Abstract
In mucosal tissues, epithelial M cells capture and transport microbes across the barrier to underlying immune cells. Previous studies suggested that high affinity ligands targeting M cells may be used to deliver mucosal vaccines; here, we show that particle composition and dispersion buffer ionic strength can independently influence their uptake in vivo. First, addition of a poloxamer 188 to nanoparticle formulations increased uptake of intranasally administered nanoparticles in vivo, but the effect was dependent on the presence of the M cell-targeting ligand. Second, solvent ionic strength is known to effect electrostatic interactions; accordingly, reduced ionic strength increased the electrostatic potential between the epithelium and the particles. Interestingly, below a critical ionic strength, intranasal particle uptake in vivo significantly was increased even when controlled for osmolarity. Similar results were obtained for uptake of bacterial particles. Surprisingly, at low ionic strength, the specific enhancement effect by the targeting peptide was negligible. Modeling of the electrostatic forces predicted that the enhancing effects of the M cell-targeting ligand only are enabled at high ionic strength, as particle electrostatic forces are reduced through Debye screening. Thus, electrostatic forces can have a dramatic effect on the in vivo M cell particle uptake independent of the action of targeting ligands. Examination of these forces will be helpful to optimizing mucosal vaccine and drug delivery.
Keywords: Bacteria, Drug Transport, Epithelium, Immunology, Ligand Binding Protein
Introduction
Most infectious diseases invade the body at mucosal sites. For the immune system to detect these pathogens, microfold cells (M cells)2 among the mucosal epithelium capture viruses and bacteria and transport them across the mucosal barrier (“transcytosis”) to underlying immune cells to induce protective immunity (1–4). One strategy for combating these infections is to induce immunity using vaccination, and some have focused on targeted delivery of vaccines to M cells on the expectation that mucosal immune responses will be more efficiently induced (5–8). The underlying expectation is that M cell uptake may be extremely selective, as many invasive pathogens such as Salmonella, Yersinia, and reovirus exploit specific mechanisms of M cell transcytosis. Proof of principle for the efficacy of M cell targeting has been provided in mouse models, using targeting molecules such as the fucose binding lectin Ulex europaeus agglutinin 1 (UEA-1) (9–11), and the sigma protein from reovirus (12, 13). Unfortunately, these molecules only are specific to mouse M cells and cannot be applied to human clinical practice.
Bioengineering approaches also have been studied but without the use of specific M cell-targeting strategies. Here, a tacit assumption is that M cell uptake may instead be relatively nonselective and may depend only on adhesive properties of the delivery vehicles. With the use of various polymer formulations and nanoemulsions, groups have been able to modulate physicochemical properties as a way to increase entry across mucosal epithelium and not necessarily through mucosal M cells (14–17). Polymer-based systems also can provide protection of therapeutic molecules from degradation (18–20). Despite the contrasts between immunological and bioengineering approaches, one relatively unexplored avenue is a bioengineering approach to mimic the invasive properties of mucosal pathogens, combining the use of both targeting and physicochemical strategies.
In a previous study, we explored the mechanism by which M cells within the mucosal immune system capture and uptake poly(lactic-co-glycolic acid) (PLGA) nanoparticles. PLGA nanoparticles were produced incorporating a recombinant fusion protein with influenza hemagglutinin (HA) fused to a C-terminal targeting peptide derived from Clostridium perfringens enterotoxin (CPE), creating a colloidal carrier for targeted delivery to mucosal M cells (21–23). These particles displayed enough of the protein and its targeting peptide on the surface to mediate enhanced delivery through M cells into mucosal lymphoid tissues in vivo, validating this approach to vaccine or drug delivery (24). In the present study, we investigated how physicochemical properties of the vaccine delivery vehicles influence the intranasal uptake of targeted PLGA nanoparticles in vivo. We found that modifications of the particle formulation can influence the ability of the targeting ligand to mediate M cell binding and uptake. In addition, the ionic properties of the dispersion buffer also can have a separate but no less influential effect on particle uptake. Such studies will provide new strategies for designing and optimizing mucosal vaccines; in addition, our results point to forces that may influence the invasive properties of mucosal pathogens, especially in the context of their activity in contaminated food and water.
EXPERIMENTAL PROCEDURES
Materials
The PLGA (poly(dl-lactide-co-glycolide) 85:15, molecular weight 50,000–75,000), polyvinyl alcohol (molecular weight 30,000–70,000, 87–90% hydrolyzed), and plx (molecular weight: 8500) were obtained from Sigma-Aldrich. 1 m HEPES, phosphate-buffered saline (PBS, 1×), SDS solution (10%) and Staphylococcus aureus (Wood strain without protein A) BioParticles®-Alexa Fluor® 488 conjugate were purchased from Invitrogen. Methylene Chloride optima®, PBS (10× ready concentrate pouches), HEPES (powder fine white crystals), d-mannitol, and sodium hydroxide were obtained from Thermo Fisher Scientific. Rhodamine 6G was obtained from Fluka® Analytical, and 16% paraformaldehyde was obtained from Electron Microscopy Sciences. Prolong Gold antifade reagent with 4′,6-diamidino-2-phenylindole was purchased from Molecular Probes, and a Pierce BCATM protein assay kit was obtained from Thermo Fisher Scientific.
Nanoparticle Preparation
PLGA nanoparticles containing targeting (HA-ts-HT-CPE) and nontargeting (HA-ts-HT) peptides were prepared from 85:15 PLGA using solvent evaporation/double emulsion (also known as water-in-oil-in-water, w/o/w) method as described in Rajapaksa et al. (24). Briefly, 4% PLGA polymer solution was prepared by dissolving 0.18 g of PLGA in 4.5 ml of methylene chloride. For labeled nanoparticles, 1 mg of rhodamine 6G was added to the PLGA solution. Then, 0.5 ml of protein solution and 0.25 ml of 2% polyvinyl alcohol solution in 10 mm HEPES buffer adjusted to pH 7.5 were added to 4.25 ml of the polymer solution and emulsified by probe sonication for 20 s (Branson® Sonifier 450 with a Duty cycle of 20% and output control of 3). The resulting emulsion (w/o) was divided into two tubes, then we added 12.5 ml of 2% polyvinyl alcohol solution to each tube and emulsified them for 30 s to obtain the final w/o/w emulsion. The final w/o/w was then combined in a 50-ml beaker and stirred for 20 h with a magnetic stirrer at 400 rpm at 4 °C to allow solvent evaporation. The nanoparticles were collected by centrifugation at 3800 rpm for 30 min and resuspended in double distilled water; the washing step was repeated three times. The nanoparticles were freeze-dried, and the final product was stored at 4 °C until used.
For plx-incorporated particles, 11.5 ml of 2% polyvinyl alcohol was added to the equally divided first w/o emulsion and sonicated for 20 s. Then, 1 ml of 12.5% plx solution (final concentration of 1%) was added to the resulting emulsion and sonicated for another 10 s to obtain the final w/o/w emulsion and processed similar to non-plx incorporated particles as described above.
Scanning Electron Microscopy
The morphology of the protein-loaded nanoparticles was visualized by scanning electron microscopy. The nanoparticles were placed on a double-sided adhesive tape attached to an aluminum stub and sputter-coated with gold/palladium beam for 2 min. The coated sampled were imaged with Philips XL30-FEG scanning electron microscopy at 10kV.
Particle Size and Polydispersity Measurements
The particle size and the polydispersity index (PDI) of the nanoparticles were measured by dynamic light scattering using a Zetasizer Nano ZS90 (Malvern Instruments). Samples of PLGA nanoparticle dispersion in PBS (1 mg/ml concentration) were placed in a disposable cuvette for size measurements. Each sample was measured three times for triplicate preparations of nanoparticles and is reported as mean ± S.D.
Determination of Total Protein Loading (%w/w) and Surface Loading
Total protein loading was estimated using a BCA assay. Approximately 5–8 mg of freeze-dried nanoparticles were accurately measured and added to 1 ml of 5% SDS in 0.1 m NaOH solution and incubated with shaking for 24 h at room temperature until a clear solution was obtained. The protein content was measured in triplicates for each sample using BCA protein assay. The protein loading (%, w/w) was expressed as the amount of protein relative to the weight of the nanoparticles assayed. In a separate experiment, blank PLGA nanoparticles were prepared, and protein loading for these “nonprotein-loaded particles” was measured using BCA assay. The protein loading calculations showed negligible interference in BCA assay by blank particles.
The surface loading of HA-ts-HT and HA-ts-HT-CPE30 in nanoparticles with and without plx also was measured. Approximately 5–8 mg of freeze-dried nanoparticles from three different experiments were measured accurately and dispersed in 1 ml of 2% SDS solution. The samples were shaken at 200 rpm at room temperature. 4 h later, samples were centrifuged at 6000 rpm for 5 min. The protein content of the supernatant was measured in triplicates using the BCA assay. Percent surface loading was calculated based on the percent protein loading for each sample.
Zeta Potential Measurements
The zeta potential of all nanoparticle formulations was measured using laser Doppler electrophoresis (Zetasizer Nano ZS90, Malvern Instruments). The nanoparticles were dispersed in different buffer solutions to obtain a 0.05% (w/v) solution. To determine the effect of the dispersion buffer ionic strength on zeta potential, the nanoparticles were dispersed in dilutions of PBS at pH 7.0 (ionic strengths: 1.5 × 10−1 m, 1.5 × 10−2, 8.5 × 10−3 m, 1.5 × 10−3 m, 1.5 × 10−4 m, and 1.5 × 10−5 m). The ionic strength (I) of the solutions were determined using the following equation: I = ½Σ(cizi2), where ci is the concentration, and zi is the charge (valence) of all significant ions in solution (25). The ionic strength of deionized water is reported as 1 × 10−7 m (25). The Debye parameter (κ) relates ionic strength (I), universal gas constant (R), absolute temperature (T), Faraday's constant (F), and permittivity of the solution (ϵ, which equals to the relative permittivity or dielectric constant ϵr multiplied by the permittivity of free space ϵo) and defined as κ2 = (1000F2)/(ϵRT)/Σcizi2. The value of κ simplifies to κ = 3.29√I (nm−1) at 25 °C. The Debye length is defined as 1/κ (25).
The zeta potentials of bacterial strains Staphylococcus aureus, Streptococcus pneumoniae, and Yersinia were measured in different ionic strength PBS solutions by dispersing 8 × 107 bacteria per 1.5 ml of solution. The zeta potential for each bacterial strain was measured in triplicate.
In Vivo Uptake Studies
For in vivo uptake studies, rhodamine 6G-labeled and HA-ts-HT- or HA-ts-HT-CPE-loaded nanoparticles with or without plx were used depending on the experiment. Under anesthesia, mice were given 20 μg of protein in 40 μl of dispersion buffer (dilutions of PBS at various ionic strengths or distilled H2O) intranasally (20 μl/nostril), and nasal-associated lymphoid tissue (NALT) was dissected after 3–5 min of nanoparticle incubation. Dissected NALT tissues were fixed in 4% paraformaldehyde with 30% sucrose in PBS, which were then stained with UEA-1 (Vector). Casein/PBS (Fisher) with a final concentration of 0.1% Tween was used for blocking and for dilutions of antibody. Tissues were post-fixed for 10 min with 4% paraformaldehyde and mounted using ProLong Gold with 4′,6-diamidino-2-phenylindole. NALT tissue was analyzed using a BD CARV II spinning disc confocal microscope, using IPLab software. Three representative images were selected from each specimen. Image deconvolution and analysis was performed using Volocity software (Volocity, version 5.3.1, Improvision Inc.). Nanoparticle uptake was measured from z-projection images from UEA-1-stained NALT taking 7850 μm2 areas where UEA-1-positive cells were evident. Nanoparticle counts were performed using Volocity software, and number of particles taken up in individual 7850 μm2 area “stamps” was plotted for three independent experiments. Five-count stamps were analyzed per image. Parameters for analysis were based on object intensity and volume. In Figs. 2 and 4, each symbol denotes the number of particles taken up in each 7850 μm2 area that was analyzed in NALT tissue sections. For comparisons of particle uptake, the uptake is represented as the means ± S.E. and a two-tailed t test was used for statistical analysis (Prism, GraphPad).
FIGURE 2.
NALT uptake of nanoparticles prepared with and without plx. A, 20 μl of nanoparticle dispersion (in PBS) was given to each nostril (20 μg of protein per animal), and the NALT tissue was dissected and stained for UEA-1. Confocal images were recorded in three different areas of the same NALT where UEA-1-positive M cells were visible. Each symbol denotes the number of particles taken up in each 7850 μm2 area that was analyzed in each NALT tissue dissected, with three mice for each condition. Uptake of fluorescent PLGA nanoparticles show increased NALT uptake when targeted protein (HA-ts-HT-CPE) is incorporated, with a further enhancement when plx is also incorporated.
FIGURE 4.
Targeted PLGA nanoparticle uptake in varying ionic strength buffers. NALT uptake of plx-incorporated targeted protein-loaded nanoparticles dispersed in PBS was compared with the uptake of targeted nanoparticles prepared with and without plx dispersed in deionized water. Each symbol denotes the number of particles taken up in each 7850 μm2 area that was analyzed in each NALT tissue dissected in three mice for each condition. PLGA nanoparticles with targeted protein show increased NALT uptake when administered in water versus buffered saline.
To control for osmolarity of low ionic strength dispersion buffer in the experiments, reduced NaCl was replaced by mannitol. NALT uptake studies were performed in a NaCl solution of 1.55 × 10−1 m ionic strength and 300 mosmol/kg, which represents the physiological strength and isosmotic low NaCl solutions of 1:20 and 1:100 dilutions of NaCl: mannitol and isosmotic 100% mannitol solution (2.9 × 10−1 m, 312 mosmol/kg). The osmolality of the solutions was measured with a Vapor Pressure Osmometer (Wescor 5500). The uptake studies were performed and analyzed as described above.
Similarly, S. aureus uptake in NALT was performed by dispersing fluorescently labeled S. aureus (1 × 109; Invitrogen) in PBS or a 0.17 × 10−3 m ionic strength dilution of PBS. The NALT was processed as described earlier, and S. aureus uptake counts were performed using Volocity software. Because of the higher rate of uptake of bacteria, counts were measured using 1600 μm2 areas, and means ± S.E. for the number of particles taken up in each was plotted for three independent experiments for each condition.
RESULTS
Development and Physiochemical Characteristics of PLGA Nanoparticles with plx
We showed previously that claudin 4-targeted protein incorporated into PLGA nanoparticles can mediate M cell-targeted delivery (24). Here, we modified the surface properties of our PLGA nanoparticles containing recombinant influenza HA fusion protein (with a C-terminal targeting peptide, CPE30, HA-ts-HT-CPE, or without, HA-ts-HT). By adding 1% plx solution in the final w/o/w emulsion, we produced nanoparticles that would incorporate plx. Plx (Poloxamer 188 or Pluronic F68) is an amphiphilic triblock co-polymer with hydrophilic polyethylene glycol chains (also known as polyethylene oxide) linked to a more hydrophobic polypropylene glycol backbone (26–29). Scanning electron microscopy images revealed that nanoparticles from all formulations were spherical with smooth surfaces (Fig. 1), similar to our previous nanoparticles (24). An increase in plx concentration of >1% resulted in formation of morphologically distorted particles with a nonuniform size distribution. All nanoparticle formulations possessed a narrow size distribution, as confirmed by polydispersity measurements (Table 1). The average hydrodynamic diameter of particles ranged from 449 to 512 nm. The average protein loading was between 1.62 and 2.16% (w/w) for all formulations with and without plx. Modification of nanoparticles with plx did not affect the surface loading of protein, where percent surface loading of protein for all formulations was between 15.25–17.77% (w/w) of total protein loading. Additionally, native gel analysis showed no sign of protein aggregation inside nanoparticles (supplemental Fig. S1). The in vitro release studies showed rather slow release of HA protein from the particles; plx particles showed slightly increased protein release but was not significantly different (supplemental Fig. S2). Physicochemical analysis showed no significant differences in particle size, polydispersity index, total and surface protein loading, and in vitro release profile between particles formulated with and without plx.
FIGURE 1.
Scanning electron microscopy of plx-incorporated PLGA nanoparticles. PLGA nanoparticles loaded with recombinant proteins (HA-ts-HT and HA-ts-HT-CPE) were prepared by a double emulsion, solvent evaporation method. The nanoparticles were placed on a double-sided adhesive tape attached to an aluminum stub and sputter coated with gold/palladium beam for 2 min. The coated samples were imaged with a Philips XL30-FEG scanning electron microscope at acceleration voltage 10 kV and magnification 20,000×. The nanoparticles show uniform shape and size.
TABLE 1.
Physical characteristics of PLGA nanoparticles
Nontargeted HA-ts-HT- and targeted HA-ts-HT-CPE-loaded nanoparticles were prepared with or without plx. The diameter of the particles in solution and P.D.I. were measured by Zetasizer Nano (ZS90) by a dynamic light scattering method. The total % protein loading (w/w) and % surface loading from total loading was measured using BCA protein assay. Control experiments revealed that there is negligible interference by blank PLGA particles on BCA assay. Percent protein (% w/w) loading for blank particles was ∼0.031%. The zeta potential in PBS at pH 7.4 was measured by Zetasizer Nano (ZS90). The particle size, % protein loading, % surface loading, and zeta potential are expressed as mean ± S.D. for three different preparations of nanoparticles.
| Sample | Particle diameter | P.D.I. | % protein loading | % surface loading from total loading | Zeta potential in PBS, pH 7.4 |
|---|---|---|---|---|---|
| nm | w/w | mV | |||
| HA-ts-HT | 464 ± 12 | 0.11 | 2.02 ± 0.39 | 15.25 ± 1.40 | −2.46 ± 0.67 |
| HA-ts-HT-CPE | 449 ± 17 | 0.08 | 2.16 ± 0.38 | 16.80 ± 1.08 | −2.34 ± 0.19 |
| HA-ts-HT + plx | 512 ± 12 | 0.11 | 1.62 ± 0.33 | 17.77 ± 4.24 | −2.08 ± 0.60 |
| HA-ts-HT-CPE + plx | 449 ± 21 | 0.10 | 1.76 ± 0.28 | 15.45 ± 1.04 | −1.66 ± 0.34 |
Surface Modification of Nanoparticles with plx and Its Effect on M Cell Uptake
Plx was incorporated into our delivery system to modify the surface properties of PLGA nanoparticles. Incorporation of polyethylene oxide is known to modify the surface charge of PLGA, change the hydrophobic/hydrophilic nature of the particles, and affect the rate of diffusion of particles through mucus (28, 30, 31). The relative surface charge of particles in colloidal dispersion can be calculated based on their mobility across an electric field, described as zeta potential. Here, zeta potential measurements showed no significant difference between particles prepared with and without plx in 1× PBS at pH 7.4. However, at this physiological dispersion condition, all nanoparticle formulations showed close-to-neutral charge, which ranged from −2.46 to −1.66 mV (Table 1).
The plx used here has a hydrophilic-lipophilic balance of 29, which is likely to make the surface of our PLGA particles more hydrophilic. Though studies suggest that hydrophobic particles are taken up better by M cells (32), it has also been shown that viscous, elastic and sticky mucus that line all mucosal tissues readily trap and eliminate hydrophobic particles as a protective measure (17). Interestingly, other studies show evidence for increased uptake of hydrophilic polyethylene glycol and plx-incorporated particles by M cells compared with classical PLGA particles (17, 30, 33). Our intranasal NALT uptake studies showed a significant increase in the uptake of plx incorporated HA-ts-HT-CPE nanoparticles when compared with non-plx incorporated HA-ts-HT-CPE particles and HA-ts-HT particles both with and without plx (Fig. 2). Addition of plx caused a 2-fold increase in targeted nanoparticle uptake but only when the targeting peptide CPE was included. Thus, the effect of plx mainly may be to increase the ability of the particle targeting peptide to interact with the mucosal epithelial membrane without specifically increasing particle adhesion.
Properties of Nanoparticle/Dispersion Buffer Colloidal System: The Influence of pH and Ionic Strength
Although the physiochemical and surface properties of nanoparticles provide vital information on polymer-based drug delivery vehicles, evaluation of particle/dispersion buffer colloidal systems may provide insight on further enhancing delivery of antigen into mucosal surfaces (34–36). Thus, we measured the zeta potential of particles dispersed in different buffer conditions by varying pH and ionic strength. Nanoparticle dispersions showed a very negative surface charge between pH 5.0–7.0, and the effect of plx was more significant in this region, making the particles less electronegative (supplemental Fig. S3). At the lowest pH (pH 2.5), all four nanoparticle dispersions showed positive zeta potentials (supplemental Fig. S3). We also evaluated the effect of ionic strength on measured zeta potential. At physiological buffer ionic strengths (1.7 × 10−1 m, pH 7.4), the zeta potential was close to zero (Table 1 and Fig. 3). However, as the ionic strength was reduced, the zeta potential values became more negative (Fig. 3). All four nanoparticle formulations possessed a very negative zeta potential (around −26 mV) at the lowest ionic strength (1.7 × 10−5 m, pH 7.4). Blank (i.e. no protein loaded) PLGA particles prepared with and without plx showed even more negative zeta potential at this ionic strength. Interestingly, incorporation of plx did not affect the particle zeta potential significantly at tested ionic strengths except for the lowest ionic strength for HA-ts-HT-CPE loaded particles.
FIGURE 3.
Nanoparticle zeta potential and ionic strength. PLGA nanoparticles were produced empty (Blank) or with recombinant proteins both with and without plx. The nanoparticles were dispersed in different buffer solutions to obtain a 0.05% (w/v) solution. The zeta potential was measured using laser Doppler electrophoresis (Zetasizer Nano ZS90). To determine the effect of dispersion buffer ionic strength on the zeta potential of nanoparticles, the nanoparticles were dispersed in dilutions of PBS at pH 7.0 (ionic strengths: 1.5 × 10−1 m, 1.5 × 10−2 m, 8.5 × 10−3 m, 1.5 × 10−3 m, 1.5 × 10−4 m, and 1.5 × 10−5 m). The zeta potentials were measured in triplicate for three different preparations of nanoparticles for each condition, and each point presents the mean ± S.D.
Effect of Dispersion Buffer Ionic Strength on M Cell Uptake of Nanoparticles
Solvent ionic strength is known to affect particle electrostatic interactions (36, 37); however, to our knowledge no study systematically has examined the influence of dispersion buffer ionic strength on in vivo M cell uptake of particles. Targeted (HA-ts-HT-CPE) nanoparticles both with and without plx, when dispersed in low ionic strength solution (1 × 10−7 m, pH 7.4), showed a significant increase in uptake compared with plx-incorporated targeted particles dispersed in physiological ionic strength (1.7 × 10−1 m, pH 7.4) (Fig. 4). Thus, at low ionic strength dispersion conditions, nanoparticles were taken up in higher numbers regardless of incorporation of plx, suggesting that additional forces may independently contribute to M cell uptake.
Because targeted particles were taken up better than non-targeted particles at physiological buffer conditions (Fig. 2) (24), we tested whether this also would apply when plx-incorporated particles were dispersed in low ionic strength buffers. First, as expected, when particles were dispersed in physiological ionic strength, HA-ts-HT-CPE particles showed significantly higher uptake compared with HA-ts-HT-loaded particles (Fig. 5A). When dispersion buffer ionic strength was reduced to 8.5 × 10−3 m, HA-ts-HT-CPE particles still were taken up in higher numbers compared with HA-ts-HT particles; however, the difference was not significant (p = 0.1833). With additional reduction in dispersion buffer ionic strength, the uptake of both HA-ts-HT- and HT-ts-HT-CPE-loaded nanoparticles increased further. Thus, at low ionic strengths, nanoparticles readily were taken up regardless of presence or absence of targeting peptide and/or incorporation of plx.
FIGURE 5.
Plx-incorporated nanoparticle uptake by NALT with different ionic strength buffers. A, PLGA nanoparticle uptake versus ionic strength and effect of targeting ligand. 20 μl of nanoparticle dispersion was given to each nostril (20 μg of protein per animal), and the NALT tissue was dissected and stained for UEA-1. Confocal images were recorded for three different areas of each whole mount NALT, where UEA-1-positive M cells were visible. Nanoparticle counts were performed using Volocity software, and the number of particles taken up in individual 7850 μm2 area stamps was analyzed for three independent experiments. Five-count stamps were analyzed per image. Parameters for analysis were based on object intensity and volume. For comparisons of particle uptake, the uptake is represented as the means ± S.E., and a two-tailed t test was used for statistical analysis (Prism; GraphPad). The results show increased uptake at lower ionic strength and loss of targeting effect at very low ionic strength. The p values at the top of the figure refer to comparisons of HA-ts-HT-CPE + plx and HA-ts-HT + plx, respectively. B, PLGA nanoparticle uptake versus ionic strength controlled for osmolarity, using mannitol as an osmolyte, showing increased uptake at lower ionic strength when controlled for osmolarity. C, confocal projection image of NALT showing UEA-1-positive M cells (green) and fluorescently labeled nanoparticles (red).
Exposure of the mucosal epithelium to low ionic strength solutions could have effects on the epithelial cells and mucus, mainly due to the hypo-osmolarity of the solution. Transient cellular swelling directly could affect particle uptake mechanisms, or epithelial damage could indirectly increase the penetration of particles. Mucosal epithelium can be exposed to a wide range of ionic conditions especially in the stomach and upper small intestine, where hypo-osmolar solutions (e.g. water) routinely are tolerated. In addition, our studies suggest that particle uptake is both active and selective, so nonspecific effects from hypo-osmolar solutions would seem less likely. However, to control for such effects, we performed a similar study using mannitol to maintain the osmolarity of the dispersion buffer. Interestingly, decreasing ionic strength still proved to have a significant effect on uptake, with the main transition occurring in the same range of ionic strength seen previously (Fig. 5, B and C). Thus, it would appear that the main effect on NALT particle uptake may be attributable to buffer ionic strength rather than osmolarity.
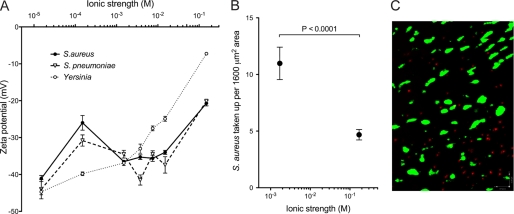
Mucosal epithelium M cells are an important entry point for many invasive pathogens, so we investigated zeta potential and M cell uptake of bacterial strains S. aureus, S. pneumoniae, and Yersinia enterocolitica (Yersinia), which are prevalent in the mucosal immune system, and shown to be taken up by M cells. At all tested ionic strengths, bacteria showed more negative zeta potential compared with PLGA nanoparticles (Fig. 6A). Interestingly, S. aureus uptake by NALT was significantly increased when S. aureus was administered in low ionic strength buffer (1.7 × 10−3 m) compared with physiological ionic strength (Fig. 6, B and C), suggesting that dispersion buffer ionic strength also may influence uptake of bacterial particles at mucosal surfaces.
FIGURE 6.
Physical characteristics and uptake of microbial particles. A, the zeta potentials of bacterial strains S. aureus, S. pneumoniae, and Yersinia were measured in different ionic strength PBS solutions by dispersing 8 × 107 bacteria/1.5 ml solution. The zeta potential for each bacterial strain was measured in triplicate. Microbial particles show higher magnitude zeta potentials (more negative) than PLGA nanoparticles across the range of ionic strength. B, S. aureus uptake versus ionic strength, showing increased uptake at low ionic strength. C, confocal projection image of NALT showing UEA-1-positive M cells (green) and fluorescently labeled bacteria (red).
DISCUSSION
In this study, we examined factors that may enhance targeting and uptake of our M cell targeted nanoparticles by modifying the surface properties and dispersion buffer conditions. In the case of plx addition to PLGA nanoparticles, we believe that the reason for increased uptake of plx-incorporated particles is due to increased penetration of these particles into the nasal mucus. It has been shown that polymers such as polyethylene glycol incorporated on the particle surface will enhance transport in mucus (33, 38, 39). Given the structural similarity between polyethylene glycol and plx, we speculate that PLGA-plx particles would penetrate the mucus layer better than non-plx PLGA particles. Csesa et al. (30) also have reported increased intranasal uptake of PLGA: plx blended particles compared with classical PLGA particles. Interestingly, plx alone could not increase particle uptake significantly without the presence of CPE targeting at physiological dispersion buffer conditions, suggesting that although plx may increase the penetration of particles, targeting was still necessary for interaction with M cells and the particles at mucosal epithelium.
Therapeutics such as mucosal vaccines normally would be expected to be administered in buffer solutions at physiological ionic strength. Under these conditions, the enhancing effects of targeting and plx were most pronounced, validating the strategy of modifying nanoparticle delivery vehicles with M cell targeting. The Debye length at 1.7 × 10−1 m ionic strength solution (i.e. physiological saline) is ∼0.74 nm, which is >5× shorter than the length of CPE30, which we estimated at 4 nm, based on the modeling studies by Kimura et al. (40). Note that the CPE peptide (30 amino acids) only has a net charge of +1 (only three charged residues: two lysines and one aspartic acid) and, thus, is not electrostatically significant. Thus, at physiological ionic strength, particles that reached the mucosal epithelial surface efficiently (perhaps in part due to plx on the surface) could be taken up by M cells with the assistance of specific short range receptor-ligand interactions (41).
Importantly, we found that the uptake of nanoparticles was even greater under low ionic strength conditions, even when controlled for buffer osmolarity. At lower ionic strength, the electrostatic potential would extend beyond the distance of influence of the CPE30-targeting peptide on the surface of the particle. (The Debye length increases by a factor of 100 when the ionic strength is reduced from 1.7 × 10−1 m to 1.7 × 10−5 m.) This may be why any enhancing effect of the targeting peptide was lost. This greater range can provide for long range interactions with mucosal membranes, although we cannot be certain whether this is due to charge interactions with the epithelial cells themselves, which are likely to be negatively charged, or with the overlying mucus layer, which could provide multiple complex charge interactions (42, 43). In any case, these results suggest that ionic strength of any dispersion buffer can be manipulated to optimize mucosal delivery of nanoparticles, relying on the long range electrostatic interactions to enhance particle-epithelium interactions (41, 44). Our results using mannitol to control buffer osmolarity suggest that this effect may also be applied to oral formulations in which a nonionic osmolyte could be used to maintain low ionic strength at least through the upper part of the small intestine (45, 46).
Our results also may have implications for microbial pathogenesis in mucosal tissues. In the case of intestinal pathogens, foodborne pathogens have the potential to infect and colonize the epithelium depending in large part on the targeting and adhesion proteins on the viruses and bacteria (44, 47). However, in the case of ingestion of contaminated fresh water, the low ionic strength will be maintained at least through the stomach and upper small intestine; ionic strength would not be normalized to blood plasma values until the jejunum (34, 48, 49). Therefore, electrostatic forces could dominate in the stomach and upper intestine under these conditions, enabling invasive organisms to rapidly interact with mucosal epithelium with forces greater than that provided by targeting or adhesive proteins alone. This effect would place particular importance on fresh water quality, especially in developing countries where diarrheal diseases are an important factor in public health.
Supplementary Material
Acknowledgments
We thank B. Anvari, J. S. Schultz, and the Center for Bioengineering at the University of California, Riverside, for use of the Malvern Zetasizer, C. Kieslich for related electrostatics calculations, P. Quinton for valuable discussions, and N. Appleby for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AI63426 and AI73689 and the Foundation for the National Institutes of Health. This work was also supported by an award from the Grand Challenges in Global Health and the Bill and Melinda Gates Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S3.
- M cells
- microfold cells
- plx
- poloxamer 188
- NALT
- nasal-associated lymphoid tissue
- PLGA
- poly(lactic-co-glycolic acid)
- HA
- hemagglutinin
- UEA-1
- Ulex europaeus agglutinin 1
- CPE
- Clostridium perfringens enterotoxin.
REFERENCES
- 1.Neutra M. R., Kozlowski P. A. (2006) Nat. Rev. Immunol. 6, 148–158 [DOI] [PubMed] [Google Scholar]
- 2.Neutra M. R., Mantis N. J., Frey A., Giannasca P. J. (1999) Semin. Immunol. 11, 171–181 [DOI] [PubMed] [Google Scholar]
- 3.Shahiwala A., Vyas T. K., Amiji M. M. (2007) Recent Pat. Drug Deliv. Formul. 1, 1–9 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H., Sekine S., Kataoka K., Pascual D. W., Maddaloni M., Kobayashi R., Kozono H., McGhee J. R., Fujihashi K. (2008) Gastroenterology 135, 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark M. A., Jepson M. A., Hirst B. H. (2001) Adv. Drug Deliv. Rev. 50, 81–106 [DOI] [PubMed] [Google Scholar]
- 6.Florence A. T. (2004) J. Drug Target. 12, 65–70 [DOI] [PubMed] [Google Scholar]
- 7.Jepson M. A., Clark M. A., Foster N., Mason C. M., Bennett M. K., Simmons N. L., Hirst B. H. (1996) J. Anat. 189, 507–516 [PMC free article] [PubMed] [Google Scholar]
- 8.Shakweh M., Ponchel G., Fattal E. (2004) Expert Opin. Drug Deliv. 1, 141–163 [DOI] [PubMed] [Google Scholar]
- 9.Foster N., Clark M. A., Jepson M. A., Hirst B. H. (1998) Vaccine 16, 536–541 [DOI] [PubMed] [Google Scholar]
- 10.Gupta P. N., Khatri K., Goyal A. K., Mishra N., Vyas S. P. (2007) J. Drug Target. 15, 701–713 [DOI] [PubMed] [Google Scholar]
- 11.Manocha M., Pal P. C., Chitralekha K. T., Thomas B. E., Tripathi V., Gupta S. D., Paranjape R., Kulkarni S., Rao D. N. (2005) Vaccine 23, 5599–5617 [DOI] [PubMed] [Google Scholar]
- 12.Helander A., Silvey K. J., Mantis N. J., Hutchings A. B., Chandran K., Lucas W. T., Nibert M. L., Neutra M. R. (2003) J. Virol. 77, 7964–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y., Wang X., Csencsits K. L., Haddad A., Walters N., Pascual D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9318–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielinska A. U., Janczak K. W., Landers J. J., Markovitz D. M., Montefiori D. C., Baker J. R., Jr. (2008) AIDS Res. Hum Retroviruses 24, 271–281 [DOI] [PubMed] [Google Scholar]
- 15.Eldridge J. H., Gilley R. M., Staas J. K., Moldoveanu Z., Meulbroek J. A., Tice T. R. (1989) Curr. Top Microbiol. Immunol. 146, 59–66 [DOI] [PubMed] [Google Scholar]
- 16.Kharenko E. A., Larionova N. I., Demina N. B. (2009) Pharm. Chem. J. 43, 200–208 [Google Scholar]
- 17.Lai S. K., Wang Y. Y., Hanes J. (2009) Adv. Drug Deliv. Rev. 61, 158–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al Haushey L., Bolzinger M. A., Bordes C., Gauvrit J. Y., Briançon S. (2007) Int. J. Pharm. 344, 16–25 [DOI] [PubMed] [Google Scholar]
- 19.Lavelle E. C., Yeh M. K., Coombes A. G., Davis S. S. (1999) Vaccine 17, 512–529 [DOI] [PubMed] [Google Scholar]
- 20.Rajapaksa T. E., Lo D. D. (2010) Curr. Immunol. Rev. 6, 29–37 [Google Scholar]
- 21.Kondoh M., Takahashi A., Fujii M., Yagi K., Watanabe Y. (2006) Biol. Pharm. Bull. 29, 1783–1789 [DOI] [PubMed] [Google Scholar]
- 22.Ling J., Liao H., Clark R., Wong M. S., Lo D. D. (2008) J. Biol. Chem. 283, 30585–30595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo D., Tynan W., Dickerson J., Scharf M., Cooper J., Byrne D., Brayden D., Higgins L., Evans C., O'Mahony D. J. (2004) Int. Immunol. 16, 91–99 [DOI] [PubMed] [Google Scholar]
- 24.Rajapaksa T. E., Stover-Hamer M., Fernandez X., Eckelhoefer H. A., Lo D. D. (2010) J. Control Release 142, 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory J. (2005) Particles in Water: Properties and Processes, Taylor & Francis, London [Google Scholar]
- 26.Besheer A., Vogel J., Glanz D., Kressler J., Groth T., Mader K. (2009) Mol. Pharm., in press [DOI] [PubMed] [Google Scholar]
- 27.Moghimi S. M., Hunter A. C. (2000) Trends Biotechnol. 18, 412–420 [DOI] [PubMed] [Google Scholar]
- 28.Ratzinger G., Länger U., Neutsch L., Pittner F., Wirth M., Gabor F. (2010) Langmuir. 26, 1855–1859 [DOI] [PubMed] [Google Scholar]
- 29.Tobío M., Nolley J., Guo Y., McIver J., Alonso M. J. (1999) Pharm. Res. 16, 682–688 [DOI] [PubMed] [Google Scholar]
- 30.Csaba N., Sánchez A., Alonso M. J. (2006) J. Control Release 113, 164–172 [DOI] [PubMed] [Google Scholar]
- 31.Santander-Ortega M. J., Csaba N., Alonso M. J., Ortega-Vinuesa J. L., Bastos-Gonzalez D. (2007) Colloid Surface A 296, 132–140 [Google Scholar]
- 32.Hillery A. M., Florence A. T. (1996) Int. J. Pharm. 132, 123–130 [Google Scholar]
- 33.Lai S. K., O'Hanlon D. E., Harrold S., Man S. T., Wang Y. Y., Cone R., Hanes J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonferoni M. C., Rossi S., Ferrari F., Stavik E., Pena-Romero A., Caramella C. (2000) AAPS Pharm. Sci. Tech. 1, E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurtovenko A. A., Lyulin S. V., Karttunen M., Vattulainen I. (2006) J Chem. Phys. 124, 094904 [DOI] [PubMed] [Google Scholar]
- 36.Ryan J. N., Gschwend P. M. (1994) J Colloid Interf. Sci. 165, 536–536 [Google Scholar]
- 37.Zhang Y., Yang M., Portney N. G., Cui D., Budak G., Ozbay E., Ozkan M., Ozkan C. S. (2008) Biomed. Microdevices. 10, 321–328 [DOI] [PubMed] [Google Scholar]
- 38.Cu Y., Saltzman W. M. (2009) Mol. Pharm. 6, 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang B. C., Dawson M., Lai S. K., Wang Y. Y., Suk J. S., Yang M., Zeitlin P., Boyle M. P., Fu J., Hanes J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19268–19273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura J., Abe H., Kamitani S., Toshima H., Fukui A., Miyake M., Kamata Y., Sugita-Konishi Y., Yamamoto S., Horiguchi Y. (2010) J. Biol. Chem. 285, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Florence A. T. (2005) Drug. Discov. Today 2, 75–81 [DOI] [PubMed] [Google Scholar]
- 42.Li Q., Logan B. E. (1999) Water Res. 33, 1090–1100 [Google Scholar]
- 43.Tsao H. K., Sheng Y. J. (2001) J Colloid Interf. Sci. 233, 124–130 [DOI] [PubMed] [Google Scholar]
- 44.Busscher H. J., van de Belt-Gritter B., Dijkstra R. J., Norde W., van der Mei H. C. (2008) Langmuir 24, 10968–10973 [DOI] [PubMed] [Google Scholar]
- 45.Doble M. A., Tola V. B., Chamberlain S. A., Cima R. R., Van Hoek A., Soybel D. I. (2002) J Gastrointest Surg. 6, 387–395 [DOI] [PubMed] [Google Scholar]
- 46.Sleator R. D., Hill C. (2002) Fems. Microbiol Rev. 26, 49–71 [DOI] [PubMed] [Google Scholar]
- 47.van Merode A. E. J., Duval J. F. L., van der Mei H. C., Busscher H. J., Krom B. P. (2008) Colloid Surface B 64, 302–306 [DOI] [PubMed] [Google Scholar]
- 48.Allen M. J., Edberg S. C., Reasoner D. J. (2004) Int. J Food Microbiol. 92, 265–274 [DOI] [PubMed] [Google Scholar]
- 49.Busscher H. J., Dijkstra R. J., Engels E., Langworthy D. E., Collias D. I., Bjorkquist D. W., Mitchell M. D., Van der Mei H. C. (2006) Environ. Sci. Technol. 40, 6799–6804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.