Abstract
Dephosphorylation and endocytic down-regulation are distinct processes that together control the signaling output of a variety of receptor tyrosine kinases (RTKs). PTP1B can directly dephosphorylate several RTKs, but it can also promote activation of downstream pathways through largely unknown mechanisms. These positive signaling functions likely contribute to the tumor-promoting effect of PTP1B in mouse cancer models. Here, we have identified STAM2, an endosomal protein involved in sorting activated RTKs for lysosomal degradation, as a substrate of PTP1B. PTP1B interacts with STAM2 at defined phosphotyrosine sites, and knockdown of PTP1B expression augments STAM2 phosphorylation. Intriguingly, manipulating the expression and phosphorylation state of STAM2 did not have a general effect on epidermal growth factor (EGF)-induced EGF receptor trafficking, degradation, or signaling. Instead, phosphorylated STAM2 specifically suppressed Akt activation, and a phosphorylation-deficient STAM2 mutant displayed prolonged localization on endosomes following EGF stimulation. These results reveal a novel link between the dephosphorylation and endocytic machinery and suggest that PTP1B can affect RTK signaling in a previously unrecognized manner.
Keywords: Endosomes, Phosphotyrosine Signaling, Receptor Endocytosis, Receptor Tyrosine Kinase, Protein-tyrosine Phosphatase (Tyrosine Phosphatase), PTP1B, STAM2
Introduction
The proper attenuation of cell surface receptor signaling following ligand stimulation is critical for cellular homeostasis. Receptor tyrosine kinases (RTKs),5 which become tyrosine-phosphorylated on their intracellular domains following activation by polypeptide growth factors, are subject to two predominant down-regulatory mechanisms: dephosphorylation, mediated by the protein-tyrosine phosphatases (PTPs), and endocytosis, comprising RTK internalization and endosomal sorting for recycling or degradation (1, 2). RTKs can control various cellular functions, including growth, metabolism, and motility, and defects in the processes that terminate RTK signaling can have profound physiological effects, including altered development and susceptibility to cancer (3–7). In addition to promoting sustained kinase activity, the tyrosine phosphorylation of RTKs provides docking sites for phosphotyrosine-binding adapter proteins. Along with numerous other effectors of downstream signaling and regulatory processes, members of the Cbl family of E3 ubiquitin ligases are recruited to a variety of RTKs (3). Cbl induces ubiquitination of RTKs on lysine residues; this modification (monoubiquitin or Lys63-linked ubiquitin chains) is distinct from the ubiquitin chains that target proteins for proteasomal degradation (Lys48-linked) and is an important determinant of the fate of activated RTKs within the cell (3).
Ligand-bound RTKs are rapidly internalized into endocytic vesicles, which then fuse to form early endosomes. At this stage, RTKs modified by ubiquitin are targeted for lysosomal degradation by a series of ESCRT (endosomal sorting complex required for transport) complexes. The ESCRT-0 complex, consisting of the ubiquitin-binding proteins STAM (signal-transducing adapter molecule) and Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate), is implicated in the initial recognition of ubiquitin moieties on endosomal RTKs (3). Targeting of the ESCRT-0 complex to endosomes involves the FYVE domain of Hrs, which binds phosphatidylinositol 3-phosphate on early endosomal membranes (8). Subsequent ESCRT complexes (ESCRT-1, -2, and -3) are progressively recruited to sites of ESCRT-0 binding and mediate internalization of RTKs into the endosomal lumen, forming multivesicular bodies (MVBs) that fuse with lysosomes (3). Importantly, activated RTKs can continue to initiate signaling cascades as they traffic in endosomes before incorporation into MVBs, and emerging evidence suggests that different pathways are preferentially activated from specific endosomal compartments (9).
In mammals, STAM is present as two isoforms, STAM1 and STAM2, which likely play largely redundant cellular functions. The two proteins share high sequence similarity, and although STAM1 and STAM2 are individually dispensable for mouse development (6, 10), double-null mice die at embryonic day 11.5 (6). Double-null mutant embryos are defective in ventral folding morphogenesis, similar to embryos lacking Hrs (7). Fibroblasts derived from STAM-deficient embryos display enlarged endosomes and impaired epidermal growth factor (EGF)-induced EGF receptor (EGFR) degradation (11), similar to the STAM2 overexpression phenotype in COS-7 cells (12). These results are consistent with several studies on the cellular function of Hrs (13), supporting a key role for the ESCRT-0 complex in the proper down-regulation of RTKs.
The STAM proteins are tyrosine-phosphorylated, directly or indirectly, by a wide range of RTKs and cytokine receptors (13). Four specific phosphorylation sites on STAM2, Tyr192, Tyr291, Tyr371, and Tyr374, have been identified in mass spectrometry studies (see PhosphoSitePlus on Web). Although their function is unknown, the regulation of one of these sites, Tyr192, has been examined in detail (15). Interestingly, the profile of STAM2 phosphorylation (comparing Tyr192 phosphorylation with overall phosphorylation) is distinct in cells stimulated with hepatocyte growth factor, platelet-derived growth factor (PDGF) and EGF (15). If tyrosine phosphorylation affects STAM function, it could allow specific RTKs to regulate the dynamics of their trafficking in endosomes and fine tune the strength of downstream signaling pathways.
Protein-tyrosine phosphatase 1B (PTP1B) has been implicated by numerous studies in the direct dephosphorylation and negative regulation of RTKs (16). PTP1B is localized to the cytoplasmic face of the endoplasmic reticulum (ER) and is believed to act on ligand-activated RTKs after endocytosis or at sites of direct contact between the ER and plasma membranes (17–19). The most well validated target of PTP1B is the insulin RTK, and increased insulin sensitivity is an important factor in the metabolic phenotype of PTP1B-null mice, which are resistant to diet-induced obesity and diabetes (20, 21). Although a number of protooncogenic RTKs, including EGFR, PDGF receptor, insulin-like growth factor 1 receptor, and Met are also PTP1B substrates (16, 22), PTP1B-null mice are paradoxically resistant to mammary tumorigenesis induced by activated mutants of the RTK ErbB2 (23, 24). These findings suggest that therapeutic inhibitors of PTP1B, already in development for metabolic disorders, could be used to treat human cancers driven by ErbB2 and possibly other RTKs. Recently, we have focused on characterizing novel non-RTK targets of PTP1B that could account for its protumorigenic effects. We identified p62Dok, a p120RasGAP regulator that inhibits Ras signaling when phosphorylated, and cortactin, an actin cytoskeletal regulator involved in tumor invasion and metastasis, as substrates of PTP1B (25, 26).
In this study, we show for the first time that PTP1B regulates the endosomal sorting machinery. Based on sequence similarity to its target site on cortactin, we identified STAM2 as a putative substrate of PTP1B. We then mapped the site of interaction of a PTP1B trapping mutant with STAM2, demonstrated that PTP1B knockdown augments STAM phosphorylation, and showed that mutation of the PTP1B target sites alters STAM2 function and localization in response to EGF. These results indicate a novel mechanism that could contribute to the tumor-suppressive effect of PTP1B inhibition.
EXPERIMENTAL PROCEDURES
Antibodies
The following commercial antibodies were used: anti-c-myc (A-14), anti-GST (Z-5), anti-c-Src (N-16), and anti-EGFR (1005; Santa Cruz Biotechnology); anti-phosphotyrosine clone 4G10 (Millipore); anti-phospho-Akt (Ser473), anti-phospho-Erk1/2 (Thr202/Tyr204), anti-Akt, anti-Erk1/2, and anti-phospho-EGFR (Tyr1068; Cell Signaling Technology); anti-PTP1B (clone 15; BD Transduction Laboratories); anti-STAM1 (Calbiochem); and anti-phospho-Src (Tyr418, Invitrogen). Additional antibodies against STAM2 (27) and Hrs (28) were described previously.
Plasmids
Mammalian expression plasmids encoding GST-tagged human PTP1B, TCPTP, and PTP-PEST (pEBG, WT, and DA mutant) (29) and myc-tagged WT human STAM2 (pEF1) (30) as well as the bacterial expression plasmids encoding Fyn (pDB1) (31) and GST-ubiquitin (mono- and diubiquitin, pGEX) (32) were described previously. STAM2 YF mutants and siRNA-resistant constructs containing four silent mutations within the STAM2 siRNA target sequence were prepared from pEF1-STAM2 WT by site-directed mutagenesis (QuikChange kit; Stratagene). For generation of pEF1-STAM1, human STAM1 was amplified by PCR from an Incyte cDNA clone (clone LIFESEQ6140828; Open Biosystems) and inserted between the SacI and XbaI sites of pEF1. The bacterial expression plasmid encoding His-tagged STAM2 was prepared by PCR amplification from pEF1-STAM2 WT and 3YF, introducing XhoI (5′) and BamHI (3′) sites for insertion into pET-15b (Novagen). To generate the GST-tagged PTP1B bacterial expression vector, a sequence encoding amino acids 1–321 of WT human PTP1B was inserted between the BamHI and EcoRI sites of pGEX-5X (GE Healthcare) by PCR subcloning. The D181A form of pGEX-PTP1B was generated by site-directed mutagenesis. Insert sequences of all constructs were confirmed by DNA sequencing. Sequences of primers used for cloning and mutagenesis are available on request.
Substrate Trapping and Immunoprecipitation
Trapping experiments to assess binding of GST-PTP1B to endogenous STAM2 or transfected, myc-tagged STAM1 or STAM2 were conducted as described previously for cortactin (26). Immunoprecipitations of STAM2 from transfected HeLa or COS-7 cells were performed using standard methods. For details, see supplemental Experimental Procedures.
Purification of Recombinant Proteins
GST, GST-PTP1B (WT and D181A), and GST-ubiquitin (mono- and diubiquitin) were expressed in Escherichia coli and purified as described (33), except proteins were not eluted from the glutathione-Sepharose beads. Instead, GST-bound beads were stored at −20 °C in a buffer containing 20 mm Tris, pH 7.4, 50% glycerol and, for PTP1B only, 5 mm dithiothreitol.
His-tagged recombinant STAM2 was purified from E. coli co-transformed with an inducible expression construct encoding Fyn (a protein-tyrosine kinase that phosphorylates STAM2) (31). For details, see supplemental Experimental Procedures.
In Vitro Binding Assays
To assess binding of recombinant PTP1B and STAM2, His-STAM2 WT or 3YF (10 ng), purified from bacteria co-expressing Fyn, was diluted to 500 μl in HNMETG containing 1% bovine serum albumin and 5 mm dithiothreitol. Glutathione-Sepharose beads (15 μl) bound to GST, GST-PTP1B WT, or GST-PTP1B DA (1 μg each) were then added and incubated with lysates for 4 h, rotating, at 4 °C. Beads were then washed six times with 500 μl of HNMETG prior to resuspension in Laemmli sample buffer, denaturation, and analysis of bound proteins by immunoblotting.
For GST-ubiquitin pulldowns, lysates from EGF-treated, myc-STAM2-expressing HeLa cells were prepared as described for immunoprecipitation experiments. For knockdown of PTP1B expression, HeLa cells, plated at 0.5 × 106 cells/6-cm dish, were transfected with PTP1B siRNA (Dharmacon human PTPN1 SMARTpool) or nontargeting siRNA at a final concentration of 100 nm using Dharmafect transfection reagent (Dharmacon) according to the manufacturer's instructions. At 24 h after transfection, cells were replated at 0.5 × 106 cells/6-cm dish and grown for an additional 36 h. Cells were then serum-starved, treated with 100 ng/ml EGF, and lysed as above. Glutathione-Sepharose beads (15 μl) bound to GST, GST-ubiquitin, or GST-diubiquitin (1 μg each) were added to 500 μg of cell lysate in a total volume of 500 μl, and mixtures were incubated for 2 h, rotating, at 4 °C. Beads were washed, and bound proteins were analyzed by immunoblotting as above.
STAM Knockdown and EGF Stimulation
HeLa cells were treated with STAM1 and STAM2 siRNA or nontargeting, scrambled siRNA as described above. siRNA duplexes against STAM2 (target sequence CTGCTCAAACTTCATATTTAA) and STAM1 (target sequence CAGCAATGATTAAGAACCTTA) were from Qiagen. After a 24-h treatment with siRNA, cells were replated at 0.25 × 106 cells/well in 6-well plates and transfected with 1 μg/well pEF-STAM2 WT or 4YF DNA (siRNA-resistant) or mock-transfected using Lipofectamine 2000 following the manufacturer's instructions. At 48 h after transfection, cells were starved 4 h in serum-free Dulbecco's modified Eagle's medium and stimulated with 10 ng/ml EGF. Cells were lysed in 300 μl of mRIPA (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate) containing 1 mm sodium orthovanadate, 50 mm sodium fluoride, and 1 × Complete protease inhibitors and analyzed by immunoblotting.
Immunofluorescence and Confocal Microscopy
HeLa cells, treated with STAM1 and STAM2 siRNA as described above, were replated at 5 × 104 cells/well in 24-well plates containing glass coverslips and transfected with 0.2 μg/well siRNA-resistant pEF1-STAM2 WT or 4YF, also as described above. At 24 h after transfection, cells were starved for 2 h in serum-free Dulbecco's modified Eagle's medium and stimulated with Alexa Fluor 555-conjugated EGF (100 ng/ml; Molecular Probes) using the “cold-load” method, essentially as described (34). Briefly, labeled EGF was diluted in cold (4 °C), serum-free Dulbecco's modified Eagle's medium and applied to coverslips for 1 h at 4 °C. Medium was then replaced with warm (37 °C) Dulbecco's modified Eagle's medium, and plates were placed in a 37 °C incubator for the indicated times. Cells were then fixed in paraformaldehyde, permeabilized with Triton X-100, stained with myc antibody and 4′,6-diamidino-2-phenylindole (DAPI), and analyzed using a confocal microscope, as described (34).
RESULTS
Association of PTP1B D181A with STAM2
Recently, we characterized the interaction of PTP1B with cortactin and found that a single residue, cortactin Tyr446, is targeted by the phosphatase with high specificity (26). The five-residue sequence upstream of this tyrosine (EPEPVpY) is found in a single additional human protein, STAM2 (Tyr291). STAM2 peptides phosphorylated at Tyr291 have been observed in mass spectrometry studies (see PhosphoSitePlus on Web), and its molecular weight closely matches an unidentified phosphoprotein found bound to PTP1B in earlier studies (26). To examine whether STAM2 is also a PTP1B substrate, we expressed in COS-7 cells GST-tagged PTP1B, either the WT enzyme or the inactive D181A substrate trapping mutant, and purified interacting proteins by GST pulldown (PD). Endogenous STAM2 was found in complex with PTP1B D181A, and the interaction was enhanced by co-expression of c-Src, a tyrosine kinase implicated in the direct phosphorylation of STAM2 (15, 35) (Fig. 1A). Transiently expressed, myc-tagged STAM2 was also readily precipitated by PTP1B-D181A, but not by trapping mutants of two closely related PTPs, TCPTP and PTP-PEST, indicating that this interaction is specific (supplemental Fig. 1). STAM2 could be detected in complex with WT PTP1B, but at much lower levels than the D181A mutant. Thus, STAM2 binding is likely transient and phosphorylation-dependent, which is common for PTP1B substrates.
FIGURE 1.
PTP1B trapping mutant binds to specific phosphotyrosine residues of STAM2. A, PTP1B D181A associates with endogenous STAM2 in intact cells. GST-PTP1B (WT or D181A) was expressed in COS-7 cells with or without c-Src. Interacting proteins were purified by GST PD and analyzed by immunoblotting (IB) along with the total cell lysates (TCL). B, STAM2/PTP1B binding site is mapped. Blots shown are as in A, except GST-PTP1B was co-expressed with myc-STAM2 (WT and YF mutants, upper panels). The results of three independent experiments were quantified (lower panel). The band density of STAM2 (myc) isolated by GST PD and in the total cell lysates were quantified by densitometry. Error bars correspond to S.E., and asterisks denote ratios significantly less than for WT STAM2 (p < 0.05, Student's t test), to which the ratios of the YF mutants were normalized. C, mutation of STAM2 Tyr291, Tyr371, and Tyr374 (3YF) blocks its interaction with PTP1B. GST-PTP1B and myc-STAM2 were expressed, and the interaction was analyzed as in B. D, PTP1B D181A and phosphorylated WT, but not 3YF, STAM2 can interact directly. Recombinant GST alone, GST-PTP1B (WT and D181A), and phosphorylated STAM2 (WT and 3YF) were purified and subjected to in vitro binding assays, and their interaction was analyzed by immunoblotting. E, STAM2 3YF displays decreased EGF-induced tyrosine phosphorylation. myc-tagged STAM2 was purified by immunoprecipitation (IP) from HeLa cells left untreated or stimulated with 100 ng/ml EGF for 15 min. F, STAM2 structure and location of PTP1B binding sites are diagrammed. STAM2 consists of N-terminal Vps27/Hrs/STAM (VHS) and ubiquitin-interacting (UI) motifs required for ubiquitin binding, a central SH3 domain, and coiled-coiled (CC), and STAM-specific (SSM) motifs implicated in Hrs binding. The positions of Tyr291, Tyr371, Tyr374, and Tyr461 are indicated.
Although the EPEPV(pY) motif on cortactin is essential for association with PTP1B (26), mutation of STAM2 Tyr291 decreased its interaction with the phosphatase by only ∼30% (Fig. 1B). Likewise, STAM1, which lacks Tyr291, still interacts with PTP1B D181A, but less efficiently (supplemental Fig. 2). To establish the target sites of PTP1B on STAM2, we prepared a series of mutants in which individual tyrosine residues were replaced by phenylalanine. In addition to Tyr291, mutation of Tyr371, Tyr374, and Tyr461 caused significantly reduced PTP1B D181A binding (Fig. 1B), and the combined mutation of Tyr291, Tyr371, and Tyr374 was sufficient to nearly eliminate the interaction (3YF mutant, Fig. 1C). Similar results were obtained with a mutant (4YF) lacking all four sites (not shown). This effect is not due to a dramatic change in overall tyrosine phosphorylation: STAM2 WT and 3YF displayed similar phosphotyrosine content when immunoprecipitated under the conditions used for GST PD experiments (supplemental Fig. 3). To establish whether the two proteins can interact directly, we purified GST-PTP1B (WT and D181A) and tyrosine-phosphorylated His-STAM2 (WT and 3YF) from E. coli (supplemental Fig. 4). The binding detected in intact cells could be reproduced in vitro using these recombinant proteins: phosphorylated, WT STAM2 was precipitated by PTP1B-D181A, and not by WT PTP1B or GST alone, and the 3YF mutation dramatically decreased this interaction (Fig. 1D).
Effect of YF Mutations on Phosphorylation and Interactions of STAM2
In HeLa cells stimulated with EGF, immunoprecipitated myc-STAM2 3YF displayed ∼50% reduced phosphotyrosine content compared with WT (Fig. 1E), confirming that together, Tyr291, Tyr371, and Tyr374 are important sites of EGF-induced tyrosine phosphorylation. These residues are located in a region of STAM2 containing the coiled-coil and STAM-specific motifs (Fig. 1F) required for Hrs binding (8), suggesting that phosphorylation could regulate formation of this complex. As expected, the Hrs·STAM2 complex was readily detected under various conditions (Fig. 1E and supplemental Fig. 3), and Hrs was the most prominent tyrosine phosphoprotein to co-precipitate with STAM2 from EGF-treated cell lysates (Fig. 1E). Nonetheless, neither the 3YF mutation nor EGF stimulation had a significant impact on STAM2 association with Hrs (Fig. 1E and supplemental Fig. 3).
A recent study suggested another potential function of STAM2 phosphorylation: NMR analysis of proteins in intact bacterial cells indicated that phosphorylation at Tyr291, Tyr371, and Tyr374 may regulate the affinity of STAM2 for ubiquitin (31). To confirm these results using an alternative method, we tested the ability of recombinant GST-tagged ubiquitin, either a single ubiquitin group or diubiquitin (2Ub; supplemental Fig. 4), to pull down STAM2 from cell extracts in vitro. The 2Ub fusion was markedly more effective at binding STAM2 than mono-ubiquitin (Fig. 2A), an effect observed previously for other ubiquitin-binding endocytic regulators, including Hrs and Eps15 (32, 36). However, the association was not affected by the STAM2 3YF mutation and occurred independently of EGF-induced tyrosine phosphorylation (Fig. 2A).
FIGURE 2.
PTP1B regulates STAM phosphorylation. A, STAM2 binds ubiquitin independently of EGF-induced tyrosine phosphorylation. Lysates from myc-STAM2 (WT and 3YF)-expressing HeLa cells, untreated or stimulated with EGF for 15 min (lower panel), were subjected to in vitro PDs with GST alone or GST-tagged ubiquitin (monoubiquitin, 1×Ub, or diubiquitin, 2×Ub). Precipitated proteins were analyzed by immunoblotting (IB, upper panel). B, PTP1B knockdown augments EGF-induced tyrosine phosphorylation of GST-diubiquitin-interacting proteins. HeLa cells treated with PTP1B-targeted or scrambled siRNA were stimulated with 100 ng/ml EGF for the indicated times. Diubiquitin-interacting proteins were purified by GST PD and analyzed by immunoblotting. The positions of STAM1/2 and Hrs on the PD P-Tyr IB are indicated. TCL, total cell lysates.
PTP1B Knockdown Augments EGF-induced Phosphorylation of ESCRT-0 Components
Among the numerous ubiquitin-binding proteins in human cells, only four prominent tyrosine phosphoproteins bind GST-2Ub in extracts from EGF-treated HeLa cells: STAM1 and STAM2, which run as a doublet of ∼65 kDa, Hrs, at ∼110 kDa, and an unidentified protein at ∼85 kDa (Fig. 2B). Therefore, we took advantage of its phosphorylation-independent binding to GST-2Ub to purify endogenous STAM2 for subsequent analysis by anti-phosphotyrosine immunoblotting. This method was substantially more effective at enriching for STAM2 than immunoprecipitation using available antibodies (data not shown).
To assess the importance of PTP1B in regulating STAM phosphorylation, we first overexpressed PTP1B in HeLa cells prior to stimulation with EGF. Under these conditions, PTP1B induces a decrease in the EGF-induced phosphorylation of several proteins, including STAM2 (supplemental Fig. 5). However, we suspected that this dramatic, nonspecific effect could simply be an artifact of the high level of PTP1B expression. Therefore, we proceeded to knock down endogenous PTP1B in these cells by siRNA. In contrast to the overexpression results, PTP1B knockdown had no apparent effect on overall phosphotyrosine content in EGF-stimulated cells, despite a >90% reduction in PTP1B protein levels (Fig. 2B, TCL P-Tyr blot). The phosphorylation of EGFR and Src, two kinases that can promote phosphorylation of STAM2, was also unaffected (supplemental Fig. 6). However, for the proteins precipitated by GST-2Ub, there was a clear difference: tyrosine phosphorylation of STAM1 and STAM2 was increased in PTP1B siRNA-treated cells at multiple time points after EGF stimulation (Fig. 2B). Unexpectedly, although we were unable to detect Hrs binding to PTP1B D181A (not shown), phosphorylation of Hrs was also increased by PTP1B knockdown (Fig. 2B). These results show that, in HeLa cells, endogenous PTP1B has an important role in regulating EGF-induced ESCRT-0 phosphorylation.
STAM Proteins Have a Specific Effect on EGF-induced Akt Activation
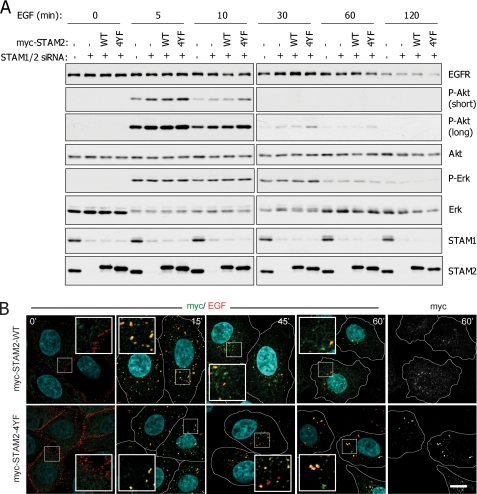
To investigate the cellular function of STAM2 phosphorylation at the sites regulated by PTP1B, we manipulated expression of the STAM proteins in HeLa cells using specific siRNAs and siRNA-resistant STAM2 expression constructs. We compared the EGF signaling response of control cells, STAM1/STAM2 siRNA-treated cells, and siRNA-treated cells reexpressing STAM2 WT or 4YF (Fig. 3A). Modulating STAM expression and mutating the STAM2 PTP1B target sites had a specific impact on EGF-induced Akt signaling. Knockdown of STAM1 and STAM2 resulted in a small but consistent increase in phospho-Akt levels at multiple time points, and although reexpression of WT STAM2 had little effect, reintroduction of STAM2 4YF resulted in further enhanced and sustained Akt phosphorylation (Fig. 3A). Conversely, there were no significant differences in the EGF-induced activation profiles of Erk or STAT3 (Fig. 3A and data not shown). Likewise, degradation of the EGFR, which was evident at 1 and 2 h after stimulation, was not affected in knockdown or reexpressing cells (Fig. 3A), despite the reported importance of the STAM proteins in degradative RTK sorting.
FIGURE 3.
Tyrosine phosphorylation of STAM2 affects its function and localization. A, tyrosine-phosphorylated STAM2 modulates EGF-induced Akt phosphorylation. HeLa cells were treated with siRNAs targeting STAM1 and STAM2 and subsequently transfected with siRNA-resistant myc-STAM2 (WT or 4YF) or mock-transfected. Control cells treated with scrambled siRNA were also mock-transfected. Cells were then stimulated with 10 ng/ml EGF for the indicated, times and total lysates were analyzed by immunoblotting. Two different exposures of the P-Akt blots are shown to avoid band saturation at early time points. B, phosphorylation-deficient STAM2 mutant displays altered localization after EGF stimulation. HeLa cells, treated with siRNAs targeting STAM1 and STAM2, were plated on coverslips and transfected with siRNA-resistant myc-STAM2 (WT or 4YF). Cells were cold-loaded with labeled EGF (100 ng/ml, red) and then transferred to 37 °C for the indicated times. Cells were then fixed, permeabilized, and stained with a myc antibody (green or white) and DAPI (blue). Results are representative of three independent experiments. Scale bar, 10 μm.
Tyrosine Phosphorylation Affects STAM2 Localization
To ascertain whether STAM2 tyrosine phosphorylation could influence its intracellular distribution or alter EGF-induced EGFR trafficking, HeLa cells expressing myc-STAM2 WT or 4YF (in which expression of endogenous STAM1 and STAM2 had been suppressed, as above) were treated with labeled EGF and analyzed by immunofluorescence and confocal microscopy. To synchronize EGFR endocytosis, cells were loaded with EGF at 4 °C, a treatment that permits EGFR·EGF complexes to accumulate in clathrin-coated pits but that does not allow internalization to proceed further (37). After the cold incubation, cells were either fixed immediately or transferred to 37 °C (to allow endocytosis to progress) and then fixed, prior to staining with anti-myc antibodies. As expected, before transfer to 37 °C, the fluorescent EGF is present exclusively at the cell periphery, whereas STAM2 (WT and 4YF) is localized to small dots in the cytoplasm (Fig. 3B). Within 15 min after transfer to 37 °C, both STAM2 WT and 4YF become localized to larger endosomes that are marked by internalized EGF (Fig. 3B). At later time points, differences in the localization patterns of the two forms of STAM2 become evident: the staining for the WT protein becomes progressively more diffuse, resembling the 0-min pattern, whereas STAM2 4YF is more persistently localized to large punctae, similar to the 15-min pattern (Fig. 3B and supplemental Fig. 7). This result suggests that tyrosine phosphorylation could be important for releasing STAM2 from these endosomal structures. Nonetheless, there were no clear differences in trafficking of the fluorescent EGF from the cell periphery (at early time points) to the perinuclear region (at late time points) in cells expressing STAM2 WT or 4YF (Fig. 3B). This correlates with the lack of effect on EGFR degradation observed in the time course experiments presented in Fig. 3A.
DISCUSSION
In this study, a series of experiments were conducted to characterize the interaction between PTP1B and STAM2 and verify its importance in cells. We showed that a trapping mutant of PTP1B can form a complex with STAM2 in intact cells, an interaction that can be reconstituted using purified proteins in vitro. Mutating specific tyrosine residues of STAM2 blocks its association with PTP1B without affecting its Hrs- or ubiquitin-binding functions, and knockdown of PTP1B expression causes hyperphosphorylation of both STAM proteins in response to EGF. Together, these results provide strong evidence that STAM2 is a genuine substrate of PTP1B.
The characterization of cortactin Tyr446 as a preferred substrate of PTP1B (26) led us to investigate the possibility that the EPEPV(pY) sequence could represent a PTP1B consensus target motif. Notably, although STAM2 shares this sequence (Tyr291) and it was identified as a candidate PTP1B substrate in this manner, this motif is not the exclusive target of PTP1B in this case. The only other proposed PTP1B consensus motif, (D/E)(pY)(pY)(R/K), based on the major target of PTP1B in the insulin receptor (38), has been used to identify PTP1B substrates including Jak2 and Tyk2 (39). Nonetheless, many PTP1B substrates do not share such conserved target sites. Although primary sequence motifs are useful for identifying PTP1B targets, other factors, including tertiary structure, localization, and non–active site-binding elements, certainly also contribute to the in vivo specificity of the phosphatase.
A large number of proteins that mediate the internalization and early endosomal sorting of RTKs are tyrosine-phosphorylated after receptor activation (see PhosphoSitePlus on Web). Although this could be due in part to their proximity to active kinase domains, phosphorylation of the endocytic machinery can certainly modify its function (40). This form of regulation could account for the differences in the endocytic dynamics of RTKs versus constitutively endocytosed, nonactivatable receptors (e.g. transferrin), even though the molecular components that mediate the two processes are similar (41). For example, the endocytic adapter protein Eps15 contains a phosphorylated tyrosine residue that, when mutated, affects internalization of the EGFR but not transferrin (40). Nonetheless, there are few other cases where a specific effect has been attributed to tyrosine phosphorylation of an endocytic regulator.
Our results indicate that tyrosine phosphorylation of STAM2 regulates both its ability to suppress EGF-induced Akt signaling and its association with EGF-positive endosomes. We were unable to uncover whether these effects on STAM2 function and localization are linked or which mechanisms could be responsible. Importantly, mutation of the PTP1B binding sites of STAM2 did not affect its interaction with two binding partners, Hrs and ubiquitin, which are believed to dictate its intracellular localization. Furthermore, expression of the mutant did not dramatically impact EGFR trafficking. Still, we suspect that STAM2 phosphorylation could still have a subtle influence on the kinetics of EGFR endocytosis that we could not detect. Maximal EGF-induced PI3K-Akt activation occurs at a postinternalization, endosomal stage (42), so a small increase in EGFR residency in this compartment could explain the signaling effects we observed. Alternatively, STAM2 phosphotyrosine residues could serve as docking sites for an unidentified modulator of Akt signaling.
A recent phosphoproteomic study showed that EGF strongly promotes differentiation of mesenchymal stem cells into osteoblasts whereas PDGF does not (43). Although the signaling networks induced by these growth factors are very similar, a key difference is the strong PI3K-Akt activation in PDGF-stimulated cells. Indeed, inhibition of PI3K permitted differentiation of these cells into osteoblasts in response to PDGF. Interestingly, STAM1 and STAM2 are among a small group of proteins that were found in phosphotyrosine-containing complexes at much higher levels in cells treated with EGF than with PDGF (43). Thus, it is tempting to speculate that, similar to the effect we observed in HeLa cells, strong STAM1/2 phosphorylation could contribute to low PI3K-Akt activity in mesenchymal stem cells and support their differentiation in response to EGF.
The relatively mild impact of STAM knockdown on EGFR down-regulation and signaling was surprising considering our current understanding of the importance of the ESCRT-0 complex in endosomal sorting. However, compared with Hrs, few studies have investigated the cellular functions of STAM1 or STAM2, and ours is the first to use an RNA interference approach to knock down both isoforms. Interestingly, the phenotype we observe after knockdown of STAM1/2 in EGF-treated HeLa cells is much less pronounced than that reported for Hrs, which causes significantly delayed EGFR degradation (44). These differences suggest that although Hrs and STAM are believed to act primarily as a complex, at least in this system, they seem to play distinct roles and are not functionally equivalent in directing RTK degradation.
Our results demonstrate a novel form of cross-talk between the RTK dephosphorylation and endocytic down-regulatory machinery. Specifically, we show that PTP1B is capable of directly regulating not only RTKs themselves but also the protein complexes that mediate their endosomal sorting. The dynamics of the interaction between ER-localized PTP1B and extracellular ligand-activated RTKs has been an important subject of research over the last decade. Different laboratories have concluded that PTP1B likely dephosphorylates RTKs after endocytosis as they traffic in close proximity to the ER (17, 18). Very recently, after completion of the current research project, a study was published which examined, by electron microscopy, the interaction of PTP1B with ligand-activated, internalized EGFR in unprecedented detail (45). In EGF-stimulated HeLa cells, expression of PTP1B D181A induced formation of extensive, direct contacts between the ER and the peripheral membrane of MVBs (45). EGFR and PTP1B were both localized to these contacts, and importantly, in cells with reduced PTP1B expression, inward vesiculation of EGFR into MVBs was dramatically impaired (45). However, the mechanism behind this MVB internalization defect was not established. The authors propose that it could involve targeting of an ESCRT protein by PTP1B, but they also provide evidence that it is not due to dephosphorylation of Hrs (45). Our data suggest that the other ESCRT-0 component, STAM, could be a prime candidate to investigate in this context in future studies.
Although the effect of STAM2 phosphorylation on EGF-induced Akt activation in HeLa cells was modest, it provides another potential explanation for the positive signaling effects of PTP1B in different cell and animal models. It has become clear in recent years that the negative regulation of RTKs and other kinases by PTP1B is tightly counterbalanced by its activation of downstream pathways. At the physiological level, this concept is illustrated by the phenotype of PTP1B-null mice, which are healthy despite the numerous RTKs known to be down-regulated by the enzyme (20, 21). At a molecular level, our laboratory and others are beginning to elucidate the substrates of PTP1B, such as p62Dok (25) and STAM2, which could allow it to promote RTK-induced signaling independently of its activity on the receptor itself.
These positive signaling functions are likely responsible for the resistance of PTP1B-null mice to specific types of cancer: PTP1B deficiency delays tumorigenesis in ErbB2-induced breast cancer (23, 24), but not in polyoma middle T-induced breast cancer (24) or p53-null lymphoma models (46). Interestingly, our laboratory detected consistently decreased phospho-Akt levels in tumors from ErbB2 transgenic mice lacking PTP1B (23). A similar effect has been reported in cultured cells: in primary PTP1B-null fibroblasts, EGF- and PDGF-induced Akt activation tended to be decreased, despite increased receptor phosphorylation (14). However, it is clear that PTP1B has remarkably pleiotrophic and cell type-specific effects on cancer cell signaling (16). PTP1B knockdown in HeLa cells has little net effect on EGF-induced EGFR, Erk, or Akt activation6 (supplemental Fig. 6), despite hyperphosphorylation of STAM1/2 and the previously reported regulation of EGFR (14) and Erk (25) by PTP1B. To understand how PTP1B-targeted therapies could benefit cancer patients, it will be useful to uncover molecular determinants of sensitivity to PTP1B inhibition. In future studies, it will be interesting to determine whether PTP1B-mediated STAM dephosphorylation could promote Akt activation specifically in forms of cancer driven by activated RTKs.
Together, the results presented here demonstrate that PTP1B can influence RTK signaling in a previously unrecognized manner. We show that PTP1B directly targets STAM2, part of the ESCRT-0 endosomal sorting complex, and we provide the first evidence that tyrosine phosphorylation affects STAM localization and function. This regulatory mechanism could impact signaling downstream of numerous cell surface receptors that are ubiquitinated and recognized by this conserved sorting machinery.
Supplementary Material
Acknowledgments
We thank Dr. Maxime Hallé for helpful discussions and for critical reading of the manuscript. We also thank Dr. Alexander Shekhtman (University at Albany) for providing the Fyn expression construct and Dr. Gergely Lukacs (McGill University) for providing the GST-ubiquitin constructs.
This work was supported by Canadian Institutes of Health Research Operating Grants MOP-62887 (to M. L. T.) and MOP-1154 (to M. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. 1–7.
M. Stuible and M. L. Tremblay, unpublished observations.
- RTK
- receptor tyrosine kinase
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- ER
- endoplasmic reticulum
- ESCRT
- endosomal sorting complex required for transport
- GST
- glutathione S-transferase
- MVB
- multivesicular body
- PD
- pulldown
- PDGF
- platelet-derived growth factor
- PI3K-Akt
- phosphoinositide 3-kinase/protein kinase A
- PTP
- protein-tyrosine phosphatase
- siRNA
- small interfering RNA
- 2Ub
- diubiquitin
- WT
- wild type.
REFERENCES
- 1.Dikic I., Giordano S. (2003) Curr. Opin. Cell Biol. 15, 128–135 [DOI] [PubMed] [Google Scholar]
- 2.Peschard P., Park M. (2007) Oncogene 26, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 3.Abella J. V., Park M. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E973–E984 [DOI] [PubMed] [Google Scholar]
- 4.Ostman A., Hellberg C., Böhmer F. D. (2006) Nat. Rev. Cancer 6, 307–320 [DOI] [PubMed] [Google Scholar]
- 5.Uetani N., Bertozzi K., Chagnon M. J., Hendriks W., Tremblay M. L., Bouchard M. (2009) J. Clin. Invest. 119, 924–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada M., Ishii N., Asao H., Murata K., Kanazawa C., Sasaki H., Sugamura K. (2002) Mol. Cell. Biol. 22, 8648–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komada M., Soriano P. (1999) Genes Dev. 13, 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuno E., Kawahata K., Okamoto A., Kitamura N., Komada M. (2004) J. Biochem. 135, 385–396 [DOI] [PubMed] [Google Scholar]
- 9.von Zastrow M., Sorkin A. (2007) Curr. Opin. Cell Biol. 19, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada M., Takeshita T., Miura S., Murata K., Kimura Y., Ishii N., Nose M., Sakagami H., Kondo H., Tashiro F., Miyazaki J. I., Sasaki H., Sugamura K. (2001) Mol. Cell. Biol. 21, 3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanazawa C., Morita E., Yamada M., Ishii N., Miura S., Asao H., Yoshimori T., Sugamura K. (2003) Biochem. Biophys. Res. Commun. 309, 848–856 [DOI] [PubMed] [Google Scholar]
- 12.Mizuno E., Kawahata K., Kato M., Kitamura N., Komada M. (2003) Mol. Biol. Cell 14, 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komada M., Kitamura N. (2005) J. Biochem. 137, 1–8 [DOI] [PubMed] [Google Scholar]
- 14.Haj F. G., Markova B., Klaman L. D., Böhmer F. D., Neel B. G. (2003) J. Biol. Chem. 278, 739–744 [DOI] [PubMed] [Google Scholar]
- 15.Row P. E., Clague M. J., Urbé S. (2005) Biochem. J. 389, 629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuible M., Doody K. M., Tremblay M. L. (2008) Cancer Metastasis Rev. 27, 215–230 [DOI] [PubMed] [Google Scholar]
- 17.Haj F. G., Verveer P. J., Squire A., Neel B. G., Bastiaens P. I. (2002) Science 295, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 18.Romsicki Y., Reece M., Gauthier J. Y., Asante-Appiah E., Kennedy B. P. (2004) J. Biol. Chem. 279, 12868–12875 [DOI] [PubMed] [Google Scholar]
- 19.Anderie I., Schulz I., Schmid A. (2007) Cell. Signal. 19, 582–592 [DOI] [PubMed] [Google Scholar]
- 20.Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., Ramachandran C., Gresser M. J., Tremblay M. L., Kennedy B. P. (1999) Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 21.Klaman L. D., Boss O., Peroni O. D., Kim J. K., Martino J. L., Zabolotny J. M., Moghal N., Lubkin M., Kim Y. B., Sharpe A. H., Stricker-Krongrad A., Shulman G. I., Neel B. G., Kahn B. B. (2000) Mol. Cell. Biol. 20, 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sangwan V., Paliouras G. N., Abella J. V., Dubé N., Monast A., Tremblay M. L., Park M. (2008) J. Biol. Chem. 283, 34374–34383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 24.Bentires-Alj M., Neel B. G. (2007) Cancer Res. 67, 2420–2424 [DOI] [PubMed] [Google Scholar]
- 25.Dubé N., Cheng A., Tremblay M. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuible M., Dubé N., Tremblay M. L. (2008) J. Biol. Chem. 283, 15740–15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 28.Hammond D. E., Carter S., McCullough J., Urbé S., Vande Woude G., Clague M. J. (2003) Mol. Biol. Cell 14, 1346–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoncic P. D., Lee-Loy A., Barber D. L., Tremblay M. L., McGlade C. J. (2002) Curr. Biol. 12, 446–453 [DOI] [PubMed] [Google Scholar]
- 30.Pandey A., Fernandez M. M., Steen H., Blagoev B., Nielsen M. M., Roche S., Mann M., Lodish H. F. (2000) J. Biol. Chem. 275, 38633–38639 [DOI] [PubMed] [Google Scholar]
- 31.Burz D. S., Shekhtman A. (2008) PLoS One 3, e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barriere H., Nemes C., Lechardeur D., Khan-Mohammad M., Fruh K., Lukacs G. L. (2006) Traffic 7, 282–297 [DOI] [PubMed] [Google Scholar]
- 33.Stuible M., Zhao L., Aubry I., Schmidt-Arras D., Böhmer F. D., Li C. J., Tremblay M. L. (2007) ChemBioChem 8, 179–186 [DOI] [PubMed] [Google Scholar]
- 34.Parachoniak C. A., Park M. (2009) J. Biol. Chem. 284, 8382–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohi O., Lehto V. P. (1998) FEBS Lett. 432, 225–227 [DOI] [PubMed] [Google Scholar]
- 36.Barriere H., Nemes C., Du K., Lukacs G. L. (2007) Mol. Biol. Cell 18, 3952–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorkina T., Huang F., Beguinot L., Sorkin A. (2002) J. Biol. Chem. 277, 27433–27441 [DOI] [PubMed] [Google Scholar]
- 38.Salmeen A., Andersen J. N., Myers M. P., Tonks N. K., Barford D. (2000) Mol. Cell 6, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 39.Myers M. P., Andersen J. N., Cheng A., Tremblay M. L., Horvath C. M., Parisien J. P., Salmeen A., Barford D., Tonks N. K. (2001) J. Biol. Chem. 276, 47771–47774 [DOI] [PubMed] [Google Scholar]
- 40.Confalonieri S., Salcini A. E., Puri C., Tacchetti C., Di Fiore P. P. (2000) J. Cell Biol. 150, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorkin A., Von Zastrow M. (2002) Nat. Rev. Mol. Cell Biol. 3, 600–614 [DOI] [PubMed] [Google Scholar]
- 42.Vieira A. V., Lamaze C., Schmid S. L. (1996) Science 274, 2086–2089 [DOI] [PubMed] [Google Scholar]
- 43.Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 44.Raiborg C., Malerød L., Pedersen N. M., Stenmark H. (2008) Exp. Cell Res. 314, 801–813 [DOI] [PubMed] [Google Scholar]
- 45.Eden E. R., White I. J., Tsapara A., Futter C. E. (2010) Nat. Cell Biol. 12, 267–272 [DOI] [PubMed] [Google Scholar]
- 46.Dubé N., Bourdeau A., Heinonen K. M., Cheng A., Loy A. L., Tremblay M. L. (2005) Cancer Res. 65, 10088–10095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.