Abstract
Hydrolytic editing activities are present in aminoacyl-tRNA synthetases possessing reduced amino acid discrimination in the synthetic reactions. Post-transfer hydrolysis of misacylated tRNA in class I editing enzymes occurs in a spatially separate domain inserted into the catalytic Rossmann fold, but the location and mechanisms of pre-transfer hydrolysis of misactivated amino acids have been uncertain. Here, we use novel kinetic approaches to distinguish among three models for pre-transfer editing by Escherichia coli isoleucyl-tRNA synthetase (IleRS). We demonstrate that tRNA-dependent hydrolysis of noncognate valyl-adenylate by IleRS is largely insensitive to mutations in the editing domain of the enzyme and that noncatalytic hydrolysis after release is too slow to account for the observed rate of clearing. Measurements of the microscopic rate constants for amino acid transfer to tRNA in IleRS and the related valyl-tRNA synthetase (ValRS) further suggest that pre-transfer editing in IleRS is an enzyme-catalyzed activity residing in the synthetic active site. In this model, the balance between pre-transfer and post-transfer editing pathways is controlled by kinetic partitioning of the noncognate aminoacyl-adenylate. Rate constants for hydrolysis and transfer of a noncognate intermediate are roughly equal in IleRS, whereas in ValRS transfer to tRNA is 200-fold faster than hydrolysis. In consequence, editing by ValRS occurs nearly exclusively by post-transfer hydrolysis in the editing domain, whereas in IleRS both pre- and post-transfer editing are important. In both enzymes, the rates of amino acid transfer to tRNA are similar for cognate and noncognate aminoacyl-adenylates, providing a significant contrast with editing DNA polymerases.
Keywords: Aminoacyl tRNA Synthesis, Aminoacyl tRNA Synthetase, Enzyme Mechanisms, Protein-Nucleic Acid Interaction, Transfer RNA (tRNA), Post-transfer Editing, Pre-transfer Editing, Hydrolytic Proofreading, Isoleucyl-tRNA Synthetase, Valyl-tRNA Synthetase
Introduction
Aminoacyl-tRNA synthetases (AARS)2 join amino acids with their cognate tRNAs in high fidelity reactions that define the genetic code (reviewed in Ref. 1). They are divided into two groups, class I and class II, based in part on mutually exclusive structural features of their respective catalytic domains. Aminoacylation by AARS occurs in a two-step reaction, in which ATP-dependent activation of the amino acid is followed by transfer of the amino acid to tRNA. The aminoacyl-adenylate (AA-AMP) intermediate, formed in the activation step, is held noncovalently in the active site. In the following transfer step, the 2′- or 3′-OH group of the terminal tRNA adenosine attacks the carbonyl carbon of the amino acid portion of the adenylate, with liberation of AMP.
Highly accurate aminoacyl-tRNA synthesis is a prerequisite for the precise transmission of genetic information and thus for cell survival. However, structural similarities among certain amino acids render some AARS unable to distinguish them in the synthetic reactions alone. For example, class I isoleucyl-tRNA synthetase (IleRS) activates the smaller valine only ∼200-fold less efficiently than cognate isoleucine (2). Class I valyl- and leucyl-tRNA synthetases (ValRS and LeuRS), as well as a number of class II AARS, also efficiently activate noncognate amino acids and transfer them to tRNA (3–5). To maintain error rates in protein biosynthesis within tolerable levels (10−3–10−4), the AARS of both classes have developed hydrolytic proofreading activities to correct initial errors in amino acid selection (reviewed in Ref. 5). Both the AA-AMP intermediate and final aminoacyl-tRNA product may be proofread to prevent translational mistakes (Fig. 1). In “pre-transfer” editing, AARS hydrolyze the noncognate AA-AMP prior to transfer of the aminoacyl moiety to the 3′-end of tRNA. Alternatively, if the noncognate amino acid is transferred to tRNA, the misacylated tRNA is hydrolyzed by “post-transfer” editing.
FIGURE 1.
A, schematic presentation of editing pathways 1–4. Pre-transfer editing occurs through enhanced dissociation (pathway 2) of noncognate aminoacyl-adenylate or its enzymatic hydrolysis (pathways 1 and 3), which may be tRNA-independent (pathway 1) or tRNA-dependent (pathway 3). After transfer, mischarged tRNA can be deacylated through post-transfer editing (pathway 4). The central pathway of the scheme (colored in black) represents amino acid activation, tRNA binding, and aminoacylation of both cognate and noncognate amino acid. Pathways described in the upper or lower part of the scheme refer only to noncognate amino acid. B, crystal structure of Staphylococcus aureus IleRS in complex with tRNA (Protein Data Bank code 1QU2 (6)). tRNA is shown in green, CP1 domain in pink, and the Rossmann fold in blue, and the rest of the protein is represented in gray. The 3′-end of tRNAIle is disordered in the crystal structure.
Class I IleRS, ValRS, and LeuRS are related by the presence of a common, large insertion (connective polypeptide 1, CP1) in the Rossmann fold catalytic domain (6–10). Examination of the IleRS, ValRS, and LeuRS CP1 domains as free-standing fragments showed that they possess hydrolytic activity toward misacylated tRNA (11–13). Analogs of misacylated tRNA were shown to bind to a threonine-rich cleft in the CP1 domains of IleRS and LeuRS, uncovering a proofreading site located 30 Å away from the synthetic site (14, 15) and pinpointing specific amino acid residues that may participate in post-transfer editing. Using a deacylation assay that measures enzymatic hydrolysis of preformed misacylated tRNA, a number of these highly conserved residues within the proofreading site were shown to be critical to post-transfer editing (15–18). Based on the IleRS:tRNAIle cocrystal structure, a model was proposed whereby post-transfer editing occurs through translocation of the flexible, single-stranded 3′-end of aminoacylated tRNA from the synthetic site to the CP1 domain editing site, whereas the tRNA body remains bound to the enzyme core (6). This model was later also supported by additional structures of ValRS and LeuRS bound to tRNA, establishing it as a feature common to all editing class I AARS possessing the large CP1 insertion (9, 19, 20).
Three distinct models for pre-transfer editing by AARS have been proposed (5). First, it has been suggested that some AARS may selectively release noncognate AA-AMP into solution for noncatalytic hydrolysis of the relatively unstable mixed anhydride linkage (21–23). Second, a hydrolytic activity within the synthetic active site was proposed based on the finding that the normally nonediting class I glutaminyl-tRNA synthetase (GlnRS) is able to hydrolyze cognate Gln-AMP when bound to tRNAGln possessing the A76 2′-deoxy modification (24). This activity has subsequently been characterized in a seryl-tRNA synthetase (SerRS) enzyme that lacks a separate editing domain (25) and in mutated forms of ProRS, ThrRS, and LeuRS in which post-transfer editing is selectively inactivated (26–30).
The third model for pre-transfer editing, first proposed for Escherichia coli IleRS, posits an active translocation of noncognate AA-AMP from the synthetic site to the CP1 domain site that catalyzes post-transfer editing (16). The model was initially based on studies that monitor rebinding of fluorescent ATP to the active site vacated by translocation of misactivated valine to the CP1 domain editing site. Justification that the misactivated valine was indeed translocated to the editing site, rather than being simply released into solution, was based on observed changes in the fluorescence when CP1 domain mutants were studied (31). Further justification was offered by mutational studies in which CP1 domain mutants fully or partially disabled for post-transfer editing were found to be inactive in overall editing as well (16, 32). Finally, crystal structures of both IleRS and LeuRS showed binding of AA-AMP analogs to the CP1 domain at a site highly overlapping with that catalyzing post-transfer editing (14, 15). A conserved aspartate appeared central to anchoring both pre-transfer and post-transfer editing substrates. These x-ray structures extended the translocation model to LeuRS as well.
The sequence of molecular events proposed for the translocation model of pre-transfer editing is highly complex. In the model, an initial misacylation event is followed by post-transfer editing hydrolysis (priming step). This is proposed to induce a conformational change, facilitating shuttling of the noncognate AA-AMP from the synthetic site to the CP1-editing site in a tRNA-dependent manner (16, 32). Multiple pre-transfer editing hydrolytic events were suggested to be possible, although the enzyme is locked in a tRNA-dependent conformation facilitating shuttling (16).
Numerous criticisms of the translocation model have been noted (14, 24, 33). First, it appears that the cognate AA-AMP, after checking at the editing site, would have to return to its starting position for the transfer to tRNA to occur (33), an apparently highly inefficient process. Second, a biological rationale for evolution of such a complex mechanism is unclear, because released misacylated AA-AMPs are not toxic to the cell. Third, the AA-AMP analogs bind to the CP1 domain in strained conformations with very low affinities, suggesting artifactual interactions in the x-ray structures arising from the structural similarity with the aminoacyl linkage (14, 19). Fourth, the ability of a free-standing CP1 domain to hydrolyze AA-AMP has not been demonstrated. Fifth, no direct kinetic evidence for intramolecular translocation of the misactivated AA-AMP is available, because no assay isolating the first order mechanistic step corresponding to this protein-RNA rearrangement has been developed for any AARS. Finally, and perhaps most importantly, there is no evidence of an intramolecular tunnel in the tRNA-bound or unliganded crystal structures of either IleRS or LeuRS, by which the translocating AA-AMP could be kept sequestered from dissociating while en route from the synthetic site to the CP1 domain.
Another disputed issue in the field has been whether tRNA is required for pre-transfer editing to proceed. It has been asserted that pre-transfer editing is in general tRNA-dependent (34), and this perspective appears to have been an organizing principle in the many studies of IleRS. However, several early studies reported tRNA-independent editing in LeuRS, IleRS, and ValRS (4, 21, 35). Recent studies confirmed tRNA-independent editing in LeuRS (28, 29). tRNA-independent editing has also been documented in class II ProRS, SerRS, and ThrRS (23, 25, 26, 30).
To distinguish more clearly whether pre-transfer editing indeed requires tRNA as cofactor, and to provide a new perspective on the three persisting models for pre-transfer editing, we have studied the proofreading reactions of the canonical IleRS and ValRS using several novel assays originally developed in our studies of GlnRS (24). In the new approach, the fates of both [32P]AMP and AA-[32P]AMP in the synthesis and hydrolysis of AA-AMP are followed through the use of [α-32P]ATP. This enables direct tracking of AMP and AA-AMP after separation by TLC. In contrast, the widely used [γ-32P]ATP-based consumption assay previously applied to IleRS and ValRS does not monitor the pre-transfer editing reaction directly. We also introduced a cold-trapping methodology to distinguish enzyme-catalyzed from nonenzymatic hydrolysis in pre-transfer editing (24). These approaches were applied in the above-mentioned studies of SerRS, ProRS, and LeuRS that demonstrated pre-transfer editing in the respective synthetic sites of those enzymes (23, 25, 26, 28, 29).
Here, we apply these approaches to the canonical E. coli IleRS and ValRS enzymes. We demonstrate that both IleRS and ValRS exhibit tRNA-independent proofreading pathways, including 3% of the overall editing. In sharp contrast to the earlier findings, we also show that CP1 domain mutants of E. coli IleRS inactive in post-transfer editing exhibit kcat/Km values for tRNA-dependent editing that are highly similar to the wild-type (WT) enzyme. Thus, tRNA-dependent pre-transfer editing by IleRS appears not to occur in the CP1 domain. The mutational studies confirm and extend previous findings showing that IleRS depends critically on pre-transfer editing, whereas ValRS preferentially uses post-transfer hydrolysis of noncognate Thr-tRNAVal. To provide a rationale for this disparate behavior, which has not previously been understood, we also introduce a further single-turnover kinetic assay to monitor the rate of the tRNA transfer step in IleRS and ValRS. Remarkably, we find that the rate of tRNA transfer is inversely correlated with the occurrence of pre-transfer editing: fast transfer of amino acid to tRNA in ValRS is coincident with a lack of pre-transfer editing, whereas slow transfer in IleRS permits hydrolysis to occur before attack by the tRNA nucleophile. These data form the basis for a model of synthetic site pre-transfer editing that features kinetic partitioning of noncognate AA-AMP between hydrolysis and tRNA transfer. We suggest that this model may be general to all editing tRNA synthetases possessing spatially separate domains dedicated to post-transfer hydrolysis.
EXPERIMENTAL PROCEDURES
Enzymes and tRNAs
WT IleRS and ValRS, and their corresponding mutants, were overexpressed in E. coli BL21 (DE3). Expression vectors encoding genes for E. coli IleRS and ValRS were used as templates for site-directed mutagenesis using QuikChange (Stratagene). Mutations were confirmed by DNA sequencing of a fragment that includes the mutation site. During the growth of E. coli cells transformed with the IleRS overexpression vector, ZnCl2 was added to a final concentration of 1 mm to ensure formation of the proper zinc-bound IleRS conformation. Both IleRS and ValRS were purified by the same procedure employing affinity chromatography on Ni+-nitrilotriacetic acid resin and were eluted with a buffer containing 200 mm imidazole. The purity of both enzymes was estimated on SDS gels as greater than 98%.
Synthetic genes for E. coli tRNAGATIle (with G1–C72 instead of WT A1–U72 sequence) and tRNATACVal, under inducible T7 promoters, were inserted between the SalI and BamHI sites of the pET3a vector, upstream of the T7 RNA polymerase terminator site. Substitution of the first tRNAIle base pair was previously used to enhance transcription by T7 RNA polymerase, and it was shown not to affect isoleucylation parameters (36). Our observations are in agreement with that study. Overexpression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 14 h at 30 °C in E. coli BL21 (DE3). High molecular weight nucleic acids were removed by precipitation with 8% polyethylene glycol in the presence of 0.4 m NaCl. Plateau aminoacylation for the unfractionated tRNA showed that the samples possess 50% tRNAIle and 80% tRNAVal. An additional purification step, yielding tRNAIle capable of aminoacylation to a level of 90%, was performed by reverse phase chromatography on a semi-preparative C4 column (Vydac), as described previously (37).
AA-AMP Synthesis Assay
The AA-AMP synthesis assay was carried out at 37 °C in a buffer containing 50 mm Hepes (pH 7.5), 20 mm MgCl2, 5 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 0.004 units/μl inorganic pyrophosphatase, and 0.5 mm [α-32P]ATP (0.01–0.1 mCi/ml). Steady-state parameters for tRNA-independent pre-transfer editing by IleRS, ValRS, and their mutants were determined by varying concentrations of noncognate valine or threonine from 0.2 to 20 and from 0.5 to 75 mm, respectively. IleRS and ValRS (WT and mutants) were present at concentrations of 2 μm and 500 nm, respectively. Steady-state parameters for tRNA-dependent proofreading were determined by varying concentrations of valine or threonine over the range 0.2–20 times the Km value. WT IleRS and ValRS were present at concentrations of 50 and 10 nm, respectively, and their corresponding mutants were present at 50 and 500 nm, respectively. Concentration of tRNAIle was 8 μm, and tRNAVal was present at 10 μm.
Reactions were initiated by addition of amino acids and were stopped by quenching 1.5 μl of the reaction mixture in 3.0 μl of 1.5 m formic acid. 1.5 μl of this mixture was then spotted onto polyethyleneimine-cellulose plates (Fluka) prewashed in water. Separation of AA-[32P]AMP, [32P]AMP, and [32P]ATP was performed by TLC in 0.1 m ammonium acetate, 5% acetic acid, followed by quantitation by phosphorimaging and kinetic analysis as described previously (24). Initial velocities obtained by time course analyses were plotted against substrate concentration and were fit to the Michaelis-Menten equation. Km and kcat values were determined directly from these plots.
Nonenzymatic Hydrolysis of AA-AMP
The stability of Val-AMP and Thr-AMP in solution was assessed as described previously (24). The AA-AMPs were produced in reactions containing 50 mm Hepes (pH 7.5), 20 mm MgCl2, 5 mm dithiothreitol, 50 μm [32P]ATP, 5 μm ValRS, and 50 mm amino acid (Val or Thr). After allowing accumulation of enzymatically synthesized AA-[32P]AMP, unlabeled ATP was added in 250- or 2500-fold molar excess. The reaction time points were taken and quenched in formic acid (1 m final concentration) at ambient temperature. AA-AMP was then separated from AMP by TLC, and signals were quantified as described above.
Aminoacylation Assay
Aminoacylation reactions were performed in a buffer containing 20 mm Hepes (pH 7.5), 100 μm EDTA, 150 mm NH4Cl, 10 μg/ml bovine serum albumin, 10 mm MgCl2, 2 mm ATP at 37 °C. In all reactions, tRNA was present at 10 μm concentration; [14C]Ile was present at 30 μm, and [14C]Val was present at 100 μm. WT IleRS and its mutants were present at 20 nm; WT ValRS, D286A, and K277P/D286A were present at 5 nm, and ValRS K277P was present at 50 nm.
Misacylated Val-tRNAIle or Thr-tRNAVal was prepared by incubating 10 μm tRNA, 30 μm [14C]valine, or [14C]threonine (50 μCi μmol−1) and 5 μm of an IleRS or ValRS deacylation-defective mutant for 45 min at 37 °C in the same buffer used for aminoacylation reactions.
Deacylation Assay
Deacylation reactions were carried out at 37 °C in a mixture containing 100 mm Hepes (pH 7.5), 20 mm MgCl2, 5 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 5–8 μm misacylated [14C]tRNA, and various concentrations of enzyme to achieve steady-state conditions (5 nm-1 μm). Reactions were quenched in 10% trichloroacetic acid, spotted on filter papers (Whatman), washed, and quantified.
Transfer Step by Chemical Quench-flow Kinetics
The transfer step was measured using AA-AMP that was preformed in situ (as described in Ref. 38) on AARS, by incubation of 20 μm IleRS D342A with 0.5 mm Ile or 5 mm Val or 40 μm ValRS D286A with 5 mm Val or 5 mm Thr, in each case in buffer containing 10 mm ATP, 20 mm Hepes (pH 7.5), 100 μm EDTA, 150 mm NH4Cl, 10 μg/ml bovine serum albumin, 10 mm MgCl2, 0.008 unit/μl inorganic pyrophosphatase at 37 °C for 30 min. Rapid quench assays were done on a KinTek RQF-3 instrument, by rapid mixing equal volumes of AARS:AA-AMP incubated in one syringe with 2 μm 32P-labeled tRNA incubated in the second syringe. Radiolabeling of the 3′ terminus of tRNA was performed using tRNA nucleotidyltransferase to exchange the endogenous A76 of tRNA with [α-32P]ATP as described previously (39, 40). Reactions were stopped with sodium acetate (pH 5.0) (final concentration of 0.4 m), and collection tubes contained SDS (final concentration of 0.1%). tRNA was degraded using P1 nuclease, and AA-AMP was separated from AMP by TLC (40). The ratio of AA-AMP to AMP is equivalent to the ratio of aminoacylated versus nonaminoacylated tRNA in the reaction. TLC plates were developed in a solution containing 100 mm ammonium acetate, 5% acetic acid. The amount of aminoacylated tRNA was plotted versus time and fit to the first order exponential equation y = Y0 + A × e−ktrans × t, where Y0 is the y intercept; A is a scaling constant; ktrans is the apparent transfer rate constant, and t is time.
RESULTS
tRNA-independent Pre-transfer Editing by Class I IleRS and ValRS
We employed a steady-state AA-AMP synthesis assay in the absence of tRNA to examine whether E. coli IleRS and ValRS are able to catalyze tRNA-independent proofreading of valine and threonine, respectively. In this assay, the generation of AMP is indicative of both enzyme-catalyzed and nonenzymatic hydrolysis. Control experiments established that neither enzyme possesses significant inherent ATPase activity, that AMP formation is not stimulated in the presence of cognate amino acids, and that insignificant levels of endogenous tRNA and AARS were present in the enzyme preparations. To exclude the possibility that tRNA-independent ATPase activity might be due to trace tRNA impurities in the enzyme preparations, RNase A was included in the AA-AMP synthesis assay. We found that the measured reaction velocities were identical.
Special controls were also taken concerning possible contamination with endogenous AARSs. If endogenous IleRS and/or ValRS were copurified with overexpressed IleRS and ValRS, both preparations should have the same amount of endogenous enzymes because both were obtained by elution with 200 mm imidazole, and approximately the same level of overexpression was achieved in each case. To test the possible presence of endogenous IleRS in the protein preparation, purified ValRS was used to measure Ile-tRNAIle formation in the presence of Ile and tRNAIle. Similarly, the presence of endogenous ValRS was tested by incubating an IleRS sample with Val and tRNAVal. In both cases, no formation of Ile-tRNAIle or Val-tRNAVal was observed using 400 nm concentrations of ValRS and IleRS, respectively. Similarly, no formation of Ile-tRNAIle was observed in preparations of ValRS D286A.
For both IleRS and ValRS, we find that AMP production in the presence of the noncognate amino acid is significantly enhanced compared with reactions containing the cognate amino acid or with control reactions lacking amino acid or enzyme (Fig. 2, A and B). Thus, in contrast to the prevailing view for IleRS (34), it is clear that this enzyme possesses readily detectable tRNA-independent pre-transfer editing activity. We also performed steady-state reactions to extract Michaelis parameters for comparison with mutant enzymes (see below). Comparative parameters for cognate reactions could not be accurately determined by this assay, because the cognate AA-AMP is stably bound, and its slow dissociation limits product accumulation.
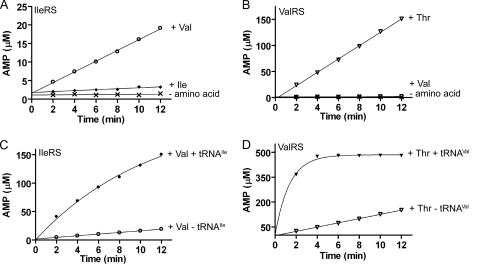
FIGURE 2.
Pre-transfer and overall editing by WT IleRS and WT ValRS. A, AMP formation by 500 nm WT IleRS in the presence of 20 mm valine and lacking tRNAIle (○). Control reactions were performed with 0.5 mm isoleucine (♦) and in the absence of amino acid (×). B, AMP formation by 500 nm WT ValRS in the presence of 80 mm threonine and lacking tRNAVal (▿). Control reactions were performed with 6 mm valine (□) and in the absence of amino acid (×). C, AMP formation by 500 nm WT IleRS with 20 mm valine and in the presence (●) or absence (○) of 8 μm tRNAIle. D, AMP formation by 500 nm WT ValRS with 80 mm threonine and in the presence (▾) or absence (▿) of 10 μm tRNAVal. C and D, high concentrations of enzymes are used to most clearly depict the effect of tRNA. Enzyme concentrations giving linear product accumulation are used to determine kcat and Km values (see “Experimental Procedures”).
Although both IleRS and ValRS steady-state reactions showed a linear increase in AMP production with time, linear accumulation of noncognate AA-AMP was observed only in IleRS reactions (see supplemental Fig. 1; supplemental Table 1). Comparison of turnover numbers for Val-AMP and AMP formation revealed that IleRS synthesizes Val-AMP 6-fold more slowly than AMP (0.009 versus 0.053 s−1, respectively; Table 1 and supplemental Table 1), suggesting enzymatic hydrolysis as a major AMP production pathway. To establish that the production of AMP is truly enzyme-catalyzed, we independently measured the nonenzymatic hydrolysis of Val-AMP and Thr-AMP in solution, under identical conditions. Reaction mixtures containing enzyme, ATP, and noncognate amino acid were incubated for 10 min to accumulate noncognate AA-AMP, as described previously (24). A large molar excess of unlabeled ATP was then added, and reaction time points were taken. These data (Table 2) reveal that nonenzymatic hydrolysis of Val-AMP and Thr-AMP is 30- and 300-fold slower than the rate of AMP formation by IleRS and ValRS, respectively, and therefore, it does not significantly contribute to the observed AMP signal. Thus, AMP formation represents a tRNA-independent intrinsic IleRS and ValRS hydrolytic activity for noncognate AA-AMP.
TABLE 1.
Steady-state parameters for AMP formation by IleRS and ValRS
The values represent the best fit value ± S.E. of at least three independent experiments.
| −tRNAa |
+tRNAb |
|||||
|---|---|---|---|---|---|---|
| Km (AA) | kcat | kcat/Km | Km (AA) | kcat | kcat/Km | |
| mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | |
| IleRS + Val | ||||||
| WT IleRS | 1.9 ± 0.3 | 0.053 ± 0.002 | 0.03 | 4.4 ± 0.9 | 1.56 ± 0.08 | 0.35 |
| IleRS T243R | 1.2 ± 0.2 | 0.063 ± 0.003 | 0.05 | 0.7 ± 0.1 | 1.04 ± 0.03 | 1.49 |
| IleRS D342A | 1.3 ± 0.2 | 0.038 ± 0.001 | 0.03 | 2.4 ± 0.5 | 0.48 ± 0.04 | 0.20 |
| IleRS T243R/D342A | 0.7 ± 0.1 | 0.043 ± 0.001 | 0.06 | 0.6 ± 0.2 | 0.29 ± 0.02 | 0.48 |
| ValRS + Thr | ||||||
| WT ValRS | 8.9 ± 2.4 | 0.37 ± 0.03 | 0.04 | 9.4 ± 1.0 | 12.9 ± 0.4 | 1.37 |
| ValRS K277P | 15.4 ± 3.0 | 0.28 ± 0.02 | 0.02 | 13.4 ± 2.2 | 0.34 ± 0.02 | 0.03 |
| ValRS D286A | 9.7 ± 1.8 | 0.28 ± 0.02 | 0.03 | 5.8 ± 1.2 | 0.28 ± 0.02 | 0.05 |
| ValRS K277P/D286A | 12.5 ± 2.0 | 0.48 ± 0.03 | 0.04 | 10.7 ± 1.8 | 0.48 ± 0.02 | 0.05 |
a WT and mutant IleRS were used at 2 μm concentration, and WT and mutant ValRS were used at 500 nm.
b WT and mutant IleRS were used at 50 nm concentration; WT ValRS was used at 10 nm, and mutant ValRS was used at 50 nm.
TABLE 2.
Steady-state rates for nonenzymatic hydrolysis
The values represent the mean ± S.D. of at least three independent experiments.
| kobs | |
|---|---|
| s−1 | |
| Val-AMP | (1.9 ± 0.7) × 10−3 |
| Thr-AMP | (1.2 ± 0.6) × 10−3 |
We speculate that tRNA-independent editing has often been unobserved because the previously often-employed assay (41), which relies on separation by filter adsorption, may be less sensitive and more error-prone than the TLC-based separation used here. This conjecture is supported by early reports of tRNA-independent editing activity by E. coli IleRS and ValRS toward cysteine (21) and by yellow lupine seed ValRS toward both cysteine and threonine (35), in each case using a TLC-based assay.
Location of the tRNA-independent Activity
The CP1 editing domains of IleRS and ValRS were mutated at several previously identified, conserved positions (16, 17, 32); the mutant enzymes were purified, and their activities in post-transfer editing were tested using a steady-state deacylation assay, with high concentrations (up to 8 μm) of preformed misacylated tRNA (Table 3). Deacylation-defective IleRS and ValRS mutants were used to prepare tRNAs misacylated to 80–90% levels. Both ValRS (D286A, K277P, and K277P/D286A) and IleRS (D342A and T243R/D342A) mutants were unable to deacylate misacylated tRNA even at high enzyme concentrations (up to 1 μm), demonstrating inactivation of the CP1 post-transfer editing site. These findings confirm prior observations for the ValRS D286A and IleRS D342A mutants (16, 17). All mutants possessed aminoacylation rates within 2-fold of the WT enzymes with the exception of ValRS K277P, which was reduced by 10-fold (Table 4).
TABLE 3.
Steady-state deacylation by IleRS and ValRS
The values represent the mean ± S.D. of at least three independent experiments.
| kobs | |
|---|---|
| s−1 | |
| IleRS + Val-tRNAIle | |
| WT IleRSa | 0.22 ± 0.04 |
| IleRS T243Ra | 0.093 ± 0.004 |
| IleRS D342Ab | 0.009 ± 0.003 |
| IleRS T243R/D342Ab | c |
| ValRS + Thr-tRNAVal | |
| WT ValRSd | 6.4 ± 0.09 |
| ValRS K277Pb | c |
| ValRS D286Ab | c |
| ValRS K277P/D286Ab | c |
a WT IleRS and the IleRS T243R mutant were each present at 100 nm concentration.
b IleRS D342A and T243R/D342A, and all ValRS mutants, were each present at 1 μm concentration.
c Activity was too low for reliable detection.
d WT ValRS was present at 5 nm concentration.
TABLE 4.
Steady-state rates for cognate aminoacylation by WT and mutant IleRS and ValRS
All substrate concentrations are saturated and therefore kobs approaches kcat. The values represent the mean ± S.D. of at least three independent experiments.
| kobs | |
|---|---|
| s−1 | |
| IleRS | |
| WT IleRSa | 0.72 ± 0.22 |
| IleRS T243Ra | 0.75 ± 0.26 |
| IleRS D342Aa | 0.55 ± 0.27 |
| IleRS T243R/D342Aa | 0.33 ± 0.14 |
| ValRS | |
| WT ValRSb | 5.15 ± 1.14 |
| ValRS K277Pc | 0.31 ± 0.09 |
| ValRS D286Ab | 2.87 ± 0.98 |
| ValRS K277P/D286Ab | 2.27 ± 0.33 |
a WT IleRS and its mutants were present at 20 nm.
b WT ValRS, D286A, and K277P/D286A were present at 5 nm.
c ValRS K277P was present at 50 nm, and it was assayed in the same conditions as the other enzymes, only concentration of Mg2+ was lowered to equal the concentration of ATP (2 mm).
tRNA-independent proofreading by mutant enzymes was measured as described above for the WT enzymes. Comparison of the kcat and Km values for AMP formation among WT and mutant enzymes revealed no significant differences in tRNA-independent pre-transfer editing by either IleRS or ValRS (Table 1). The steady-state kinetic constants for Val-AMP formation by IleRS mutants were also very similar to the WT enzyme (supplemental Table 1). These data demonstrate that tRNA-independent pre-transfer editing is unaffected by mutations within the CP1-editing site. Crystallographic analysis had suggested that the fully conserved aspartate (Asp-342 in IleRS and Asp-286 in ValRS) anchors both pre- and post-transfer editing substrates for hydrolytic attack within the CP1 domain (14, 15, 17). Insensitivity of the pre-transfer editing rate to mutation of this key residue strongly suggests that tRNA-independent pre-transfer editing by IleRS and ValRS does not occur in the CP1 post-transfer editing domain. Because the translocation model (16) presupposes the presence of tRNA to facilitate shuttling of noncognate AA-AMP from the synthetic to the editing site, this demonstration of tRNA-independent editing also shows that the model cannot account for all pre-transfer editing by either IleRS or ValRS.
tRNA-dependent Editing by IleRS and ValRS
The presence of aminoacylable tRNA in the AA-AMP synthesis assay enables the use of this approach to measure overall editing, because misacylation and post-transfer editing then occur in parallel with pre-transfer editing. The AMP generated will originate from both the pre- and/or post-transfer editing steps, and it is not possible to distinguish these processes in the WT enzymes. We find that the presence of tRNAIle or tRNAVal generates substantial increases in IleRS- and ValRS-stimulated AMP production relative to conditions deprived of tRNA (Fig. 2, C and D, and Table 1), in agreement with previous work (3, 42). At the level of kcat/Km, the presence of tRNA increases AMP synthesis rates by 12-fold for IleRS and by 30-fold for ValRS (Table 1).
Mutational analysis reveals significant differences in the mechanism of tRNA-dependent proofreading by IleRS and ValRS. Cognate tRNAIle strongly stimulates AMP production by deacylation-defective IleRS mutants (D342A and T243R/D342A); kcat values in the presence of tRNA are 12- and 7-fold higher than in its absence (0.48 s−1 versus 0.038 s−1 and 0.29 s−1 versus 0.043 s−1, respectively; see Table 1). This represents the first direct experimental evidence of a tRNA-dependent pre-transfer pathway in IleRS, because earlier work inferred pre-transfer editing as the major IleRS pathway based solely on kinetic modeling of the WT enzyme reactions (2). The magnitude of stimulation by tRNAIle in these mutants parallels the stimulation observed in WT IleRS. Thus, as observed for tRNA-independent editing, tRNA-dependent pre-transfer editing by IleRS is also not greatly affected by mutations in the CP1 post-transfer editing site.
In sharp contrast, tRNAVal did not stimulate an increase in AMP production by deacylation-defective ValRS mutants (Table 1). Considered independently, these data suggest either that ValRS lacks measurable tRNA-dependent pre-transfer editing activity or that this activity occurs within the CP1 domain. However, early transient kinetic studies clearly demonstrated post-transfer editing as a major pathway for ValRS (3). Our data now suggest further that tRNA-dependent pre-transfer editing makes no measurable contribution to error correction in this enzyme. It is informative to view these findings in light of recent data showing that E. coli LeuRS possesses efficient tRNA-dependent pre-transfer editing (28). The absence of pre-transfer editing in ValRS shows that it employs a significantly different proofreading mechanism as compared with IleRS and LeuRS, which share the common property of using both activities to exclude noncognate aminoacyl-tRNA formation.
Robust tRNA-dependent Pre-transfer Editing in IleRS CP1 Domain Mutants
Although the IleRS D342A and T243R/D342A mutants retain WT kcat/Km values for tRNA-dependent pre-transfer editing, the individual kcat and Km values for the mutants are each decreased (Table 1). It appears then that key residues in the IleRS CP1 domain, although dispensable for high levels of tRNA-dependent pre-transfer editing, nonetheless are capable of modulating the activity. We suggest that such modulation arises because, in the enzyme-tRNA complex, the nonaminoacylated 3′-acceptor end is able to access both the synthetic and editing sites. Thus, mutants in CP1 may modulate this equilibrium and produce modest compensating differences in the Michaelis parameters. However, because the kcat/Km value is insensitive to the presence of the key Asp-342 residue in CP1, and because D342A is also fully inactive in post-transfer editing (Table 3), it is highly unlikely that the robust tRNA-dependent pre-transfer activity resides in CP1. Thus, these data provide evidence against the translocation model for tRNA-dependent pre-transfer editing by IleRS.
By contrast, earlier work on IleRS D342A reported lack of nearly all editing activity, and this was taken as evidence for CP1-based pre-transfer editing (16, 17). Although a simple explanation for the observed discrepancy is not evident, it is possible that the prior use of an indirect assay monitoring 32PPi formation after adsorption of [γ-32P]ATP on activated charcoal may have contributed to it (15, 16, 41). The insensitivity of that assay is also evident based on its failure to detect tRNA-independent editing, as described above. In contrast, our approach measures pre-transfer editing directly. Confidence in our measurements also arises from the rigorous determination of Michaelis parameters for all WT and mutant pre-transfer editing reactions (Table 1). In contrast, the previous mutational experiments drew conclusions based entirely on the qualitative comparison of reaction time courses. Our work highlights the hazards of relying on such data and underscores the importance of rigorous determination of kinetic constants as a basis for proposing detailed models of enzyme function.
IleRS T243R Mutant
As described above for the IleRS CP1 domain mutants D342A and T243R/D342A, the IleRS T243R single mutant also retains tRNA-independent editing at a level identical to the WT enzyme (Table 1). Furthermore, T243R also exhibits similar stimulation of AMP production by cognate tRNAIle; kcat in the presence of tRNA is 16-fold higher than kcat in its absence (1.04 s−1 versus 0.063 s−1; see Table 1). However, unlike D342A and T243R/D342A, this mutant IleRS retains 40% of the WT post-transfer editing activity (Table 3). Thus, AMP formation by T243R in the presence of tRNA includes contributions from both pathways. Previously, it was reported that T243R was substantially decreased in overall editing while retaining WT levels of post-transfer activity (32); based on this, it was suggested that Thr-243 is a key component of a CP1-based pre-transfer editing site. However, we find only a very modest drop in overall editing (kcat 1.04 s−1 for T243R versus 1.56 s−1 for WT IleRS; see Table 1) in the context of a small (2.5-fold) deficit in post-transfer editing. Clearly, T243R retains highly robust tRNA-dependent pre-transfer editing activity. Our data again do not support a direct catalytic role of the CP1 domain in noncognate AA-AMP hydrolysis.
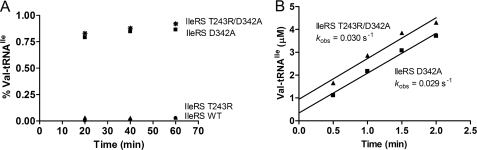
We also note that IleRS D342A and IleRS T243R/D342A accumulate Val-tRNAIle to levels identical to Ile-tRNAIle accumulation by WT IleRS, albeit with lower catalytic rate constants (Fig. 3). In contrast, IleRS T243R, which retains post-transfer editing, does not significantly accumulate Val-tRNAIle (Fig. 3). Clearly, IleRS requires both pathways to achieve high accuracy in Ile-tRNAIle formation. A similar finding was recently reported for LeuRS (28).
FIGURE 3.
A, plateau aminoacylation of tRNAIle with Val using IleRS WT (●), T243R (▴), D342A (■), and T243R/D342A (*). tRNA was 5 μm; [14C]Val was 100 μm, and enzymes were 5 μm. B, steady-state aminoacylation of tRNAIle with Val using IleRS D342A (■) and T243R/D342A (▴). tRNA was 10 μm; [14C]Val was 100 μm, and enzymes were 1 μm.
Transfer of Noncognate Amino Acid to Cognate tRNA
To examine whether the rate of noncognate aminoacyl transfer may influence partitioning of tRNA-dependent editing between the pre- and post-transfer pathways, rapid chemical quench experiments were performed using the deacylation- defective ValRS D286A and IleRS D342A mutants. Single turnover experiments were done by rapidly forming AARS:AA-AMP (20–40 μm) in situ and then immediately adding 32P-labeled cognate tRNA (2 μm). Because the rates of the ATP/PPi exchange reactions for noncognate substrates are very high (supplemental Table 2), AA-AMP will be rapidly reformed (from ATP and amino acid) after pre-transfer hydrolysis. Therefore, this approach permits measuring the rate constant for transfer of the amino acid to tRNA (ktrans; representing either the chemical reaction or a closely linked conformational change (43)) in the context of simultaneous pre-transfer editing.
ValRS D286A rapidly transfers both valine and threonine to tRNAVal (39 and 55 s−1, respectively; see Fig. 4B) although transfer of isoleucine and valine to tRNAIle by IleRS D342A occurs much more slowly (0.7 and 0.4 s−1, respectively; Fig. 4A). Transfer rate constants that we measured toward cognate substrates are very similar for the WT enzymes (46 ± 4 s−1 for ValRS and 1.1 ± 0.1 s−1 for IleRS) and correspond to previously published values (38, 44). For IleRS and ValRS, however, the extent to which pre-transfer editing is utilized is inversely related to the rate of the transfer step; slow transfer by IleRS permits kinetic partitioning toward Val-AMP hydrolysis. In contrast, ValRS transfers threonine rapidly to tRNAVal, and therefore, it must rely on post-transfer editing to ensure fidelity.
FIGURE 4.
Single turnover aminoacyl transfer of cognate and noncognate amino acids by IleRS D342A and ValRS D286A. A, transfer of Ile (■) and Val (▴) by IleRS D342A. tRNAIle was present at 1 μm, and IleRS D342A:AA-AMP was present at 10 μm. B, transfer of Val (■) and Thr (▴) by ValRS D286A. tRNAVal was present at 1 μm and ValRS D286A:AA-AMP was present at 20 μm.
DISCUSSION
Pre-transfer Editing in the Synthetic Active Site of IleRS
We have evaluated the three models for pre-transfer editing hydrolysis by a set of kinetic experiments on the canonical E. coli IleRS enzyme. Addition of excess unlabeled ATP to a tRNA-independent editing reaction has shown that the rate of solution hydrolysis is much too low to account for the observed rate of AMP accumulation (Table 2). This eliminates the selective release mechanism. Next, comparison of kcat/Km values between WT enzymes and their corresponding deacylation-defective mutants shows that inactivation of the CP1 hydrolytic site does not affect the efficiency of pre-transfer editing (Table 1). This provides strong evidence against the translocation model. Finally, transient kinetics was used to demonstrate that rate constants for tRNA-dependent pre-transfer editing are inversely correlated with that for transfer to tRNA. This provides the basis for a synthetic site model, in which kinetic competition between water and the tRNA A76 2′-hydroxyl group determines the balance between pre-transfer and post-transfer editing.
The exclusion of the translocation model in IleRS, however, is not fully definitive because the formal possibility exists that a different and so far unrecognized hydrolytic site exists elsewhere on the CP1 domain. We consider this possibility to be very unlikely, for two reasons. First, structure-based sequence alignments of the IleRS, ValRS, and LeuRS CP1 domains and inspection of the crystal structures does not reveal an evident alternative active site. Such a site has never been proposed; indeed, the crystallographic data relied on by the translocation model indicated highly overlapping binding sites for pre-transfer and post-transfer analogs (14, 15). Second, deletion of the entire E. coli LeuRS CP1 domain does not eliminate tRNA-dependent editing of Ile-AMP (27). Therefore, for this homologous enzyme that also utilizes both pre-transfer and post-transfer editing, and for which deacylation-defective CP1 domain point mutants have also been shown to hydrolyze noncognate AA-AMP using the direct assay employed here (28), exclusion of the translocation model appears to be nearly certain.
An important general implication of synthetic site pre-transfer editing is that lack of a distinct editing domain does not necessarily imply that a tRNA synthetase has no capacity to hydrolyze noncognate AA-AMP. The clear examples are yeast SerRS and Methanococcus jannaschii and human ProRS. Each of these class II enzymes that naturally lacks an editing domain has been shown to possess tRNA-independent pre-transfer editing (25, 26). Another possible example is the mitochondrial LeuRS enzyme that possesses significant sequence differences in its editing domain (45). The apparent absence of editing in this case might be in part rationalized based on a less stringent need for highly specific coding in the limited mitochondrial proteome. However, a synthetic site pre-transfer activity, even of limited efficiency, would offer improved fidelity despite the absence of post-transfer hydrolysis. Possibly, the kinetic approaches described here might detect and quantitate such activity even though it was not clearly evident in ATP consumption assays (45). In general, more detailed kinetic studies of editing enzymes in distinct cell compartments is of interest given the recent demonstration of cell-specific differences in translational quality control (46).
We also demonstrated a robust tRNA-independent pre-transfer editing activity in the context of deacylation-defective IleRS mutants (Table 1). These data further support the synthetic site model and also demonstrate that pre-transfer editing does not require a priming post-transfer hydrolytic event, as demanded by the translocation model. tRNA-independent pre-transfer editing has been recently assigned to the synthetic active site of class I LeuRSs from Aquifex aeolicus and E. coli and class II ProRS and ThrRS from E. coli (26, 28, 30). Therefore, as with synthetic site tRNA-dependent pre-transfer editing, a weaker tRNA-independent activity also may be a general feature of many of the editing AARS.
Kinetic Partitioning of AA-AMP within the Synthetic Site Determines the Balance between Pre- and Post-transfer Editing Pathways
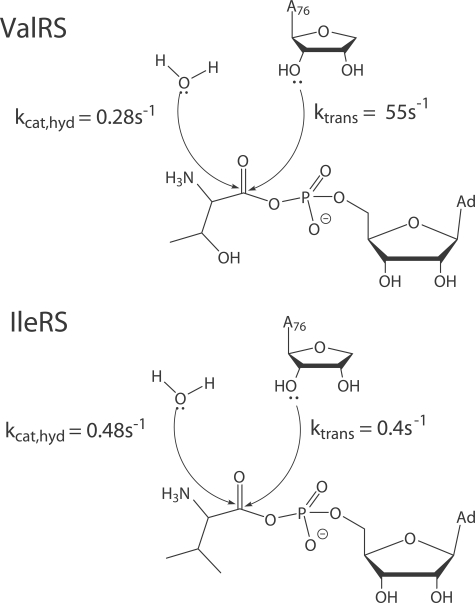
Our experimental data provide the basis for a new model for how editing is partitioned between pre-transfer and post-transfer reactions (Fig. 5). In both IleRS and ValRS, noncognate AA-AMP is formed rapidly, with kcat for ATP-PPi exchange similar to that of the cognate reaction (supplemental Table 2). During binding and/or activation of the noncognate substrate, the presumed decreased rigidity of the enzyme-substrate complex would allow penetration of a hydrolytic water molecule that is otherwise excluded (24). This provides the basis for synthetic site pre-transfer hydrolysis. However, this reaction occurs in competition with transfer to tRNA. Kinetic partitioning of the noncognate intermediate thus occurs, as the tRNA A76 2′-ribose hydroxyl competes with water for nucleophilic attack on the carbonyl carbon of the adenylate mixed anhydride.
FIGURE 5.
Active site partitioning of noncognate AA-AMP within the synthetic site. Fast transfer of threonine to tRNAVal predominates in the synthetic site of ValRS. In contrast, water competes efficiently with the tRNA for nucleophilic attack on carbonyl carbon atom of Val-AMP in IleRS.
Our pre-steady state data show dramatic differences in the behavior of IleRS and ValRS, which are consistent with the mutational studies described above and with earlier work that also drew a clear distinction between the two enzymes (2, 3). Fast transfer of threonine to tRNAVal does not allow significant partitioning of Thr-AMP toward hydrolysis; there is little to no pre-transfer editing in ValRS. Thus, ValRS relies almost entirely on discrimination after transfer, i.e. on post-transfer editing in the CP1 domain. In contrast, water can compete more efficiently with the tRNA transfer step in IleRS, thereby increasing the contribution of pre-transfer editing in that enzyme. In support of this model, comparison of kcat for tRNA-dependent hydrolysis by deacylation-defective IleRS mutants (Table 1), with the microscopic rate constant (ktrans; Fig. 4) for the transfer of amino acid to tRNA, shows that both processes occur on a similar time scale. Misacylated Val-tRNAIle is still synthesized at a significant rate, requiring a further post-transfer editing step as well.
The significance of these pre-steady state measurements is that they offer a clear explanation for the different reliance of IleRS and ValRS on the two editing reactions. It is now evident that the essential difference between the two enzymes is the extremely fast transfer rate constant in ValRS, as compared with the slow transfer in IleRS. Comparisons with the transfer rates in other tRNA synthetases show that the behavior of IleRS is most unusual, because fast tRNA transfer is commonly observed among AARS whether or not they also possess editing activities (44, 47, 48).
Similar observations have been made for editing of serine by E. coli ThrRS (30). This class II enzyme edits Ser-AMP in a tRNA-independent manner within the synthetic catalytic core. In the presence of tRNAThr, fast transfer and efficient post-transfer editing within the N-terminal editing domain takes place. Under these conditions, pre-transfer editing of serine is negligible. However, slowing of the transfer step by a designed mutation increases the relative contribution of pre-transfer editing, establishing an inverse correlation between rates of aminoacyl transfer and pre-transfer editing, as we have observed by comparing IleRS with ValRS. Thus, kinetic partitioning of AA-AMP within the synthetic site may determine the balance between pre- and post-transfer editing in AARS of both classes.
Stimulation of Synthetic Site Pre-transfer Editing by tRNA
The structural origins of tRNA-dependent stimulation of pre-transfer editing remain as an important area for future research. One possibility is that the presence of the A76 ribose in the vicinity of the adenylate increases ordering of the active site solvent structure to better stabilize a water nucleophile. In ValRS, the very fast transfer of amino acid to tRNA may imply that the A76 2′-OH group is well positioned for in-line attack, although the roughly 100-fold lower transfer rate constant in IleRS indicates that the reactive moieties may be juxtaposed less precisely. However, because the tRNA 3′-end is likely sampling different conformations and making interactions with protein in both active sites, acceleration of pre-transfer editing might also arise from a tRNA-induced change in the conformation of the synthetic active site. It has been suggested that initial tRNA binding positions the 3′-CCA terminus in the editing site of class I editing enzymes (49, 50). If this is the case, tRNA enhancement of synthetic site pre-transfer editing may indeed occur without binding adjacent to the AA-AMP. The observed large difference in ktrans between IleRS and ValRS (Fig. 4) might then represent different translocation rates of the respective nonaminoacylated tRNA 3′-ends from editing to synthetic sites, rather than the rate of the chemical step directly. A variety of approaches, including mutational analysis of synthetic site amino acids, development of an approach to directly measure the first-order translocations of the tRNA 3′-end in either direction, structural studies of the enzymes with tRNA bound in the synthetic active site, and the use of burst kinetics to follow the first round of editing (30),3 will likely be essential to shed further light on this important question.
More detailed kinetic studies will also be needed to quantitate precisely how total editing is partitioned between synthetic site pre-transfer and editing site post-transfer reactions. It was suggested for LeuRS that such partitioning might be adequately determined by comparing observed steady-state rate constants (kobs) for AMP formation in the WT enzyme and in a mutant incapable of post-transfer editing (29). However, because the tRNA participates in both pathways and samples both active sites, any mutation at either site could have pleiotropic effects. Indeed, our steady-state data show that mutations in the IleRS CP1 domain influence both Km and kcat values for tRNA-dependent Val-AMP hydrolysis. We suggest that measurement of the mechanistic constant for AA-AMP hydrolysis by pre-steady state kinetics, and comparison of this first-order constant with that for tRNA transfer, may provide the sought for quantitative measure of partitioning between the two pathways. Comparison of the kcat,hyd with kcat is unsuitable for this purpose (Fig. 5), because the measured kcat, hyd represents only a lower limit on the true value of khyd.
Distinct Mechanisms to Achieve Specificity in tRNA Synthetases and DNA Polymerases
DNA polymerases also employ hydrolytic proofreading within a distant editing domain, in a reaction that parallels post-transfer editing by AARS. However, significant contrasts between DNA polymerases and AARSs are observed during processing of the noncognate substrates within the synthetic active sites (49, 51). In polymerases, initial formation of an enzyme-DNA complex with an incoming noncognate dNTP is followed by rapid reversal of the induced fit conformational change needed to fully assemble the active site. A noncognate dNTP is also misaligned with respect to catalytic residues; formation of the phosphodiester bond proceeds 1000-fold more slowly than with the cognate dNTP. Together, the slow chemical step and reversible conformational change greatly enhance dissociation of the noncognate dNTP. It was further suggested that this utilization of induced fit (during the mismatch recognition) to slow catalysis and promote noncognate substrate release may be widely used to increase selectivity (51). However, our data for class I AARS clearly show that cognate and noncognate amino acids are each transferred to tRNA with similar rates under saturating conditions. Thus, misalignment of reactive groups in the chemical step is not a component of amino acid selectivity by ValRS and IleRS.
Absence of specificity at the tRNA transfer step appears not to be confined to class I AARS, because the class II ThrRS transfers noncognate serine and cognate threonine to tRNAThr with similar rates (30). Establishing the extent to which this phenomenon may be general to all editing AARS will require similar measurements on other enzymes.
The double-sieve model for editing AARS proposes that the synthetic site functions to reject larger amino acids on steric grounds, although a second editing site excludes cognate amino acids based on size and hydrophobicity (52). It appears that this classic model, which relies on rather rigid pre-formed sieves, must be expanded further to account for a distinct editing activity that is assembled in the synthetic site during the recognition process. Thus, the first “coarse” sieve not only excludes larger noncognate amino acids but also functions to hydrolyze the AA-AMP formed with smaller amino acids. Cellular requirements to operate the synthetic site at a sufficiently high rate to maintain protein synthesis may have limited the efficiency of this pre-transfer editing sieve. This could have provided an evolutionary driving force for the addition of the separate post-transfer editing domain to a primordial editing tRNA synthetase that may have carried out proofreading using synthetic site pre-transfer hydrolysis alone.
Supplementary Material
Acknowledgments
We thank Ya-Ming Hou (Thomas Jefferson University, Philadelphia) for the IleRS and ValRS overexpression plasmids and Ivana Weygand-Durasevic (University of Zagreb, Croatia) for access to research facilities. We are indebted to Boris Lenhard for critical reading of the manuscript. I. G.-S. attended the EMBO Practical Course: Transient Kinetic Methods Applied to Biological Macromolecules (UK, 2008), and this training helped her in developing transient kinetic methods at the University of Zagreb, Croatia.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R03TW008024 (to J. J. P. and I. G.-S.) and GM63713 (to J. J. P.). This work was also supported by the Ministry of Science, Education, and Sports of the Republic of Croatia Grant 119-0982913-1358 (to Ivana Weygand-Durasevic).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
M. Dulic, N. Cvetesic, J. Perona, and I. Gruic-Sovulj, unpublished results.
- AARS
- aminoacyl-tRNA synthetase
- AA-AMP
- aminoacyl-adenylate
- IleRS
- isoleucyl-tRNA synthetase
- ValRS
- valyl-tRNA synthetase
- LeuRS
- leucyl-tRNA synthetase
- SerRS
- seryl-tRNA synthetase
- ProRS
- prolyl-tRNA synthetase
- ThrRS
- threonyl-tRNA synthetase
- WT
- wild type.
REFERENCES
- 1.Ibba M., Soll D. (2000) Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2.Fersht A. R. (1977) Biochemistry 16, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 3.Fersht A. R., Kaethner M. M. (1976) Biochemistry 15, 3342–3346 [DOI] [PubMed] [Google Scholar]
- 4.Englisch S., Englisch U., von der Haar F., Cramer F. (1986) Nucleic Acids Res. 14, 7529–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling J., Reynolds N., Ibba M. (2009) Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 6.Silvian L. F., Wang J., Steitz T. A. (1999) Science 285, 1074–1077 [PubMed] [Google Scholar]
- 7.O'Donoghue P., Luthey-Schulten Z. (2003) Microbiol. Mol. Biol. Rev. 67, 550–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. (1998) Science 280, 578–582 [DOI] [PubMed] [Google Scholar]
- 9.Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. (2000) Cell 103, 793–803 [DOI] [PubMed] [Google Scholar]
- 10.Cusack S., Yaremchuk A., Tukalo M. (2000) EMBO J. 19, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin L., Hale S. P., Schimmel P. (1996) Nature 384, 33–34 [DOI] [PubMed] [Google Scholar]
- 12.Zhao M. W., Zhu B., Hao R., Xu M. G., Eriani G., Wang E. D. (2005) EMBO J. 24, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betha A. K., Williams A. M., Martinis S. A. (2007) Biochemistry 46, 6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 15.Fukunaga R., Yokoyama S. (2006) J. Mol. Biol. 359, 901–912 [DOI] [PubMed] [Google Scholar]
- 16.Bishop A. C., Nomanbhoy T. K., Schimmel P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukunaga R., Yokoyama S. (2005) J. Biol. Chem. 280, 29937–29945 [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson T. L., Nomanbhoy T. K., Schimmel P. (2000) Biochemistry 39, 8180–8186 [DOI] [PubMed] [Google Scholar]
- 19.Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 20.Fukunaga R., Yokoyama S. (2005) Nat. Struct. Mol. Biol. 12, 915–922 [DOI] [PubMed] [Google Scholar]
- 21.Jakubowski H., Fersht A. R. (1981) Nucleic Acids Res. 9, 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopfield J. J. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., Musier-Forsyth K. (2006) J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 24.Gruic-Sovulj I., Uter N., Bullock T., Perona J. J. (2005) J. Biol. Chem. 280, 23978–23986 [DOI] [PubMed] [Google Scholar]
- 25.Gruic-Sovulj I., Rokov-Plavec J., Weygand-Durasevic I. (2007) FEBS Lett. 581, 5110–5114 [DOI] [PubMed] [Google Scholar]
- 26.Splan K. E., Ignatov M. E., Musier-Forsyth K. (2008) J. Biol. Chem. 283, 7128–7134 [DOI] [PubMed] [Google Scholar]
- 27.Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan M., Zhu B., Zhou X. L., He R., Chen X., Eriani G., Wang E. D. (2010) J. Biol. Chem. 285, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu B., Yao P., Tan M., Eriani G., Wang E. D. (2009) J. Biol. Chem. 284, 3418–3424 [DOI] [PubMed] [Google Scholar]
- 30.Minajigi A., Francklyn C. S. (2010) J. Biol. Chem. 285, 23810–23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nomanbhoy T. K., Hendrickson T. L., Schimmel P. (1999) Mol. Cell 4, 519–528 [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson T. L., Nomanbhoy T. K., de Crécy-Lagard V., Fukai S., Nureki O., Yokoyama S., Schimmel P. (2002) Mol. Cell 9, 353–362 [DOI] [PubMed] [Google Scholar]
- 33.Dock-Bregeon A. C., Rees B., Torres-Larios A., Bey G., Caillet J., Moras D. (2004) Mol. Cell 16, 375–386 [DOI] [PubMed] [Google Scholar]
- 34.Schimmel P., Schmidt E. (1995) Trends Biochem. Sci. 20, 1–2 [DOI] [PubMed] [Google Scholar]
- 35.Jakubowski H. (1980) Biochemistry 19, 5071–5078 [DOI] [PubMed] [Google Scholar]
- 36.Nureki O., Niimi T., Muramatsu T., Kanno H., Kohno T., Florentz C., Giegé R., Yokoyama S. (1994) J. Mol. Biol. 236, 710–724 [DOI] [PubMed] [Google Scholar]
- 37.Salazar J. C., Ambrogelly A., Crain P. F., McCloskey J. A., Söll D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fersht A. R., Kaethner M. M. (1976) Biochemistry 15, 818–823 [DOI] [PubMed] [Google Scholar]
- 39.Gruic-Sovulj I., Dulic M., Jaric J., Cvetesic N., Majsec K., Weygand-Durasevic I. (2010) Croat. Chem. Acta, in press [Google Scholar]
- 40.Wolfson A. D., Uhlenbeck O. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hale S. P., Schimmel P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2755–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldwin A. N., Berg P. (1966) J. Biol. Chem. 241, 839–845 [PubMed] [Google Scholar]
- 43.Johnson K. A. (1992) The Enzymes (Sigman D. S. ed) 3rd Ed., pp. 2–61, Academic Press, Inc., San Diego [Google Scholar]
- 44.Zhang C. M., Perona J. J., Ryu K., Francklyn C., Hou Y. M. (2006) J. Mol. Biol. 361, 300–311 [DOI] [PubMed] [Google Scholar]
- 45.Lue S. W., Kelley S. O. (2005) Biochemistry 44, 3010–3016 [DOI] [PubMed] [Google Scholar]
- 46.Reynolds N. M., Ling J., Roy H., Banerjee R., Repasky S. E., Hamel P., Ibba M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4063–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uter N. T., Perona J. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14396–14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minajigi A., Francklyn C. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17748–17753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francklyn C. S. (2008) Biochemistry 47, 11695–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., Alley M. R. (2007) Science 316, 1759–1761 [DOI] [PubMed] [Google Scholar]
- 51.Johnson K. A. (2010) Biochim. Biophys. Acta 1804, 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fersht A. R. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, pp. 384–389, W. H. Freeman & Co., New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.