Abstract
Epicatechin gallate (ECg) sensitizes methicillin-resistant Staphylococcus aureus (MRSA) to oxacillin and other β-lactam agents; it also reduces the secretion of virulence-associated proteins, prevents biofilm formation, and induces gross morphological changes in MRSA cells without compromising the growth rate. MRSA is resistant to oxacillin because of the presence of penicillin-binding protein 2a (PBP2a), which allows peptidoglycan synthesis to continue after oxacillin-mediated acylation of native PBPs. We show that ECg binds predominantly to the cytoplasmic membrane (CM), initially decreasing the fluidity of the bilayer, and induces changes in gene expression indicative of an attempt to preserve and repair a compromised cell wall. On further incubation, the CM is reorganized; the amount of lysylphosphatidylglycerol is markedly reduced, with a concomitant increase in phosphatidylglycerol, and the proportion of branched chain fatty acids increases, resulting in a more fluid structure. We found no evidence that ECg modulates the enzymatic activity of PBP2a through direct binding to the protein but determined that PBP2 is delocalized from the FtsZ-anchored cell wall biosynthetic machinery at the septal division site following intercalation into the CM. We argue that many features of the ECg-induced phenotype can be explained by changes in the fluid dynamics of the CM.
Keywords: Antibiotics, Bacteria, Membrane Bilayer, Membrane Lipids, Microarray, Staphylococcus aureus, Catechin Gallates, Penicillin-binding Protein, Peptidoglycan
Introduction
Staphylococcus aureus is an opportunistic pathogen responsible for hospital- and community-acquired infections that extend from localized cutaneous lesions to life-threatening conditions such as sepsis and infective endocarditis (1, 2). Staphylococcal infections are becoming increasingly difficult to treat because of the rapid emergence of multidrug resistance. In particular, the acquisition, evolution, and horizontal dissemination of genes conferring resistance to β-lactam antibiotics have eroded the capacity to employ these agents for effective chemotherapy (3, 4). Although new agents such as linezolid, daptomycin, and tigecycline have been developed in response to evolving resistance in Gram-positive pathogens and are proving highly effective (5), resistance to these drugs is emerging (6, 7) and is likely to increase as their clinical use increases. There is a continuing need for new treatments for these infections, in particular those employing agents that suppress or abrogate the emergence of resistance (8). The utility of antibiotics made less potent by the evolution of resistance could be restored by compounds with the capacity to reverse antibiotic resistance during the course of infection; such modifying agents, that generate new phenotypes rather than kill the target bacteria, may have the potential to reduce the emergence of resistance to drug-modifier combinations (9).
In this context, we are examining the therapeutic anti-staphylococcal potential of (−)-epicatechin gallate (ECg),4 a polyphenol found in abundance in green tea (10). Galloyl catechins have negligible intrinsic antibacterial activity but reduce penicillin-binding protein 2a (PBP2a)-mediated β-lactam resistance (11); they also disrupt the secretion of virulence-related proteins (9) and prevent the formation of biofilms (12, 13). In addition, they promote cell wall thickening and cell aggregation without affecting the rate or extent of growth in culture (12, 14). ECg, the most potent galloyl catechin (10, 11), does not suppress the transcription of mecA or the production of its product PBP2a (12), and it is clear that restoration of susceptibility of methicillin-resistant S. aureus (MRSA) to β-lactam antibiotics is due to a more complex and as yet incompletely defined mechanism. There is some evidence that ECg-mediated modification of staphylococci is related to the capacity of galloyl catechins to partition into the cytoplasmic membrane (CM). ECg penetrates the lipid palisade of unilamellar vesicles of phosphatidylcholine or phosphatidylethanolamine, increasing the lipid order of the fluid bilayers (15–18). ECg and other catechins bind to MRSA during the mid-logarithmic phase of growth to a degree that reflects their capacity to intercalate into phosphatidylcholine and phosphatidylethanolamine bilayers (19). However, S. aureus elaborates an unusual CM comprising three major phospholipids (20–22): negatively charged phosphatidylglycerol (PG) and cardiolipin in addition to positively charged lysyl-PG (LPG), with evidence for asymmetric distribution across the outer and inner CM leaflets (21). The intercalation of catechins into the CM of viable staphylococci has yet to be demonstrated.
ECg-mediated perturbation of orderly cell division, cell wall turnover, and cell separation may be related to changes in the secretion of autolysins from the staphylococcal cell; ECg-grown cells retained autolysins within the thickened cell wall, probably in a predominantly inactive form, with greatly reduced amounts released into the growth medium (12). Activity and retention of these enzymes, key players in cell separation and peptidoglycan turnover, are strongly influenced by the charge characteristics of the cell wall; growth in the presence of ECg decreases the net positive charge of the cell surface (9, 23), in part by reducing the degree of d-alanylation of wall teichoic acids associated with peptidoglycan (23). An increased negative surface charge leads to electrostatic repulsion from negatively charged surfaces and in all likelihood accounts for the biofilm inhibiting properties of ECg (24). Further, ECg modulates the levels of PBPs 1 and 3 in the staphylococcal CM, resulting in a 5–10% reduction in peptidoglycan cross-linking without compromising cell integrity (12) and is responsible for the less dense appearance of the wall and the greater packed cell volume of ECg-treated bacteria (23).
Although these changes explain some aspects of the complex ECg-induced MRSA phenotype, they are unlikely to account for the profound changes in β-lactam susceptibility engendered by ECg; moderate concentrations elicit complete restoration of oxacillin sensitivity (11, 19), and we contend that such a large consequence can only be achieved by direct interference with the underlying antibiotic resistance machinery of the cell. In dividing cells, PBP2-mediated transglycosylation and transpeptidation reactions occur predominantly at the division septum (25). The PBP2 substrate within extending peptidoglycan chains is restricted to the septum; PBP2 molecules are recruited to this site by a process requiring the presence of FtsZ, a protein that directly or indirectly anchors the cell wall synthetic machinery to the site of cell division (26). β-Lactam antibiotics bind covalently to the transpeptidase active site of PBP2 and abrogate catalytic activity, preventing formation of pentaglycine cross-bridges and, in β-lactam-susceptible strains, delocalizing PBP2 from the septum (26). The acquisition of PBP2a allows transpeptidation to occur in the presence of β-lactam agents (27); in its native conformation, PBP2a has a very low affinity for β-lactams because of the closed conformation of its active site (28). PBP2 and PBP2a function cooperatively in the presence of oxacillin, possibly as a multienzyme complex, to ensure orderly peptidoglycan synthesis at the septum with no delocalization of PBP2 (26). Intercalation of ECg into the CM of dividing MRSA may interfere with the traffic of membrane-anchored PBPs; in addition, exposure of PBP2a to ECg may result in a conformational change leading to the opening of the active site from the closed conformation and the concomitant loss of resistance. In this study, we investigated binding of ECg to the CM, the effect of ECg on PBP2 localization, and its capacity to alter the conformation of PBP2a. Our data show that ECg penetrates the staphylococcal CM, initially decreasing membrane fluidity; the targeted bacteria respond by elaborating a more mobile and more negatively charged bilayer. ECg does not appear to interact with PBP2a or alter its interactions with β-lactams but delocalizes PBP2 from the septum; we suggest that delocalization of PBP2 from the site of cell division is primarily responsible for galloyl catechin-mediated sensitization of MRSA strains to β-lactam antibiotics.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Reagents
Epidemic MRSA isolate EMRSA-16 was from a clinical sample obtained at the Royal Free Hospital (London, UK). Methicillin-susceptible S. aureus strain ATCC29213 was obtained from the American Type Culture Collection; vancomycin-intermediate-resistant S. aureus (VISA) Mu50 was isolated from a Japanese patient in 1997. Methicillin-susceptible S. aureus strain LH607 is a protein A-deficient mutant constructed in the background of 8325-4 by inactivation of the spa gene using a tetracycline cassette. COL spa− was obtained by transduction of the inactivated spa gene from LH607 into COL. The homogeneous MRSA strain COLpPBP2-31, derived from COL, carries a gfp-pbp2 fusion (26). RUSA130 (29) is a COL mutant in which PBP2 has been inactivated by insertion of Tn551 (EryR) into the transpeptidase-encoding region of pbp2; this strain was the gift of A. Tomasz (Rockefeller University, New York, NY). The minimum inhibitory concentration (MIC) of S. aureus strains was determined as described previously (11). ECg and (−)-[4(n)-3H] ECg were gifts from Mitsui Norin (Tokyo, Japan); the radiolabeled compound (specific activity, 481 GBq/mmol; radioactive concentration, 74.0 MBq/ml), was prepared from sodium boro[3H]hydride by Amersham Biosciences using a proprietary method developed by Amersham Biosciences. PG and cardiolipin were obtained from Sigma-Aldrich; LPG was produced by custom synthesis (Avanti Polar Lipids Inc., Delfzijl, The Netherlands). The bacteria were grown with aeration at 37 °C in Mueller-Hinton (MH) broth (Oxoid Ltd., Basingstoke, UK). Erythromycin (10 μg/ml) and tetracycline (5 μg/ml) were added as necessary. ECg was dissolved in 50% ethanol and added to MH broth to give a final concentration of 12.5 μg/ml.
Microarray Analysis
The BμG@S SAv1.1.0 microarray has been described elsewhere (30) and contains PCR products representing all predicted open reading frames from the initial seven S. aureus genome sequencing projects. Total RNA extracts were prepared as described previously (31). EMRSA-16 was grown in the presence of ECg until the A600 reached 0.7 (logarithmic phase) or 3.0 (stationary phase). Hybridizations were performed as described (31). Hybridization data were analyzed using an Affymetrix 428 scanner and then quantified using Bluefuse for Microarrays 3.5 software (BlueGnome, Cambridge, UK). Data analysis was performed in GeneSpring GX 7.3 (Agilent Technologies, Santa Clara, CA) using median-normalized Cy5/Cy3 ratio intensities for three biological replicates as described previously (31). Fully annotated microarray data have been deposited in BμG@Sbase (accession number BUGS-97) and ArrayExpress (accession number E-BUGS-97). The modulation of selected genes by ECg was confirmed using quantitative reverse transcription-PCR as described previously (31).
Localization of [3H]ECg in S. aureus
EMRSA-16 was grown in MH broth containing ECg and [3H]ECg (3.7 KBq/ml); the cells were harvested by centrifugation once the A600 had reached 0.6–0.8 and washed twice with cold Buffer A (0.05 m Tris-HCl, pH 7.5, containing 100 mm NaCl and 20 mm MgCl2·6H2O). Washed cells were suspended in Buffer A containing 50% (w/v) sucrose and converted to protoplasts by incubation with lysostaphin (200 μg/ml) for 2 h at 37 °C with gentle swirling. Control cells were incubated without lysostaphin. The protoplasts were harvested by centrifugation (38,000 × g, 1 h, 4 °C), and the supernatant was retained for determination of radioactivity. The protoplasts were then lysed by dilution in excess of 50 mm phosphate buffer, pH 6.6, containing 10 mm MgCl2·6H2O, 5 μg/ml DNase, and 5 μg/ml RNase. The lysates were incubated for 15 min at 37 °C with shaking (200 rpm). EDTA was added to a final concentration of 10 mm, and the lysates were incubated for a further 15 min; MgCl2·6H2O was added to give a final concentration of 15 mm, and residual whole cells and cell debris were removed by centrifugation (2,000 × g, 4 °C, 30 min). The membrane fraction was harvested by centrifugation (130,000 × g, 1 h, 4 °C), and radioactivity in the pellet and supernatant was measured. The values obtained from parallel control experiments omitting the cell wall hydrolase lysostaphin were subtracted from values using lysostaphin-treated cells to compensate for nonspecific [3H]ECg release.
Phospholipid Analysis
To determine the nature of the head groups of CM phospholipids, 100-ml cultures of EMRSA-16 were grown in MH broth containing [32P]orthophosphate (18.5 KBq/ml). Phospholipids were extracted during mid-logarithmic phase (A600 = 0.7) according to Bligh and Dyer (32) and separated as described by Ames (33) with the exception that high performance TLC glass plates were used (coated with Silica Gel 60 F254, 100 × 100 mm; Merck). The first dimension was developed in chloroform, methanol, 7 n ammonia (60:35:5, v/v/v), and the second was developed in chloroform, methanol, glacial acetic acid (65:25:8, v/v/v). Radioactive spots were visualized by autoradiography, identified by comparison with commercial standards or by MS/MS, and quantified using a Bio-Rad Molecular Imager FX. Fatty acid esters were hydrolyzed with MeOH, 20 m NaOH (19:1) for 30 min at 45 °C. Two volumes of CH2Cl2 and 1 volume of water were then added. The samples were mixed and centrifuged, and the organic layer was collected and evaporated. TMS derivatives were obtained using 50 μl of acetonitrile and 50 μl of N,O-bis(trimethylsilyl)trifluoroacetamide (30 min, 80 °C). The derivatives were separated by gas chromatography and identified by MS using combined gas chromatography-MS comprising an Agilent HP HP6890 series gas chromatography with HP7683 series injector interfaced directly to an Agilent HP5973 mass selective detector (mass range, 10–700 m/z; scan acquisition mode); a J & W DB-5ms capillary column (40 m length, 250.0 μm diameter, 0.25-μm film thickness) was installed, and an oven temperature gradient of 80–280 °C was employed.
Cell Membrane Labeling and Fluorescence Anisotropy Measurements
The membrane fluidity of mid-logarithmic EMRSA-16 was determined by measuring the fluorescence polarization of the probe 1,6-diphenyl-1,3,5-hexatriene (DPH) inserted into the CM as previously described (34). Fluorescence polarization was measured using a thermostat-controlled PerkinElmer Life Sciences LS55 luminescence spectrometer. The excitation and emission wavelengths for the DPH probe were 358 and 428 nm, respectively. The data were analyzed using FL WINLAB software program, version 4.00.02.
Epifluorescence Microscopy
The bacteria were grown in MH broth to A600 of 0.3; ECg (12.5 μg/ml), oxacillin (4 μg/ml), or ECg + oxacillin (12.5 and 4 μg/ml) were added to aliquots and incubated (37 °C; 1 h). For phase contrast and fluorescence microscopy, live cells were examined using an Axi Observer.Z1 inverted microscope (Zeiss) on glass slides coated with 1% agarose in PBS. Image acquisition was undertaken with a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ) and Metamorph versus 7.5 software (Molecular Devices, Uckfield, UK).
Immunofluorescence Microscopy
ECg-, oxacillin-, and ECg/oxacillin-treated bacteria were fixed and examined by fluorescence microscopy as described (25). Lysostaphin-treated cells were washed with PBS, air-dried, rehydrated with PBS, blocked with 2% bovine serum albumin for 15 min and incubated overnight at 4 °C with the primary antibody against Bacillus subtilis FtsZ added in consecutive 2-fold dilutions over the range 1/800–1/6,400. The cells were washed with PBS and incubated in the dark for 1 h with anti-rabbit IgG fluorescein isothiocyanate conjugate diluted 1/500 in 2% bovine serum albumin before further washing. Vectashield mounting medium (Vector Laboratories, Peterborough, UK) containing 1 μg/ml 4′,6-diamidino-2-phenylindole was added before visualization by fluorescence microscopy.
PBP Extraction and Profiling
CMs were prepared from logarithmic phase cultures, and PBPs were released from membrane vesicles with 2% (v/v) Triton X-100 as described previously (12). Following exposure of PBP preparations (10 μg of protein) to ECg for 15 min at 37 °C, PBPs were selectively labeled by preincubation with 25 μm Bocillin FL (Invitrogen) for 10 min at 37 °C. The proteins were then separated by 10% SDS-PAGE, and PBPs were detected by fluorography using the Bio-Rad FX imager.
Interactions with PBP2a
The capacity of ECg to bind PBP2a was determined. Recombinant PBP2a was expressed and purified as described earlier (35, 36). We ascertained that the protein was soluble during all conditions of assay. Binding studies were conducted with β-lactams (Bocillin FL or benzyl[14C]penicillin; specific activity of 59 mCi/mmol; Amersham Biosciences) as acylating agents or [3H]ECg. The aliquots were collected at different time intervals, proteins were separated by 10% SDS-PAGE, and the acyl-enzyme complex was detected by fluorimetry (Gel Doc 2000; Bio-Rad) or scintillation counting of gel slices. The kinetic parameters for interactions of nitrocefin with PBP2a (acylation and deacylation studies) were measured as previously described (35). Intrinsic tryptophan fluorescence of PBP2a was assayed using an excitation wavelength of 295 nm (PerkinElmer Life Sciences LS55 spectrofluorimeter).
RESULTS
Effect of ECg on Antibiotic Resistance
Incorporation of 12.5 μg/ml ECg into the assay reduced the MIC of EMRSA-16 from 512 to 1 μg/ml and Mu50 from 256 to 0.5 μg/ml (n = 3). The compound had no effect on the vancomycin MIC of vancomycin-susceptible EMRSA-16 (0.5 μg/ml; n = 6) or VISA isolate Mu50 (2 μg/ml; n = 6).
ECg-induced Changes in Gene Expression
Transcriptional profiling using DNA microarrays can shed light on mechanisms of drug action by providing a genome-wide view of the cellular response to bioactive agents. Because gene expression in S. aureus is markedly influenced by population density and environmental cues (37), we determined transcriptional profiles of mid-logarithmic and stationary phase ECg-containing cultures. Statistically significant 2-fold or greater changes were found for 103 up-regulated and 166 down-regulated genes following growth of EMRSA-16 in mid-logarithmic culture in comparison with bacteria cultured in ECg-free medium. After growth with ECg to stationary phase, 102 genes were up-regulated and 102 were down-regulated compared with culture in the absence of the galloyl catechin.
Genes up-regulated by growth to mid-logarithmic phase in ECg were involved in cation transport and binding (fhuABD and sstABCD), transport and metabolism of amino acids (e.g. MRSA252 ORFs SAR0119–0127) and riboflavin (ribADEH), electron transport (cydAB), β-lactamase expression (blaIR1Z), shape determination (e.g. mreC), and pathogenesis (e.g. fnbA and isaA). Most prominent was the induction of genes belonging to the general cell wall stress stimulon (Table 1), a group of genes that are induced following treatment with cell wall active antibiotics such as vancomycin, oxacillin, d-cycloserine, and Bacitracin (38–40) and with CM depolarizing agents such as daptomycin (41) and cationic defense peptides (42). Many genes of the stimulon are under the control of the two component signal transduction system vraRS, a positive regulator of peptidoglycan synthesis (39), initially shown to be up-regulated by exposure to vancomycin (39) and overexpressed in VISA clinical isolates in comparison with the isogenic vancomycin-susceptible parent (40). ECg increased transcription of the response regulator vraR and the sensor histidine kinase vraS (Table 1) and some, but not all, genes in the regulon controlled by vraRS (39); thus, sgtB and fmt, involved in cell envelope biogenesis, pbpB (encoding PBP2), yvqF, tcaA (implicated in teicoplainin resistance), prsA (protease maturation), and N315 ORFs SA2220, SA1703, SA1712, SA2103, SA2221, and SA2343 were up-regulated following exposure to ECg (Table 1). These genes belong to the “core” cell wall stress stimulon, defined as genes common to the four initial studies defining the stimulon (40); genes from this core group that were not up-regulated by ECg included murZ, involved in cell wall and membrane biogenesis, the serine protease encoding htrA, and SA2297. Some stimulon-related genes of unknown function were highly up-regulated by ECg, in particular vraX, annotated on the basis of its up-regulation by imipenem, and SA1477 (24.4- and 11.9-fold respectively; Table 1). The profile presented in Table 1 provides strong evidence that EMRSA-16 responds to ECg by taking steps to preserve and/or repair the cell wall and CM compromised by exposure to this agent (43).
TABLE 1.
Genes with increased expression of at least 2-fold following exposure to ECg, compared with cell wall stress stimulon up-regulation determined in other studies
| MRSA252a | N315b | Gene | Product or putative function | ECgc | VISAd | Vancomycind | Oxacillin bacitracin d-cycloserinee | Daptomycinf | Functional category |
|---|---|---|---|---|---|---|---|---|---|
| SAR1340 | SA1166 | thrB | Homoserine kinase | 2.48 | Amino acid biosynthesis | ||||
| SAR1408 | SA1228 | dapB | Dihydropicolinate synthase | 2.66 | Amino acid biosynthesis | ||||
| SAR1339 | SA1165 | thrC | Threonine synthase | 3.06 | Amino acid biosynthesis | ||||
| SAR1407 | SA1227 | dapA | Homoserine dehydrogenase | 3.76 | Amino acid biosynthesis | ||||
| SAR1024 | SA0903 | Conserved hypothetical protein | 2.33 | 3.6 | Up | Carbohydrate transport and metabolism | |||
| SAR2522 | SA2220 | Glycerate kinase | 2.88 | Up | 14.8 | Carbohydrate transport and metabolism | |||
| SAR1374 | SA1195 | msrR | Reductase regulator | 2.84 | 4.8 | 2.5 | Transcription | ||
| SAR2394 | SA2103 | Transcriptional regulator | 3.2 | 2.7 | 3.4 | 14 | Regulation/transcription | ||
| SAR2585 | SA2296 | Transcriptional regulator, MerR family | 2.18 | Regulatory functions | |||||
| SAR1974 | SA1700 | vraR | Response regulator | 4.80 | 2.5 | 8.7 | 10.39 | Signal transduction mechanism | |
| SAR1975 | SA1701 | vraS | Sensor histidine kinase | 8.05 | 2.5 | 8.7 | 10.4 | 9.57 | Signal transduction mechanism |
| SAR1976 | SA1702 | yvpF | Conserved hypothetical protein | 6.57 | 2.60 | 8.1 | 7.1 | Function unknown | |
| SAR1977 | SA1703 | Hypothetical protein | 6.8 | 3.1 | Up | Function unknown | |||
| SAR0204 | SA0205 | Peptidase domain protein | 2 | 4.5 | Up | Cell envelope biogenesis | |||
| SAR0273 | SA0265 | lytM | Peptidoglycan hydrolase | 2.58 | 4.8 | 2.2 | Cell envelope biogenesis | ||
| SAR1030 | SA0909 | fmt | Autolysis/methicillin resistance | 4.11 | Up | Up | 4.93 | Cell envelope biogenesis | |
| SAR1964 | SA1691 | sgtB | Transglycosylase domain protein | 5.47 | 2.9 | 10.6 | 8.10 | Cell envelope biogenesis | |
| SAR0251 | SA0244 | Teichoic acid biosynthesis protein | 2.19 | 2.90 | Cell envelope biogenesis | ||||
| SAR2442 | SA2146 | tcaA | TcaA protein | 3.73 | 4.1 | Up | 3.03 | Cell envelope | |
| SAR1461 | SA1283 | pbp2 | Penicillin binding protein 2 | 2.21 | Cell envelope | ||||
| SAR2212 | SA1926 | murZ | UDP-GlcNAc-carboxytransferase | 4.14 | Cell envelope | ||||
| SAR1932 | SA1659 | prsA | Protease maturation protein | 4.77 | 3.2 | 12 | 7.7 | 9.59 | Protein turnover and chaperones |
| SAR1805 | SA1549 | htrA | Serine protease HtrA | 2.74 | 6.8 | 11 | 3.06 | Protein turnover and chaperones | |
| SAR0528 | SA0483 | clpC | ATP-dependent Clp protease, ClpC | 3.98 | Protein turnover and chaperones | ||||
| SAR0938 | SA0835 | clpB | ATP-dependent Clp protease, CLpB | 3.87 | Protein turnover and chaperones | ||||
| SAR1658 | SA1410 | grpE | Heat shock protein GrpE | 3.28 | Protein folding and stabilization | ||||
| SAR2117 | SA1837 | groES | Chaperonin, 10 kDa | 2.84 | Protein folding and stabilization | ||||
| SAR0513 | SA0470 | Chaperonin, 33 kDa | 2.15 | Protein folding and stabilization | |||||
| SAR1230 | SA1097 | hslU | Heat shock protein HslVU | 2.12 | Protein folding and stabilization | ||||
| SAR2475 | SA2175 | heat shock protein Hsp20 | 3.08 | Protein folding and stabilization | |||||
| SAR2064 | SA1777 | Hypothetical protein | 2.2 | 2.3 | General function | ||||
| SAR2388 | SA2097 | Secretory antigen precursor | 2.04 | 4.8 | General function | ||||
| SAR0925 | SA0824 | Conserved hypothetical protein | 3.19 | 2.1 | 5.8 | Function unknown | |||
| SAR2690 | SA2405 | betA/cudB | Choline dehydrogenase | 5.94 | Up | Cell motility and secretion | |||
| SAR0927 | SA0826 | spsB | Type-1 signal peptidase 1B | 2.39 | 4.7 | Up | 2.56 | Cell motility and secretion | |
| SAR1851 | SA1587 | ribA | Riboflavin biosynthesis protein | 5.92 | 2.9 | Coenzyme metabolism | |||
| SAR0580 | SA0533 | vraA | Long chain fatty acid CoA ligase | Lipid metabolism | |||||
| SAR0581 | SA0534 | vraB | Acetyl-CoA acetyltransferase | 2.7 | Lipid metabolism | ||||
| SAR0582 | SA0535 | vraC | Hypothetical protein | 5.3 | Function unknown | ||||
| SAR0583 | SA0536 | vraY | Hypothetical protein | 2.9 | Function unknown | ||||
| SAR0584 | vraX | Hypothetical protein | 24.2 | 22.7 | 224.7 | 26.6 | Function unknown | ||
| SAR0645 | SA0591 | Hypothetical protein | 3.7 | 5.3 | Up | Up | Function unknown | ||
| SAR2509 | SA2207 | hlgA | γ-Hemolysin, component A | 10.18 | Toxin production and resistance | ||||
| SAR2770 | SA2480 | drp35 | drP35 | 8.87 | Toxin production and resistance | ||||
| SAR2511 | SA2209 | hlgB | γ-Hemolysin, component B | 5.04 | Toxin production and resistance | ||||
| SAR2510 | SA2208 | hlgC | g-Hemolysin, component C | 3.52 | Toxin production and resistance | ||||
| SAR1035 | SA0914 | Similar to chitinase B | 3.26 | 2.2 | 10.6 | 5.5 | |||
| SAR1730 | SA1476 | Hypothetical protein | 6.95 | 4 | 20.9 | ||||
| SAR1731 | SA1477 | Hypothetical protein | 11.94 | 5.2 | 27.6 | ||||
| SAR1987 | SA1712 | Hypothetical protein | 2.04 | 2 | Up | Up | |||
| SAR2065 | SA1778 | Hypothetical protein | 2.4 | 2.2 | |||||
| SAR2184 | SA1898 | SceD protein | 4.45 | 10.5 | |||||
| SAR2407 | SA2113 | Hypothetical protein | 4.99 | 3.8 | 10.4 | ||||
| SAR2523 | SA2221 | Hypothetical protein | 5.16 | 2.4 | 9.5 | 18.7 | |||
| SAR2636 | SA2343 | Hypothetical protein | 8.28 | 84.4 | Up | Up | |||
| SAR0599 | SA0550 | Hypothetical protein | 2.2 | Up | |||||
| SAR2771 | SA2481 | Hypothetical protein | 4.58 | 3.4 | 5.7 | ||||
| SAR2586 | SA2297 | GTP-pyrophosphokinase | 2.0 | 8.6 | Up |
a MRSA252 ORF identifiers for the BμG@S SAv1.1.0 microarray used in this study (30). ORF identifiers in bold form the genes of the core cell wall stress stimulon as defined by McAleese et al. (40).
b N315 ORF identifiers for the S. aureus N315 microarray described by McAleese et al. (40). ORF identifiers in bold form the genes of the core cell wall stress stimulon as defined by McAleese et al. (40).
c Data from this study with S. aureus EMRSA-16.
d Up-regulation of cell wall stress stimulon genes induced by spontaneous mutation of a clinical S. aureus isolate to VISA and by exposure to suprainhibitory concentrations of vancomycin (40).
e Up-regulated genes after exposure of S. aureus RN450 to these agents (38).
f Up-regulated genes after exposure of S. aureus ATCC 29213 to suprainhibitory concentrations of daptomycin (41).
EMRSA-16 genes down-regulated by ECg in mid-logarithmic cultures fell into a number of functional categories, including genes encoding osmotic shock proteins, membrane and exported proteins and lipoproteins, genes involved in nitrate/nitrite, amino acid and carbohydrate metabolism, and protein and ion transport. In addition, genes of the agr regulon (agrABCDI and agrIII), controlling the expression of proteins involved in pathogenesis, were strongly down-regulated. Gene transcriptional analysis following growth in the presence of ECg to stationary phase gave no evidence that the cell wall stress stimulon was maintained into this phase of the growth cycle.
Microarrays were validated using quantitative reverse transcription-PCR on selected genes. There was excellent correlation between quantitative reverse transcription-PCR and microarray data for the 11 genes described in supplemental Fig. S1 (R2 = 0.9448; p = 0.0000121).
Subcellular Localization of ECg
Prior to production of protoplasts, the cells grown in the presence of [3H]ECg were washed in buffer; in a typical experiment, 2–4% of total radioactivity was recovered in the wash fraction. Nonspecific release of radioactivity was determined by processing ECg-grown bacteria as described under “Experimental Procedures” with the exception that lysostaphin was omitted from the first incubation step and the values obtained were subtracted from the values obtained for the test samples. Supernatants obtained following lysostaphin digestion represented [3H]ECg firmly bound to the cell wall and constituted 31% of total recovered radioactivity. After lysis of protoplasts in hypotonic buffer and collection of membranes by ultracentrifugation, the supernatant represented the intracellular fraction and contained 10.9% of recovered 3H; 58.1% of total recovered radioactivity was found in the membrane fraction. The values obtained were the means of four experiments.
CM Changes Induced by ECg
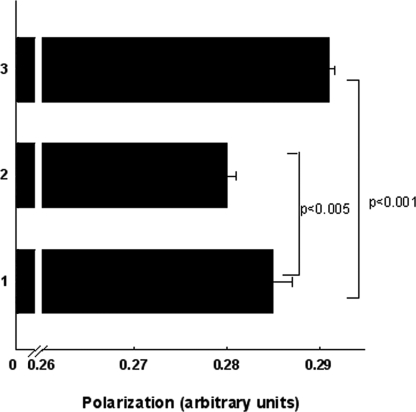
Intercalation of ECg into the CM will impact on the physical properties of the bilayer (15). We therefore measured the effect of ECg on membrane fluidity using DPH; this hydrophobic fluorescent probe associates with the lipophilic tails of phospholipids without disturbing bilayer structure (44). A reduction in the freedom of rotation of the probe will be reflected in increased emission of light in the same plane as the excited light and a reduction in emission of light at 90° to the plane of excitation. There is therefore an inverse relationship between the amount of polarized light emitted and the fluidity of the membrane. When ECg was added to DPH-loaded mid-logarithmic cultures of EMRSA-16, there was an immediate, significant (p < 0.005) increase in membrane probe polarization, indicating a reduction in fluidity (Fig. 1). When bacteria were grown to mid-logarithmic phase in the presence of ECg prior to the addition of DPH, there was significant (p < 0.001) reduction in probe polarization, indicative of a more fluid bilayer.
FIGURE 1.
Membrane fluidity of MRSA-16 grown in the absence of ECg (bar 1), grown to mid-logarithmic phase in 12.5 μg/ml of ECg (bar 2), and grown to mid-logarithmic phase in ECg-free MH broth with the galloyl catechin added and polarization determined immediately (bar 3). The fluorescent probe DPH was incorporated into the CM as described under “Experimental Procedures.” The bars represent the means ± S.D. (n = 15), and the levels of significance were determined using paired Student's t test.
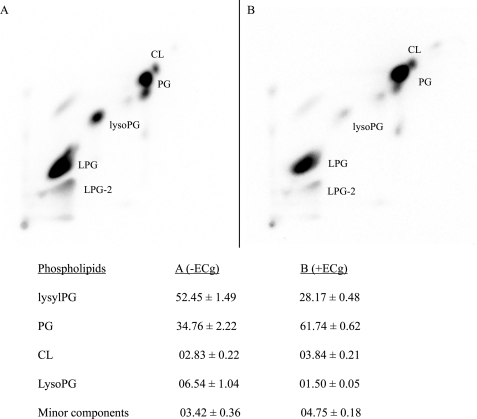
The polarization data strongly suggest that the bacteria respond to binding of ECg by altering the composition of the CM. We therefore investigated the composition of phospholipids extracted from ECg-grown and conventionally grown EMRSA-16 cells. Two-dimensional TLC revealed up to 10 components in membranes from both cultures (Fig. 2); five spots represented minor components, each constituting less than 1% of total phospholipids, and were not investigated further. Growth to mid-logarithmic phase in ECg reduced the content of the most prominent phospholipid, LPG, from 52.5 to 28.7%; there was a concomitant increase in PG from 34.8 to 61.7%, an increase in cardiolipin from 2.8 to 3.8%, and a significant reduction in lysophosphatidylglycerol from 6.5 to 1.5%. Fatty acid moieties of these phospholipids ranged in length from C10 to C21 with the most abundant being C16:0, C18:0, C19:0, and C20:0 (Table 2). As previously reported for S. aureus (45, 46), EMRSA-16 membranes contained a low percentage of unsaturated fatty acids and a relative abundance of branched chain fatty acids. There was no significant difference between C-even and C-odd chain lengths of mid-logarithmic phase (A600 = 0.7) cultures grown in MH broth or in broth supplemented with ECg (even number of C atoms: 69.5 and 68.4%, respectively) and only minor variations in fatty acid chain length between control and ECg-grown cultures (Table 2). However, ECg-grown cultures contained a higher complement of branched chain fatty acids compared with control cells (22.5% compared with 15.5%); branched fatty acids are known to increase the fluidity of the CM because of their inability to form crystalline structures (47). The proportions of both iso and anteiso fatty acids were increased by ECg; the latter confer greater fluidizing properties on membranes than iso fatty acids because they disturb packing order to a greater extent (46). Because CM properties are known to be influenced by the growth phase of the culture (46), we determined fatty acid composition in early logarithmic phase; whereas the proportion of branched chain fatty acids in the control culture declined from 21.5% after 1 h growth to 18.4% after 2 h and 15.5% after 3 h, the proportion did not change when EMRSA-16 was grown in the presence of ECg (22.0% at 1 h, 22.4% at 2 h, and 22.5% at 3 h).
FIGURE 2.
Two-dimensional TLC analysis of membrane lipids extracted from MRSA-16 grown to mid-logarithmic phase (A600 = 0. 7) in the absence (A) or presence (B) of 12.5 μg/ml of ECg. The growth medium (MH broth) contained 18.5 KBq/ml of [32P]orthophosphate. The spots were visualized by autoradiography, and their intensity was quantified using a Bio-Rad Molecular Imager FX; the data are presented in the bottom panel. CL, cardiolipin; lysoPG, lysophosphatidylglycerol. The LPG total is the sum of values obtained from spots LPG and LPG-2. Each of the minor phospholipid components from ECg-free and ECg-containing cultures constituted less than 1% of total lipid extracted. The values given are the percentages of total lipid extracted ± S.D. (n = 9).
TABLE 2.
Fatty acid composition of phospholipids extracted from EMRSA-16 following growth to mid-logarithmic phase (grown to A600 = 0.7; 3 h from broth inoculation) in ECg (12.5 μg/ml)
| Fatty acid | MH broth | MH broth + ECg |
|---|---|---|
| C10:0 | 0.48 ± 0.07 | 0.65 ± 0.05 |
| C11:0 | 0.23 ± 0.04 | 0.39 ± 0.05 |
| C12:0 | 0.62 ± 0.07 | 0.54 ± 0.07 |
| isoC14:0 | 0.56 ± 0.07 | 0.65 ± 0.02 |
| C14:0 | 8.19 ± 0.72 | 4.84 ± 0.50 |
| isoC15:0 | 2.33 ± 0.09 | 3.36 ± 0.30 |
| anteisoC15:0 | 3.31 ± 0.15 | 4.82 ± 0.28 |
| C15:0 | 3.27 ± 0.50 | 1.80 ± 0.05 |
| isoC16:0 | 0.86 ± 0.07 | 0.97 ± 0.06 |
| C16:0 | 13.42 ± 0.44 | 13.84 ± 0.55 |
| isoC17 | 1.52 ± 0.03 | 2.09 ± 0.12 |
| anteisoC17:0 | 2.25 ± 0.10 | 3.64 ± 0.08 |
| C17:0 | 3.01 ± 0.17 | 2.32 ± 0.14 |
| C18:1,9 cis ol | 2.49 ± 0.46 | 3.92 ± 0.61 |
| C18:0 | 24.61 ± 1.34 | 28.09 ± 1.13 |
| isoC19:0 | 1.24 ± 0.10 | 1.57 ± 0.03 |
| anteisoC19:0 | 0.95 ± 0.07 | 1.47 ± 0.17 |
| C19:0 | 9.82 ± 0.30 | 7.35 ± 0.45 |
| C20:0 | 18.23 ± 1.79 | 14.91 ± 1.26 |
| C21:0 | 2.62 ± 0.18 | 2.78 ± 0.16 |
| % Branched (br) | 15.51 | 22.49 |
| % Non-branched (n-br) | 84.49 | 77.51 |
| Ratio (br/n-br) | 0.18 | 0.29 |
Effect of ECg on PBP2 Recruitment to the Septum
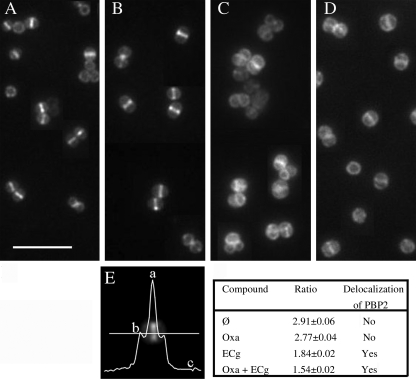
The septum is the main site of cell wall synthesis in S. aureus and requires the presence of FtsZ; this key cell division protein directly or indirectly recruits PBP2 to the septum during the growth cycle (25). In strain COL spa−, FtsZ was found to be localized at the septum of cells grown to mid-logarithmic phase using anti-FtsZ antiserum and fluorescence microscopy (data not shown). We also calculated the amount of GFP-PBP2 fluorescence at the septum compared with the nonseptal membrane in dividing cells of the MRSA strain COLpPBP2-31 (Fig. 3). When grown in the absence of ECg, the GFP-PBP2 fusion was localized at the septum, indicating that PBP2 was retained to the site of cell division by the FtsZ-containing machinery (Fig. 3A); oxacillin at a concentration of 4 μg/ml had no effect on the localization of the fusion in this MRSA strain (Fig. 3B). Exposure to ECg did not affect the localization of FtsZ at the septum of COL spa− cells (data not shown) but was found to delocalize the GFP-PBP2 fusion in COLpPBP2-31 (Fig. 3C); the effect was even more pronounced in the presence of ECg and oxacillin (Fig. 3D). This methodology could not be used to localize PBP2a in the membrane because it has not proved possible to engineer MRSA producing functional GFP-PBP2a fusions.5
FIGURE 3.
Location of PBP2 in MRSA strain COLpPBP2-31 as determined by fluorescence microscopy. This strain carries a gfp-pbp2 fusion. The bacteria were visualized after growth in MH broth (A) and in broth containing 4 μg/ml oxacillin (B), 12.5 μg/ml ECg (C), and 4 μg/ml oxacillin and 12.5 μg/ml ECg (D). The extent of localization of PBP2 at the septum was determined by calculating the ratio (a − c/b − c), where a stands for fluorescence found at the septum, b stands for fluorescence at the lateral membrane, and c stands for background fluorescence (E). The values are the means ± S.D. from measurement of fluorescence ratios of 200 cells in each sample. A ratio (a − c/b − c) greater than 2 indicates localization at the septum, where two membranes are present, whereas a ratio of 2 or less than two indicates delocalization (59).
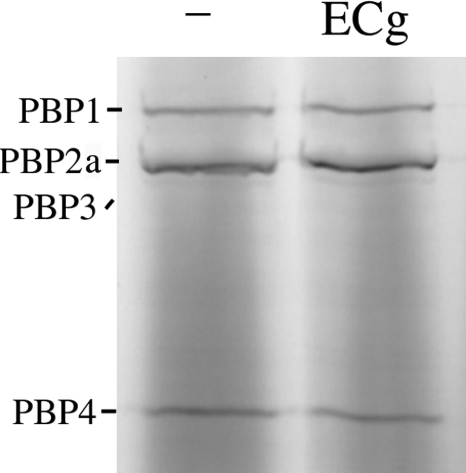
Binding of ECg to PBPs
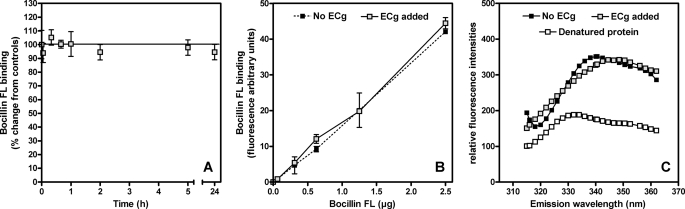
We investigated the possibility that ECg binds to PBP2a and either blocks access to the active site or alters its structure. PBPs were prepared from membranes recovered from RUSA130; Tn551 insertion into the transpeptidase domain of PBP2 produces a truncated form of the protein and allows resolution of PBP2a, which migrates in a virtually identical fashion to PBP2 in polyacrylamide gels (48). ECg was incubated with the membrane extracts, and the mixture was probed with Bocillin FL. At a concentration (12.5 μg/ml) that effects complete transition from β-lactam resistance to full susceptibility in MRSA strains, ECg had no effect on the capacity of Bocillin FL to bind to PBP2a (Fig. 4).
FIGURE 4.
Binding of the fluorescent penicillin derivative Bocillin FL to PBPs recovered from RUSA130 and exposed to 12.5 μg/ml ECg. This strain produces a truncated form of PBP2 and enables the detection of PBP2a, which migrates in gels in an identical fashion to native PBP2. After incubation with ECg, the samples were labeled with 25 μm Bocillin FL and separated by 10% SDS-PAGE, and the proteins were detected by fluorography. −, no ECg. Truncated PBP2 has very low affinity for β-lactam agents (29, 48) and does not bind sufficient Bocillin FL for visualization on the gel.
We then undertook studies with purified PBP2a; we determined the affinities of PBP2a for penicillin in competition studies with ECg (12.5 μg/ml). In all assays, no differences in penicillin-PBP2a binding were found even if (i) the time of interaction between ECg and PBP2a was increased up to 24 h (using both Bocillin FL (Fig. 5A) or benzyl[14C]penicillin); (ii) the concentration of Bocillin in the assay was lowered in stepwise fashion to below the level of detection in the assay (Fig. 5B); or (iii) the concentration of ECg was increased (maximal concentration tested, 50 μg/ml). PBP2a was then incubated in the presence of increasing concentrations of [3H]ECg (0.8–3.5 μg) over 1 h, but in contrast to 14C-benzylpenicillin, no evidence for complex formation between ECg and PBP2a was found. These experiments were complemented by determination of the kinetic parameters for interaction between nitrocefin (a cephalosporin showing a distinctive change in the adsorption spectrum upon cleavage of the amide bond in the β-lactam ring by enzyme acylation) and PBP2a (see Ref 35 for experimental design). As previously described, the minimal interaction scheme assumes binding of nitrocefin (N) to the enzyme in an initial noncovalent complex (EN), which proceeds to acylate the active site serine of PBP2a and gives rise to the covalent complex E-N. This covalent complex will ultimately undergo hydrolytic deacylation. These studies indicate that preincubation of the protein with ECg (12.5 μg/ml; 1 h) did not modify either the dissociation constant for the formation of the preacylation complex (Kd = ∼220 μm), the PBP2a acylation rate constant (k2 = ∼2 × 103 s−1), or the deacylation rate constant (k3 = ∼3.7 × 106 s−1). Examination of PBP2a folding by fluorescence spectroscopy of tryptophan residues showed no differences between untreated and ECg-treated PBP2a (Fig. 5C) compared with denatured protein (1 h; 60 °C).
FIGURE 5.
β-Lactam binding (Aand B) and fluorescence assay (C) of PBP2a in 25 mm HEPES, 1 m NaCl (pH 7. 0). A, binding of the fluorescent penicillin derivative Bocillin FL to PBP2a (5 μm). PBP2a was incubated for up to 24 h in 12.5 μg/ml ECg. The samples were then incubated for 25 min in the presence of 1.25 μg of Bocillin FL and prepared for fluorescence measurement. The results were expressed as percentages of change from controls (no ECg added). The values are the means ± S.D. B, binding of increasing concentrations of Bocillin FL to PBP2a (5 μm). PBP2a was incubated over 1 h in 12.5 μg/ml ECg. Binding was then measured after 25 min in the presence of increasing concentrations of Bocillin FL. The values are the means ± S.D. C, tryptophan fluorescence emission spectra of PBP2a (0.18 μg/ml). The pretreatments of PBP2a are as follows: ECg added (1 h, 12.5 μg/ml) and denatured protein (1 h, 60 °C). We have previously determined (36) that the Bocillin FL concentrations employed in these experiments enable the detection of changes in the affinity of PBP2a for β-lactam antibiotics.
DISCUSSION
Exposure of multidrug-resistant isolates of S. aureus to ECg elicits phenotypic changes that are likely to reduce the capacity of these pathogens to produce severe systemic infection. ECg compromises biofilm formation, markedly suppresses the secretion of key virulence determinants such as α-toxin and coagulase and, perhaps most significantly, abrogates resistance to β-lactam antibiotics (9, 10). These alterations are accompanied by a marked change in the appearance of cells when viewed by light or electron microscopy but not by a reduction in growth rate. Galloyl catechins also reduce halotolerance in staphylococci, probably by interfering with Na+-specific antiporter systems in the CM (49), and the interrelatedness of these components of the ECg-induced phenotype is emphasized by the capacity of NaCl to prevent biofilm formation in much the same way as ECg. The health promoting properties of catechins have attracted much attention and have been largely attributed to their antioxidative, free radical scavenging activities (50), although they also modulate the physical structure of phospholipid model bilayers and, potentially, cell membranes (15–18). Many proteins involved in the regulation and maintenance of bacterial growth and metabolism reside in the CM and require a lipid environment to function; in addition, proteins embedded in the CM facilitate intracellular and intercellular signaling. Thus, modulation of the physical properties of the CM may be responsible for the generation of the complex phenotype elicited by incubation with ECg.
Galloyl catechins, particularly those in the epi (cis) configuration such as ECg and (−)-epigallocatechin gallate, partition into the phospholipid palisade, with their relative affinities for membranes reflected in n-octanol-saline partition coefficients (51), because of the presence of an exposed hydrophobic membrane-perturbing domain (10). We sought clues to the molecular processes involved in ECg-mediated β-lactam sensitization by examining changes in gene expression following EMRSA-16 exposure to ECg. A significant proportion (38 of 103) of genes up-regulated in the logarithmic phase formed part of the cell wall stress stimulon associated with responses of staphylococci to cell wall-active antibiotics, although genes of this stimulon are also up-regulated by daptomycin. This lipopeptide antibiotic functions primarily through disruption of CM function (52). Many of these stress response genes are under the control of the two-component system VraRS that positively regulates cell wall biosynthesis; orderly wall synthesis is known to be perturbed following ECg exposure (12, 23). A comparison of cell wall stress stimulon genes modulated by cell wall and membrane antibiotics (Table 1) reveals that the pattern of up-regulation induced by ECg, with no antibiotic activity, is distinct from but more comparable with that engendered by the cell wall biosynthesis inhibitors vancomycin and oxacillin than by daptomycin. Thus, ECg may have cell wall and membrane targets, or it may disrupt orderly cell wall assembly in a nonlethal fashion by perturbing the lipid environment of membrane-bound enzymes involved in peptidoglycan biosynthesis. Interestingly, although the ECg-induced stress response pattern is strikingly similar to those genes overexpressed in VISA clinical isolates (40), ECg had no effect on MRSA or VISA vancomycin susceptibility.
Direct evidence for interaction with the CM and cell wall was obtained following exposure of EMRSA-16 to radiolabeled ECg. Most (58%) of the label was found in the CM fraction with a smaller proportion (31%) associated with the cell wall. ECg intercalated rapidly into the CM as evidenced by an immediate increase in polarization of the spin probe on exposure of DPH-loaded bacteria to ECg, indicating an increase in the lipid order of the CM. ECg is known to interact stably with hydrocarbon chains in the bilayer of phospholipid model membranes, forcing more tightly packed acyl chains (15). It is likely that these abrupt changes will have a significant impact on metabolic processes associated with the CM, and we present evidence that the targeted bacteria respond by radically altering the composition and biophysical properties of the membrane; there is an increase in the proportion of branched to nonbranched fatty acids within phospholipids leading to “overcompensation” in response to ECg-mediated perturbation. These changes may improve the capacity of CM-associated cell wall biosynthetic enzymes to continue to function in the presence of ECg, albeit in suboptimal fashion. The proportion of ECg that bound to the cell wall is significantly large but not unexpected, because entry into and egress from the staphylococcal cell of hydrophilic molecules is to a large extent controlled by a complex continuum of positive and negative charges within the wall matrix (24) and is likely to reflect loosely bound ECg. We previously demonstrated that exogenous peptidoglycan had no effect on the capacity of ECg to reduce the MIC of oxacillin (12), suggesting that binding is transient and does not blockade sites within the cell wall that influence the sensitization mechanism.
The coupling of lysine to PG to yield LPG was markedly reduced by growth in the presence of ECg. Whether this is a bacterial response to CM intercalation or an inhibition of LPG conjugation by ECg is currently unclear; lysine is attached to PG by MprF, a CM-anchored protein that also translocates LPG to the outer leaflet of the CM, where its positive charge profile provides protection against host cationic antibacterial peptides and daptomycin (53). Thus, mprF mutants are less virulent when assessed in vitro and in vivo (54), and MprF has been proposed as a novel target for anti-staphylococcal chemotherapeutic drug development (53). Thus, ECg may elicit reductions in virulence potential over and above that caused by large decreases in the secretion of enzymes and toxins (9, 55) by reduction of the proportion of LPG in the membrane. In biological milieu, ECg forms complexes with monovalent and divalent metal ions (56) and thus carries an overall positive charge; a reduction in LPG at the CM surface could increase the interaction of ECg-metal ion complexes with the phospholipid palisade caused by decreased electrostatic repulsion. LPG head groups in all likelihood contribute to the complex charge matrix at or near the staphylococcal surface, and a reduced complement of LPG-related positive charges probably contribute, along with reduced wall teichoic acid d-alanylation (23), to a more negatively charged surface. Such ECg-modified bacteria are likely to lose their capacity to form biofilms on artificial surfaces because of electrostatic repulsion (12).
In staphylococci, the new cell wall is synthesized mainly, if not exclusively, at the septum and is dependent on the formation of an FtsZ ring that ensures the cell wall biosynthetic machinery remains localized at the septation site throughout the cell division cycle (57, 58). In MRSA, the functional cooperation between PBP2 and PBP2a is maintained in the presence of β-lactam agents, growth and cell division proceed in orderly fashion and PBP2, and presumably PBP2a are found at the septum (26). In this study we show that ECg delocalizes PBP2 from the septum, and this is likely to contribute to the gross morphological changes that ECg induces in the absence of β-lactam agents. MRSA strains are sensitized to oxacillin even when PBP2a is constitutively expressed (11), suggesting either that PBP2a is also delocalized from the septum or that PBP2 and PBP2a are functionally decoupled, and PBP2a is no longer able to compensate for the loss of PBP2 transpeptidase activity in the presence of oxacillin. Delocalization of PBP2 would abrogate peptidoglycan chain extension at the site of cell division, an event with significant consequences on cell replication (57).
We have no evidence that ECg engenders greater physical separation of these proteins in the membrane or that the changes in the fluid dynamics of the membrane are responsible for the decoupling. In fact, there is little or no evidence in the literature that PBP2 and PBP2a form a close physical association within the membrane, although intuitively one would expect the functional cooperation between the two proteins to be manifest through formation of such a complex. The division site appears to be enriched for certain phospholipids (58), suggesting that the lipid composition at the septum plays a role in the assembly and maintenance of the biosynthetic machinery; ECg-induced rearrangement of the membrane bilayer may therefore contribute to PBP delocalization by inducing a suboptimal lipid environment for these membrane-anchored proteins.
There is evidence that PBP2 recruitment to the septum is at least partially dependent on recognition of the substrate, because covalent modification of the active site with β-lactams or blocking of the d-Ala-d-Ala terminus of the peptide stem of peptidoglycan with vancomycin results in delocalization (26, 58). It has been suggested (58) that β-lactam-mediated inactivation of the transpeptidase domain of PBP2 allows PBP2a, which has a low affinity for β-lactams, to bind effectively to peptidoglycan (its substrate) at the septum. This in turn allows the recruitment of PBP2 to ensure continued transglycosylation and transpeptidation in the presence of the antibiotic. Such a process is clearly disrupted by ECg and may be related to gross changes to the lipid environment as discussed above. It could also occur, however, if ECg bound directly to PBP2a and induced a conformational change that allowed acylation of the active site, as occurs at low pH (36). We therefore examined the interaction of ECg with PBP2a; incubation of PBPs with ECg did not alter their capacity to bind Bocillin FL and had no impact on kinetic properties (as measured by acylation and deacylation studies) or on the folded structure of PBP2a (as determined by tryptophan fluorescence spectroscopy). We think it likely, therefore, that changes in the lipid composition of the CM provide an environment that is incompatible with maintenance of the biosynthetic machinery at the division site.
In summary, we provide evidence that ECg binds to the CM of MRSA and penetrates deep into the hydrophobic region of the bilayer, leading to tighter acyl chain packing, a reduction in membrane fluidity, and nonlethal perturbation of cell wall synthesis. The bacteria recover by reconfiguring the membrane; a reduction in LPG, likely to affect virulence, is accompanied by an increase in branched chain fatty acid moieties of the phospholipids, resulting in the elaboration of a bilayer with higher overall fluidity than that found in nontreated cells. We propose that these gross changes in membrane fluid dynamics uncouple at least some components from the peptidoglycan biosynthetic machinery at the septum, resulting in delocalization of PBP2 and eventually also of PBP2a or abrogating the functional cooperation between PBP2 and PBP2a that allows cell wall synthesis to proceed normally in the presence of β-lactam agents. Decoupling does not appear to involve direct interactions between ECg and PBP2a. A smaller proportion of ECg can be recovered from the cell wall, and it is possible that ECg binds to nascent peptidoglycan in the wall and uncouples PBP2/PBP2a from substrate, a hypothesis that requires elucidation.
Supplementary Material
Acknowledgments
We acknowledge the Bacterial Microarray Group at St. George's for supply of the microarray and the Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. We are grateful to Elena Lastochkin for purification of PBP2a and Yukihiko Hara for supply of [3H]ECg. We thank Anna Przyborowska (King's College London) for undertaking the fatty acid analyses and Kersti Karu (London School of Pharmacy) for MS identification of lysophosphatidylglycerol. We thank Jeff Errington (University of Newcastle) for the FtsZ antibody.
This work was supported by Research Grant G0600004 from the Medical Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
M. G. Pinho, unpublished data.
- ECg
- (−)-epicatechin gallate
- PBP
- penicillin-binding protein
- MRSA
- methicillin-resistant S. aureus
- VISA
- vancomycin intermediate-resistant S. aureus
- MIC
- minimum inhibitory concentration
- CM
- cytoplasmic membrane
- PG
- phosphatidylglycerol
- LPG
- lysylphosphatidylglycerol
- MS
- mass spectrometry
- DPH
- 1,6-diphenyl-1,3,5-hexatriene
- MH
- Mueller-Hinton
- PBS
- phosphate-buffered saline
- ORF
- open reading frame.
REFERENCES
- 1.Benfield T., Espersen F., Frimodt-Møller N., Jensen A. G., Larsen A. R., Pallesen L. V., Skov R., Westh H., Skinhøj P. (2007) Clin. Microbiol. Infect. 13, 257–263 [DOI] [PubMed] [Google Scholar]
- 2.Deurenberg R. H., Stobberingh E. E. (2008) Infect. Genet. Evol. 8, 747–763 [DOI] [PubMed] [Google Scholar]
- 3.Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., Fridkin S. K. (2007) J. Am. Med. Assoc. 298, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 4.Gordon R. J., Lowy F. D. (2008) Clin. Infect. Dis. 46, S350–S359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eliopoulos G. M. (2009) J. Infect. 59, S17–S24 [DOI] [PubMed] [Google Scholar]
- 6.Shakil S., Akram M., Khan A. U. (2008) J. Chemother. 20, 411–419 [DOI] [PubMed] [Google Scholar]
- 7.Sorlozano A., Gutierrez J., Martinez T., Yuste M. E., Perez-Lopez J. A., Vindel A., Guillen J., Boquete T. (2010) Eur. J. Clin. Microbiol. Infect. Dis. 29, 73–80 [DOI] [PubMed] [Google Scholar]
- 8.Martínez J. L., Baquero F., Andersson D. I. (2007) Nat. Rev. Microbiol. 5, 958–965 [DOI] [PubMed] [Google Scholar]
- 9.Taylor P. W., Bernal P., Zelmer A. (2009) Antibiotic Resistance: Causes and Risk Factors, Mechanisms and Alternatives, pp. 43–78, Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 10.Taylor P. W., Hamilton-Miller J. M., Stapleton P. D. (2005) Food Sci. Technol. Bull. 2, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stapleton P. D., Shah S., Anderson J. C., Hara Y., Hamilton-Miller J. M., Taylor P. W. (2004) Int. J. Antimicrob. Agents 23, 462–467 [DOI] [PubMed] [Google Scholar]
- 12.Stapleton P. D., Shah S., Ehlert K., Hara Y., Taylor P. W. (2007) Microbiology 153, 2093–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco A. R., Sudano-Roccaro A., Spoto G. C., Nostro A., Rusciano D. (2005) Antimicrob. Agents Chemother. 49, 4339–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton-Miller J. M., Shah S. (1999) FEMS Microbiol. Lett. 176, 463–469 [DOI] [PubMed] [Google Scholar]
- 15.Caturla N., Vera-Samper E., Villalaín J., Mateo C. R., Micol V. (2003) Free Rad. Biol. Med. 34, 648–662 [DOI] [PubMed] [Google Scholar]
- 16.Kajiya K., Kumazawa S., Nakayama T. (2001) Biosci. Biotechnol. Biochem. 65, 2638–2643 [DOI] [PubMed] [Google Scholar]
- 17.Kajiya K., Kumazawa S., Nakayama T. (2002) Biosci. Biotechnol. Biochem. 66, 2330–2335 [DOI] [PubMed] [Google Scholar]
- 18.Kumazawa S., Kajiya K., Naito A., Saito H., Tuzi S., Tanio M., Suzuki M., Nanjo F., Suzuki E., Nakayama T. (2004) Biosci. Biotechnol. Biochem. 68, 1743–1747 [DOI] [PubMed] [Google Scholar]
- 19.Stapleton P. D., Shah S., Hara Y., Taylor P. W. (2006) Antimicrob. Agents Chemother. 50, 752–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Short S. A., White D. C. (1971) J. Bacteriol. 108, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay K., Whitmire W., Xiong Y. Q., Molden J., Jones T., Peschel A., Staubitz P., Adler-Moore J., McNamara P. J., Proctor R. A., Yeaman M. R., Bayer A. S. (2007) Microbiology 153, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 22.Haest C. W., de Gier J., op den Kamp J. A., Bartels P., van Deenen L. L. (1972) Biochim. Biophys. Acta 255, 720–733 [DOI] [PubMed] [Google Scholar]
- 23.Bernal P., Zloh M., Taylor P. W. (2009) J. Antimicrob. Chemother. 63, 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuhaus F. C., Baddiley J. (2003) Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinho M. G., Errington J. (2003) Mol. Microbiol. 50, 871–881 [DOI] [PubMed] [Google Scholar]
- 26.Pinho M. G., Errington J. (2005) Mol. Microbiol. 55, 799–807 [DOI] [PubMed] [Google Scholar]
- 27.Pinho M. G., de Lencastre H., Tomasz A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuda C., Hesek D., Lee M., Morio K., Nowak T., Mobashery S. (2005) J. Am. Chem. Soc. 127, 2056–2057 [DOI] [PubMed] [Google Scholar]
- 29.Pinho M. G., Ludovice A. M., Wu S., de Lencastre H. (1997) Microb. Drug Res. 3, 409–413 [DOI] [PubMed] [Google Scholar]
- 30.Witney A. A., Marsden G. L., Holden M. T., Stabler R. A., Husain S. E., Vass J. K., Butcher P. D., Hinds J., Lindsay J. A. (2005) Appl. Environ. Microbiol. 71, 7504–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle M., Feuerbaum E. A., Fox K. R., Hinds J., Thurston D. E., Taylor P. W. (2009) J. Antimicrob. Chemother. 64, 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 33.Ames G. F. (1968) J. Bacteriol. 95, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernal P., Muñoz-Rojas J., Hurtado A., Ramos J. L., Segura A. (2007) Environ. Microbiol. 9, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 35.Fuda C., Suvorov M., Vakulenko S. B., Mobashery S. (2004) J. Biol. Chem. 279, 40802–40806 [DOI] [PubMed] [Google Scholar]
- 36.Lemaire S., Fuda C., Van Bambeke F., Tulkens P. M., Mobashery S. (2008) J. Biol. Chem. 283, 12769–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick R. P. (2003) Mol. Microbiol. 48, 1429–1449 [DOI] [PubMed] [Google Scholar]
- 38.Utaida S., Dunman P. M., Macapagal D., Murphy E., Projan S. J., Singh V. K., Jayaswal R. K., Wilkinson B. J. (2003) Microbiology 149, 2719–2732 [DOI] [PubMed] [Google Scholar]
- 39.Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003) Mol. Microbiol. 49, 807–821 [DOI] [PubMed] [Google Scholar]
- 40.McAleese F., Wu S. W., Sieradzki K., Dunman P., Murphy E., Projan S., Tomasz A. (2006) J. Bacteriol. 188, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthaiyan A., Silverman J. A., Jayaswal R. K., Wilkinson B. J. (2008) Antimicrob. Agents Chemother. 52, 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sass V., Pag U., Tossi A., Bierbaum G., Sahl H. G. (2008) Int. J. Med. Microbiol. 298, 619–633 [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson B. J., Muthaiyan A., Jayaswal K. (2005) Curr. Med. Chem. 4, 259–276 [Google Scholar]
- 44.Trevors J. T. (2003) J. Biochem. Biophys. Methods 57, 87–103 [DOI] [PubMed] [Google Scholar]
- 45.Lennarz W. J. (1966) Adv. Lipid Res. 4, 175–225 [DOI] [PubMed] [Google Scholar]
- 46.Denich T. J., Beaudette L. A., Lee H., Trevors J. T. (2003) J. Microbiol. Methods 52, 149–182 [DOI] [PubMed] [Google Scholar]
- 47.Sutton G. C., Quinn P. J., Russell N. J. (1990) Biochem Soc. Trans. 18, 950. [DOI] [PubMed] [Google Scholar]
- 48.Chung M., Antignac A., Kim C., Tomasz A. (2008) Antimicrob. Agents Chemother. 52, 2709–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stapleton P. D., Gettert J., Taylor P. W. (2006) Int. J. Food Microbiol. 111, 276–279 [DOI] [PubMed] [Google Scholar]
- 50.Babu P. V., Liu D. (2008) Curr. Med. Chem. 15, 1840–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto T., Kumazawa S., Nanjo F., Hara Y., Nakayama T. (1999) Biosci. Biotechnol. Biochem. 63, 2252–2255 [DOI] [PubMed] [Google Scholar]
- 52.Baltz R. H. (2009) Curr. Opin. Chem. Biol. 13, 144–151 [DOI] [PubMed] [Google Scholar]
- 53.Ernst C. M., Staubitz P., Mishra N. N., Yang S. J., Hornig G., Kalbacher H., Bayer A. S., Kraus D., Peschel A. (2009) PLoS Path. 5, e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peschel A., Jack R. W., Otto M., Collins L. V., Staubitz P., Nicholson G., Kalbacher H., Nieuwenhuizen W. F., Jung G., Tarkowski A., van Kessel K. P., van Strijp J. A. (2001) J. Exp. Med. 193, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah S., Stapleton P. D., Taylor P. W. (2008) Lett. Appl. Microbiol. 46, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumamoto M., Sonda T., Nagayama K., Tabata M. (2001) Biosci. Biotechnol. Biochem. 65, 126–132 [DOI] [PubMed] [Google Scholar]
- 57.Scheffers D. J., Pinho M. G. (2005) Microbiol. Mol. Biol. Rev. 69, 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zapun A., Vernet T., Pinho M. G. (2008) FEMS Microbiol. Rev. 32, 345–360 [DOI] [PubMed] [Google Scholar]
- 59.Pereira P. M., Filipe S. R., Tomasz A., Pinho M. G. (2007) Antimicrob. Agents Chemother. 51, 3627–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.