Abstract
Human immunodeficiency virus, type 1 (HIV-1) envelope glycoprotein (gp120/gp41) plays a critical role in virus infection and pathogenesis. Three of the six monoclonal antibodies considered to have broadly neutralizing activities (2F5, 4E10, and Z13e1) bind to the membrane-proximal external region (MPER) of gp41. This makes the MPER a desirable template for developing immunogens that can elicit antibodies with properties similar to these monoclonal antibodies, with a long term goal of developing antigens that could serve as novel HIV vaccines. In order to provide a structural basis for rational antigen design, an MPER construct, HR1-54Q, was generated for x-ray crystallographic and x-ray footprinting studies to provide both high resolution atomic coordinates and verification of the solution state of the antigen, respectively. The crystal structure of HR1-54Q reveals a trimeric, coiled-coil six-helical bundle, which probably represents a postfusion form of gp41. The MPER portion extends from HR2 in continuation of a slightly bent long helix and is relatively flexible. The structures observed for the 2F5 and 4E10 epitopes agree well with existing structural data, and enzyme-linked immunosorbent assays indicate that the antigen binds well to antibodies that recognize the above epitopes. Hydroxyl radical-mediated protein footprinting of the antigen in solution reveals specifically protected and accessible regions consistent with the predictions based on the trimeric structure from the crystallographic data. Overall, the HR1-54Q antigen, as characterized by crystallography and footprinting, represents a postfusion, trimeric form of HIV gp41, and its structure provides a rational basis for gp41 antigen design suitable for HIV vaccine development.
Keywords: Antigen, Glycoprotein, HIV, Protein Structure, X-ray Crystallography, X-ray Footprinting, Vaccine Design
Introduction
The HIV2 glycoprotein is initially expressed as the precursor protein gp160, which is post-translationally cleaved into two non-covalently associated proteins: gp120, the receptor-binding protein that targets the CD4 receptor, and gp41, which mediates the fusion of the viral and host cellular membranes (1, 2). The native, prefusion form of the gp120-gp41 complex is thought to be a trimer comprising three gp120 subunits and three membrane-anchored gp41 subunits and is in a metastable conformation with the heavily glycosylated gp120 shielding gp41 (3, 4). Upon binding of gp120 to CD4 and a co-receptor (CCR5 or CXCR4), gp41 undergoes a large conformational change to form the prehairpin fusion intermediate in which the N-terminal and C-terminal helices separate from each other and the fusion peptide extends toward the host membrane (1, 5). This ultimately leads to the fusion of the viral and host membranes and leaves a postfusion form of gp41 with a structure of hairpins in a trimer.
Several crystal structures of HIV-1 and simian immunodeficiency virus gp41 have been reported (6–9). All structures contain the gp41 core, which is composed of the two heptad repeat regions, the N-terminal helical heptad repeat (HR1) and the C-terminal helical heptad repeat (HR2). The largest structure, GCN4/gp41, has a trimeric GCN4 leucine zipper fused to the N-terminal segment of gp41 core in place of the fusion domain (8). The smallest structure is a thermostable gp41 subdomain, N34/C28, connected by a 6-residue flexible linker (6). A third structure, N36/C34, is of the same length as the simian immunodeficiency virus gp41 core (7, 9). All of the molecules fold into a six-stranded helical bundle in which three N-terminal helices (HR1) form an interior, trimeric coiled-coil. Three C-terminal helices (HR2) pack into hydrophobic grooves on the surface of the HR1 coiled-coil in anti-parallel fashion. All of the crystal structures of HIV/simian immunodeficiency virus gp41 cores are similar and represent postfusion conformations of gp41. However, the high resolution structure of a longer, intact gp41 with the membrane-proximal external region (MPER) has not been solved.
Four broadly neutralizing monoclonal antibodies (bnmAbs) that target gp41 have been characterized. These include 2F5, 4E10, Z13e1, and m44 (10–13). The bnmAb m44 interacts with a conformational epitope located in the HR2 and the neighboring loop region upstream. The other three antibodies all target the MPER of gp41 between HR2 and the transmembrane domain. The MPER of gp41 encompasses ∼30 residues, ending at Lys683 (mutated to Gln in HR1-54Q; see Fig. 1), immediately preceding the transmembrane domain. All three bnmAbs targeting the MPER bind to highly conserved contiguous epitopes in the MPER. The immune system's recognition of these epitopes is not well understood; it may occur during the fusion process, in the fusion intermediate form of gp41, or prior to the fusion process, in the prefusion form of gp41. NMR and EPR studies of 2F5, 4E10, and Z13e1 epitope peptides in a membrane-mimetic environment suggest that these bnmAbs disrupt MPER fusogenic functions by preventing requested motion or correct orientation of MPER (14, 15). The 2F5 epitope is centered on 662ELDKWA667, whereas 4E10 and Z13e1 recognize the overlapping epitope 670WNWFDITNW678 (12, 16, 17). Crystal structures of Fab domains of the bnmAbs in complex with peptides representing all three of these epitopes have been reported (18–21). The peptides interact with the complementarity-determining regions of the mAbs and are in mostly helical conformations.
FIGURE 1.
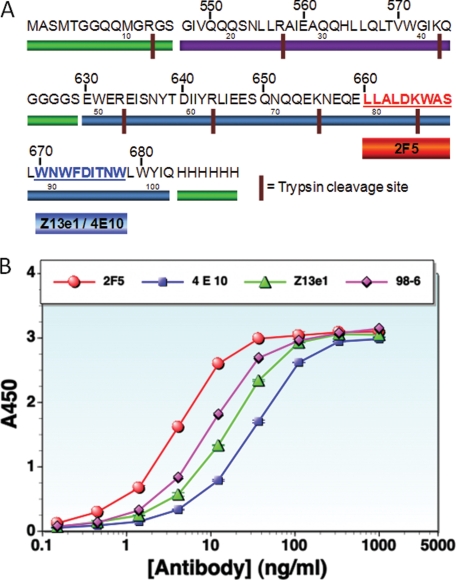
A, sequence of the HR1-54Q construct. Green bars mark the processing tags and GGGGS linker; purple and blue bars indicate the gp41 HR1 and HR2+MPER sequences, respectively. B, ELISAs of HR1-54Q with relevant antibodies: 2F5 (red circles), 4E10 (purple squares), Z13e1 (green triangles), and 98-6 (pink diamonds). 98-6 is the antibody that binds to the six-helix bundle.
The three-dimensional structure of MPER has been studied previously by NMR spectroscopy using a synthetic peptide in the presence of dodecylphosphocholine micelles and also by x-ray crystallography using a fusion peptide with a GCN4 leucine zipper (10, 22). Recently, the crystal structure of a longer MPER construct that includes HR2 (HR2/MPER, residues 630–683) has been published, and the tryptophan-rich MPER region has a helical conformation in these structures (23). In the crystal structure of the GCN4 fusion peptide, the MPER forms a parallel, trimeric helical coiled-coil structure, probably induced by the fused GCN4 leucine zipper. In the gp41 HR2/MPER construct, the molecule associates with itself to form a dimeric, asymmetric anti-parallel coiled-coil in the crystal structure. So far, the structure of the MPER has not been observed within an intact gp41 protein containing the gp41 core. Here we present a gp41 structure consisting of HR1, HR2, and the MPER, termed HR1-54Q, in an effort to characterize the MPER structural properties within an intact gp41 protein. We also characterize the solution structural properties of the antigen using techniques of structural mass spectrometry, providing a novel characterization of the physiological antigen structure. These structural data will assist in the design of immunogens intended to elicit neutralizing responses for ultimate vaccine development.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The HR1 fragment was amplified by PCR from Mcon6gp160 (46). The sense primer was 5′-CCATGGATCCGGCATCGTGCAGCAG-3′, and the antisense primer was 5′-CCATGGATCCTCCTCCTCCCTGCTTGATGCCCCACAC-3′. The amplified HR1 fragment was digested by BamHI and then inserted into the pET-gp41-54Q3 BamHI site to yield pET-HR1-54Q. The orientation was identified by PCR, and the sequence was confirmed by sequencing. Protein expression and purification was performed according to the method of Penn-Nicholson et al. (24) with slight modifications. For gp41-HR1-54Q expression, Escherchia coli T7 Express IysY/Iq (New England Biolabs) was transformed with pET-gp41-HR1-54Q and cultured overnight at 37 °C in superbroth containing ampicillin (50 μg/ml). Cells were diluted 1:100 in fresh superbroth and cultured to 1.0 A600 at 37 °C. Protein expression was then induced with 1 mm isopropyl-β-d-thiogalactopyranoside, and cells were cultured to 5.0 A600. Cells were harvested by centrifugation at 5000 rpm for 20 min in a Sorvall Superspeed centrifuge. The pellet was washed and lysed in phosphate-buffered saline (PBS) by sonication using a Branson digital sonifier. Following centrifugation at 10,000 rpm (HB4 rotor) for 20 min, the pellet containing inclusion bodies was solubilized in PBS containing 8 m urea and sonicated. Insoluble debris was removed by centrifugation at 10,000 rpm for 20 min, and soluble proteins were bound to Ni2+-nitrilotriacetic acid resin (Qiagen) by mixing on an end to end shaker overnight at 4 °C. The mixture was loaded onto a column, and the protein was renatured through serial incubations with 10 bed volumes of PBS containing a decreasing step gradient of urea at 8, 6, 4, 3, 2, 1, and 0 m. The column was washed with PBS containing 20 mm imidazole, and the protein was eluted with PBS containing 250 mm of imidazole. After dialysis against 20 mm HEPES, 50 mm NaCl (pH 8.0), the protein was subjected to structural analysis.
Crystallization
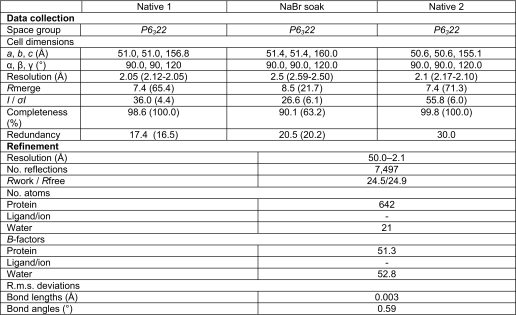
The protein was crystallized by sitting drop vapor diffusion at 18 °C. HR1-54Q (2 μl of 25 mg/ml) was mixed with solution 1 μl of the reservoir solution and 1 μl of Silver Bullets kit solution 93 (Hampton Research) and equilibrated against 0.2 ml of the reservoir solution containing 100 mm BisTris, pH 6.5, 200 mm sodium acetate, and 45% 2-methyl-2,4-pentanediol. Hexagonal plate-shaped crystals appeared in 10 days and grew to the maximum size of 0.1 × 0.1 × 0.03 mm3. Diffraction from these crystals is consistent with the space group P6322 (a = b = 51 Å, c = 156 Å), with a monomer in the asymmetric unit (50% solvent content).
X-ray Data Collection and Processing
X-ray diffraction data were collected on an ADSC 315 detector using synchrotron radiation at beamline X29A at the National Synchrotron Light Source. The native data set used for molecular replacement was collected from crystals flash-cooled in liquid N2 directly from the crystallization drops. For the bromide derivative, HR1-54Q crystals were transferred to 25 μl of mother liquid containing 1 m sodium bromide and soaked for 120 s prior to flash cooling. The data set for the single anomalous dispersion phasing experiment was collected at the bromine peak wavelength (0.9195 Å). The data sets used for final structure refinement were collected from a crystal with dehydration treatment. The seal of the crystallization well was opened, and 2 μl of mother liquid was added to the drop. The drop was then left to dry in the air for 40 min before flash cooling of the crystals from the drop. All x-ray data were reduced using the HKL suite (25).
Structure Determination and Refinement
The structure of HR1-54Q was solved by combining phases from molecular replacement using the gp41 core structure as the search model (Protein Data Bank accession code 1AIK) and a bromine single anomalous dispersion experiment using the PHENIX suite (26). Residues 15–86 with all side chains were built automatically. Refinement using PHENIX reduced the Rfree to ∼33%, and densities for the C-terminal MPER regions beyond 86 (corresponding to gp41 sequence residue Ala667) were poor, indicating that the MPER was flexible. Dehydration treatment of the crystals provides a reduction of Rfree of 4% and allows building of an additional 8 residues in MPER. Models were built using the program COOT (27). The final model includes residues 13–94 and a total of 23 water molecules, with Rcryst and Rfree of 24.5 and 24.9%, respectively. The model displays excellent stereochemistry, with 99% of the residues in the most favored region and 1% in additionally allowed regions of a Ramachandran plot by PROCHECK (28). Surface areas were calculated with the SURFACE program in the CCP4 suite (29). Data collection and structure refinement statistics are shown in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
Synchrotron Hydroxyl Radical Footprinting
Samples were thawed on ice and diluted to 5 μm HR1-54Q in 10 mm sodium phosphate, 50 mm NaCl buffer, pH 8.0. Diluted samples were kept at 4 °C prior to the experiments and used within 12 h of thawing. Exposure conditions were predetermined by following the dose-dependent degradation of the fluorescent compound Alexa 488 (Invitrogen) in the presence of the sample in the sample buffer (30). Three sets of samples (50-μl injection volume) were exposed to a mirror-focused (31) synchrotron x-ray beam (5.5-milliradian angle, focus value of 6) at the X28C beamline of the National Synchrotron Light Source at Brookhaven National Laboratory for 0–20 ms. The exposure time of the samples was controlled via flow rate through the flow cell in the KinTek (Austin, TX) stopped-flow apparatus (30). Oxidation was quenched by the flow of sample from the KinTek into the collection vessel containing methionine amide (final concentration of 10 mm). The samples were then flash-frozen in liquid nitrogen and stored at −80 °C until digestion. Irradiated protein samples were digested with sequencing grade modified trypsin (Promega, Madison, WI) at an enzyme/protein ratio of 1:20 (w/w) at 37 °C overnight; the digestion reaction was terminated by freezing the sample. The resulting peptides (∼1 pmol) were loaded onto a 300 μm inner diameter × 5-mm C18, Acclaim PepMap nano-reverse phase trapping column to preconcentrate and wash away excess salts using a U 3000 nano-high pressure liquid chromatograph (Dionex, Sunnyvale, CA). The loading flow rate was set to a 25 μl/min, with 0.1% formic acid (pH 2.9) as the loading solvent. Reverse phase separation was performed on a 75-μm inner diameter × 15-cm C18, Acclaim PepMap nanoseparation column using nanoseparation system U 3000 (Dionex). Peptide separation was accomplished using buffer A (100% water and 0.1% formic acid) and buffer B (20% water, 80% acetonitrile, and 0.04% formic acid). Proteolytic peptide mixtures eluted from the column with a 2%/min acetonitrile gradient were introduced into an LTQ FT ULTRA mass spectrometer (ThermoFisher Scientific, Waltham, MA) equipped with a nanospray ion source and using a needle voltage of 2.4 kV. MS and tandem MS spectra were acquired in the positive ion mode; the acquisition cycle included a full scan recorded in the FT analyzer at resolution R = 100,000, followed by CID MS/MS of the eight most intense peptide ions in the LTQ analyzer. MS/MS spectra of the peptide mixtures were searched for modifications of the tryptic peptides from the HR1-54Q construct using the ProtMap software (32). In addition, detected MS/MS mass spectral data for modified peptides were manually interpreted and correlated with hypothetical MS/MS spectra predicted for the proteolysis products of the HR1-54Q construct with the aid of the ProteinProspector (University of California, San Francisco, CA) algorithm. The detected total ion currents were utilized to determine the extent of oxidation by separate quantitation of the unmodified proteolytic peptides and their radiolytic products by dividing the peak area corresponding to the modified peptide by that of the total peptide (modified and unmodified) (33). Levels of modification versus exposure time, normalized to the beam current for each experiment, were plotted (supplemental Fig. 1) and fitted with a first-order exponential (34) via χ2 minimization to determine the rate constant. Solvent-accessible surface area was calculated using the “surface” function in the CCP4 package (29). The Protein Data Bank structure 3K9A solved here was used for monomer calculations; the trimeric model for accessibility analysis was constructed by the application of crystallographic symmetry in the program COOT (27).
ELISA
Thirty ng of HR1-54Q was coated onto each well in Nunc immunosobent 96-well plates using coating buffer (150 mm Na2CO3, 350 mm NaHCO3, 30 mm NaN3, pH 9.6) at 4 °C overnight. Subsequently, the coated wells were treated with blocking buffer (2.5% skim milk, 25% fetal bovine serum, 1× PBS, pH 7.5) at 37 °C for 1 h and then washed four times with the washing buffer (0.1% Tween 20 in PBS). Serially diluted mAbs 2F5, 4E10, Z13e1, and 98-6 (obtained from the National Institutes of Health AIDS Reserach and Reagent Program) were added to the treated wells and incubated for 2 h at 37 °C in 100 μl of blocking buffer. The loaded wells were washed five times with the washing buffer, followed by incubation with the secondary antibody, goat anti-human IgG horseradish peroxidase (Pierce), at 1:3000 dilution at 37 °C for 1 h. The wells were then washed five more times with the washing buffer and developed by adding 100 μl of TMB horseradish peroxidase-substrate (Bio-Rad) for 10 min. The reaction was stopped with the addition of 50 μl of 2 n H2SO4. Plates were read on a microplate reader (Molecular Devices) at 450 nm.
RESULTS AND DISCUSSION
Design of HR1-54Q
HR1-54Q was designed to include epitopes relevant to an HIV-based immunogen and to facilitate protein crystallization for elucidation of the high resolution structure of gp41 with MPER (Fig. 1A). HR1-54Q consists of a total of 108 amino acids, including an N-terminal T7 expression tag (14 residues, MASMTGGQQMGRGS), HR1 (29 residues, corresponding to gp41 residues Gly547–Gln575), a short linker (5 residues, GGGGS), HR2/MPER (54 residues, corresponding to gp41 residues Glu630–Gln683, with Gln substituting for Lys at position 683), and a C-terminal His6 tag (Fig. 1). The expression tags appear to increase the solubility of the protein and were not cleaved after purification. Shorter versions of gp41-54Q constructs without HR1 were also produced; however, crystallization attempts of these constructs were unsuccessful even with extensive crystallization screening. Examination of previous gp41 core structures shows that HR1/HR2 of gp41 forms a six-helix bundle structure, which might provide a stable framework to stabilize MPER. The length of HR1 in HR1-54Q was chosen carefully to fulfill criteria related to both antigen design and crystallization. The HR1 contains only 29 residues from the sequence spanning Gly547–Gln575 and is shorter than full-length HR1 by 7 residues. Because the MPER is of primary interest for our immunogen design, we required that the relevant epitopes should be solvent-exposed to generate a maximum immune response. The shorter HR1 is expected to minimize interactions with the MPER, leaving it exposed. The C-terminal end of HR1 was chosen based on the examination of the previous gp41 core structures, which have a distance of ∼20 Å from the ending residues of HR1 to the starting residues of HR2. A short GGGGS linker was selected to connect the two ends without distortion of the hairpin structure. We also selected a short linker in an effort to minimize the addition of immunodominant regions other than the MPER. HR1-54Q binds to antibodies specific for a six-helix bundle in ELISAs (Fig. 1B), suggesting that the six-helix bundle in HR1-54Q is intact even with the shortened HR1.
Overall Structure of HR1-54Q
The crystal structure of HR1-54Q was determined to 2.1 Å (Table 1 and Fig. 2). Residues 13–94, corresponding to two residues (GS) from the N-terminal expression tag, gp41 residues 547–575 (HR1), the GGGGS linker, and gp41 residues 630–675 (HR2+MPER), were ordered in the electron density maps, although the last 8 residues were fitted only at the level of the backbone atoms due to weak side chain densities. The first 12 residues of the N-terminal expression tag were not ordered as well as the last 8 residues of MPER and the C-terminal His6 tag. The crystallographic asymmetric unit of HR1-54Q contains a monomer. The functional gp41 is a homotrimer, and trimeric HR1-54Q can be generated from the monomer in the asymmetric unit using crystallographic symmetry operators corresponding to space group P6322. The monomers of HR1-54Q consist of a 2-helical hairpin structure with the MPER extending from the C terminus in a slightly bent helical conformation. The trimer of HR1-54Q forms a tightly packed six-helix bundle with the HR1 helices forming the trimer interface. The surface area of each HR1-54Q monomer is ∼6600 Å2, and the total buried surface area in the trimeric interface is ∼6040 Å2. The residues that line the trimer interface are all from HR1, including Ile548, Gln552, Leu555, and Ile559, and involve mostly van der Waals interactions. The average B-factor of the gp41 core region is 45.8 Å2, and the average B factor of the MPER(662–675) region is 63.4 Å2, indicating that the MPER region is more flexible in the structure.
FIGURE 2.
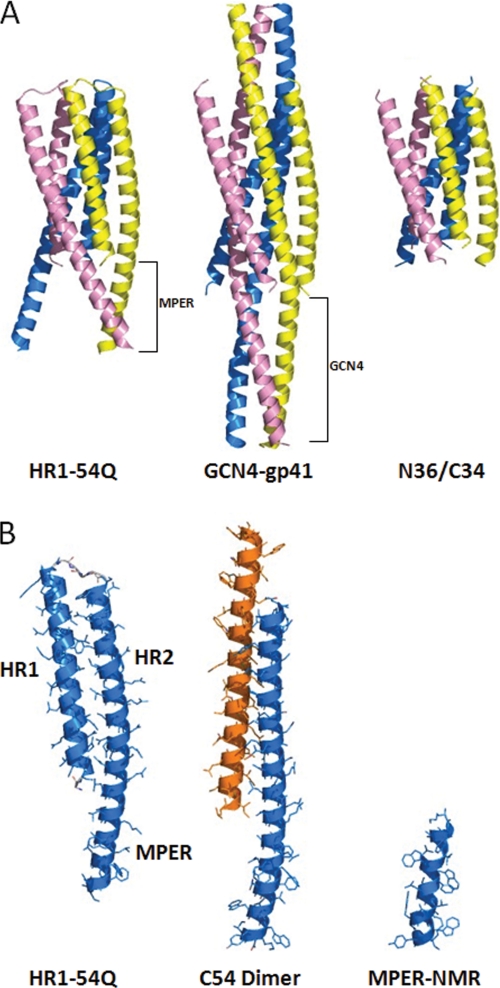
Structure of HR1-54Q and comparison with other core and MPER structures. A, trimeric structures of HR1-54Q (left), GCN4-gp41 fusion (middle; Protein Data Bank entry 1ENV), and gp41 N36/C34 core (right; Protein Data Bank entry 1AIK). HR1-54Q includes gp41 sequence 547–575 and 630–683 (the last eight residues not shown in the structure). GCN4-gp41 fusion includes gp41 sequences 541–581 and 624–665. N36/C34 core includes gp41 sequences 546–581 and 628–661. B, monomer of HR1-54Q (left), C54 dimer (middle; Protein Data Bank entry 3GWO), and gp41 MPER in docecylphosphocholine micelles (right; Protein Data Bank entry 1JAU).
GP41 Core in HR1-54Q
The previous crystal structures of the gp41 core are all synthetic peptides (HR1/HR2) refolded in solution prior to crystallization and represent gp41 in the postfusion conformation. The two representative core structures are 1AIK (a structure of HIV gp41 peptides, N36/C34, with HR1 (residues 546–581) and HR2 (residues 628–661)) and 1ENV (the largest HIV gp41 core structure, with a leucine zipper GCN4 fused onto HR1 (41 residues, 541–581) and HR2 (40 residues, 624–663)). The HR1/HR2 portion of HR1-54Q overlaps well with the two previous core structures, with an r.m.s. deviation of 0.5 Å for 60 Cα atoms (Fig. 2A). The trimeric structures are also similar for HR1-54Q and the other core structures, with the three HR1 helices located at the trimer interface and the HR2 helices packed at the outside in the hydrophobic groves. The shorter HR1 in HR1-54Q apparently does not alter the six-helix bundle structure, which provides a secure structural frame to stabilize the extended MPER.
MPER in HR1-54Q
The MPER in HR1-54Q extends from HR2 in a slightly bent helical conformation. The MPER in the crystal structure does not interact with itself and is fully exposed to solvent. The previous MPER structures of various lengths are shown in Fig. 2. In Fig. 2B, a 19-residue MPER (residues 665–683 of gp41) in docecylphosphocholine micelles solved by NMR spectroscopy (35) is illustrated. This structure begins in the middle of the 2F5 epitope but does include 4E10 and Z13e1 epitopes. The MPER in the membrane-mimetic environment is also in a helical conformation, which superimposes well with the MPER in HR1-54Q, with an r.m.s. deviation of 1.5 Å for 11 Cα atoms. The other two previous MPER structures are high resolution crystal structures (22, 23). One is an MPER peptide (residues 662–683) fused with a C-terminal GCN4 leucine zipper. In this structure, the MPER is in a helical conformation and forms a parallel triple-stranded coiled-coil. In the fusion peptide structure, the binding epitopes for both 2F5 and 4E10 are buried in the trimer interface; therefore, antibodies fail to bind to the fusion peptide, and this structure should not be considered native (22).
The newly published gp41 dimerization domain (C54) is the largest MPER structure published so far (23). It contains residues 630–683; the structure is a long helix that associates with itself, forming an anti-parallel coiled-coil. The C54 monomer overlaps reasonably well with the HR2/MPER regions in HR1-54Q, with an r.m.s. deviation of 2.0 Å, for 45 Cα atoms. It appears that the head-to-tail dimerization interface of C54 mimics the interactions between HR1 and HR2 observed in HR1-54Q. Interestingly, it does not mimic the HR1/HR2 pair in the same molecule but rather HR1 and HR2 from the neighboring molecule in the trimer. The r.m.s. deviation is 3.5 Å, for a total of 67 Cα atoms, including 22 Cα atoms from HR1 and 45 Cα atoms from HR2/MPER. Some of the interactions observed in the C54 dimer interface match closely, probably by coincidence, with the interactions between HR1 and HR2 in the gp41 core. For example, the interactions formed by Glu657 and Gln650 from the two C54 monomers match with those formed by Glu657 (HR2) and Gln550 (HR1) in HR1-54Q. Also, the interactions formed by Gln653 and Gln653 from the two C54 monomers match mostly with those formed by Gln653 (HR2) and Gln552 (HR1) in HR1-54Q. The binding epitopes 2F5 and 4E10 are exposed in both the C54 dimer and in HR1-54Q; however, HR1-54Q appears to possess a more physiologically relevant, trimeric conformation.
2F5/4E10 Binding Epitopes in HR1-54Q
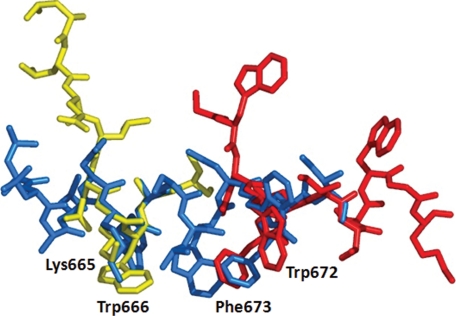
The MPER in HR1-54Q encompasses the complete 2F5 binding epitope. 2F5 is a monoclonal antibody with potent and broadly neutralizing activities against HIV-1, with 662ELDKWA667 as the binding epitope core. Crystal structures of 2F5 in complex with gp41 epitope peptides of various lengths have been determined (19, 20). The epitope core peptide forms a slightly distorted type 1 β-turn in all reported crystal structures. Residues located at the C terminus to the ELDKWA epitope core do not closely interact with the antibody. The three key residues that fit into the binding groove and that have the largest buried surface area are Asp664, Lys665, and Trp666. Fig. 3 shows superposition of the peptide 659ELLELDKWASLW670 from the 2F5 Fab domain complex onto the MPER from HR1-54Q. The last two C-terminal residues in the peptide were not ordered in the complex and were not included. The N-terminal part of the peptide is extended and does not overlap well with the MPER. However, the core piece, 664DKWA667, overlaps reasonably well with the unbound MPER structure in HR1-54Q, and the key residues, Lys665 and Trp666, in binding to the bnmAbs superimpose quite well in these structures.
FIGURE 3.
Structure comparison of HR1-54Q with 2F5 and 4E10 epitope peptides. The MPER from HR1-54Q is shown in blue. The binding epitope from the 2F5 complex is shown in yellow (Protein Data Bank entry 3D0V), and the binding epitope from the 4E10 complex is shown in red (Protein Data Bank entry 1TZG).
4E10 is the broadest neutralizing mAb discovered to date, with neutralizing activities against most HIV-1 clades. The 4E10 binding eiptope core is centered on 670WNWFDITNW678. The bound peptide adopts a helical conformation in which the key residues, Trp672, Phe673, and Ile675, are located on the same face of the helix. Because the MPER in HR1-54Q also has a helical conformation although the last 8 residues were disordered in the crystal structure, the binding epitope superimposed very well all the way to Ile675. The key residues with the largest buried surface area, Trp672 and Phe673, are in similar conformations in the two structures. The N-terminal end of the 4E10 binding epitope opens up, deviating from the helical conformation. It does not overlap well with the MPER in HR1-54Q; however, the MPER in HR1-54Q is quite flexible and will be able to adopt the bound conformation easily.
HR1-54Q is designed as an immunogen candidate to induce immune responses similar to 2F5 or 4E10/Z13e1. There has been great interest in designing MPER-based immunogens capable of eliciting broadly neutralizing antibodies similar to 2F5 and 4E10 (36); however, unstructured MPER peptides have not been successful candidates. It appears that the conformational state of the MPER is critical for eliciting 2F5 or 4E10/Z13e1 type immune responses. Several structural characteristics required for induction of 2F5- or 4E10/Z13e1-like antibodies include the correct conformation and exposure to solvent that both triggers the immune system and generates antibodies that appropriately recognize gp41. HR1-54Q is an intact MPER-containing protein that appears to possess the characteristics mentioned above. HR1-54Q indeed binds tightly to both 2F5 and 4E10 in ELISAs and is shown to have the proper antigenicity expected from the design. Preliminary immunogenicity analysis using animal models challenged by HR1-54Q showed that HR1-54Q induced extensive antibody response, but the breadth of the neutralization capabilities of the induced antibodies were mediocre (data not shown).
The CDR H3 of the two broadly neutralizing antibodies, 2F5 and 4E10, is crucial for neutralization (37–40). The CDR H3 loop of 4E10 has a hydrophobic surface facing away from the peptide epitope. The hydrophobic residues from this surface interact with the viral membrane to position the antibody to capture the exposed MPER in the transient fusion intermediate of gp41 (37, 40). A lipid component in an immunogen is perhaps required for induction of 4E10- and 2F5-like antibody responses. For the design of a future MPER-based immunogen, the transmembrane domain of gp41 and a lipid component may have to be included to mimic all of the structural characteristics required for neutralization.
Trimeric Structure in Solution Revealed by Footprinting
In order to investigate the solution state of the HR1-54Q construct, synchrotron hydroxyl radical modification of the protein, with analysis by mass spectrometry, was performed. The method is well understood to provide a sensitive probe of protein side chain accessibility, and thus residues predicted to be buried in the monomer structure as well as those involved in oligomeric contacts should be protected from covalent modification by hydroxyl radicals. In the experiment, proteins are exposed to reagents that covalently modify side chains and then digested with proteolytic enzymes; the fragments are examined for overall mass shifts indicating modification, and tandem MS is used to identify the specific sites of modification (41–43). Trypsin digestion of the protein followed by liquid chromatography-coupled MS yielded ∼78% coverage of HR1-54Q amino acids, including all residues from the N terminus to Lys665. The C-terminal peptide, containing the His6 tag, did not appear in the spectra in quantities high enough to permit detection of modifications. Analysis of the MS data revealed that the N-terminal methionine was lost during processing and that some acetylation of the N terminus occurred. The footprinting results, which are expressed as the rate of modification as a function of x-ray exposure and the specific sites observed to be modified, are summarized in Table 2 (see also supplemental Fig. 1) and were compared with side chain solvent accessibility calculations for the HR1-54Q crystal structure in monomeric and trimeric forms.
TABLE 2.
Footprinting data and solvent accessibility calculation summary
Verified modified residues are underlined. Error values shown for modification rates are S.D.
The relative reactivity of amino acid side chains is one of the two factors that determine whether it is seen to be modified in a footprinting experiment; the second is its solvent accessibility. Within HR1-54Q, the Met, Trp, Tyr, His, Leu, Ile, Arg, and Glu residues were of interest because they are frequently observed to be modified; their relative reactivity has been observed to be as follows: Met > Trp > Tyr > His > Leu, Ile > Arg > Glu (34, 42, 44, 45). Table 2 lists all peptides observed in the experiment and identifies the modified residues (where observed). The peptide overlapping with the N-terminal expression tag has a high rate of modification, and both methionine residues are seen to be modified. This region has no observable electron density in the crystal structure, indicating both solvent accessibility and significant flexibility in this peptide. The HR1 peptide 547–557 has a very low rate of modification, consistent with only Arg557 exposed in the trimer, whereas the Ile and Leu residues are buried. On the other hand, HR1 peptide 558–574 has a very high rate of modification, consistent with the predicted solvent accessibility for the highly reactive Trp571 that is confirmed to be modified in the tandem MS. In contrast, Trp631 in HR2 (peptide 575, 629–633) is predicted not to be solvent-accessible in the trimer, and this peptide was not observed to be modified, consistent with the prediction. Peptide 634–644 within HR2 has several reactive residues modified and an overall modification rate of ∼5 s−1; Tyr638 and Tyr643 are predicted to be somewhat solvent-exposed in the trimer and are confirmed as modified. Peptide 645–655 within HR2 has a very low overall rate of modification, consistent with the presence of relatively unreactive residues that are somewhat solvent-accessible. Last, peptide 656–665, which overlaps with the MPER, has several relatively unreactive residues seen to be modified. Leu660, which is exposed in the predicted monomer (103 Å2) but relatively buried in the trimer (38 Å2), is seen not to be modified, whereas the adjacent Leu661, which is predicted to be accessible in the trimer (118 Å2) is seen to be modified. Leu663 and Lys665 are also seen to be modified, further supporting our structural model for HR1-54Q.
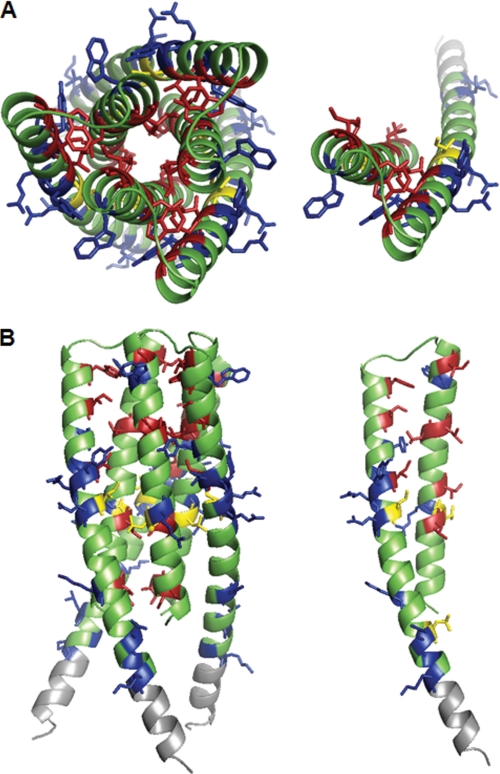
A visual representation of the footprinting results, mapped onto the crystal structure, is shown in Fig. 4, with color coding corresponding to the solvent accessibility of probe residues as experimentally determined via footprinting (red for protected, blue for solvent-exposed). As described above, the footprinting data are consistent with the trimeric structure, with nearly universal agreement between the data and the solvent accessibility as calculated for the crystal structure in this state. Red residues are buried in interface regions in the trimeric form, whereas in the monomeric form they would be quite exposed and are expected to be modified. In addition, the probe residues calculated to be significantly solvent-exposed in the trimeric form are all modified. Although higher order oligomers may be present in small quantities, the accessibility (and reactivity) of residues on the exterior of the trimer indicates that this is the predominant structure present in solution. There are several residues that do not conform to expected values (shown in yellow in Fig. 4); in all cases, these are residues for which tandem mass spectrometry was unable to verify the exact amino acid modified within the peptide. Tandem MS spectra indicated oxidative modification (+16) of Arg557 at a given retention time in the chromatogram and either Leu556 or Arg557 (slightly ambiguous spectrum) at another retention time. Taken in the context of the overall results, it is not unlikely that the second spectrum represents modification of Arg557 at a different position on the side chain, which can explain the slight change in retention time. In the case of Ile646 and Glu647, the spectrum is ambiguous, and the amount of modification is quite low; the majority of modification of peptide Leu645–Lys655 occurs as oxidative modification of Lys655. Lys and Arg are not as reactive as many other amino acids; thus, modification of these residues is relatively uncommon and signifies considerable solvent accessibility. It is perhaps unsurprising to find that Lys665 falls into this category. Although there are no highly reactive amino acids within the portion of the 2F5 binding region covered, three residues, Leu661, Leu663, and Lys665, exhibit clear modification, indicating that the 2F5 binding site is quite exposed in solution. Interestingly, Leu660 is not exposed; this is also predicted by the trimeric structure.
FIGURE 4.
Footprinting data mapped onto the HR1-54Q structure. Blue, modified probe residues. Red residues are unmodified probes, yellow shows probe residues for which modification was not verifiable, green indicates residues that are not easily modified, and gray residues are those for which no mass spectrometry signal was identified. A, top view of trimeric (left) and monomeric (right) HR1-54Q. B, side view of trimeric (left) and monomeric (right) HR1-54Q.
CONCLUSION
Our construct of HR1-54Q provided a high resolution crystal structure that included HR1, HR2, and the MPER region. A predicted trimeric structure can be generated through typical symmetry operation applied to the HR1-54Q monomer. The solution form of the protein exhibited binding to the three major broadly neutralizing antibodies to HIV as well as to an antibody that recognizes the six-helix bundle. Protein footprinting confirmed the global and local features of the trimeric structure predicted by crystallography. This antigen provides a basis for rational design of HIV vaccine candidates.
Supplementary Material
Acknowledgment
Data for this study were collected at beamlines X29A and X28C of the National Synchrotron Light Source, Brookhaven National Laboratory. Financial support for the operation of these beamlines comes principally from NIBIB, National Institutes of Health, Grant P30-EB-09998, and the office of Basic Energy Sciences of the United States Department of Energy.
This work was supported, in whole or in part, by National Institutes of Health Grant P01-AI-074286.
The atomic coordinates and structure factors (code 3K9A) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
D. P. Han, H. Habte, Y. Qin, and M. W. Cho, unpublished data.
- HIV
- human immunodeficiency virus
- MPER
- membrane-proximal external region
- mAb
- monoclonal antibody
- bnmAb
- broadly neutralizing monoclonal antibody
- PBS
- phosphate-buffered saline
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MS
- mass spectrometry
- r.m.s.
- root mean square
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Gallo S. A., Finnegan C. M., Viard M., Raviv Y., Dimitrov A., Rawat S. S., Puri A., Durell S., Blumenthal R. (2003) Biochim. Biophys. Acta. 1614, 36–50 [DOI] [PubMed] [Google Scholar]
- 2.Hunter E., Swanstrom R. (1990) Curr. Top. Microbiol. Immunol. 157, 187–253 [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. (2008) Nature 455, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J. D., Grisé H., Ofek G. A., Taylor K. A., Roux K. H. (2006) Nature 441, 847–852 [DOI] [PubMed] [Google Scholar]
- 5.Eckert D. M., Kim P. S. (2001) Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 6.Tan K., Liu J., Wang J., Shen S., Lu M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12303–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malashkevich V. N., Chan D. C., Chutkowski C. T., Kim P. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9134–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. (1997) Nature 387, 426–430 [DOI] [PubMed] [Google Scholar]
- 9.Chan D. C., Fass D., Berger J. M., Kim P. S. (1997) Cell 89, 263–273 [DOI] [PubMed] [Google Scholar]
- 10.Saphire E. O., Parren P. W., Pantophlet R., Zwick M. B., Morris G. M., Rudd P. M., Dwek R. A., Stanfield R. L., Burton D. R., Wilson I. A. (2001) Science 293, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 11.Zwick M. B., Jensen R., Church S., Wang M., Stiegler G., Kunert R., Katinger H., Burton D. R. (2005) J. Virol. 79, 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwick M. B., Labrijn A. F., Wang M., Spenlehauer C., Saphire E. O., Binley J. M., Moore J. P., Stiegler G., Katinger H., Burton D. R., Parren P. W. (2001) J. Virol. 75, 10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H., Tudor D., Alfsen A., Labrosse B., Clavel F., Bomsel M. (2008) Retrovirology 5, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L., Sun Z. Y., Coleman K. E., Zwick M. B., Gach J. S., Wang J. H., Reinherz E. L., Wagner G., Kim M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z. Y., Oh K. J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J. H., Wagner G., Reinherz E. L. (2008) Immunity 28, 52–63 [DOI] [PubMed] [Google Scholar]
- 16.Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Rüker F., Katinger H. (1993) J. Virol. 67, 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., Katinger H. (2001) AIDS Res. Hum. Retroviruses 17, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 18.Cardoso R. M., Zwick M. B., Stanfield R. L., Kunert R., Binley J. M., Katinger H., Burton D. R., Wilson I. A. (2005) Immunity 22, 163–173 [DOI] [PubMed] [Google Scholar]
- 19.Julien J. P., Bryson S., Nieva J. L., Pai E. F. (2008) J. Mol. Biol. 384, 377–392 [DOI] [PubMed] [Google Scholar]
- 20.Ofek G., Tang M., Sambor A., Katinger H., Mascola J. R., Wyatt R., Kwong P. D. (2004) J. Virol. 78, 10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejchal R., Gach J. S., Brunel F. M., Cardoso R. M., Stanfield R. L., Dawson P. E., Burton D. R., Zwick M. B., Wilson I. A. (2009) J. Virol. 83, 8451–8462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., Deng Y., Dey A. K., Moore J. P., Lu M. (2009) Biochemistry 48, 2915–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Deng Y., Li Q., Dey A. K., Moore J. P., Lu M. (2010) J. Virol. 84, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penn-Nicholson A., Han D. P., Kim S. J., Park H., Ansari R., Montefiori D. C., Cho M. W. (2008) Virology 372, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta. Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R. A. (2001) Nucleic Acids Res. 29, 221–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30.Gupta S., Sullivan M., Toomey J., Kiselar J., Chance M. R. (2007) J. Synchrotron Radiat. 14, 233–243 [DOI] [PubMed] [Google Scholar]
- 31.Sullivan M. R., Rekhi S., Bohon J., Gupta S., Abel D., Toomey J., Chance M. R. (2008) Rev. Sci. Instrum. 79, 025101 [DOI] [PubMed] [Google Scholar]
- 32.Kaur P., Kiselar J. G., Chance M. R. (2009) Anal. Chem. 81, 8141–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiselar J. G., Maleknia S. D., Sullivan M., Downard K. M., Chance M. R. (2002) Int. J. Radiat. Biol. 78, 101–114 [DOI] [PubMed] [Google Scholar]
- 34.Xu G., Kiselar J., He Q., Chance M. R. (2005) Anal. Chem. 77, 3029–3037 [DOI] [PubMed] [Google Scholar]
- 35.Schibli D. J., Montelaro R. C., Vogel H. J. (2001) Biochemistry 40, 9570–9578 [DOI] [PubMed] [Google Scholar]
- 36.Burton D. R., Desrosiers R. C., Doms R. W., Koff W. C., Kwong P. D., Moore J. P., Nabel G. J., Sodroski J., Wilson I. A., Wyatt R. T. (2004) Nat. Immunol. 5, 233–236 [DOI] [PubMed] [Google Scholar]
- 37.Alam S. M., Morelli M., Dennison S. M., Liao H. X., Zhang R., Xia S. M., Rits-Volloch S., Sun L., Harrison S. C., Haynes B. F., Chen B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julien J. P., Huarte N., Maeso R., Taneva S. G., Cunningham A., Nieva J. L., Pai E. F. (2010) J. Virol. 84, 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ofek G., McKee K., Yang Y., Yang Z. Y., Skinner J., Guenaga F. J., Wyatt R., Zwick M. B., Nabel G. J., Mascola J. R., Kwong P. D. (2010) J. Virol. 84, 2955–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer E. M., Leaman D. P., Zwick M. B., McMichael A. J., Burton D. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan J. Q., Almo S. C., Chance M. R. (2004) Acc. Chem. Res. 37, 221–229 [DOI] [PubMed] [Google Scholar]
- 42.Guan J. Q., Chance M. R. (2005) Trends Biochem. Sci. 30, 583–592 [DOI] [PubMed] [Google Scholar]
- 43.Konermann L., Stocks B. B., Pan Y., Tong X. (2009) Mass Spectrom. Rev. Early View, 1–23 [DOI] [PubMed] [Google Scholar]
- 44.Xu G., Chance M. R. (2005) Anal. Chem. 77, 4549–4555 [DOI] [PubMed] [Google Scholar]
- 45.Xu G., Chance M. R. (2004) Anal. Chem. 76, 1213–1221 [DOI] [PubMed] [Google Scholar]
- 46.Gao F., Weaver E. A., Lu Z., Li Y., Liao H. X., Ma B., Alam S. M., Scearce R. M., Sutherland L. L., Yu J. S., Decker J. M., Shaw G. M., Montefiori D. C., Korber B. T., Hahn B. H., Haynes B. F. (2005) J. Virol. 79, 1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.