Abstract
β-Catenin is a bifunctional protein participating in both cell adhesion and canonical Wnt signaling. In cell adhesion, it bridges the transmembrane cadherin and the actin-binding protein α-catenin and is essential for adherens junction formation, whereas in canonical Wnt signaling, it shuttles between the cytosol and nucleus and functions as an essential transcriptional activator. Schmidtea mediterranea β-catenin-1 was identified as a determinant of antero-posterior polarity during body regeneration by mediating Wnt signaling. Here we show that S. mediterranea β-catenin-2 is specifically expressed in epithelial cells in the gut and pharynx, where it has a putative role in mediating cell adhesion. We show evidence that planarian β-catenin-1 and -2 have distinct biochemical properties. β-Catenin-1 can interact with the components of the canonical Wnt signaling pathway but not with α-catenin, whereas β-catenin-2 interacts with cell adhesion molecules, including E-cadherin and α-catenin, but not with Wnt signaling components. Consistent with their specific function, β-catenin-1 is a potent transcriptional activator, whereas β-catenin-2 has no transcriptional activity. Protein sequence alignment also indicates that the planarian β-catenin-1 and -2 retain distinct critical residues and motifs, which are in agreement with the differences in their biochemical properties. At last, phylogenetic analysis reveals a probable Platyhelminthes- specific structural and functional segregation from which the monofunctional β-catenins evolved. Our results thus identify the first two monofunctional β-catenins in metazoans.
Keywords: Beta-Catenin, Cell Adhesion, Evolution, Signal Transduction, Wnt Pathway, Planarian
Introduction
β-Catenin was originally identified as a component of cell adhesion complexes, linking the cytoplasmic tail of cadherins with α-catenin (1). α-Catenin, in turn, binds and bundles actin filaments through its C-terminal domain (2, 3). In this way, β-catenin serves as a key protein linkage between the cadherin cytoplasmic domain and the actin cytoskeleton (4, 5). In addition, β-catenin also functions as a key component of the canonical Wnt signaling pathway, which regulates a variety of cellular processes (6–8). In the absence of Wnt ligands, a “degradation complex” forms, comprising CK1α (casein kinase 1α), Axin, GSK3β (glycogen synthase kinase 3β), and APC (adenomatous polyposis coli). Steady-state levels of β-catenin are maintained at a low level by phosphorylation, ubiquitination, and 26 S proteasome-mediated degradation (9, 10). Upon Wnt stimulation, β-catenin is stabilized and translocated into the nucleus, where it binds to transcription factors of the TCF4/LEF family and regulates target gene expression (7, 8).
Structurally, β-catenin protein contains 12 armadillo repeats, which form a conserved central superhelix with a positively charged groove that is critical for its interaction with the majority of binding partners, such as Axin, APC, TCF, and E-cadherin (11–16). The N- and C-terminal regions are less conserved and are supposed to be structurally flexible. The N terminus contains two α-catenin binding domains and one DSG motif (DpSGΦXpS, Φ represents a hydrophobic amino acid, and X represents any amino acid), in which the two phosphorylated serines (pS) could be recognized by the E3 ubiquitin ligase complex, and this causes the ubiquitination and degradation of β-catenin (10, 17, 18). The C terminus contains the transactivation domain and recruits multiple transcriptional coactivators and inhibitors (19, 20). In the extreme C terminus, there is a PDZ binding motif that has been found to participate in cell adhesion and establish cell polarity in epithelial cells (21).
Vertebrates have two closely related β-catenin homologs, β-catenin and plakoglobin (also named γ-catenin). Both of them have a similar structure and function in adherens junctions through interaction with E-cadherin and α-catenin (22, 23). In addition, plakoglobin is specifically involved in desmosome function. Both plakoglobin and β-catenin are able to interact with the major Wnt signaling components, including APC, Axin, and TCF/LEF (24). Moreover, both can directly activate transcription when fused to the Gal4 DNA-binding domain or the HMG box (DNA binding domain) from LEF1 (19, 25). Unlike vertebrates, Drosophila melanogaster has only one β-catenin homolog, armadillo, which functions in both cell adhesion and canonical Wnt signaling (26). In Caenorhabditis elegans, however, at least three divergent β-catenin homologs, BAR-1, WRM-1, and HMP-2, have been identified. Among them, WRM-1 mediates Wnt signaling via the TCF homologue POP-1, and HMP-2 functions in cell adhesion via binding to the cadherin homologue HMR-1 (27). Loss-of-function studies indicate that each of the C. elegans β-catenins has distinct functions, yet overexpression of WRM-1 or HMP-2 can rescue the BAR-1 mutant phenotype (28). Furthermore, all of them can directly activate transcription when fused to the Gal4 DNA-binding domain in yeast, suggesting that all of the C. elegans β-catenins have retained their functional redundancy, remaining bifunctional (28).
In the planarian Schmidtea mediterranea, two full-length β-catenin homologs, β-catenin-1 and -2 (Smed-β-catenin-1/2) have been reported (29–31). RNAi of β-catenin-1 resulted in an anteriorized phenotype during regeneration and in intact animals. In addition, RNAi of Smed-dishevelled, Smed-evi/wls, and Smed-WntP-1 caused a similar phenotype (30, 32, 33), and RNAi of apc, the negative regulator of the canonical Wnt signaling, caused the opposite phenotype, the double-tail worms (30). These results demonstrated that, similar to antero-posterior (A-P) axis determination in vertebrate embryos, A-P patterning in planarians is regulated by canonical Wnt signaling and that Smed-β-catenin-1 is involved in transducing the Wnt signal. In contrast, RNAi of Smed-β-catenin-2 did not reveal any obvious abnormality (30, 31). Furthermore, overexpression of Smed-β-catenin-1 but not -2 induced a secondary body axis in Xenopus embryos, a characteristic indication of Wnt signal activation, suggesting that planarian β-catenins have distinct signaling activities (31). Here we show that S. mediterranea β-catenin-2 is localized to cell junctions in the epithelia of the gut and pharynx. Biochemical analysis shows that Smed-β-catenin-1 functions specifically in Wnt signaling and Smed-β-catenin-2 in cell adhesion. This complete functional segregation is accompanied by a corresponding structural diversification. Sequence alignment and phylogenetic analysis further reveal a unique evolutionary subfunctionalization of β-catenin genes in two planarian species and possibly also in the parasitic flatworm Schistosoma mansoni.
EXPERIMENTAL PROCEDURES
Planarians
The planarians used belong to an asexual race of S. mediterranea collected from Montjuïc (Barcelona, Spain) and maintained as described elsewhere (34).
Plasmids
To generate the LEF1-HMG fusion constructs, the cDNA fragment encoding amino acids 268–394 of mouse LEF1 was amplified by PCR and inserted into the pcDNA3-Myc vector at EcoRI/BamHI sites to obtain LEF1-HMG. The cDNAs encoding full-length Smed-β-catenins or their C terminus (Smed-β-cat1, amino acids 740–954; Smed-β-cat2, amino acids 508–701) were inserted into LEF1-HMG at EcoRI/KpnI sites. The fragment encoding amino acids 958–1287 of human APC was cloned into the vector pcDNA3-FLAG, and this partial APC was used to detect interaction with β-catenins. To generate the construct of Smed-β-cat1FL-2N, the N terminus of Smed-β-cat2(1–71) was inserted in front of full-length Smed-β-cat1. To create the mutants of F253D, F293D, and F253D/F293D, Phe242 and Phe282 of Smed-β-catenin-1 were mutated to aspartate separately or simultaneously (corresponding to human β-catenin Phe253 and Phe293). All constructs were verified by sequencing.
Cell Culture and Luciferase Reporter Assay
HEK293T and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Plasmids were transfected in triplicate with Vigofect transfection reagent (Vigorous). Firefly and Renilla luciferase activities were measured 36 h after transfection using the Dual-Luc assay kit (Promega). TOPflash luciferase activity was normalized to that of Renilla. The following amounts of reporter plasmid were added per well: Super-TOPflash (1 ng) and pRL-TK (0.5 ng). PCS2+ DNA was used to adjust the total DNA amount to 150 ng/well.
Antibodies
anti-FLAG (M2) was from Sigma; anti-Myc and anti-hemagglutinin were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The polyclonal antibody against Smed-β-catenin-2 was produced against the last 14 amino acids of Smed-β-catenin-2 protein (AC EU082825) by Invitrogen. It was used at a 1:2000 dilution. Secondary antibodies were anti-mouse or anti-rabbit IgG-horseradish peroxidase and highly cross-absorbed Alexa Fluor 568-conjugated goat anti-mouse IgG (Molecular Probes) for immunodetection of Smed-b-catenin-2 in whole mount immunostainings.
Immunostaining of Mammalian Cells
HEK293T and HeLa cells were grown on glass coverslips. Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min. After washing in PBS twice for 5 min, cells were blocked with 5% bovine serum albumin in PBS for 30 min and incubated with primary antibody for 1 h. Then the samples were washed three times for 10 min in PBS and then incubated with fluorophore-conjugated secondary antibody, followed by washing three times for 10 min in PBS and staining with 4′,6-diamidino-2-phenylindole (Roche Applied Science) for 2 min. Finally, cells were mounted in Prolong Gold antifade reagent (Invitrogen) and stored at 4 °C before imaging. Images were recorded using a Zeiss LSM710 confocal microscope.
Co-immunoprecipitation Assay and Western Blot
HEK293T cells were seeded into 6-well plates and transfected with plasmid DNA samples in the following day. Forty-eight hours after transfection, cells were lysed and then sonicated in 400 μl of lysis buffer/well (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, pH 8.0, 1% Triton X-100) containing protease inhibitor mixture (Roche Applied Science) at 4 °C. After centrifugation at 14,000 rpm, 4 °C, for 15 min, 40 μl of supernatant was mixed with 8 μl of 6× SDS-loading buffer and treated at 95 °C for 2 min. The remaining supernatant was incubated with FLAG-M2 beads (Sigma) or other antibodies (anti-Myc or anti-hemagglutinin) at 4 °C for 4 h and followed by incubation with protein A/G-Sepharose (Santa Cruz Biotechnology, Inc.) overnight. The beads were than washed at least three times with lysis buffer at 4 °C for 5 min each, and bound proteins were eluted with 40 μl of 2× SDS loading buffer at 95 °C for 5 min. When testing the interactions between β-catenins with β-Trcp (β-transducin repeat-containing protein), 25 mm ALLN (Sigma) was added to the culture medium 4 h before harvesting the cells, and phosphatase inhibitors (50 mm NaF, 1 mm sodium vanadate) were added to the lysis buffer. Total lysates and immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblot with specific antibodies.
Whole-mount Immunostaining in Planarians
Immunostaining was carried out as described previously (35). After washing, staining with 4′,6-diamidino-2-phenylindole (Roche Applied Science) was performed overnight, and animals were mounted in Prolong Gold antifade reagent (Invitrogen) and stored at 4 °C before imaging. Images were recorded using a Leica SPII confocal microscope.
RNAi Silencing
RNAi analysis was performed by double-stranded RNA microinjection. Smed-β-catenin-2 double-stranded RNA was synthesized by in vitro transcription (Roche Applied Science). Planarians were injected with around 50 ng of double-stranded RNA for 3 days, and on the next day they were amputated pre- and postpharyngeally, and the head, trunk, and tail pieces were allowed to regenerate. All animals where fixed with Carnoy solution at 20 days of regeneration. Control animals were injected with water.
Phylogenetic Analysis
The full-length sequences of all β-catenin proteins were downloaded from NCBI and aligned using the ClustalW program. The armadillo repeats were identified with SMART (available from the EMBL Web site). Phylogenetic trees were constructed using the minimum evolution method with bootstrap 1000 replications using the Mega 3.1 program. Gaps were completely deleted. Accession numbers were as follows: Homo sapiens, NP_002221.1, NP_001895.1; Mus musculus, NP_034723.1, AAA37280.1; Xenopus laevis, AAA49931.1; Danio rerio, AAH47815.1, NP_571252.1; Xenopus tropicalis, AAI35470.1; D. melanogaster, NP_476666.1; Nasonia vitripennis, XP_001603109.1; Pharhyale hawaiensis, ACH92925; Aplysia californica, AAY29569.1; Cerebratulus lacteus, ABY21456.1; Platynereis dumerilii, ABQ85061.1; Chaetopterus variopedatus, AAL49497.1; Dugesia japonica, BAD93243.1; Hydra magnipapillata, AAC47137.1; S. mediterranea, ABW79875.1, ABW79874.1; S. mansoni, XM_002573746.1 (Sm-β-catenin-1; see supplemental Fig. 2 for Sm-β-catenin-2); Dictyostelium discoideum, XP_646234.
RESULTS
Smed-β-catenin-2 Is Localized to the Epithelial Cell Membrane of the Gut, Pharynx, and Epidermis
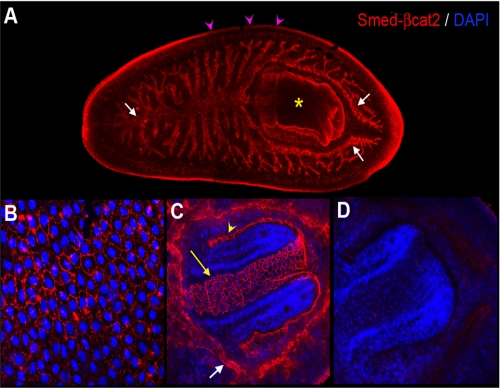
Smed-β-catenin-2 mRNA is mostly expressed in the digestive tissue (31). In order to detect the endogenous protein, we developed a polyclonal rabbit antibody against the last 14 amino acids of Smed-β-catenin-2. Immunohistochemistry using this antibody revealed staining of the gut branches, pharynx, and epidermal cells (Fig. 1, A–C). Higher magnification images showed that Smed-β-catenin-2 protein was clearly and specifically localized in the cell membrane of the epithelial cells of the epidermis (Fig. 1B) and pharynx (Fig. 1C). Note that both the inner and outer epithelial layers of the pharynx were labeled, whereas the nearby muscle cells were not stained (Fig. 1C and supplemental Movie 1). Higher magnification images of the gut indicated that the antibody labeled tubular structures (Fig. 1C), suggesting that Smed-β-catenin-2 protein is also strongly expressed in the cell membrane of the gut epithelial cells. Immunostaining of Smed-β-catenin-2 RNAi-treated animals with the anti Smed-β-catenin-2 antibody demonstrated the specificity of the antibody labeling (Fig. 1D). Taken together, these results suggest that Smed-β-catenin-2 corresponds to the β-catenin responsible for mediating cell adhesion and is mostly localized to areas of cell-cell contact in the epithelia.
FIGURE 1.
Immunostaining of planarians with anti-Smed-β-catenin-2 antibody demonstrates its location in the epithelial cell membrane of different tissues. A, immunodetection of Smed-β-catenin-2 protein in the three gut branches (white arrows), the pharynx (yellow asterisk), and the epidermis (pink arrowheads) of planarians. B, detection of Smed-β-catenin-2 protein in the cell membrane of the epidermal cells. C, detection of Smed-β-catenin-2 protein in the cell membrane of the gut cells (white arrow) and the pharynx (yellow arrow and arrowhead indicate the inner and outer pharynx epidermal cells, respectively). D, Smed-β-catenin-2 is completely absent after silencing the corresponding gene by RNAi. All images correspond to z projections of confocal sections. DAPI, 4′,6-diamidino-2-phenylindole.
Smed-β-catenin-2 Interacts with the Cadherin Complex
It has been reported that Smed-β-catenin-1 acts in the canonical Wnt/β-catenin signaling pathway to regulate A-P axis determination in planarians (29–31). Our immunohistochemical observations suggested that, in contrast, Smed-β-catenin-2 is involved in cell adhesion. To explore the biochemical differences between these two β-catenin proteins, we compared their signal transduction capacity and cell adhesion activities in mammalian cells and Xenopus embryos. In these experiments, human or Xenopus β-catenin were used as positive controls because they are functional in both contexts.
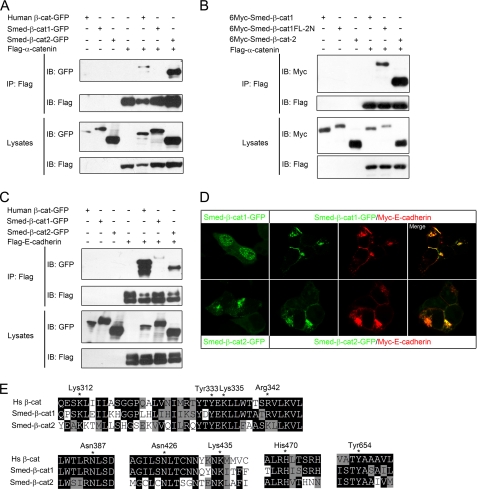
Previous studies have shown that β-catenin mediates cell adhesion by bridging cadherin and α-catenin, which further links to actin fibers (4, 5). Amino acid sequence analysis clearly indicated that Smed-β-catenin-2 contains the two conserved α-catenin-binding domains in the N terminus, whereas Smed-β-catenin-1 does not (supplemental Fig. 1) (31). As predicted, when co-transfected in HEK293T cells, Smed-β-catenin-2 was co-immunoprecipitated with α-catenin, whereas Smed-β-catenin-1 was not (Fig. 2A). Moreover, when these two α-catenin-binding domains of Smed-β-catenin-2 were transplanted to the N terminus of Smed-β-catenin-1, the resulting fusion protein (Smed-β-catenin-1FL-2N) regained α-catenin binding (Fig. 2B). Interestingly, however, when E-cadherin was co-expressed with the two Smed-β-catenins, it could interact with both, albeit to a slightly lesser extent with β-catenin-1 (Fig. 2C). We also found that the subcellular localization of the two planarian β-catenins were different. When overexpressed in HEK293T cells, Smed-β-catenin-1 localized mainly to the nucleus, where Smed-β-catenin2 was never observed. In contrast, Smed-β-catenin-2 was largely localized to the cytosol and weakly at the cell membrane (Fig. 2D, left panels). Furthermore, overexpressed E-cadherin was able to recruit both Smed-β-catenins to the cell membrane (Fig. 2D), indicating that both β-catenins are able to interact with E-cadherin.
FIGURE 2.
Smed-β-catenin-2 interacts with the cadherin complex. A, HEK293T cells were transfected with the indicated plasmids, and lysates were immunoprecipitated (IP) with FLAG-M2 beads. Western blotting (IB) with anti-GFP revealed co-IP of Smed-β-catenin-2 but not Smed-β-catenin-1 following FLAG-M2 immunoprecipitation, indicating that only Smed-β-catenin-2 interacts with α-catenin. B, co-IP experiments in HEK293T cells indicated that the transplantation of the α-catenin-binding domain into Smed-β-catenin-1 rescued its interaction with α-catenin. C, co-IP with E-cadherin reveals an interaction with both Smed-β-catenin-1 and -2. D, localization of Smed-β-catenins alone or co-expressed with E-cadherin in transfected HEK293T cells. E, alignments of stretches of amino acid sequences from human β-catenin and Smed-β-catenin-1 and -2 that are important for β-catenin/cadherin interaction. Essential residues for the interaction are indicated by stars and numbered in human β-catenin.
Because the three-dimensional structure of both human and zebrafish β-catenin has been resolved and the armadillo repeats of Smed-β-catenins are quite conserved (supplemental Fig. 1), we performed three-dimensional modeling. The predicted three-dimensional structures of Smed-β-catenin-1 and -2 both have very typical β-catenin backbones (data not shown), confirming the overall conservation of the protein family. Previous studies have revealed that within the central region of the protein, the armadillo repeat 4–9 in human β-catenin constitutes a core interacting platform for cadherin binding, in which Lys312 and Lys435 form the critical salt bridges with E-cadherin, and Asn387, Asn426, and His470 form the hydrogen bonds to strengthen the interaction (13). In addition, Arg342, Tyr333, and Lys335 from armadillo repeats 3 and 4 also form salt bridges with phosphoserine residues in E-cadherin (13). Tyr654 is also critical, such that β-catenin/E-cadherin binding would be 6 times weaker when it was phosphorylated (13). According to the predicted three-dimensional structure, we aligned the stretches of amino acid sequences that contain the critical residues for β-catenin/cadherin interaction (Fig. 2E). Consistent with the biochemical results, all of the mentioned residues were found to be conserved in both planarian β-catenins. The conservation of cadherin binding residues in planarian Smed-β-catenin-2 confirms that it is linked to cell adhesion. The conservation of these residues in Smed-β-catenin-1 is probably due to the fact that many of these residues are also essential for β-catenin to interact with TCF, APC, and Axin (see below). This may explain why Smed-β-catenin-1 still retains the capacity to interact with E-cadherin. These results suggested that Smed-β-catenin-2 is able to fulfill the adhesion function by interacting with both cadherin and α-catenin, whereas, despite retaining E-cadherin binding capacity, Smed-β-catenin-1 is incapable of functioning in cell adhesion due to a lack of α-catenin binding.
Smed-β-catenin-1 Can Directly Activate Canonical Wnt Signaling
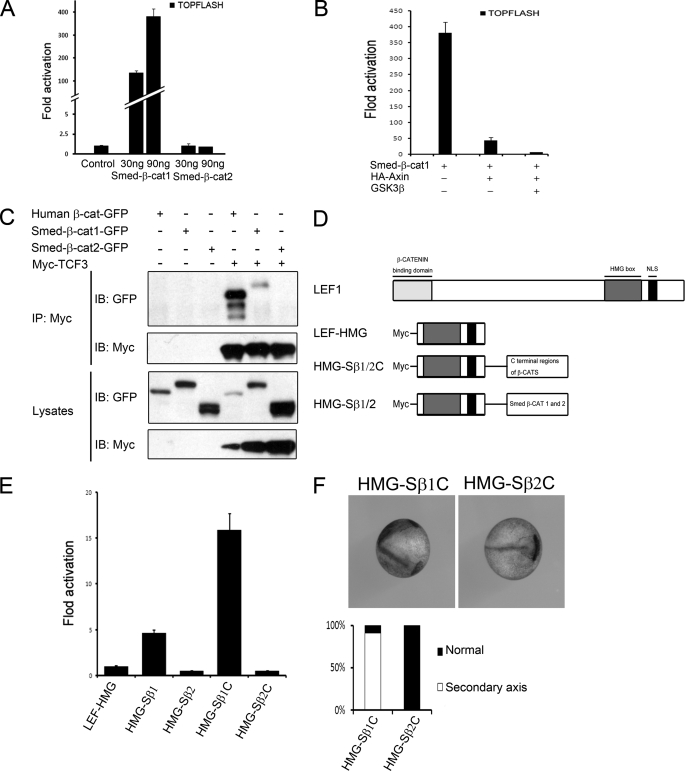
Loss-of-function studies indicate that Smed-β-catenin-1 is required for A-P axis determination (29–31), whereas the expression pattern of Smed-β-catenin-2 suggests a role in cell adhesion in planarian. These findings are inconsistent with a bifunctional β-catenin as in most metazoans. As shown above, Smed-β-catenin-1 is unable to mediate cell adhesion due to lack of α-catenin binding. Because vertebrate plakoglobin and C. elegans HMP2, both of which are involved in cell adhesion alone in vivo, still retain signal transduction ability (28), we addressed whether Smed-β-catenin-2 is able to directly trigger canonical Wnt signaling. Smed-β-catenin-1 but not -2 was reported to induce a secondary axis in Xenopus embryos (31). Here we used the Super-TOPflash reporter, a Wnt-responsive element-driven luciferase expression plasmid, to detect Wnt signaling in cultured mammalian cells. The reporter was significantly activated when increasing doses of Smed-β-catenin-1 were co-transfected, whereas Smed-β-catenin-2 had no affect at all (Fig. 3A). This reporter activation by Smed-β-catenin-1 was inhibited by co-transfection of Axin and further by Axin and GSK3β together (Fig. 3B). Furthermore, the FOPflash reporter, a control reporter construct that contains mutant TCF binding sites, was not activated by Smed-β-catenin-1 (data not shown). These results indicate that Smed-β-catenin-1 is able to specifically activate canonical Wnt signaling in mammalian cells, whereas Smed-β-catenin-2 is inactive. Interestingly, these findings also confirm that cultured mammalian cells can be used to characterize planarian genes, at least for their signaling activities.
FIGURE 3.
Only Smed-β-catenin-1 can activate canonical Wnt signaling in mammalian cells and Xenopus embryos. A, TOPflash reporter assay following co-transfection of HEK293T cells with 30 or 90 ng of Smed β-catenins and reporter plasmids. Only Smed-β-catenin-1 activated Wnt/β-catenin signaling. B, TOPflash reporter assay following co-transfection of HEK293T cells with 90 ng of Smed-β-catenin-1, 20 ng of Axin, and 10 ng of GSK3β plasmids. Axin cotransfection inhibited Smed-β-catenin-1 activation of the reporter. This inhibition was strengthened by GSK3β transfection. C, co-IP assays with TCF indicate an interaction with Smed-β-catenin-1 but not Smed-β-catenin-2. D, schematic structure of LEF1, its DNA binding domain (HMG box), and different fusion proteins with the full-length or C-terminal domains of Smed-β-catenins. E, TOPflash reporter assay in HEK293T cells. Both full-length and C-terminal Smed-β-catenin-1 in the fusion proteins activated the reporter, but no transactivation was observed with that of Smed-β-catenin-2. F, HMG-Smed-β-catenin1-C induces a secondary axis in Xenopus embryos, whereas no effect is observed with HMG-Smed-β-catenin-2-C (top). The percentages of embryos with the indicated phenotypes are shown below (n = 30). IB, immunoblot.
Because the transcriptional activity of β-catenin is largely mediated by the LEF/TCF family of transcriptional factors (7), we further tested the interaction of Smed-β-catenin-1/2 with TCF3. Co-immunoprecipitation (co-IP) experiments indicated that Smed-β-catenin-1 was able to interact with Xenopus TCF3, but Smed-β-catenin-2 was not (Fig. 3C). This result suggests that Smed-β-catenin-1 but not -2 has the potential to bind and function via a TCF transcription factor. Although an S. mediterranea TCF homolog has not been reported, several homologs are found in silico in the genome (data not shown).
Binding to LEF/TCF transcription factors is a prerequisite for β-catenin to regulate target gene expression. The capacity to mediate direct transcriptional activation is also fundamental. To address whether Smed-β-catenins are able to activate transcription directly, we fused either full-length or the C-terminal domain of β-catenins with the DNA-binding domain of Xenopus LEF1 (Fig. 3D), an approach to bypass the β-catenin/TCF interaction. This method has been used successfully to demonstrate transactivation of vertebrate β-catenin and plakoglobin (25). As shown in Fig. 3E, the fusion proteins with full-length or C-terminal Smed-β-catenin-1 both activated Wnt-responsive reporter expression, but no such effects were observed with Smed-β-catenin-2. Similar results were obtained with injected Xenopus embryos, in which Smed-β-catenin-1 fusion proteins but not those from Smed-β-catenin-2 induced secondary axis (Fig. 3F). These results suggest that, like the bifunctional β-catenin homologues, Smed-β-catenin-1 also contains a transactivation domain in its C terminus, whereas neither C-terminal nor full-length Smed-β-catenin-2 has transactivation ability, even when it is forced to bind to the Wnt-responsive DNA element. Considering its in vivo expression pattern and biochemical properties, we conclude that Smed-β-catenin-2 is monofunctional and involved specifically in cell adhesion.
Smed-β-catenin-1 Is Regulated by Axin-APC-GSK3β-mediated Degradation
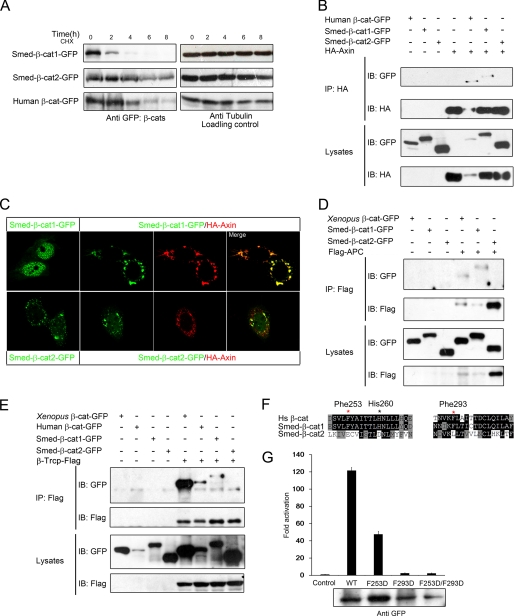
It is well documented that the Wnt signal-mediating β-catenin is under the control of the degradation complex composed of APC-Axin-GSK3β and that activation of Wnt signaling occurs through stabilization of β-catenin by disturbing the formation and function of the degradation complex. We therefore measured the stability of overexpressed Smed-β-catenins in HEK293T cells. As shown in Fig. 4A, Smed-β-catenin-1 was quickly degraded, whereas Smed-β-catenin-2 was much more stable when protein synthesis was blocked by cycloheximide. This suggested that Smed-β-catenin-1 but not -2 was recognized and phosphorylated by the endogenous APC-Axin-GSK3β complex and subsequently ubiquitinated via E3 ligase β-Trcp and degraded by the proteasome. To confirm this hypothesis, we carried out co-IP experiments to assess the interaction of the two Smed-β-catenins with Axin, APC, and β-Trcp. As shown in Fig. 4B, co-IP with Axin revealed an interaction with Smed-β-catenin-1 but not Smed-β-catenin-2. Furthermore, Axin coexpression led to relocation of overexpressed Smed-β-catenin-1 from the nucleus to the cytosol in transfected HeLa cells (Fig. 4C), similar to human β-catenin (data not shown). Smed-β-catenin-2 was cytosolic alone and did not colocalize with Axin when coexpressed (Fig. 4C). Similar binding patterns were also observed between Smed-β-catenins and both APC and β-Trcp (Fig. 4, D and E, and Table 1), suggesting that Smed-β-catenin-1 is captured and degraded in a similar manner to human β-catenin, whereas Smed-β-catenin-2 has escaped from this regulation.
FIGURE 4.
Only Smed-β-catenin-1 is captured by the degradation complex. A, the Smed-β-catenins showed different stability in HEK293T cells. HEK293T cells were transfected as indicated. Thirty-two hours after transfection, cycloheximide (CHX) was added to the culture medium at a final concentration of 100 μg/ml. Cell lysates were collected every 2 h thereafter and analyzed by Western blot. Whereas Smed-β-catenin-2 remained relatively stable for up to 8 h, Smed-β-catenin-1 was hardly detectable after only 4 h. B, co-IP assays with Axin reveal an interaction with Smed-β-catenin-1 but not Smed-β-catenin-2. C, localization of Smed-β-catenins alone or co-expressed with Axin in HeLa cells. D and E, co-IP with APC and β-Trcp reveals an interaction with Smed-β-catenin-1 but not Smed-β-catenin-2. Human APC fragment (amino acids 958–1287) and full-length β-Trcp were used in the assays. Human and Xenopus β-catenins were used as positive controls. F, alignments of the amino acid sequences from human β-catenin and Smed-β-catenins. Critical residues for β-catenin to interact with Axin, APC, and TCF are shown. Red star, important residues in the hydrophobic pocket; black star, residue for hydrogen bond. G, Super-TOPflash reporter assay following co-transfection of HEK293T cells with reporter plasmids and 90 ng of wild type Smed-β-catenin-1 or its mutations. The protein expression level is shown below. IB, immunoblot; HA, hemagglutinin.
TABLE 1.
The interaction pattern of Smed-β-catenins with the cell adhesion complex and the Wnt signaling components
| Smed-β-cat-1 | Smed-β-cat-2 | Human β-cat | |
|---|---|---|---|
| E-cadherin | + | + | + |
| α-Catenin | − | + | + |
| TCF | + | − | + |
| Axin | + | − | + |
| APC | + | − | + |
| β-Trcp | + | − | + |
According to the three-dimensional structures, human β-catenin uses the same core platform to interact with TCF and cadherin (12, 13). As shown in Fig. 2E, these residues are conserved in both Smed-β-catenin-1 and -2. In addition, a hydrophobic pocket in armadillo repeats 3 and 4, which contains Phe253 and Phe293, is also critical for β-catenin to interact with APC, Axin, and TCF but dispensable for cadherin binding (12). Sequence alignment indicated that the hydrophobic pocket, including the corresponding Phe253, Phe293, and some of the surrounding residues, is highly conserved in Smed-β-catenin-1 but not in Smed-β-catenin-2 (Fig. 4F). The corresponding residue of His260, which forms a critical hydrogen bond with Axin in human β-catenin, is also conserved only in Smed-β-catenin-1. To confirm the importance of the hydrophobic pocket of Smed-β-catenin-1 in Wnt signal activation, we mutated the two phenylalanine residues (Phe253 and Phe293; in Smed-β-catenin-1, Phe242 and Phe282, respectively) to aspartate separately or simultaneously (F253D, F293D, and F253D/F293D). In a Wnt-responsive reporter assay, Smed-β-catenin-1(F253D) showed much reduced activity, and the F293D mutant showed almost no activity (Fig. 4G). Consistently, the double mutation resulted in nearly complete loss of Wnt signaling activation (Fig. 4G), indicating that the hydrophobic pocket is essential for Smed-β-catenin-1 to transduce Wnt signaling. These analyses thus reveal that only Smed-β-catenin-1 preserves all of the essential residues for binding to APC, Axin, and TCF and therefore its ability to participate in Wnt signaling. Smed-β-catenin-2 has lost the critical hydrophobic pocket and subsequently its interaction with APC, Axin, and TCF (Table 2).
TABLE 2.
Summary of the domain composition of Smed-β-catenins
| Smed-β-cat-1 | Smed-β-cat-2 | Human β-cat | |
|---|---|---|---|
| Phosphorylation sites | + | − | + |
| α-Catenin binding domains | − | + | + |
| Hydrophobic pocket | + | − | + |
| Transactivation ability | + | − | + |
| PDZ binding domain | − | + | + |
Evolutionary Analysis of β-Catenin Proteins
Evidence suggests that β-catenin is a very ancient protein, because it plays an important role during fruiting body formation under starvation in the unicellular amoeba D. discoideum (36). Here it functions not only as a mediator of adherens junctions in the stalk cells of the fruiting body but also as a regulator of gene expression and spore cell differentiation in a GSK3-dependent manner. Sequence analysis indicated that Dictyostelium β-catenin contains both the potential GSK3 phosphorylation sites and the α-catenin binding motifs, in agreement with its bifunctional nature (36) (supplemental Fig. 1). The existence of a bifunctional β-catenin in unicellular amoeba suggests that the β-catenins in higher organisms are likely to have a bifunctional origin (36, 37).
Our results indicated, unexpectedly, that Smed-β-catenins are monofunctional. In order to further explore the evolutionary relationship, we compared the amino acid sequences of β-catenin from other Platyhelminthes. In D. japonica, one β-catenin gene (Dj-β-catenin) has been characterized (38), and another has recently been reported without full sequence information (39). In addition to freshwater planarians, the parasite S. mansoni is also a flatworm, and its genome sequence was reported recently (40). One β-catenin homolog was annotated by genomic analysis (40) (XM_002573746.1), and we refer to it as S. mansoni β-catenin-1 (Sm-β-catenin-1). Furthermore, we recognized that the putative Rab11 (XM_002570009.1) was misannotated, and its 3′ region encodes another β-catenin homolog of the parasite (we refer to this as Sm-β-catenin-2, and its amino acid sequence is shown in supplemental Fig. 2). Because the N-terminal sequence of Sm-β-catenin-2 is not complete (∼50 amino acids are missing), we included it in the phylogenetic analysis but not the alignment.
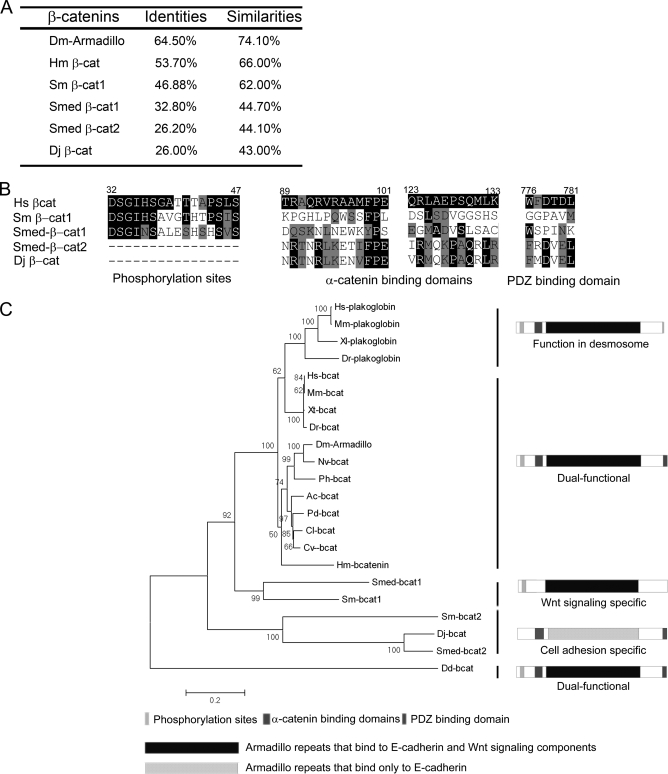
Overall sequence comparison indicated that the planarian and Schistosoma β-catenins all have above 40% similarity with human β-catenin (Fig. 5A), suggesting that they are indeed β-catenin homologs. The armadillo repeats are highly conserved in all of these β-catenin proteins, indicating a very similar and robust backbone structure, whereas the N- and C-terminal sequences are relatively divergent (supplemental Fig. 1). Two elements are easily recognizable in the N-terminal sequences, namely the potential GSK3 phosphorylation sites and the α-catenin binding motifs (Fig. 5B and supplemental Fig. 1). The phosphorylation sites (specifically, the stretch of Ser/Thr residues) are conserved in Smed-β-catenin-1 and Sm-β-catenin-1 but are missing in Smed-β-catenin-2 and Dj-β-catenin (which should be renamed Dj-β-catenin-2) (Fig. 5B and supplemental Fig. 1). In contrast, the α-catenin binding motifs are conserved in Smed-β-catenin-2 and Dj-β-catenin but not in Sm-β-catenin-1 (Fig. 5B and supplemental Fig. 1). In the C-terminal end, a conserved PDZ binding domain functioning in cell adhesion is observed in Smed-β-catenin-2 and Dj-β-catenin but not in Smed-β-catenin-1 or Sm-β-catenin-1 (Fig. 5B and supplemental Fig. 1). These results suggest that, in addition to Smed-β-catenins, β-catenins from D. japonica and S. mansoni are also likely to be monofunctional, participating in either Wnt signaling or cell adhesion but not both.
FIGURE 5.
Evolutionary analysis of β-catenin proteins. A, identity and similarity comparisons of β-catenins from different species with human β-catenin. B, alignments of the subdomains of β-catenins from human (Hs) and other species. Dm, D. melanogaster; Hm, H. magnipapillata; Sm, S. mansoni; Smed, S. mediterranea; Dj, D. japonica. The alignment with full-length β-catenin proteins is shown in supplemental Fig. 1. C, phylogenetic analysis of β-catenin and plakoglobin homologs from 17 species. Confidence values are shown at the main nodes. Black bars and schematic diagrams indicate different types of β-catenin homologs. Mm, M. musculus; Xl, X. laevis; Dr, D. rerio; Xt, X. tropicalis; Nv, N. vitripennis; Ph, P. hawaiensis; Ac, A. californica; Cl, C. lacteus; Pd, P. dumerilii; Cv, C. variopedatus; Dd, D. discoideum.
Phylogenetic analysis of most of the β-catenin proteins from metazoans (C. elegans β-catenins were not included because we believe that they have undergone a different subfunctionalization; see “Discussion”) suggested four evolutionary branches from a bifunctional origin (Fig. 5C). Plakoglobin is the result of a vertebrate-specific diversification that mediates desmosome adhesion in vivo but retains Wnt signaling activity. β-Catenins from all vertebrate and most of the invertebrate species analyzed so far grouped nicely together. Smed-β-catenin-1 and Sm-β-catenin-1 comprised the third branch, which represents the Wnt signaling-specific β-catenin. Finally, Smed-β-catenin-2, Dj-β-catenin, and Sm-β-catenin-2 together comprised the fourth branch, which represents the putative cell adhesion-specific protein. Both the sequence alignment and the phylogenetic tree supported the hypothesis that these monofunctional β-catenins evolved from a bifunctional ancestor and arose via structural and functional segregation.
DISCUSSION
Putative Role of β-Catenin-2 in Planarian Cell Adhesion
In vertebrates and most invertebrates, β-catenin functions in both Wnt signaling and cell adhesion. In agreement with its bifunctional nature, there is only one β-catenin homolog in most of the invertebrates studied (e.g. armadillo in Drosophila) (26). In vertebrates, β-catenin is involved in both Wnt signaling and adherens junctions, and its closely related protein, plakoglobin, is specifically required for cell adhesion (22, 23). It was shown recently that there are at least two β-catenin homologs in planarians (31). β-Catenin-1 is involved in a canonical Wnt signaling pathway that controls antero-posterior axis polarity, but β-catenin-2 is apparently not involved because RNAi causes no A-P axis defects. The expression of the two β-catenins differs significantly. β-Catenin-1 expression is ubiquitous, with a stronger signal detected in the central nervous system, whereas β-catenin-2 mRNA is mostly detected in the digestive system (31). The localization of β-catenin-1 protein is currently unknown because no specific antibodies are available. However, its role as a transducer of Wnt signaling suggests that it is likely to translocate to the nucleus. Our results show that β-catenin-2 protein is detected at the epithelial cell border of the gut, pharynx, and epidermis. The immunostaining signal is specific because it is consistent with the mRNA expression pattern, and the signal is eliminated by RNAi. The localization of β-catenin-2 strongly suggests that it is cell membrane-associated and likely to be involved in cell-cell adhesion of the epithelia. Confocal microscopy revealed no nuclear localization of β-catenin-2 protein, further confirming its cell adhesion-specific role. Differential expression of the signaling-specific β-catenin-1 and the cell adhesion-specific β-catenin-2 also suggests that the two genes have adopted their own regulatory elements to control their spatial and temporal expression patterns. It is interesting to note that the expression of β-catenin-2 is tissue-specific, raising the possibility of more β-catenin homologs that mediate cell adhesion in other planarian tissues. Consistent with this possibility, we have identified fragments of two different putative β-catenin homologs in the genome data base of S. mediterranea (data not shown). This could partly explain the apparent absence of behavioral or morphologic phenotypes in Smed-β-cat-2 RNAi animals despite a complete absence of Smed-β-catenin-2.
Wnt Signaling-specific Versus Cell Adhesion-specific β-Catenins
Both armadillo and vertebrate β-catenin are bifunctional. Plakoglobin functions in the desmosome in vivo, but it is able to trigger Wnt signaling when overexpressed (22, 24, 25). The C. elegans homologs BAR-1, WRM-1, and HMP-2 have distinct biological functions; however, all of them have transcriptional activity (27, 28), suggesting that they are also bifunctional. β-Catenin proteins are bound by completely different proteins and are distributed at distinct subcellular locations according to whether they are involved in cell adhesion or cell signaling. This raises the question of how the same protein can fulfill two independent cellular functions. Structurally, β-catenin can be divided into five functional portions: the N-terminal phosphorylation region, the two α-catenin binding motifs, the central armadillo repeats, the C-terminal transactivation domain, and the PDZ binding motif. The armadillo repeats, as the backbone of the β-catenin protein, are required for both cell adhesion and signaling because they contain the interacting surface for multiple β-catenin-binding proteins, including E-cadherin, APC, Axin, and TCF (11). The phosphorylation region and the transactivation domain are necessary for its signaling function but dispensable for cell adhesion (13, 16). In contrast, the α-catenin binding motifs and the PDZ binding domain are important for cell adhesion but not necessary for signaling (10, 11). Therefore, a simple Wnt signaling-specific β-catenin would in principle be composed of the phosphorylation region, the armadillo repeats, and the transactivation domain, whereas a cell adhesion-specific β-catenin would consist of the α-catenin-binding motifs, the armadillo repeats, and the PDZ binding domain. This is indeed the case in each of the two planarian β-catenins. In β-catenin-1, the α-catenin binding motif and the PDZ binding domain are missing, whereas in β-catenin-2, the phosphorylation region is missing, and the transactivation domain is non-functional. Thus, our results suggest that, at least in terms of their domain structure, the two planarian β-catenins are simplified monofunctional versions of the ancient bifunctional protein (see Table 1). It remains unclear why the C terminus of β-catenin-2 is transcriptionally inactive. A possible explanation is that it has lost its interaction with general transcriptional activators like CBP/P300.
The central core structure involving 12 armadillo repeats provides the binding surface for most β-catenin partners. A series of studies addressing the co-crystal structure of β-catenin with each of its main binding partners revealed a shared binding groove and identified important residues in the interacting surface (11–16). In principle, β-catenin protein is able to interact with E-cadherin, APC, Axin, and TCF via the same binding surface, providing the molecular basis for the bifunctional nature of β-catenin. In particular, it has been noted that β-catenin interacts with cadherin, APC, and TCF in a very similar manner (11, 15). However, a closer look at the individual residues or the subbinding surfaces, especially through structure-based mutagenesis, has revealed that binding of β-catenin to each of these partners can be uncoupled. For example, mutation of Phe253 or Phe293 abolished TCF binding without affecting cadherin interaction (12). In planarian β-catenin-1, the armadillo repeats region has preserved its essential residues to interact with APC, Axin, and TCF, whereas in β-catenin-2, it has lost its specific interacting pocket for these partners, thereby abolishing the Wnt signaling function (for protein interactions, see Table 2). Importantly, these types of mutations have occurred naturally during evolution, further confirming that the structure of the armadillo repeats can be diversified to uncouple binding to its multiple partners. These results thus suggested that the central backbone can be further classified into two types: the armadillo repeats that bind to E-cadherin only and the armadillo repeats that bind to E-cadherin as well as the Wnt components.
Our results reveal a basis for monofunctionalization of two planarian β-catenins arising from a bifunctional ancestor protein through diversification of armadillo repeats combined with distinct N and C termini. To our knowledge, this is the first demonstration of truly Wnt signaling-specific and cell adhesion- specific β-catenins in a metazoan.
Structural and Functional Segregation of β-Catenin Proteins during Evolution
It is rare that the same protein has two independent functions in two different subcellular locations. How such bifunctionality evolved in β-catenin is unclear. Proteins containing armadillo repeats have been identified in yeast and plants, but they do not appear to be β-catenin homologs (37). The unicellular amoeba D. discoideum is so far the simplest animal that contains a typical β-catenin, Aardvark (Aar) (36). Aar has been shown to function in both cell adhesion and cell signaling leading to regulation of gene expression. Sequence analysis also confirmed that Aar contains all of the necessary elements for its dual function (36). In contrast, our biochemical results indicate that planarian β-catenins are monofunctional. The amino acid sequence comparison confirmed that each of the monofunctional β-catenin proteins has a structure that is necessary and sufficient for the single activity it performs. Phylogenetic analysis further revealed that a similar monofunctionalization process might also have occurred in D. japonica and S. mansoni, another two species from the phylum Platyhelminthes. Further investigation is needed to verify whether such a structural and functional segregation is specific to the Platyhelminthes. We thus hypothesize that planarian (and possibly Platyhelminthes) β-catenin-1 and -2 originated by a gene duplication event from an ancestral, bifunctional β-catenin gene. Each of the duplicated copies adopted distinct regulatory elements for tissue-specific expression and was freed from one of the dual functions. This subfunctionalization would allow numerous residues and motifs to avoid strong selection and therefore undergo sequence diversification. The functionally partitioned β-catenins would then be likely to evolve much faster than their ancestor to fulfill their single functions.
β-Catenin subfunctionalization is not unique to flatworms. Plakoglobin is one example that originated from an ancient β-catenin homolog and specified into a desmosome junction protein (22, 23). In C. elegans, the situation is more complicated because at least three β-catenin-like proteins have roles in different biological processes (27). Functional characterization and sequence analysis have suggested that both the structure and function of the C. elegans β-catenins are just partially segregated. It seems that in the roundworms β-catenin has taken a different approach for subfunctionalization. It is still possible that other examples of β-catenin segregation are waiting to be identified. One unique feature of planaria is their ability to reproduce both sexually and asexually, via a striking regenerative capacity that allows the formation of complete animals from any small fragment of their body. Whether cellular plasticity has any impact on gene subfunctionalization will be an interesting question to address.
Supplementary Material
Acknowledgments
We thank Drs. Randy Moon, Junlin Teng, Tianhui Hu, Ye-Guang Chen, and R. M. Kypta for reagents and Dr. J. L. Gomez-Skarmeta for providing the Smed-β-cat1/2-GFP constructs. We also thank Dr. Zhongde Ye for technical assistance with confocal imaging experiments, Drs. Qinghua Tao and Wen Wang for helpful discussions, and Dr. I. Patten for advice on English style.
This work was supported by National Natural Science Foundation of China Grant 30670225 and Major Science Programs of China Grant 2006CB943402 (to W. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie 1 and Figs. 1 and 2.
- TCF
- T-cell factor
- LEF
- lymphoid enhancer-binding factor
- β-RNAi
- RNA interference
- A-P
- antero-posterior
- PBS
- phosphate-buffered saline
- IP
- immunoprecipitation
- GFP
- green fluorescent protein.
REFERENCES
- 1.Kemler R. (1993) Trends Genet. 9, 317–321 [DOI] [PubMed] [Google Scholar]
- 2.Rimm D. L., Koslov E. R., Kebriaei P., Cianci C. D., Morrow J. S. (1995) Proc. Natl. Acad. Sci. 92, 8813–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokutta S., Drees F., Takai Y., Nelson W. J., Weis W. I. (2002) J. Biol. Chem. 277, 18868–18874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson W. J., Nusse R. (2004) Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamora C., Fuchs E. (2002) Nat. Cell Biol. 4, 101–108 [DOI] [PubMed] [Google Scholar]
- 6.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 7.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 8.MacDonald B. T., Tamai K., He X. (2009) Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimelman D., Xu W. (2006) Oncogene 25, 7482–7491 [DOI] [PubMed] [Google Scholar]
- 10.Hart M., Concordet J. P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., Polakis P. (1999) Curr. Biol. 25, 207–210 [DOI] [PubMed] [Google Scholar]
- 11.Huber A. H., Nelson W. J., Weis W. I. (1997) Cell 90, 871–882 [DOI] [PubMed] [Google Scholar]
- 12.Graham T. A., Weaver C., Mao F., Kimelman D., Xu W. (2000) Cell 103, 885–896 [DOI] [PubMed] [Google Scholar]
- 13.Huber A. H., Weis W. I. (2001) Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 14.Xing Y., Clements W. K., Kimelman D., Xu W. (2003) Genes Dev. 17, 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y., Clements W. K., Le Trong I., Hinds T. R., Stenkamp R., Kimelman D., Xu W. (2004) Mol. Cell 15, 523–533 [DOI] [PubMed] [Google Scholar]
- 16.Xing Y., Takemaru K., Liu J., Berndt J. D., Zheng J. J., Moon R. T., Xu W. (2008) Structure 16, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C., Kato Y., Zhang Z., Do V. M., Yankner B. A., He X. (1999) Proc. Natl. Acad. Sci. 96, 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokutta S., Weis W. I. (2000) Mol. Cell 5, 533–543 [DOI] [PubMed] [Google Scholar]
- 19.Vleminckx K., Kemler R., Hecht A. (1999) Mech. Dev. 81, 65–74 [DOI] [PubMed] [Google Scholar]
- 20.Städeli R., Hoffmans R., Basler K. (2006) Curr. Biol. 16, 378–385 [DOI] [PubMed] [Google Scholar]
- 21.Perego C., Vanoni C., Massari S., Longhi R., Pietrini G. (2000) EMBO J. 19, 3978–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butz S., Stappert J., Weissig H., Kemler R. (1992) Science 257, 1142–1144 [DOI] [PubMed] [Google Scholar]
- 23.Peifer M., McCrea P. D., Green K. J., Wieschaus E., Gumbiner B. M. (1992) J. Cell Biol. 118, 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Ze'ev A., Geiger B. (1998) Curr. Opin. Cell Biol. 10, 629–639 [DOI] [PubMed] [Google Scholar]
- 25.Simcha I., Shtutman M., Salomon D., Zhurinsky J., Sadot E., Geiger B., Ben-Ze'ev A. (1998) J. Cell Biol. 141, 1433–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orsulic S., Peifer M. (1996) J. Cell Biol. 134, 1283–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korswagen H. C., Herman M. A., Clevers H. C. (2000) Nature 406, 527–532 [DOI] [PubMed] [Google Scholar]
- 28.Natarajan L., Witwer N. E., Eisenmann D. M. (2001) Genetics 159, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen C. P., Reddien P. W. (2008) Science 319, 327–330 [DOI] [PubMed] [Google Scholar]
- 30.Gurley K. A., Rink J. C., Sánchez Alvarado A. (2008) Science 319, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglesias M., Gomez-Skarmeta J. L., Saló E., Adell T. (2008) Development 135, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 32.Adell T., Salò E., Boutros M., Bartscherer K. (2009) Development 136, 905–910 [DOI] [PubMed] [Google Scholar]
- 33.Petersen C. P., Reddien P. W. (2009) Proc. Natl. Acad. Sci. 106, 17061–17066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molina M. D., Saló E., Cebrià F. (2007) Dev. Biol. 311, 79–94 [DOI] [PubMed] [Google Scholar]
- 35.Cebrià F., Newmark P. A. (2005) Development 132, 3691–3703 [DOI] [PubMed] [Google Scholar]
- 36.Grimson M. J., Coates J. C., Reynolds J. P., Shipman M., Blanton R. L., Harwood A. J. (2000) Nature 408, 727–731 [DOI] [PubMed] [Google Scholar]
- 37.Schneider S. Q., Finnerty J. R., Martindale M. Q. (2003) J. Exp. Zool. B Mol. Dev. Evol. 295, 25–44 [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi C., Saito Y., Ogawa K., Agata K. (2007) Dev. Biol. 306, 714–724 [DOI] [PubMed] [Google Scholar]
- 39.Oviedo N. J., Morokuma J., Walentek P., Kema I. P., Gu M. B., Ahn J. M., Hwang J. S., Gojobori T., Levin M. (2010) Dev. Biol. 339, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berriman M., Haas B. J., LoVerde P. T., Wilson R. A., Dillon G. P., Cerqueira G. C., Mashiyama S. T., Al-Lazikani B., Andrade L. F., Ashton P. D., Aslett M. A., Bartholomeu D. C., Blandin G., Caffrey C. R., Coghlan A., Coulson R., Day T. A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J. A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D. A., Lacerda D., Macedo C. D., McVeigh P., Ning Z., Oliveira G., Overington J. P., Parkhill J., Pertea M., Pierce R. J., Protasio A. V., Quail M. A., Rajandream M. A., Rogers J., Sajid M., Salzberg S. L., Stanke M., Tivey A. R., White O., Williams D. L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C. M., Barrell B. G., El-Sayed N. M. (2009) Nature 460, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.