Abstract
The epidermal growth factor (EGF) receptor is a tyrosine kinase that dimerizes in response to ligand binding. Ligand-induced dimerization of the extracellular domain of the receptor promotes formation of an asymmetric dimer of the intracellular kinase domains, leading to stimulation of the tyrosine kinase activity of the receptor. We recently monitored ligand-promoted conformational changes within the EGF receptor in real time using luciferase fragment complementation imaging and showed that there was significant movement of the C-terminal tail of the EGF receptor following EGF binding (Yang, K. S., Ilagan, M. X. G., Piwnica-Worms, D., and Pike, L. J. (2009) J. Biol. Chem. 284, 7474–7482). To investigate the structural basis for this conformational change, we analyzed the effect of several mutations on the kinase activity and luciferase fragment complementation activity of the EGF receptor. Mutation of Asp-960 and Glu-961, two residues at the beginning of the C-terminal tail, to alanine resulted in a marked enhancement of EGF-stimulated kinase activity as well as enhanced downstream signaling. The side chain of Asp-960 interacts with that of Ser-787. Mutation of Ser-787 to Phe, which precludes this interaction, also leads to enhanced receptor kinase activity. Our data are consistent with the hypothesis that Asp-960/Glu-961 represents a hinge or fulcrum for the movement of the C-terminal tail of the EGF receptor. Mutation of these residues destabilizes this hinge, facilitating the formation of the activating asymmetric dimer and leading to enhanced receptor signaling.
Keywords: Hormone Receptors, Protein Kinases, Receptor Structure-Function, Receptor-Tyrosine Kinase, Signal Transduction, EGF Receptor
Introduction
The epidermal growth factor (EGF)2 receptor is a classical receptor-tyrosine kinase composed of an extracellular ligand binding domain and an intracellular tyrosine kinase domain (1). In its inactive state, the EGF receptor is thought to exist in membranes as a monomer (2, 3), although substantial evidence suggests that it also forms inactive “pre-dimers” (4–8). Binding of EGF induces a major conformational change in the extracellular domain of the receptor allowing the formation of back-to-back receptor dimers (9–11). This leads to the activation of the intracellular tyrosine kinase domain.
Recent studies have provided insight into the mechanism through which receptor dimerization leads to activation of the tyrosine kinase. Zhang et al. (12) reported that the EGF receptor kinase domain crystallizes as an asymmetric dimer in which the C-lobe of one kinase (the activator kinase) interacts with the N-lobe of the second kinase (the receiver kinase). Through mutational analysis, they demonstrated that the receiver kinase becomes activated as a result of asymmetric dimer formation. Subsequently, Thiel et al. (13) presented evidence that the intracellular juxtamembrane domain was involved in this allosteric activation of the EGF receptor kinase domain. The role of the juxtamembrane domain in dimer formation was clarified by Brewer et al. (14) who reported the crystal structure of an EGF receptor asymmetric kinase dimer that contained essentially all of the juxtamembrane domain. Their structure shows that the juxtamembrane domain of the receiver kinase forms a cradle or latch around the C-lobe of the activator kinase. Mutagenesis studies confirmed the importance of residues in the intracellular juxtamembrane domain for stabilization of the activating asymmetric kinase dimer.
Jura et al. (15) noted a similar latch between the activator and receiver kinase in a previously reported crystal structure of the related ErbB4 kinase domain (16). By assaying several soluble, truncated forms of the intracellular kinase domain, they verified the importance of the juxtamembrane domain in kinase activation. Thus, it appears likely that tyrosine kinase activation in all ErbB family receptors occurs via formation of asymmetric dimers and involves interactions between the intracellular juxtamembrane domain of one monomer and residues in the C-lobe of the other kinase monomer.
In their study, Jura et al. (15) also reported the crystal structure of a symmetric EGF receptor kinase dimer in which the kinase monomers are present in a head-to-head arrangement. They noted that the C-terminal residues in the symmetric dimer structure occupy the same position on the C-lobe of the kinase as residues in the juxtamembrane latch that stabilize the activating asymmetric dimer. Thus, for the activating asymmetric dimer to form, the AP-2 helix and the C-terminal tail must be displaced from their position in the symmetric dimer.
We have recently used luciferase fragment complementation imaging in living cells to identify conformational changes in the EGF receptor that occur in response to ligand binding (8). Our data suggest that full-length EGF receptors exist as inactive pre-dimers in which the C-terminal tails of the two receptor monomers are in close proximity. Ligand binding induces a conformational change that leads to separation of the C-terminal tails. Over time, this effect is reversed, and the receptors again adopt a conformation in which the C-terminal tails are close together.
To investigate the structural basis of this conformational change, we made several mutations in the C-terminal tail of the EGF receptor, including the replacement of Asp-960 and Glu-961 with Ala. This mutation results in an EGF receptor kinase that is significantly more active than the wild-type receptor. Mutation of Ser-787, with which Asp-960 interacts, leads to a similar phenotype. Our data are consistent with the hypothesis that Asp-960/Glu-961 represents a hinge or fulcrum for the movement of the C-terminal tail. Destabilization of this hinge by mutation of Asp-960/Glu-961 or Ser-787 facilitates displacement of the C-terminal tail by the juxtamembrane latch sequences, allowing easier conversion of the inactive symmetric dimer to the active asymmetric dimer.
EXPERIMENTAL PROCEDURES
Reagents
Murine EGF was purchased from Biomedical Technologies, Inc. and was dissolved in sterile water. U0126 (EMD Chemicals) was dissolved in DMSO. Doxycycline was from Clontech and was dissolved in sterile water. d-Luciferin (Biosynth) was dissolved in PBS and coelenterizine (Sigma) was dissolved in ethanol. The MAP kinase and phosphospecific Thr-669 antibodies were from Upstate Biotechnology. The phosphospecific MAP kinase antibody was from Promega and the phosphotyrosine (PY20) antibody was from BD Biosciences. A mixture of EGF receptor antibodies from Cell Signaling and Santa Cruz Biotechnology was used to detect the EGF receptor. Pervanadate was generated by mixing equal concentrations of sodium orthovanadate and H2O2.
DNA Constructs
The wild-type EGF receptor construct was subcloned from pcDNA3.1(−) (Invitrogen) into pcDNA5/FRT (Invitrogen) using the NheI and HindIII restriction enzyme sites. The wild-type EGFR-NLuc/CLuc constructs were described previously (8)). Using the wild-type EGF receptor (pcDNA3.1(−)) as the template, QuikChange site-directed mutagenesis (Stratagene) was used to generate the D950A, D960A, E961A, D960A/E961A, and S787F mutants. Mutants were digested with the BstEII and HindIII restriction enzyme sites and were ligated into the wild-type EGF receptor construct (pcDNA5/FRT) cut with the same enzymes. All mutant constructs were sequenced in their entirety.
The D950A/D960A/E961A triple mutant was generated using QuikChange site-directed mutagenesis with the D960A/E961A-EGF receptor mutant (pcDNA3.1(−)) as the template. The D950A/D960A/E961A-EGF receptor pcDNA3.1(−) construct was subcloned into the wild-type pcDNA5/FRT construct using the BstEII and HindIII restriction sites and was sequenced.
QuikChange site-directed mutagenesis was used to make the T669A-EGF receptor construct with the wild-type EGF receptor (pcDNA5/FRT) as the template. The mutant was digested with NheI and BstEII and was ligated into the wild-type EGF receptor pcDNA5/FRT plasmid cut with the same enzymes. The single point mutation was verified by sequencing. The T669A-CLuc construct was made using QuikChange site-directed mutagenesis with the EGFR-CLuc (pcDNA6/V5-HisB) construct (8) as template. The product was re-ligated into the same vector using the NheI and BsiWI restriction enzyme sites. T669A-NLuc was generated by restriction enzyme digest of the EGFR-NLuc pcDNA3.1TOPO construct with BstEII and KpnI, followed by ligation into the T669A-EGF receptor pcDNA5/FRT construct.
The D960A/E961A-CLuc fusion construct was made by QuikChange site-directed mutagenesis of EGFR-CLuc pcDNA6/V5-HisB (8). cDNA was digested with BstEII and BsiWI and was re-ligated into the EGFR-CLuc pcDNA6/V5-HisB construct cut with the same enzymes. D960A/E961A-NLuc was made by digesting the D960A/E961A-CLuc construct with NheI and BsiWI, followed by ligation into the EGFR-NLuc pBI-Tet construct. The D960A/E961A-NLuc construct was then subcloned into the pcDNA5/FRT vector using the BstEII and EcoRV restriction enzyme sites.
Cell Lines
Flp-In CHO cells (Invitrogen) were co-transfected with pOG44 (Invitrogen) and the wild-type or mutant EGF receptors (pcDNA5/FRT) using Lipofectamine 2000 (Invitrogen). Stable clones were isolated by selection in 600 μg/ml hygromycin (InvivoGen). Cells were maintained in F-12 containing 10% FetalPlex, 1000 μg/ml penicillin/streptomycin, and 100 μg/ml hygromycin. CHO-K1 Tet-On cells were grown in DMEM containing 10% FetalPlex, 1000 μg/ml penicillin/streptomycin, and 200 μg/ml G418. The EGFR-NLuc/EGFR-CLuc, T669A-NLuc/T669A-CLuc, and D960A/E961A-NLuc/D960A/E961A-CLuc constructs were transiently transfected into CHO-K1 Tet-On cells 24 h prior to luciferase fragment complementation imaging using Lipofectamine 2000. Cells stably expressing EGFR-NLuc and EGFR-CLuc were maintained as previously described (8). Transfection efficiency was determined by co-transfection of Renilla luciferase (pRLuc-N1, Packard Bioscience).
Kinase Activation and Western Blotting
CHO cells were grown to confluence in 35-mm dishes. Cells were serum-starved in F-12 containing 1 mg/ml BSA for 2 h. Cells were washed twice in ice-cold phosphate-buffered saline and were then scraped into RIPA buffer (150 mm NaCl, 10 mm Tris pH 7.2, 0.1% sodium dodecylsulfate, 1% Triton X-100, 17 mm deoxycholate, and 2.7 mm EDTA) containing 20 mm p-nitrophenyl phosphate, 1 mm sodium orthovanadate, and protease inhibitors. Equal amounts of protein (BCA assay, Pierce) were loaded onto a 9% SDS-polyacrylamide gel and then transferred to polyvinylidene difluoride (Millipore). Western blots were blocked for 1 h in TBST/10% nonfat milk. The blots were incubated in primary antibody for 1 h (overnight at 4 °C for EGF receptor and phospho-Thr-669 antibodies), washed in TBST/0.1% BSA, incubated in secondary antibody for 45 min and washed three times in TBST/0.1% BSA. Western blots were detected using the Enhanced Chemiluminescent reagent from GE Healthcare.
Cell Migration Assay
CHO cells were grown to confluence in 35-mm dishes and serum-starved for 24 h in F-12 medium containing 0.5% fetal bovine serum. Monolayers were wounded with a sterile pipette tip, washed with Hanks balanced salt solution, photographed, and the media replaced with F-12 containing 0.5% fetal bovine serum without or with 1 nm EGF. Cultures were incubated for an additional 24 h, with photographs taken 12 h and 24 h post-EGF addition on a Zeiss LSM 510 ConfoCor 2 microscope.
Luciferase Complementation Imaging
Cells were plated 48 h prior to use at 5 × 103 cells per well in growth media in a black wall 96-well plate. On the day of the assay, cells were serum-starved for 2 h and then incubated for 20 min in 175 μl of DMEM without phenol red, containing 1 mg/ml BSA, 25 mm Hepes (pH 7.2), and 0.6 mg/ml d-luciferin at 37 °C. To establish a baseline, cell radiance (photons/second/cm2/sr) was measured using a cooled CCD camera and imaging system at 37 °C (IVIS 50; Caliper) (30 s exposure; binning, 8; no filter; f-stop, 1; field of view, 12 cm). EGF was added in a volume of 25 μl in the same media (DMEM, 1 mg/ml BSA, 25 mm Hepes (pH 7.2), 0.6 mg/ml d-luciferin). Radiance was measured sequentially as described above. Transfection efficiency was assessed by monitoring Renilla luciferase expression. Media was replaced on cells with DMEM without phenol red containing 1 mg/ml BSA, 25 mm Hepes, (pH 7.2), and 400 nm coelenterazine. Radiance was immediately measured as described above except the filter was set to <510.
Data Analysis
Data were collected in quadruplicate for each condition. A flat-field correction was done to correct for differences in the baseline photon flux. Light production expressed as photon flux (photons/sec) was determined from regions-of-interest defined over wells using LIVINGIMAGE (Xenogen) and IGOR (Wavemetrics) software. Changes in photon flux were calculated by subtracting values observed in untreated cells from those of EGF-treated cells. Standard errors were determined using the formula for unpooled standard error.
RESULTS
Involvement of Asp-960 and Glu-961 in the Control of EGF Receptor Kinase Activity
Our previous studies on ligand-induced conformational changes in the EGF receptor suggested that there was movement of both the kinase domain and the C-terminal tail (8). In most EGF receptor kinase domain structures, ∼10–20 residues downstream of position 961 are missing from the structure (12, 17–20). This suggests that this is a highly flexible region that might alter its position upon ligand binding. Residue 960 is an aspartic acid, and residue 961 is a glutamic acid. At least one acidic residue at either of these positions is conserved in all four ErbB family members (supplemental Table S1).
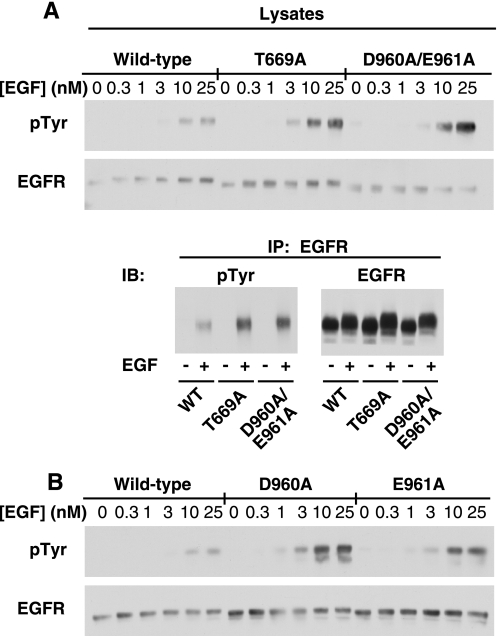
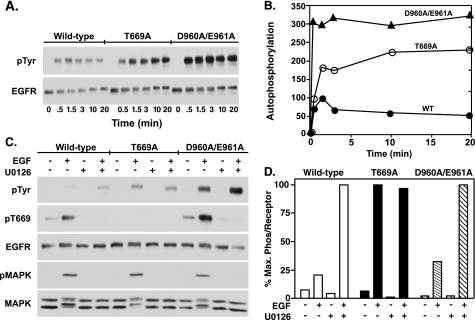
To determine whether Asp-960 and/or Glu-961 were involved in EGF-induced conformational changes in the EGF receptor, the double point mutant, D960A/E961A-EGF receptor was constructed. The mutant was expressed in CHO cells, and its ability to undergo EGF-stimulated receptor autophosphorylation was assessed. For comparison, cells expressing the T669A-EGF receptor were also analyzed. Thr-669 is phosphorylated by MAP kinase (21), a modification that leads to desensitization of the receptor (22). Mutation of this residue blocks MAP kinase-dependent desensitization and enhances the activity of the kinase (22). Fig. 1A shows the dose response to EGF in cells expressing similar levels of the wild type, T669A- and D960A/E961A-EGF receptors. EGF stimulated the phosphorylation of the receptor in all cell lines. As expected, autophosphorylation of the EGF receptor was substantially higher (∼4-fold) in cells expressing the T669A-EGF receptor, consistent with the hypothesis that it fails to undergo MAP kinase-dependent desensitization. Interestingly, the D960A/E961A-EGF receptor was also more highly phosphorylated (∼6-fold) than the wild type-EGF receptor. Immunoprecipitation of the EGF receptor confirmed that the bands phosphorylated in the cell lysates were indeed the EGF receptor (Fig. 1A, lower panel).
FIGURE 1.
Autophosphorylation of the wild-type, T669A, and D960A/E961A EGF receptors. A, CHO cells stably expressing the wild type, T669A-, or D960A/E961A-EGF receptor were plated in 6-well dishes 48 h prior to assay. Cells were incubated in medium containing 1 mg/ml BSA for 2 h before assay. The indicated concentrations of EGF were then added to cells for 5 min at 37 °C. Upper panel, cells were lysed in RIPA buffer, and equal amounts of the protein lysates were run on a 9% SDS-PAGE gel. Western blot analysis was performed with the indicated antibodies. Lower panel, lysates from cells treated without or with 25 nm EGF for 5 min were immunoprecipitated with an anti-EGF receptor antibody and analyzed by SDS-polyacrylamide gel electrophoresis. Western blot analysis was then performed with an anti-phosphotyrosine antibody or an anti-EGF receptor antibody. B, identical protocol was applied to CHO cells stably expressing the wild-type, D960A-, or E961A-EGF receptors.
To determine which residue contributed most to the observed phenotype, the D960A- and E961A-EGF receptor single point mutants were constructed and stably expressed in CHO cells. The effect of EGF on the activity of cells expressing wild type, D960A- or E961A-EGF receptors is shown in Fig. 1B. As before, treatment of cells expressing the wild-type EGF receptor led to tyrosine phosphorylation of the receptor. Stimulation of cells expressing either the D960A-EGF receptor or the E961A-EGF receptor with EGF resulted in increased levels of receptor autophosphorylation as compared with the wild-type receptor. However, autophosphorylation was somewhat higher in the D960A-EGF receptor than in the E961A-EGF receptor. This suggests that while both residues are important, Asp-960 makes a larger contribution to the phenotype than does Glu-961. Because both residues appear to contribute to the phenotype, all further experiments were carried out with the D960A/E961A double point mutant.
Downstream Signaling in the EGF Receptor Mutants
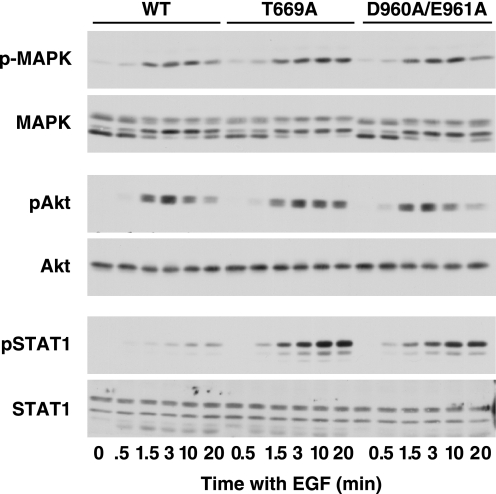
Cells expressing the wild-type-, T669A-, or D960A/E961A-EGF receptors were stimulated with 25 nm EGF for increasing lengths of time and then analyzed for the effect of the mutations on downstream signaling. As shown in Fig. 2, EGF stimulated MAP kinase activity to a similar extent in all three cell lines. Likewise, the EGF-stimulated activation of Akt was similar in all three lines. However, phosphorylation of Stat1 was markedly enhanced in cells expressing the T669A- and D960A/E961A-EGF receptors as compared with the wild-type receptor.
FIGURE 2.
Downstream signaling through the wild-type, T669A-, and D960/E961A-EGF receptors. CHO cells stably expressing the wild-type, T669A-, or D960A/E961A-EGF receptors were plated in 6-well dishes 48 h prior to assay. Cells were treated with 25 nm EGF for the times indicated, then lysed in RIPA buffer, and analyzed by Western blotting with the indicated antibody.
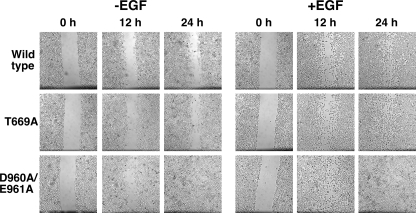
The data in Fig. 3 show the effect of the EGF receptor mutations on the migration of cells into a scratch wound in a confluent monolayer of cells plated on fibronectin-coated dishes. Integrins are known to induce some of their biological effects by triggering the activation of the EGF receptor (23–25). Thus, an activated form of the EGF receptor would be expected to enhance cell migration both in the absence and presence of EGF. In the absence of EGF, cells expressing either wild-type or T669A-EGF receptors, showed relatively modest migration into the wound by 12 h, which was still incompletely filled by 24 h. By contrast, in cells expressing the D960A/E961A-EGF receptor, the scratch had significantly narrowed by 12 h and numerous cells had migrated into the center of the wound. By 24 h, the scratch was completely closed.
FIGURE 3.
Migration of cells expressing the wild-type, T669A-, and D960/E961A-EGF receptors. CHO cells stably expressing the wild type, T669A, or D960A/E961A-EGF receptors were plated in 6-well dishes and grown to confluency. Monolayers were wounded using a sterile pipette tip, and the media replaced with F12 containing 0.5% fetal bovine serum in the absence or presence of 1 nm EGF. Cultures were returned to the incubator and photographed every 12 h as described under “Experimental Procedures.”
Addition of EGF stimulated significant migration in all cell lines. However, cells expressing the D960A/E961A-EGF receptor showed a greater degree of wound closure after both 12 h and 24 h than did cells expressing either wild-type or T669A-EGF receptor. These data demonstrate that the D960A/E961A mutation results in a receptor with enhanced the ability to activate down stream signaling pathways and cellular level responses.
Luciferase Fragment Complementation Imaging of EGF Receptor Mutants
We have previously used luciferase fragment complementation imaging to visualize conformational changes in the EGF receptor that occur in response to hormone binding (8). In this system, the N-terminal fragment of luciferase (NLuc) or the C-terminal fragment of luciferase (CLuc) are fused to the C terminus of the full-length EGF receptor. Complementation occurs when the fragments are in close proximity to each other and can reconstitute an active luciferase. Complementation declines when the fragments move apart from each other. The assay is done in intact cells under physiological conditions allowing real-time analysis of conformational changes in the EGF receptor.
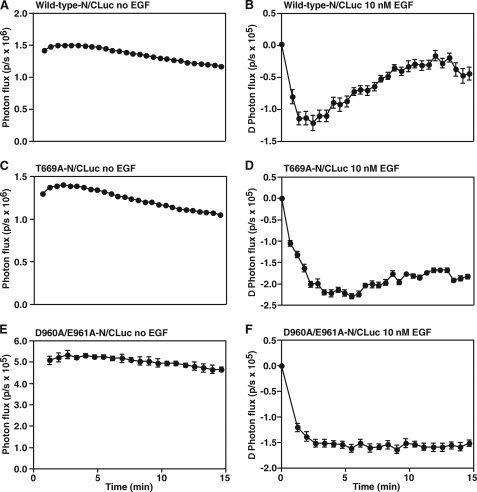
To compare the EGF-induced conformational changes in wild-type, T669A-, and D960A/E961A-EGF receptors, luciferase fragments were fused to the C termini of each receptor and the constructs stably expressed in CHO cells. Fig. 4, A and B show the results of a control experiment in which cells expressing the wild-type EGF receptor were stimulated with EGF. As previously reported (8), a significant basal luciferase activity was observed, suggesting the presence of preformed dimers in unstimulated cells. Addition of EGF led to a rapid decrease in luciferase activity followed by a slow recovery back to baseline levels of light production.
FIGURE 4.
Luciferase fragment complementation imaging of cells expressing wild-type, T669A-, or D960A/E961A-EGF receptors. CHO cells were plated in 96-well plates 48 h prior to assay. Twenty-four h prior to imaging, cells were transiently transfected with cDNA encoding the wild type-, T669A-, or D960A/E961A-EGF receptor NLuc and CLuc luciferase fusion constructs. On the day of imaging, cells were incubated in medium containing 1 mg/ml BSA for 2 h followed by incubation in 0.6 mg/ml d-luciferin for 20 min. A, C, E, photon flux (photons/sec; p/s) in the absence of EGF. B, D, F, change in photon flux following addition of 10 nm EGF. The change in photon flux was calculated by subtracting the average photon flux in the absence of EGF from the photon flux observed in the presence of EGF. Data represent the average of four independent replicates, and the standard error is shown.
The results of luciferase fragment complementation imaging in the T669A-EGF receptor mutant are shown in Fig. 4, C and D. Like the wild-type receptor, the T669A-EGF receptor showed substantial basal luciferase activity. Similarly, addition of EGF induced a rapid decrease in luciferase activity (Fig. 4D). However, in contrast to what was seen with the wild-type EGF receptor, there was no recovery phase. Luciferase activity declined and did not regain its initial level. As shown in Fig. 4, E and F, the D960A/E961A-EGF receptor mutant exhibited the same pattern of luciferase fragment complementation as that seen in the T669A-EGF receptor. Specifically, EGF stimulated a rapid decline in luciferase activity, but there was no recovery back to baseline levels.
The D960/E961 Mutation Is Independent of MAP Kinase-induced Desensitization of the EGF Receptor
These data suggest that the T669A- and D960A/E961A-EGF receptors share a common characteristic that leads to a similar pattern of ligand-induced changes in luciferase complementation that are different from those observed for the wild-type receptor. To further compare these two mutants, a time course of EGF-stimulated phosphorylation was performed. The data in Fig. 5A show that phosphorylation of the wild-type EGF receptor peaked ∼2 min after the addition of EGF and declined slightly thereafter. By contrast, maximal EGF-stimulated autophosphorylation of the T669A- and D960A/E961A-EGF receptors was ∼3-fold higher than that observed in the wild type receptor and was maintained at that high level over the 20 min time course (quantitated in Fig. 5B). Thus, in addition to similar behavior in the luciferase complementation assay, the two mutant receptors also share the characteristic of persistently elevated levels of receptor phosphorylation.
FIGURE 5.
Autophosphorylation of wild-type, T669A-, and D960A/E961A-EGF receptors. CHO cells stably expressing the wild type, T669A, or D960A/E961A-EGF receptors were plated in 6-well dishes 48 h prior to assay. A, cells were treated with 25 nm EGF for the times indicated, then lysed in RIPA buffer, and analyzed by Western blotting with an anti-phosphotyrosine or an anti-EGF receptor antibody. B, anti-phosphotyrosine blots from A were quantitated using Image J, and the level of EGF receptor autophosphorylation was normalized to the maximum phosphorylation observed in cells expressing wild-type receptors. C, cells were incubated for 20 min at 37 °C without or with 10 μm U0126, followed by stimulation with 1 nm EGF for 5 min. Cells were immediately lysed in RIPA buffer and equal amounts of protein lysates were separated on an SDS-PAGE gel. Western blotting was performed with the indicated antibodies. D, anti-phosphotyrosine and anti-EGF receptor Western blots from A were quantitated using Image J and the level of EGF receptor autophosphorylation was plotted as the percent maximum phosphorylation per unit receptor for each type of receptor.
The modest level of EGF receptor autophosphorylation in the wild-type receptor is due in part to the MAP kinase-dependent desensitization of the receptor (22, 26). This reduces EGF-stimulated kinase activity and leads to lower net receptor phosphorylation due to dephosphorylation of the receptor by protein tyrosine phosphatases. Because the D960A/E961A-EGF receptor showed a phenotype similar to that of the T669A mutant, it was possible that the double point mutation might in some way inhibit the ability of MAP kinase to phosphorylate the EGF receptor on Thr-669 and desensitize its kinase activity. To assess this possibility, cells expressing wild-type, T669A-, or D960A/E961A-EGF receptors were treated without or with the MEK inhibitor, U0126, prior to stimulation by EGF to block the activation of MAP kinase.
In the wild-type EGF receptor, stimulation with EGF resulted in receptor autophosphorylation as well as phosphorylation of the receptor on Thr-669 (Fig. 5C). Pretreatment with U0126 abolished phosphorylation of the receptor on Thr-669 and was associated with a 5-fold increase in EGF receptor autophosphorylation (quantitated in Fig. 5D). These data are consistent with the interpretation that phosphorylation of the EGF receptor by MAP kinase decreases activation of the EGF receptor kinase and indicate that this effect can be abolished by inhibiting MAP kinase activity.
As expected, the T669A-EGF receptor was not phosphorylated at residue 669 and EGF-stimulated receptor autophosphorylation was higher than in the wild-type receptor (Fig. 5C). Consistent with the absence of phosphorylation at residue 669, pretreatment of cells expressing the T669A-EGF receptor with U0126 had no effect on the level of receptor autophosphorylation. Thus, the enhanced tyrosine kinase activity of this receptor can be ascribed to the ablation of MAP kinase-dependent phosphorylation of the receptor.
In contrast to the T669A-EGF receptor, the D960A/E961A-EGF receptor was extensively phosphorylated at Thr-669 (Fig. 5C). Despite the heavy phosphorylation at Thr-669, autophosphorylation of the D960A/E961A-EGF receptor was substantially higher than was seen in the wild type EGF receptor. Inhibition of Thr-669 phosphorylation by U0126 was associated with a further ∼3-fold increase in the level of autophosphorylation of the EGF receptor. These observations indicate that the D960A/E961A-EGF receptor is still subject to MAP kinase-dependent desensitization and that the double point mutation enhances EGF receptor kinase activity by a mechanism other than ablation of MAP kinase-dependent desensitization of the receptor.
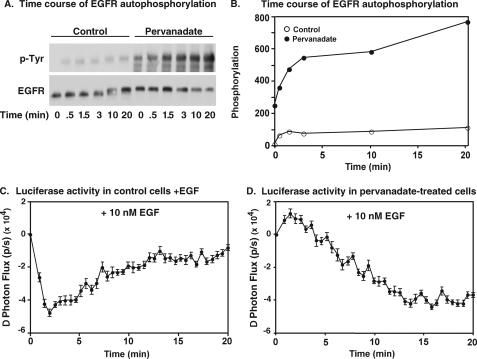
Our previous studies demonstrated that the initial decline in luciferase complementation following EGF stimulation requires the tyrosine kinase activity of the receptor (8). To determine whether elevated EGF receptor autophosphorylation was associated with changes in our luciferase fragment complementation assay, cells expressing wild-type EGF receptors fused to N-Luc and C-Luc fragments were pretreated without or with sodium pervanadate to inhibit phosphotyrosine protein phosphatases and increase receptor autophosphorylation prior to assaying for EGF-stimulated luciferase activity.
Fig. 6, A and B show the effect of pervanadate treatment on EGF receptor autophosphorylation. In control cells, EGF stimulated a modest level of receptor autophosphorylation. By contrast, in cells pretreated with pervanadate, a significant level receptor phosphorylation was seen even in the absence of EGF. Stimulation with EGF led to a further slow and persistent rise in receptor autophosphorylation to a level that was nearly 8-fold higher than that seen at the peak of phosphorylation in control cells.
FIGURE 6.
Effect of pervanadate on luciferase activity in the wild-type EGF receptor. A, CHO cells stably expressing wild-type EGF receptors were preincubated with 200 μm sodium pervanadate for 20 min and then 10 nm EGF was added for the times indicated. Cells were immediately lysed in RIPA buffer, and equal amounts of protein lysates were separated on an SDS-PAGE gel. Western blotting was performed with the indicated antibodies. B, anti-phosphotyrosine Western blots from A were quantitated using Image J, and the level of EGF receptor autophosphorylation was normalized to the maximum phosphorylation observed in cells expressing wild-type receptors. C and D, cells stably expressing EGFR-NLuc and EGFR-CLuc were plated in 96-well plates 48 h prior to assay. On the day of imaging, cells were pretreated for 20 min with 0.6 mg/ml d-luciferin in the absence or presence of 200 μm sodium pervanadate. 10 nm EGF was then added to the cells, and the change in photon flux was monitored over time in untreated cells (C) or pervanadate-treated cells (D).
With respect to luciferase fragment complementation, the control cells (Fig. 6C) exhibited the typical biphasic pattern of luciferase complementation. EGF stimulated a rapid decrease in luciferase activity followed by a return to baseline. In the pervanadate-treated cells, EGF triggered a slow decline in luciferase activity but there was no recovery of complementation (Fig. 6D). The fact that elevated phosphorylation of the EGF receptor in pervanadate-treated cells is associated with a persistent decline in luciferase complementation is consistent with the hypothesis that the decrease in luciferase activity is related to phosphorylation of the receptor. The absence of the initial rapid decline of luciferase activity after the addition of EGF to pervanadate-treated cells is likely due to the fact that the EGF receptor was extensively autophosphorylated prior to the addition of EGF to these cells (Fig. 6, A and B). These data suggest that the similarity of EGF-stimulated conformational changes in the T669A- and D960A/E961A-EGF receptors visualized by luciferase fragment complementation imaging probably reflects the extent to which they have been phosphorylated.
The D960A/E961A Mutation Facilitates Asymmetric Dimer Formation
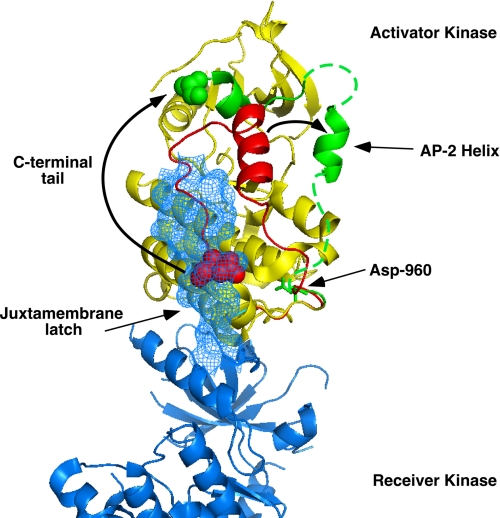
Because kinase activation is dependent upon asymmetric dimer formation, we examined the structure of the EGF receptor asymmetric dimer (14) to determine whether Asp-960 or Glu-961 could contribute to the formation of this structure. Fig. 7 shows the structure of the asymmetric dimer with the activator kinase in yellow and the receiver kinase in blue. The extreme C-terminal sequences, including the AP-2 helix (residues 969–978) are shown in green. A monomer from the inactive, symmetric kinase dimer (15) was aligned with the (yellow) activator kinase from the asymmetric dimer (14). Most of two structures align quite well (not shown). However, as shown in the figure, the AP-2 helix and C-terminal tail sequences in the inactive, symmetric form of the kinase (red residues) occupy a distinctly different position from these segments in the active, asymmetric form of the kinase (green residues).
FIGURE 7.
Structure of the EGF receptor kinase asymmetric dimer. The asymmetric dimer structure of Brewer et al. (14) was used (PDB code 3GOP). The activator kinase is shown in yellow and the receiver kinase in blue. The AP-2 helix and C terminus of the activator kinase is shown in green. A monomer from the inactive symmetric dimer structure of Jura et al. (15) (PDB code 3GT8) was aligned with the activator kinase of the asymmetric dimer using PyMol. The AP-2 helix and the C terminus of the inactive symmetric kinase monomer are shown in red. The position of the juxtamembrane latch made by sequences N-terminal to the receiver kinase domain is shown in blue mesh.
In the inactive form of the kinase (red), the AP-2 helix lies essentially parallel to the long axis of the asymmetric dimer. It is connected to its C-terminal tail by a loop such that the helix and the C-terminal tail run roughly parallel to each other. By contrast, in the active asymmetric dimer (green), the AP-2 helix is swung away from the body of the kinase domain and the C-terminal tail runs roughly perpendicular to this helix. There is an ∼18 Å shift in the position of the AP-2 helix and its attached C-terminal sequences.
As noted by Jura et al. (15), the C-terminal residues in the symmetric kinase dimer (red) occupy the same position as the juxtamembrane latch made by N-terminal sequences in the receiver kinase (blue mesh in Fig. 7). Thus, for the activating asymmetric dimer to form, the C-terminal tail of the activator kinase must be displaced by the N-terminal juxtamembrane strand of the receiver kinase. This can be achieved by rotating the strand containing the AP-2 helix around a hinge point at the position of Asp-960 in the activator kinase.
In both the symmetric and asymmetric dimers, the side chain of Asp-960 is within hydrogen bonding distance of the side chain of Ser-787 (supplemental Fig. S1). This interaction is likely to stabilize the position of Asp-960, restricting the ability of the strand containing the AP-2 helix and the C-terminal tail to rotate out to allow formation of the juxtamembrane latch. Loss of the Asp-960 side chain would abolish this stabilizing interaction, facilitating the displacement of the C-terminal tail of the activator kinase by the juxtamembrane sequences of the receiver kinase. This would improve the activator function of the D960A/E961A receptor, leading to an increase in tyrosine kinase activity.
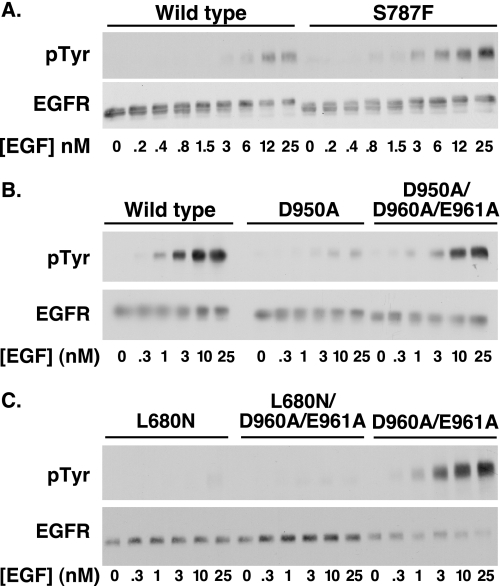
If the enhanced kinase activity of the D960A/E961A-EGF receptor is due to the release of the polar contacts that stabilize the position of Asp-960, then mutation of Ser-787 should also enhance EGF receptor kinase activity as it would eliminate the interaction of Asp-960 with the serine side chain. Na et al. (27) have reported the presence of a Ser-787 to Phe mutation in the EGF receptor of a patient with non-small cell lung carcinoma. This suggests that Phe could be an activating mutation at this position. To determine whether mutation of Ser-787 reproduces the effect of the D960A/E961A mutation, we constructed the S787F-EGF receptor and stably expressed it in CHO cells. Fig. 8A compares the dose response to EGF in cells expressing wild type and S787F-EGF receptors. As can be seen from the figure, the S787F-EGF receptor exhibited a markedly elevated level of receptor autophosphorylation, consistent with the hypothesis that interactions mediated by Ser-787 play an important role in limiting the level of activation of the EGF receptor kinase.
FIGURE 8.
The D950A/D960A mutation enhances the activator activity of the EGF receptor. A, CHO cells stably expressing wild type or S787F-EGF receptors were treated with the indicated concentrations of EGF for 2 min at 37 °C. The cells were lysed in RIPA buffer, and aliquots of the lysates containing equal amounts of protein were separated on an SDS-PAGE gel. Western blot analysis was performed with the indicated antibodies. B, CHO cells stably expressing the wild type-, D950A-, or D950A/D960A/E961A-EGF receptors were treated with the indicated concentrations of EGF for 2 min at 37 °C after which the cells were lysed in RIPA buffer. Aliquots of the lysates containing equal amounts of protein were separated on an SDS-PAGE gel. Western blot analysis was performed with the indicated antibodies. C, CHO cells stably expressing the L680N-, L680N/D960A/E961A-, or D960A/E961A-EGF receptors were treated with the indicated concentrations of EGF for 2 min at 37 °C after which the cells were lysed in RIPA buffer. Aliquots of the lysates containing equal amounts of protein were separated on an SDS-PAGE gel. Western blot analysis was performed with the indicated antibodies.
Our data suggest that D960/E961 control the activator function of the EGF receptor. If the D960A/E961A-EGF receptor has enhanced activator activity, then the mutation would be expected to counteract the effects of a different mutation that compromises the activator function of the EGF receptor. Jura et al. (15) observed that mutation of Asp-950 to Ala resulted in an EGF receptor with greatly reduced autophosphorylation in cell-based assays. Asp-950 is present on the activator side of the asymmetric dimer interface. It forms a hydrogen bond with Ser-671 in the juxtamembrane domain of the receiver kinase that stabilizes the asymmetric dimer (14).
To determine whether mutation of Asp-960/Glu-961 can compensate for the defect in the D950A-EGF receptor, the D950A single mutant and the D950A/D960A/E961A triple mutant EGF receptors were constructed and stably expressed in CHO cells. Fig. 8B compares the dose response to EGF in cells expressing wild-type, D950A, and the D950A/D960A/E961A triple mutant EGF receptors. As can be seen from the figure, substitution of Asp-950 with alanine markedly impaired the ability of EGF to stimulate autophosphorylation of its receptor. This is consistent with the observations of Jura et al. (15). This inhibition was largely overcome when the D950A mutation was combined with the D960A/E961A mutation in the triple mutant. Thus, the D960A/E961A mutation can compensate for a mutation that compromises the activator function of a kinase monomer.
Leu-680 is in the N-lobe of the receiver kinase. Substitution of the Leu-680 with Asn impairs the ability of the receiver kinase to be activated. If the D960A/E961A mutation increases kinase activity because it enhances the activator function of the kinase, it should be unable to compensate for the effect of a mutation that compromises the receiver function of the kinase domain. To test this prediction, the D960A/E961A mutation was combined with the L680N mutation and the triple mutant stably expressed in CHO cells. As shown in Fig. 8C, the L680N-EGF receptor shows negligible EGF-stimulated tyrosine kinase activity. By contrast, the D960A/E961A-EGF receptor exhibits strong EGF-stimulated receptor autophosphorylation. Combining all three mutations results in the production of a receptor that shows almost no ligand-stimulated receptor autophosphorylation. Thus, the D960A/E961A mutation cannot overcome the block in receiver function introduced by the L680N mutation. Together, these findings are consistent with the conclusion that the D960A/E961A mutation enhances the activator function of the EGF receptor.
DISCUSSION
These studies were undertaken in an effort to provide a structural explanation for the conformational changes in the EGF receptor that we documented previously using luciferase fragment complementation imaging (8). In those studies (as here), we observed significant basal luciferase activity suggesting that the luciferase fragments were in close proximity to each other, possibly due to the formation of an inactive pre-dimer. Addition of EGF led to a decrease in luciferase activity indicating that the two luciferase fragments become separated from each other. As EGF is known to induce the formation of receptor dimers, the data suggest that a conformational change that separates the two C termini is responsible for the decrease in luciferase activity. The decrease in luciferase activity is not due to degradation of the receptor/luciferase fusion protein because luciferase complementation returns to starting levels over time. Thus, the receptor ultimately must adopt a conformation that brings the two C termini back into proximity to permit enzyme complementation.
The data reported here and the recent structures of the EGF receptor kinase in an apparently inactive, symmetric dimer (15) and an active, asymmetric dimer (14) provide insight into the structural underpinnings of these conformational changes. Alignment of the structures of the inactive, symmetric dimer (15) and the active, asymmetric dimer (14) suggest that the AP-2 helix rotates away from the kinase core when the enzyme is converted from the symmetric to the asymmetric dimer. The C-terminal residues are also significantly re-positioned upon formation of the activating, asymmetric dimer. This movement of the C-terminal tail seems to have been completed in the recent ErbB3 kinase structure (28). As shown in supplemental Fig. S2, where the ErbB3 kinase (magenta in the figure) was aligned with the activator monomer of the asymmetric dimer, the C-terminal residues of the ErbB3 kinase are rotated around Asp-960 essentially 180° from their position in the EGF receptor. The C-terminal tail extends down into the proximity of the receiver kinase and would thus be available for phosphorylation. This is exactly what is predicted to occur upon allosteric activation of the EGF receptor kinase (12).
Our mutational analysis suggests that this movement of the AP-2 helix and the C-terminal tail is restricted by an interaction between Asp-960 and Ser-787 in the kinase core. Breaking this contact allows easier movement of the AP-2 helix, thereby facilitating formation of the asymmetric dimer. Loss of this interaction effectively makes the D960A/E961A-EGF receptor kinase a super activator. This is reflected in the ability of the D960A/E961A-EGF receptor to mediate enhanced receptor autophosphorylation, Stat 1 phosphorylation and cell migration.
It is unclear from the structure how the E961A mutation gives rise to the activated phenotype. However, as can be seen in supplemental Fig. S1, the side chain of Gln-952 is relatively close to that of Glu-961 and, given the apparent flexibility of this portion of the structure, it is possible that a hydrogen bond between these two side chains contributes to the stabilization of the Asp-960 hinge.
Both Ser-787 and Asp-960 are conserved from insects to man in the EGF receptor, ErbB2 and ErbB4 (supplemental Table S1). However, in all ErbB3 proteins, Ser-787 is replaced by a proline residue. Furthermore, Gln-952, which may contribute to the stabilization of the Asp-960 hinge, is conserved in most ErbB receptors but is a proline in ErbB3. As a result of these changes, the C-terminal tail of ErbB3 receptors would be more readily displaced by the juxtamembrane latch and ErbB3 would therefore be predicted to have enhanced activator activity as compared with the EGF receptor.
It is noteworthy that Thr-669, the site of MAP kinase phosphorylation that diminishes the receiver function of the EGF receptor, is conserved in the EGF receptor, ErbB2 and ErbB4 but is an aspartic acid in ErbB3. As shown by Brewer et al. (14), the Thr → Asp substitution significantly reduces the kinase activity of the soluble, intracellular domain of the EGF receptor, presumably by inhibiting the receiver function of the asymmetric dimer. Together, these observations suggest that ErbB3 has evolved in such a way that its receiver function is suppressed while its activator function is enhanced. This would enable this kinase-impaired receptor (29) to selectively adopt the activator position in an asymmetric dimer, a position in which it is most effective in signaling.
In summary, our data are consistent with a model in which Asp-960/Glu-961 serves as a hinge point for the movement of the AP-2 helix and C-terminal tail that occurs in the transition from the inactive to the active form of the kinase. This movement may be the basis for the EGF-stimulated changes in luciferase complementation observed in full length EGF receptors (8). Mutations in Asp-960, Glu-961, and the residues with which they interact, appear to enhance the activator function of the EGF receptor and, in the case of Ser-787, can give rise to an oncogenic version of this receptor (27).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM064491 (to L. J. P.) and P50 CA94056 (to D. P. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- EGF
- epidermal growth factor
- BSA
- bovine serum albumin
- RIPA
- radioimmune precipitation assay buffer.
REFERENCES
- 1.Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J., Downward J., Mayes E. L. V., Whittle N., Waterfield M. D., Seeburg P. H. (1984) Nature 309, 418–425 [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y., Schlessinger J. (1987) Biochemistry 26, 1443–1451 [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y., Schlessinger J. (1987) Biochemistry 26, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 4.Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., Burgess A. W. (2005) J. Biol. Chem. 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 5.Moriki T., Maruyama H., Maruyama I. N. (2001) J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 6.Yu X., Sharma K. D., Takahashi T., Iwamoto R., Mekada E. (2002) Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao R. H., Maruyama I. N. (2008) J. Cell Sci. 121, 3207–3217 [DOI] [PubMed] [Google Scholar]
- 8.Yang K. S., Ilagan M. X., Piwnica-Worms D., Pike L. J. (2009) J. Biol. Chem. 284, 7474–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson K. M., Berger M. B., Mendrola J. M., Cho H. S., Leahy D. J., Lemmon M. A. (2003) Mol. Cell 11, 507–517 [DOI] [PubMed] [Google Scholar]
- 10.Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 11.Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J. H., Saito K., Sakamoto A., Inoue M., Shirouzu M., Yokoyama S. (2002) Cell 110, 775–787 [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 13.Thiel K. W., Carpenter G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19238–19243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., Carpenter G. (2009) Mol. Cell 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood E. R., Shewchuk L. M., Ellis B., Brignola P., Brashear R. L., Carerro T. R., Dickerson S. H., Dickson H. D., Donaldson K. H., Gaul M., Griffin R. J., Hassell A. M., Keith B., Mullin R., Petrov K. G., Reno M. J., Rusnak D. W., Tadepalli S. M., Ulrich J. C., Wagner C. D., Vanderwall D. E., Waterson A. G., Williams J. D., White W. L., Uehling D. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2773–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair J. A., Rauh D., Kung C., Yun C. H., Fan Q. W., Rode H., Zhang C., Eck M. J., Weiss W. A., Shokat K. M. (2007) Nat. Chem. Biol. 3, 229–238 [DOI] [PubMed] [Google Scholar]
- 18.Stamos J., Sliwkowski M. X., Eigenbrot C. (2002) J. Biol. Chem. 277, 46265–46272 [DOI] [PubMed] [Google Scholar]
- 19.Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., Alligood K. J., Rusnak D. W., Gilmer T. M., Shewchuk L. (2004) Cancer Res. 64, 6652–6659 [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., Kuriyan J. (2007) Nature 450, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Northwood I. C., Gonzalez F. A., Wartmann M., Raden D. L., Davis R. J. (1991) J. Biol. Chem. 266, 15266–15276 [PubMed] [Google Scholar]
- 22.Li X., Huang Y., Jiang J., Frank S. J. (2008) Cell. Signal. 20, 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcoux N., Vuori K. (2003) Oncogene 22, 6100–6106 [DOI] [PubMed] [Google Scholar]
- 24.Moro L., Dolce L., Cabodi S., Bergatto E., Boeri, Erba E., Smeriglio M., Turco E., Retta S. F., Biuffrida M. G., Venturino M., Godovac-Zimmermann J., Conti A., Schaefer E., Beguinot L., Tacchetti C., Gaggini P., Silengo L., Tarone G., Defilippi P. (2002) J. Biol. Chem. 277, 9405–9414 [DOI] [PubMed] [Google Scholar]
- 25.Moro L., Venturino M., Bozzo C., Silengo L., Altruda F., Beguinot L., Tarone G., Defilippi P. (1998) EMBO J. 17, 6622–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heisermann G. J., Wiley H. S., Walsh B. J., Ingraham H. A., Fiol C. J., Gill G. N. (1990) J. Biol. Chem. 265, 12820–12827 [PubMed] [Google Scholar]
- 27.Na I. I., Rho J. K., Choi Y. J., Kim C. H., Park J. H., Koh J. S., Ryoo B. Y., Yang S. H., Lee J. C. (2007) Lung Cancer 57, 96–102 [DOI] [PubMed] [Google Scholar]
- 28.Jura N., Shan Y., Cao X., Shaw D. E., Kuriyan J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi F., Telesco S. E., Liu Y., Radhakrishnan R., Lemmon M. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7692–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.