Abstract
Membrane proteins constitute 20–30% of all proteins encoded by the genome of various organisms. Large amounts of purified proteins are required for activity and crystallization attempts. Thus, there is an unmet need for a heterologous membrane protein overexpression system for purification, crystallization, and activity determination. We developed a combinatorial method for overexpressing and purifying membrane proteins using Escherichia coli. This method utilizes short hydrophilic bacterial proteins, YaiN and YbeL, fused to the ends of the membrane proteins to serve as facilitating factors for expression and purification. Fourteen prokaryotic and mammalian membrane proteins were expressed using this system. Moderate to high expression was obtained for most proteins, and detergent solubilization combined with a short purification process produced stable, monodispersed membrane proteins. Five of the mammalian membrane proteins, overexpressed using our system, were reconstituted into liposomes and exhibited transport activity comparable with the native transporters.
Keywords: Gene Expression, Mammal, Membrane, Membrane Proteins, Reconstitution of Membrane Transporters, Overexpression
Introduction
Membrane proteins play key roles in fundamental biological processes in all living organisms. They catalyze the specific transport of ions and metabolites across the cell membrane, play a crucial role in energy exchanges, and act as signal receptors for receiving and transducing signals across the membrane (1). Therefore, understanding the mechanisms of action of membrane proteins is crucial for advances in basic and pharmacological research.
Determination of a three-dimensional structure is an important tool for understanding the function and mechanism of action of a protein. There are four main methods for determining three-dimensional structure: x-ray diffraction, nuclear magnetic resonance, atomic force microscopy, and electron diffraction. X-ray diffraction, based on protein crystallization, is currently the most widely used method (2). However, only a small number of high resolution three-dimensional structures of membrane proteins have been solved, most of them from bacteria. One reason for this dearth of information is that it is difficult to obtain a sufficient amount of the protein of interest for crystallography because membrane proteins are usually present at minute levels in natural biological membranes (3). Hence, structural studies of membrane proteins usually require overexpression of the membrane protein in question.
For soluble proteins, many different overexpression systems are currently available (4), with the Escherichia coli overexpression system the most prevalent (5, 6). Over the last decade, methods have been developed to enhance overexpression in E. coli. In particular, the use of fusion proteins (7–9) has become increasingly common: genetic fusion of the protein of interest to the N or C termini of other proteins can yield higher expression, increase solubility, and simplify detection and affinity purification of the overexpressed products. Examples of proteins that are commonly used for construction fusion proteins are maltose-binding protein (8–9), glutathione S-transferase (10), NuSA (11), and thioredoxin (12). However, most of the proteins successfully overexpressed as fusion proteins are soluble proteins, and there have only been a few reports of overexpressed fusion membrane proteins (13–16). For example, the G protein-coupled receptor, neurotensin receptor, was actively expressed in E. coli with maltose-binding protein fused to its N terminus and thioredoxin fused to its C terminus. Because G protein-coupled receptors have their N terminus at the periplasm, maltose-binding protein, with its own signal peptide, assists efficient translocation across the E. coli cytoplasmic membrane, positioning the N terminus of the receptor in the periplasm (16). But, most membrane proteins show a strong preference for the N terminus in the cytoplasm (17).
Therefore, few better fusion partners for membrane protein overexpression have been developed. Zhang et al. (18) used fusion with a set of short, nonglobular, negatively charged peptides to overexpress the adenovirus receptor immunoglobulin variable-type domains successfully. Roosild et al. (19) used MISTIC, a short, nonglobular Bacillus subtilis integral membrane protein as a fusion domain for overexpression of 15 membrane proteins in E. coli. Rasmussen et al. (20) fused the T4 lysozyme in place of the third intracellular loop of the β2-adrenergic receptor and facilitated its crystallization into diffracting crystals (20).
However, all of these successful fusion systems used one fusion cassette. Here, we report the development of a combinatorial fusion system for overexpression of membrane proteins in E. coli. Our combinatorial system comprises two fusion domains; fusion with either the N or C terminus of the gene of interest, or both, allows us to create eight expression cassettes for a given protein and increases the chances that membrane protein overexpression will be successful. The fusion domains are two short hydrophilic bacterial proteins YaiN and YbeL, about 100 amino acids long, which, when fused to the ends of the membrane protein, serve as facilitating factors for its expression and purification. These two bacterial domains were selected to serve as a fusion domain because of their hydrophilic nature and nonglobular α-helical predicted structure (see supplemental Fig. 1). The YaiN fusion domain belongs to the E. coli operon frm, whose likely function is the degradation of formaldehyde (21). The YbeL fusion domain contains the first 120 amino acid of the E. coli YbeL gene that has no known function (22).
We expressed 14 prokaryote and eukaryote membrane proteins using this overexpression system, obtaining a high to moderate expression level. The proteins were solubilized using mild detergent, indicating that they were inserted into the membrane rather than in inclusion bodies. We also developed a simple purification process that resulted in stable, monodispersed membrane proteins that were amenable to crystallization. In addition, we observed transport uptake activity of five different proteins from the SLC17 family that were overexpressed in our system and reconstituted into liposomes: ATP transport activity of the human vesicular nucleotide transporter, glutamate transport activity of the rat vesicular glutamate transporter, aspartate and sialic acid transport activities of the human and mouse Sialinl transporter (hSialin and mSialin) and Pi transport activity of the mouse sodium-dependent phosphate transporter. Thus, our method may be a useful alternative system for overexpression and biochemical and structural characterization of membrane proteins that would otherwise be difficult or impossible to study (23).
EXPERIMENTAL PROCEDURES
Cloning
Cloning of the nine cassette combinations (see Fig. 1), cloning of membrane proteins into the nine combinations, cloning of MISTIC, and cloning of the TEV2 protease site are described in detail in the supplemental Methods.
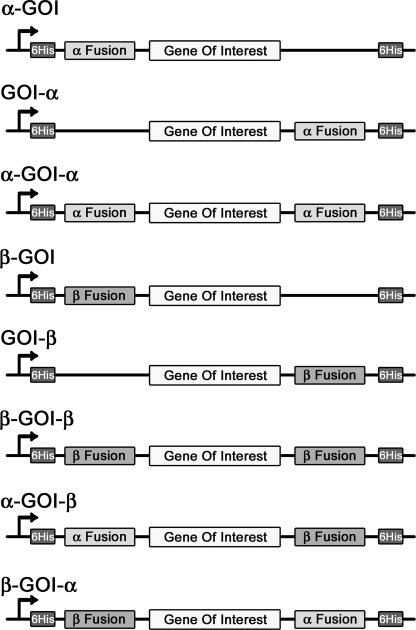
FIGURE 1.
Schematic representation of the eight inducible pET-28a(+) expression cassettes. Each construct produces a fusion protein by adding bacterial protein domains to the membrane protein at the N terminus, the C terminus, or to both termini, together with two His6 tags. GOI, gene of interest.
Protein Overexpression
E. coli C43 (DE3) cells were transformed with expression vectors and grown in 2% LB (Difco) plus 20 μg/ml kanamycin sulfate, for 16 h at 37 °C. TB medium (Terrific Broth; Sigma) plus 20 μg/ml kanamycin sulfate was inoculated with 1/1000 transformed C43 culture and grown at 37 °C. After the culture reached an A600 of 0.6–0.8, it was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and incubated for 16 h at 18 °C. The induced cell cultures were harvested and lysed until homogeneity in solubilization buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 10 mm KCl, 2 mm phenylmethylsulfonyl fluoride). Cells were crushed using French press at 15,000 pounds/square inch (SLM Instruments), centrifuged at 5,856 × g at 4 °C for 5 min (to remove large inclusion bodies and cell debris), and the supernatant was carefully separated from the pellet and then centrifuged at 150,000 × g for 1 h at 4 °C (Ti70 rotor; Beckman). The membrane-containing pellet was isolated and resuspended in solubilization buffer. Protein concentration was determined using the Bradford assay, and all samples were diluted to 10 mg/ml. Samples were analyzed by Western blotting. Western blot conditions are described in the supplemental Methods.
Detergent Solubilization and Protein Purification
Membranes (10 mg/ml) were treated with 2% detergent obtained from Anatrace (stock solution was 10%, solubilized in double-distilled water) and incubated for 10–60 min at 0 °C. The unsoluble membranes were removed by ultracentrifugation (Ti70 rotor) at 150,000 × g at 4 °C for 1 h, and the supernatant containing the fusion proteins was isolated, diluted twice, and passed through a 10–15-ml nickel-NTA (Qiagen) affinity column (Bio-Rad Econo-Column low pressure chromatography), equilibrated with washing buffer containing 20 mm imidazole, 100 mm NaCl, 10 mm KCl, 20 mm Tris, pH 8. After protein binding, the column was washed with washing buffer containing 0.02–0.1% detergent. If needed, detergent was exchanged by washing with 10 column volumes of washing buffer containing 0.02–0.1% different detergent, and 0.05–0.1 mg/ml phosphatidylethanolamine (PE; Sigma) or a mixture of 0.05:0.016 mg/ml cholesterol/PE (Sigma). For lipid preparation, 40 mg of PE was suspended in 1 ml of washing buffer by sonication in bath sonicator until homogeneity. 60 mg of cholesterol was suspended in 1 ml of 20 mg/ml PE by sonication in bath sonicator until homogeneity. The lipid aliquots (40 μl, 3:1 mg/ml for cholesterol/PE) were frozen (−80 °C) and prior to use were fast thawed and immediately diluted (60 times for cholesterol/PE) in washing buffer containing a detergent. The protein was eluted with buffer containing 300 mm imidazole, 100 mm NaCl, 10 mm KCl, 20 mm Tris, pH 7, and 0.02–0.1% new detergent. Selected fractions were concentrated by adding 10% polyethylene glycol 6000 (Hampton Research) to each fraction, incubating for 10 min at 0 °C, and centrifuging at 11,950 × g at 4 °C for 10 min. The pellet was resuspended in buffer containing 20 mm Tris, pH 7.5, 0.02–0.1 detergent. For further purification, a 120-ml size-exclusion gel-filtration column (HiLoad 16/60, Superdex 200 preparation grade) was equilibrated with buffer containing 20 mm Tris, pH 7.5, 70 mm NaCl, 10 mm KCl, and 0.02–0.05% detergent. The flow rate was 1 ml/min.
Reconstitution and Transport Assay
Reconstitution of purified SLAC17 protein into liposomes and transport assay were carried out as described (26–28). For more details, see supplemental Methods.
Oligomerization Assays
A purified and concentrated (∼10–20 μg) sample of fusion protein was loaded onto a 7–60% sucrose gradient containing 20 mm Tris, pH 7.5, and 0.02–0.1% detergent. The gradient was centrifuged in an SW40 rotor at 170,000 × g for 14–16 h. Fractions were collected from the bottom of the tube.
RESULTS
Construction of Eight Fusion Protein Expression Cassettes using Two Bacterial Fusion Domains
Using two bacterial protein domains, we designed eight expression cassettes for each protein utilizing the pET-28a(+) expression vector. The two bacterial protein fusion domain genes were YaiN (21) (98 amino acids), which we called α, and YbeL (22) (120 amino acids), which we called β. The cassettes were designed to fuse α or β to the N terminus, the C terminus, or to both termini of the gene encoding each membrane protein (Fig. 1). Construction of the expression cassettes is described in detail in supplemental Table 1.
Selection of Membrane Proteins for Expression
We have concentrated our efforts on overexpressing multiple transmembrane proteins such as transporters. Therefore, 14 prokaryotic and eukaryotic multiple transmembrane proteins were chosen for cloning into the eight expression cassettes: 12 transporters, one channel and one enzyme (for additional information, see supplemental Table 2). In addition, because many membrane proteins have a strong preference for the N terminus in the cytoplasm (17), all of the membrane proteins we chose have a cytoplasmic N terminus. We have not yet tested proteins with periplasmic N terminus, such as G protein-coupled receptors.
Estimation of Membrane Protein Overexpression
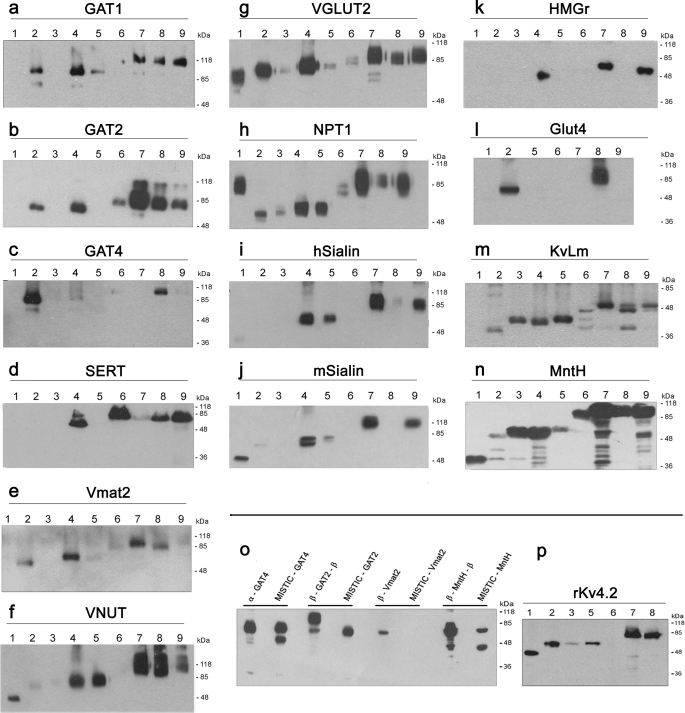
Following the genetic cloning of the selected membrane proteins into the eight overexpression cassettes, small scale overexpression experiments were performed. As a control, a ninth construct was prepared for each protein tested, with the gene cloned into the pET-28a(+) vector for expression without any fusion domains. Expression levels were analyzed by Western blotting (Fig. 2, a–n). For each membrane protein, at least two of the combinations yielded high levels of expression that were markedly higher than that of the control clone; in some cases, we observed high yields even when there was no detectable expression using the control construct. Note that in these experiments, the proteins are not purified yet, and inclusion bodies are not fully removed, yielding multiple bands on the SDS-PAGE. However, these impurities are being mostly removed in the large scale purification process that follows. We also compared the currently reported expression system with the MISTIC method (19). As shown in Fig. 2o, whereas similar results were obtained with the expression of GAT4 and GAT2, Vmat2 was not expressed at all by the MISTIC method and was highly expressed by our system. In addition, the prokaryotic E. coli membrane protein MntH exhibited higher expression in our system. Furthermore, we have shown in Fig. 2p that the Rattus norvegicus Kv4.2 (24) potassium channel, which could not be overexpressed with the MISTIC fusion protein (19), was highly expressed in our system.
FIGURE 2.
Western blot analysis of the small scale overexpression tests. Small scale overexpression of GAT1 (a), GAT2 (b), GAT4 (c), SERT (d), Vmat2 (e), VNUT (f), VGLUT2 (g), NPT1 (h), hSialin (i), mSialin (j), HMGR (k), GLUT4 (l), KvLm (m), and MntH (n) is shown. Numbers indicate the specific construct for each protein. GOI, gene of interest. Lanes 1, GOI in control vector; lanes 2, α-GOI; lanes 3, GOI-α; lanes 4, β-GOI; lanes 5, GOI-β; lanes 6, α-GOI-α; lanes 7, β-GOI-β; lanes 8, α-GOI-β; lanes 9, β-GOI-α. o, Western blot analysis of overexpressed membrane proteins fused either to the α/β combinations or the MISTIC protein. Three eukaryotic membrane proteins (GAT4, GAT2, and Vmat2) and one prokaryote E. coli protein (MntH) were cloned downstream of the MISTIC gene. The MISTIC fusion proteins were tested for overexpression alongside the best overexpressed clone of each gene in the α/β fusion system. The calculated molecular masses of the overexpressed fusion proteins in kDa, left to right, are 84.5, 89, 99, 85.7, 72, 71.8, 76.4, and 63. p, Western blot analysis of small scale overexpression test of rKv4.2. Numbers indicate the specific fusion construct as described above. For all Western blot analyses, samples containing 5 μg of proteins were separated by SDS-PAGE, transferred to a membrane, probed with an anti-His6 antibody, and detected using ECL. Aggregates at the top of the gel, typical for polytopic membrane proteins, are not shown.
Optimization of Fusion Protein Solubilization
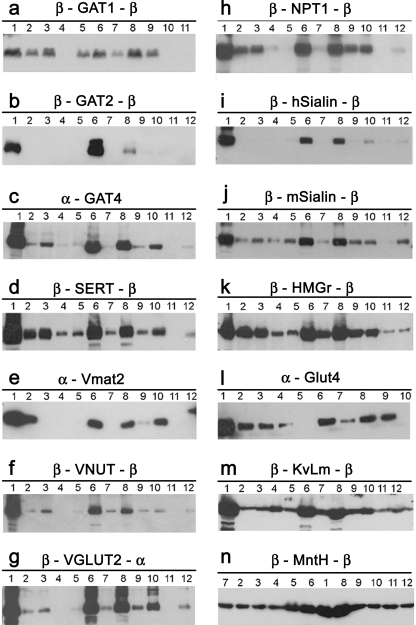
For each membrane protein, the overexpressing combinations were tested to determined whether the proteins could be solubilized from the membrane. Note that if an overexpressed protein is sequestered in inclusion bodies, mild detergents will not solubilize it. However, if it is inserted into the membrane, treatment with mild detergent will solubilize it. We tested 11 relatively mild detergents, using the detergent sodium-lauryl-sarcosine as a positive control due to its strong ionic properties. Most of the detergents tested have previously been used successfully in membrane protein crystallography. Fig. 3 shows Western blot analysis of the solubilization test for one combination from each membrane protein tested. As can be seen, every overexpressed membrane protein could be solubilized by at least one of the mild detergents, which mean that considerable amounts of the membrane proteins were not in inclusion bodies. Similar results were observed for most of the additional overexpressing combinations of each membrane protein. However, as expected, the fusion domains did not eliminate inclusion body formation, and further investigation is required to decrease the fusion body formation and to improve the intact membrane protein amounts.
FIGURE 3.
Western blot analysis of detergent solubilization of the fusion membrane proteins. Fusion proteins (∼10–20 μg) were tested for solubilization using 2% solutions of 12 detergents: β-GAT1-β (a), β-GAT2-β (b), α-GAT4 (c), β-SERT-β (d), α-Vmat2 (e), β-VNUT (f), β-VGLUT2-α (g), β-NPT1-β (h), β-hSialin-β (i), β-mSialin-β (j), β-HMGR-β (k), α-GLUT4 (l), β-KvLm-β (m), and β-MntH-β (n). The soluble fractions after ultracentrifugation were subjected to Western blot analysis using an anti-His6 antibody and ECL. Numbers indicate the detergent used: lanes 1, sodium-lauryl-sarcosine; lanes 2, DDM; lanes 3, n-dodecyl-β-d-thiomaltoside (DTM); lanes 4, lauryl dimethylamine oxide; lanes 5, ChapsO; lanes 6, Fos-choline-16/14; lanes 7, sucrose monododecanoate; lanes 8, Cyclofos-7; lanes 9, Cymal-7; lanes 10, sodium cholate; lanes 11, n-octyl-β-d-glucoside; lanes 12, Triton X-100. Aggregates at the top of the gel, typical for polytopic membrane proteins, are not shown.
Scale-up and Purification of the Overexpressed Proteins
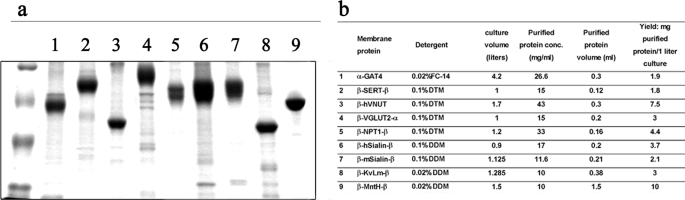
For each selected combination, large scale expression experiments (9-liter E. coli culture) were conducted, and purification was done by standard protein separation techniques. To obtain large protein amounts, the initial purification step, solubilization from the membrane, was done mostly by Fos-choline, although milder detergents were able to solubilize most of the membrane proteins tested. Solubilizations from the membrane with milder detergents have not yet been tested. In the following purification step, the Fos-choline detergent was exchanged upon the Ni-NTA affinity column, and we were able to incorporate lipids, including cholesterol (GAT1–4, SERT, NPT1, Sialinl) during this process. Because the E. coli membrane dose not contains cholesterol, incorporation of cholesterol to the solubilized membrane proteins can potentially become a crucial step for proper folding and activity experiments. In addition, detergent exchange can dramatically increase the possibilities for manipulating crystallization conditions (25). For some proteins, an additional purification step was done by a size-exclusion gel-filtration column, and as shown in supplemental Fig. 2, the solubilized membrane proteins maintained their stability and solubility through the gel-filtration purification, which enabled separation and purification of inhomogenities. Obviously, when two peaks were obtained, we examined both in SDS-PAGE Coomassie Brilliant Blue staining and chose the most purified and homogenic peak for further testing. We were able to purify each overexpressed membrane protein, obtaining exceptionally high amounts of pure protein for almost all membrane proteins tested (purification yields are in Fig. 4b). Fig. 4a shows SDS-PAGE Coomassie Brilliant Blue staining analyses of nine purified membrane proteins.
FIGURE 4.
Purification of the overexpressed fusion membrane proteins. Solubilized membrane proteins were applied to a Ni-NTA affinity column. If necessary, detergent exchanges were performed before elution in the presence of 0.05–0.1 mg/ml lipids (PE or PE + cholesterol). The relevant eluted Ni-NTA fractions were concentrated and subjected to size-exclusion gel-filtration chromatography. Selected gel-filtration column fractions were concentrated by polyethylene glycol precipitation and resuspended in crystallization buffer containing 0.02–0.1% detergent. a, Coomassie Brilliant Blue staining of the purified and concentrated proteins: lane 1, α-GAT4 solubilized in Fos-choline-14; lane 2, β-SERT-β solubilized in Fos-choline-14 followed by detergent exchange to DTM + PE + cholesterol; lane 3, β-VNUT solubilized in Fos-choline-14 followed by detergent exchange to DTM + PE; lane 4, β-VGLUT2-β solubilized in Fos-choline-14 followed by detergent exchange to DTM; lane 5, β-NPT1-β solubilized in Fos-choline-14 followed by detergent exchange to DTM + PE + cholesterol; lane 6, β-hSialin-β solubilized in Fos-choline-14 followed by detergent exchange to DDM + PE + cholesterol; lane 7, β-mSialin-β solubilized in Fos-choline-14 followed by detergent exchange to DDM + PE + cholesterol; lane 8, β-KvLm-β solubilized in Fos-choline-16 followed by detergent exchange to DDM; lane 9, β-MntH-β solubilized in DDM. The amounts of purified proteins loaded on the gel are 5–10 μg. Aggregates at the top of the gel, typical for polytopic membrane proteins, are not shown. Molecular mass marker sizes, top to bottom, are 118, 85, 48, 36, 26, and 20 kDa. b, purification yields of the overexpressed membrane proteins. Yields (in milligrams of purified protein/liter of E. coli culture) were calculated by dividing the final amount of purified protein (in milligrams) by the volume (in liters) of the E. coli culture used.
Stability and Oligomerization State of the Purified Membrane Proteins
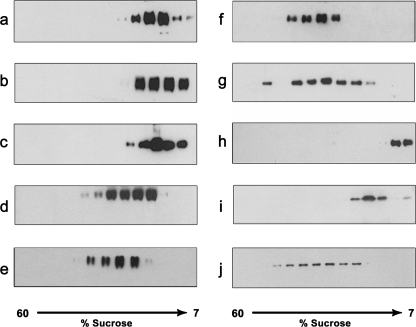
We next used linear sucrose gradient to determine the stability and oligomerization state of the purified membrane proteins (excluding the proteins VNUT, VGLUT2, NPT1, and Vmat). In this method, analysis of the protein distribution along the sucrose gradient reveals whether the protein is stably monodispersed or is in aggregates. As shown in Fig. 5, the overexpressed proteins sedimented in distinct peaks. This experiment suggests that regardless of their oligomeric state (monomer, dimer, trimer, or tetramer), the purified membrane proteins are monodispersed, not aggregated.
FIGURE 5.
Oligomerization state of the purified fusion membrane proteins. The purified proteins were loaded on a 7–60% linear sucrose gradient and centrifuged for 15 h. Fractions were collected and analyzed by Western blotting using an anti-His6 antibody and ECL. a, β-GAT1-β in 0.1% DDM (exchanged from Cyclofos-7); b, β-GAT2-β in 0.1% Fos-choline-16; c, α-GAT4 in 0.02% Fos-choline-16; d, β-SERT-β in 0.02% DTM (exchanged from Fos-choline-14); e, β-hSialin-β in 0.1% DDM (exchanged from Fos-choline-14; f, β-mSialin-β in 0.1% DDM (exchanged from Fos-choline-14); g, β-HMGR-β in 0.02% DDM; h, α-GLUT4 in 0.02% Fos-choline-16; i, β-KvLm-β in 0.02% DDM (exchanged from Fos-choline-16); j, β-MntH-β in 0.02% DDM.
In Vitro Activity Tests
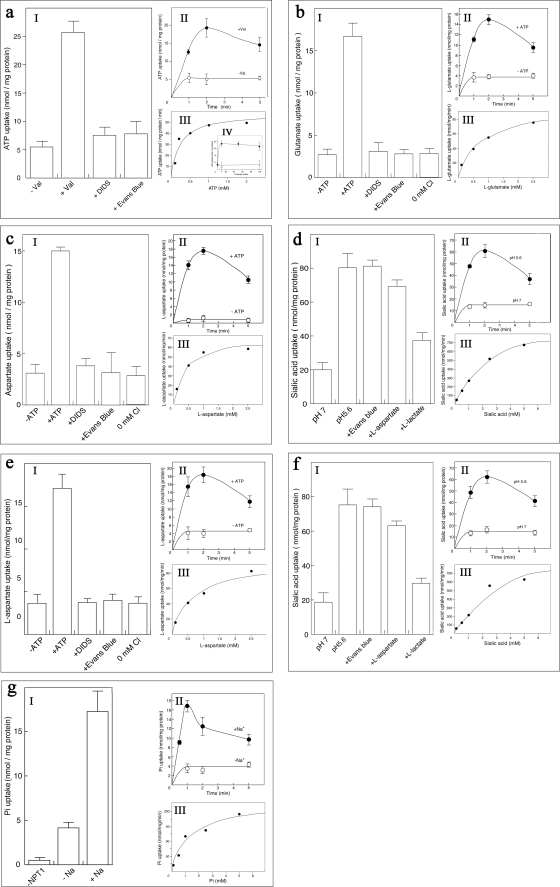
To support the notion that membrane proteins overexpressed in our system are properly folded and can maintain their activity, we reconstituted five of the mammalian SLC17 transporters overexpressed in our system and conducted specific uptake experiments: human vesicular nucleotide transporter (hVNUT) fused to the β domain at its N terminus, rat vesicular glutamate transporter-2 (rVGLUT2) fused to the β domain at its N terminus and to the α domain at its C terminus, human and mouse sialic acid transporters (hSialinl, mSialinl), each fused to the β domain at both N and C termini and mouse Na-Pi transporter-1 (mNPT1), fused to the β domain at both N and C termini. We have tested their kinetic properties and whether the reconstituted transporters maintained their previously reported features (26–28), namely, inhibition by diisothiocyanatostilbene-2,2′-disulfonate (DIDS) and Evans blue and Cl− dependence of the transport activity. The purified β-hVNUT (Fig. 4a, lane 3) was reconstituted into liposomes, and uptake of [α-32P]ATP in proteoliposomes was measured. When an internal inside positive Δψ was established (by a K+-diffusion potential through the addition of valinomycin), the proteoliposomes took up [α-32P]ATP, which was markedly inhibited by DIDS and Evans blue (Fig. 6aI). In addition, the proteoliposomes took up ATP in a time-dependent manner (Fig. 6aII), and the valinomycin-evoked ATP uptake exhibited dose dependence with Km and Vmax values of 0.2 mm and 55.7 nmol/min per mg of protein, respectively (Fig. 6aIII). Cl− dependence was also measured, and as previously reported (26), the presence of Cl− was an absolute requirement for the ATP transport activity (Fig. 6aIV). The purified protein β-VGLUT2-α (Fig. 4a, lane 4) was co-reconstituted with purified bacterial F-ATPase into liposomes. F-ATPase is required to establish a stable Δψ by active proton pumping, which will serve as the driving force for l-glutamate uptake (27). Upon addition of ATP, the proteoliposomes facilitated l-[2,3-3H]glutamate uptake, which was markedly inhibited by DIDS and Evans blue and required Cl− for the transport activity (Fig. 6bI). In addition, the proteoliposomes took up l-glutamate in a time-dependent manner (Fig. 6bII) and the l-glutamate uptake exhibited dose dependence with Km and Vmax values of 0.63 mm and 86.4 nmol/min per mg of protein, respectively (Fig. 6bIII). The purified human and mouse sialic acid transporters, β-hSialinl-β and β-mSialinl-β (Fig. 4a, lanes 6 and 7), which were purified in the presence of cholesterol, were co-reconstituted with bacterial F-ATPase into liposomes to generate a stable Δψ which will facilitate the uptake of l-aspartate (28). Upon addition of ATP, the proteoliposomes facilitated l-[2,3-3H] aspartate uptake, which was markedly inhibited by DIDS and Evans blue and required Cl− for the transport activity (Fig. 6, c and eI). In addition, the proteoliposomes took up l-aspartate in a time-dependent manner (Fig. 6, c and eII), and the l-aspartate uptake exhibited dose dependence with Km and Vmax values of 0.39 mm and 65.1 nmol/min per mg of protein, respectively for hSialin and 0.6 mm and 88.7 nmol/min per mg, respectively, for mSialin (Fig. 6, c and eIII). In addition, we have reconstituted β-hSialin-β and β-mSialin-β into liposomes without F-ATPase and measured pH-dependent [6-3H]sialic acid co-transport activity. Upon establishing a pH gradient, the proteoliposomes facilitated [6-3H]sialic acid uptake, which, as expected (28), was not inhibited by Evans blue or l-aspartate, but was inhibited by l-lactate (Fig. 6, d and fI). In addition, the proteoliposomes took up sialic acid in a time-dependent manner (Fig. 6, d and fII), and the sialic acid uptake exhibited dose dependence with Km and Vmax values of 1.38 mm and 721 nmol/min per mg of protein, respectively, for hSialin and 2.15 mm and 809 nmol/min per mg, respectively, for mSialin (Fig. 6, d and fIII). The purified protein β-NPT1-β, which was purified in the presence of cholesterol (Fig. 4a, lane 5), was reconstituted into liposomes and Na+-dependent uptake of [32P]Na2HPO4 into the proteoliposomes was measured. When a Na+ gradient was imposed in the proteoliposomes, Pi uptake was observed and was not present in liposomes lacking the purified β-NPT1-β (Fig. 6gI). In addition, the proteoliposomes took up Pi in a time-dependent manner (Fig. 6gII), and the Pi uptake exhibited dose dependence with Km and Vmax values of 1.18 mm and 228.9 nmol/min per mg of protein, respectively (Fig. 6gIII). We have further reconstituted NPT1 with E. coli lipid instead of asolectin (identical liposomes concentrations were used), but essentially, the same transport activity was detected as that of NPT1 reconstituted with asolectin liposomes (data not shown). Therefore, E. coli lipid can be used for reconstitution as asolectin.
FIGURE 6.
Uptake activity of the reconstituted proteins into proteoliposomes. I, specific substrate uptake activities in the presence or absence of appropriate driving force, including inhibition of uptake activities (by either 2 μm DIDS, 1 μm Evans blue, 5 mm l-aspartate, or 5 mm l-lactate) and Cl− dependence (IV for VNUT). II, time course of uptake activity. Proteoliposomes were suspended in reaction buffer containing radiolabeled substrate in the presence (filled circles) or absence (open circles) of the appropriate driving force. Samples were assayed at the indicated times. III, dose dependence of uptake activity. Upon the addition of radiolabeled substrate at the listed concentrations, samples were assayed after 1 min (0.5 min for NPT1). a, [α-32P]ATP uptake activity of β-VNUT; b, l-[2,3-3H]glutamate uptake by β-VGLUT2-α; c, l-[2,3-3H]aspartate uptake activity of β-hSialin-β; d, H+/[6-3H]sialic acid co-transport uptake activity of β-hSialin-β; e, l-[2,3-3H]aspartate uptake activity of β-mSialin-β; f, H+/[6-3H]sialic acid co-transport uptake activity of β-mSialin-β; g, Pi uptake activity by β-NPT1-β.
Digestion of Fusion Proteins with Proteolytic Enzymes
After confirmation that the overexpressed membrane fusion proteins were stable, monodispersed, and correctly folded, we tested three of the overexpressed proteins for stability after removal of the bacterial fusion domains. To do this, we cloned the TEV protease site into the eight expression vectors so that one or both fusion domains could be removed (supplemental Fig. 3f). In β-MntH-β, after a 15-h digestion with TEV, only one of the β-fusion domains was removed; we hypothesized that the other β-domain was inaccessible due to folding (supplemental Fig. 3a). Supporting this idea, TEV digestion of another combination, β-MntH, resulted in ∼98% removal of β (supplemental Fig. 3b). For α-GAT4, TEV proteolysis yielded ∼98% removal of the α-fusion domain (supplemental Fig. 3c). In all of these experiments, the resulting membrane proteins were stable through size-exclusion chromatography and could be concentrated in crystallization buffers containing a low detergent concentration. In addition, we performed nonspecific (trypsin) proteolysis of β-KvLm-β (in 0.02% n-dodecyl-β-d-maltoside; DDM) for 15 h. After proteolysis, we observed a single strong band at about 28 kDa, which corresponds well to the predicted size of KvLm (supplemental Fig. 3d). Furthermore, we applied the purified trypsin-digested KvLm on a linear sucrose gradient and observed that KvLm was monodispersed (supplemental Fig. 3e).
DISCUSSION
There have been many reports of methods for overexpressing soluble fusion proteins in E. coli (7, 9). In contrast, membrane proteins have presented a much bigger challenge in terms of developing a general but reliable overexpression system. The combinatorial system for expression and purification of membrane proteins described in this report represents a step forward in the effort to achieve this important goal.
Using this expression system, we achieved overexpression of all tested proteins, namely 12 mammalian and 2 prokaryotic membrane proteins. The overexpression levels using our constructs were markedly higher than expression from the control clones (the membrane protein with no fusion domains), which generally expressed the protein poorly or not at all (Fig. 2). Furthermore, all of the membrane proteins tested were overexpressed using at least two of the overexpression combinations. Using eight cassettes for overexpression dramatically increases the chances that the membrane protein will be able to be crystallized and maintain its activity: The location (N terminus, C terminus, or both) and type (α, or β, or both) of the fusion domains in each cassette affect the solubility, stability, and homogeneity of the expressed membrane protein. In contrast to other expression systems, here the use of eight expression cassettes allows optimization of the expression of individual proteins, each of which has unique properties. For example, GAT4 was highly overexpressed as α-GAT4 (Fig. 2c, lane 2), whereas all the other GABA neurotransmitters transporters (GAT1 and GAT2) were highly expressed as fusion proteins with two fusion domains: α-GAT-β, β-GAT-α or β-GAT-β, where GAT is GAT1-2 (Fig. 2, a and b, lanes 7–9).
Overexpression of membrane proteins as fusion proteins with α- or β-domains reduced accumulating in inclusion bodies. In addition, all of the overexpressed membrane proteins tested were solubilized using mild detergents that are suitable for activity and crystallization experiments (Fig. 3). This suggests that a large portion of the membrane proteins overexpressed using this system localize to the E. coli membrane rather than being sequestered in inclusion bodies. Indeed, the overexpressed proteins could be purified using standard purification techniques (Fig. 4 and supplemental Fig. 2), yielding larger quantities of purified and stable membrane proteins than is usually possible. For example, we obtained 1.8–7.5 mg of pure mammalian protein/liter of E. coli culture, and 3–10 mg of pure prokaryote protein/liter of E. coli culture (Fig. 4b).
To obtain high protein quantities, most of the proteins were solubilized from the membrane by the Fos-choline detergents. However, because only one membrane protein was crystallized using detergent from the Fos-choline family, MscS (29), and because Fos-choline detergents can solubilize and stabilize misfolded proteins, it was later exchanged into milder detergent in the presence of lipids. Such detergent exchanges separate the folded proteins from the misfolded ones (aggregates and not eluted together with the soluble proteins) and more importantly, enable the incorporation of lipids that are very important for activity and crystallization experiments. The detergent type is key for crystal formation, and the lipid composition can dramatically affect the stability, homogeneity, and activity of the membrane protein (30). For example, plasma membrane proteins such as the Na+/Cl− neurotransmitter transporters (GAT1,2,4 and SERT) and NPT1 require cholesterol for their activity (3, 30). Cholesterol was added to the lipids during the purification of these proteins.
All of the tested purified membrane proteins could be purified into homogenic monodispersed form as judged by their migration as a narrow band during sucrose gradient centrifugation (Fig. 5) and behavior in gel-filtration chromatography (supplemental Fig. 2). These results indicate that the overexpressed membrane proteins are folded and are not in aggregates or inclusion bodies. Furthermore, the observation that three membrane proteins, MntH, KvLm, and GAT4, could maintain their solubility and distinct oligomeric state after the removal of the fusion domains (supplemental Fig. 3) suggests that the membrane proteins expressed using our system are correctly folded to some extent.
Proteoliposomes containing the purified transporters β-hVNUT, β-VGLUT2-α, β-hSialin-β, β-mSialin-β, and β-NPT1-β actively took up their specific substrates by using their appropriate driving force (Fig. 6). The uptake properties of the reconstituted transporters were similar to that of the previously reported transporters (26–28). For example, the uptake activities were absolutely dependent on the presence of Cl− and were highly inhibited by DIDS and Evans blue (Fig. 6I). In addition, the transporters exhibited similar kinetic properties to the previously reported values (Table 1), with differences that are probably consequences of differences in expression host, namely, E. coli versus eukaryotic cell lines. These results support our proposal that the overexpressed membrane proteins in our system are correctly folded and can maintain their native activity.
TABLE 1.
Kinetic parameters of SLC17 proteins overexpressed in the α/β-fusion system and insects cells
SLC17 proteins fused to the α/β-fusion proteins were overexpressed, purified, and reconstituted as described in the text. SLC17 proteins overexpressed in the baculovirus-insect cells system were purified and reconstituted as described in Refs. 26–28. Reconstitution and uptake experiments conditions were identical for proteins overexpressed in both systems. NT, not tested; NP, not published.
| Membrane protein | Organism | Substrate | Ref. | Kinetic parameters of SLC17 portions overexpressed in insect cells using baculovirus system |
Kinetic parameters of SLC17 portions overexpressed in E. coli using the α/β-fusion system |
||
|---|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | ||||
| mm | nmol/min per mg protein | mm | nmol/min per mg protein | ||||
| VNUT | Human | ATP | 26 | 0.8 | 138 | 0.2 | 55.7 |
| VGLUT | Rat | Glutamate | 27 | 4.8 | 120 | 0.63 | 86.4 |
| Sialin | Mouse | Aspartate | 28 | 0.62 | 142 | 0.6 | 88.7 |
| Sialic acid | 28 | 1.5 | 1,000 | 2.15 | 809 | ||
| Human | Aspartate | NT | NT | 0.39 | 65.1 | ||
| Sialic acid | NT | NT | 1.38 | 721 | |||
| NPT1 | Mouse | Pi | NP | 6.3 | 134 | 1.18 | 228.9 |
Taken together, these observations show that our overexpression system can produce membrane proteins that are folded, soluble, stable, and active. Therefore, membrane proteins overexpressed and purified in our system can be used for biological activity and crystallization experiments. This promising new system may allow great progress to be made in structure-function studies of membrane proteins, although obviously, the expression system described in this paper will not provide a solution for all kinds of membrane proteins. Even if 5% of membrane proteins are amenable for expression in this system, it will significantly advance the molecular study of membrane proteins.
Supplementary Material
Acknowledgments
We thank Dr. Takaaki Miyaji for help with the reconstitution experiments and Dr. Maurice Montal for the cDNA of KvLm, Dr. Gary Rudnick for the cDNA of hSERT, Dr. Shimon Schuldiner for the cDNA of rVmat2, Dr. Joseph Roitelman for the cDNA of HMGR, Dr. Amira Klip for the cDNA of Glut4, Dr. Senyon Choe for the cDNA of MISTIC, and Dr. Lily Yeh Jan for cDNA of rKv4.2.
This work was supported in part by Grant 3-6790 from the Japan-Israel Scientific Research cooperation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2, Figs. 1–3, Methods, and additional references.
- TEV
- tobacco etch virus
- DDM
- n-dodecyl-β-d-maltoside
- DIDS
- diisothiocyanatostilbene-2,2′-disulfonate
- DTM
- n-dodecyl-β-d-thiomaltoside
- NTA
- nitrilotriacetic acid
- PE
- phosphatidylethanolamine.
REFERENCES
- 1.Michel H. (1983) Trends Biochem. Sci. 8, 56–59 [Google Scholar]
- 2.Caffrey M. (2003) J. Struct. Biol. 142, 108–132 [DOI] [PubMed] [Google Scholar]
- 3.Tate C. G. (2001) FEBS Lett. 504, 94–98 [DOI] [PubMed] [Google Scholar]
- 4.Rai M., Padh H. (2001) Curr. Sci. 80, 1121–1128 [Google Scholar]
- 5.Makrides S. C. (1996) Microbiol. Rev. 60, 512–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quick M., Wright E. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 8597–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckwith J. (2000) Methods Enzymol. 326, 3–7 [DOI] [PubMed] [Google Scholar]
- 8.Kapust R. B., Waugh D. S. (1999) Protein Sci. 8, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pryor K. D., Leiting B. (1997) Protein Expr. Purif. 10, 309–319 [DOI] [PubMed] [Google Scholar]
- 10.Zhan Y., Song X., Zhou G. W. (2001) Gene 281, 1–9 [DOI] [PubMed] [Google Scholar]
- 11.Davis G. D., Elisee C., Newham D. M., Harrison R. G. (1999) Biotechnol. Bioeng. 65, 382–388 [PubMed] [Google Scholar]
- 12.LaVallie E. R., Lu Z., Diblasio-Smith E. A., Collins-Racie L. A., McCoy J. M. (2000) Methods Enzymol. 326, 322–340 [DOI] [PubMed] [Google Scholar]
- 13.Laage R., Langosch D. (2001) Traffic 2, 99–104 [DOI] [PubMed] [Google Scholar]
- 14.Begum R. R., Newbold R. J., Whitford D. J. (2000) Chromatogr. B Biomed. Sci. Appl. 737, 119–130 [DOI] [PubMed] [Google Scholar]
- 15.Drew D., Slotboom D. J., Friso G., Reda T., Genevaux P., Rapp M., Meindl-Beinker N. M., Lambert W., Lerch M., Daley D. O., Van Wijk K. J., Hirst J., Kunji E., De Gier J. W. (2005) Protein Sci. 14, 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White J. F., Trinh L. B., Shiloach J., Grisshammer R. (2004) FEBS Lett. 564, 289–293 [DOI] [PubMed] [Google Scholar]
- 17.Daley D. O., Rapp M., Granseth E., Melén K., Drew D., von Heijne G. (2005) Science 308, 1321–1323 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y. B., Howitt J., McCorkle S., Lawrence P., Springer K., Freimuth P. (2004) Protein Expr. Purif. 36, 207–216 [DOI] [PubMed] [Google Scholar]
- 19.Roosild T. P., Greenwald J., Vega M., Castronovo S., Riek R., Choe S. (2005) Science 307, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 21.Herring C. D., Blattner F. R. (2004) J. Bacteriol. 186, 6714–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perna N. T., Plunkett G., 3rd, Burland V., Mau B., Glasner J. D., Rose D. J., Mayhew G. F., Evans P. S., Gregor J., Kirkpatrick H. A., Pósfai G., Hackett J., Klink S., Boutin A., Shao Y., Miller L., Grotbeck E. J., Davis N. W., Lim A., Dimalanta E. T., Potamousis K. D., Apodaca J., Anantharaman T. S., Lin J., Yen G., Schwartz D. C., Welch R. A., Blattner F. R. (2001) Nature 409, 529–533 [DOI] [PubMed] [Google Scholar]
- 23.Serrano-Vega M. J., Magnani F., Shibata Y., Tate C. G. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng M., Tsaur M. L., Jan Y. N., Jan L. Y. (1992) Neuron 9, 271–284 [DOI] [PubMed] [Google Scholar]
- 25.Seddon A. M., Curnow P., Booth P. J. (2004) Biochim. Biophys. Acta 1666, 105–117 [DOI] [PubMed] [Google Scholar]
- 26.Rudnick G. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 5949–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juge N., Yoshida Y., Yatsushiro S., Omote H., Moriyama Y. (2006) J. Biol. Chem. 281, 39499–39506 [DOI] [PubMed] [Google Scholar]
- 28.Miyaji T., Echigo N., Hiasa M., Senoh S., Omote H., Moriyama Y. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 11720–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bass R. B., Strop P., Barclay M., Rees D. C. (2002) Science 298, 1582–1587 [DOI] [PubMed] [Google Scholar]
- 30.Shouffani A., Kanner B. I. (1990) J. Biol. Chem. 265, 6002–6008 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.