Summary
Chilo iridescent virus (CIV) is a large (~1850 Å diameter) insect virus with an icosahedral, T=147 capsid, a dsDNA genome, and an internal lipid membrane. The structure of CIV was determined to 13 Å resolution by means of cryo-electron microscopy (cryoEM) and three-dimensional image reconstruction. A homology model of P50, the CIV major capsid protein (MCP), was built based on its amino acid sequence and the structure of the homologous Paramecium bursaria chlorella virus 1 (PBCV-1) Vp54 MCP. This model was fitted into the cryoEM density for each of the 25 trimeric CIV capsomers per icosahedral asymmetric unit. A difference map, in which the fitted CIV MCP capsomers were subtracted from the CIV cryoEM reconstruction, showed that there are at least three different types of minor capsid proteins associated with the capsomers outside the lipid membrane. “Finger” proteins are situated at many, but not all, of the spaces between three adjacent capsomers within each trisymmetron, and “zip” proteins are situated between sets of three adjacent capsomers at the boundary between neighboring trisymmetrons and pentasymmetrons. Based on the results of segmentation and density correlations, there are at least eight finger proteins, and three dimeric and two monomeric zip proteins in one asymmetric unit of the CIV capsid. These minor proteins appear to stabilize the virus by acting as intercapsomer cross-links. One transmembrane “anchor” protein per icosahedral asymmetric unit, which extends from beneath one of the capsomers in the pentasymmetron to the internal leaflet of the lipid membrane, may provide additional stabilization for the capsid. These results are consistent with the observations for other large, icosahedral dsDNA viruses that also utilize minor capsid proteins for stabilization and determining their assembly.
Keywords: large DNA virus, cryo-electron microscopy, 3D image reconstruction, enveloped virus, minor capsid proteins
Introduction
The nucleocytoplasmic large DNA viruses (NCLDV) include the Asfarviridae, Baculoviridae, Iridoviridae, Phycodnaviridae, Poxviridae, and the new family of giant Mimiviridae. All of these viruses have double-stranded DNA genomes ranging in size from ~130 to 1200 kbp, and all but the families Baculoviridae and Poxviridae have virions with icosahedral capsids. Many genes of these viruses show significant sequence similarities among themselves and to their host homologs.1,2 This suggests that their viral genomes are constituted from a mosaic of genes derived from a variety of hosts during evolution.3 Such large genomes account for most of the functions of a living cell and, hence, raise questions about the boundaries that distinguish these viruses from living organisms.
Structural studies of icosahedral NCLDVs include the 7,500 Å diameter Mimivirus, which infects amoeba;4 the 2,200 Å diameter Phaeocystis pouchetii virus (PpV01), which infects and lyses haptophytes;5 the 1,900 Å diameter Paramecium bursaria chlorella virus type 1 (PBCV-1: family Phycodnaviridae), which infects unicellular, eukaryotic, chlorella-like green algae;6-8 and the 1,850 Å diameter Chilo iridescent virus (CIV: family Iridoviridae), which infects the rice stem borer insect.6 All these viruses have an outer protein capsid comprised of arrays of pseudo-hexagonal, but actually trimeric, capsomers organized into 20 equilateral trisymmetrons and 12 pentagonal pentasymmetrons.9 Inside the outer protein layer there is at least one, 40 Å-thick, lipid membrane bilayer that envelopes the genome and other viral proteins. The lipid membrane adopts an icosahedral morphology that roughly follows the contour defined by the outer layer of capsomers. The principal difference in the capsids of these large dsDNA viruses lies in the number and arrangement of capsomers that make up the trisymmetrons and pentasymmetrons, as is defined by their triangulation number.10 Capsomers within a trisymmetron all pack in essentially the same orientation, which differs by ~60° from the capsomers in neighboring trisymmetrons. This opposing arrangement creates cleavage planes at the trisymmetron boundaries. A consequence of the approximately 60° rotation between the trimeric capsomers on either side of trisymmetron boundaries is to introduce a series of successive radial, local dyads that relate the capsomers in neighboring trisymmetrons and result in alternate long and short inter particle distances.8
The structure of the capsomers in these large dsDNA viruses consists of three monomers related by 3-fold symmetry. Each monomer consists of two “jelly-roll” domains11 that occur in succession along the polypeptide chain, placed so as to generate pseudo-hexagonal symmetry. Similarly, pseudo-hexameric capsomers occur in smaller icosahedral dsDNA viruses such as adenovirus,12,13 the lipid containing phage PRD1,13,14 the marine phage PM2,15 Bam35,16 as well as small, non-enveloped RNA viruses7 such as Cowpea Mosaic virus17 and picornaviruses,11 but none of these have capsomers organized into trisymmetrons and pentasymmetrons. These similarities suggest that the capsids of these icosahedral dsDNA viruses have evolved from a primordial double jelly-roll structure that forms pseudo-hexagonal capsomers.7,18 The jelly-roll structure itself may be even more ancient as it also occurs in the capsid proteins of small icosahedral viruses such as the single-stranded DNA parvoviruses19 and bacteriophage phiX174.20
CIV has a wide host range that includes leafhoppers and some arthropods and has been touted as a potential pest control agent.21 The densely packed, crystalline arrays of virions in CIV-infected cells produce a characteristic blue to purple iridescence. The CIV virion (~1×109 Daltons) encapsidates a 212,482 bp, dsDNA genome within its fiber-covered, icosahedral shell.21,22 The CIV genome accounts for about 12% of the particle mass. Similar to other iridoviruses such as frog virus 3 (FV3)23 and fish lymphocystis disease virus24 and also the T-even bacteriophages (Myoviridae),25 the CIV genome is circularly permuted and has about 12% genome redundancy.26 CIV virions contain more than 30 structural proteins. P50 (51.4 kDa, 467 amino acids), the MCP, accounts for almost half of the total protein mass.27,28 It has ~21% sequence identity to Vp54, the MCP of PBCV-1.18 The location and organization of all other structural proteins are unknown. The CIV virion contains 9% lipid, sufficient to form a continuous internal lipid bilayer. The viral membrane is abundant in phosphatidylinositol and diglycerides and has a composition that distinguishes it from the host cell, indicating that CIV synthesizes its own bilayer.29
An earlier cryoEM and three-dimensional (3D) image reconstruction study of CIV at 26 Å resolution showed that the protein shell has a maximum diameter of 1850 Å and consists of 12 pentavalent pentamers and 1460 hexavalent trimers6 arranged with T=147 (h = 7, k = 7) icosahedral, quasi-equivalent symmetry.6 Each of the 12 pentasymmetrons consists of one pentameric complex at its center (vertex) surrounded by 30 P50 trimers. The asymmetric unit of the 5-fold symmetric pentasymmetron consists of one monomer from the pentamer and six P50 trimers, with the trimers adopting two distinct orientations differing by ~60° rotation about their 3-fold axes. Each of the 20, 3-fold symmetric trisymmetrons consists of 55 capsomers, all of which are aligned with a similar rotational orientation. The asymmetric unit of the trisymmetron consists of 18 1/3 capsomers, with the trimer at each icosahedral 3-fold axis contributing one monomer to each unit.
Structural and sequence similarities among the MCPs of icosahedral dsDNA viruses such as PpV01, PBCV-1, CIV, PRD1, adenovirus, and Bam35 suggest that the MCPs of these viruses originated from a common ancestor.7,18,30 However, differences of the structure and arrangement of minor capsid proteins of PRD1, adenovirus, and Bam35 indicate that the assembly pathways of these viruses evolved separately.30 For example, PRD1 and Bam35 contain extended “tape-measure” proteins that bridge the icosahedral 5-fold axes and roughly trace the 2-fold edges of the icosahedral faces and, hence, probably dictate virion size during assembly. Here we report a 3D cryo-reconstruction study of CIV in which features not apparent in an earlier study at 26 Å resolution,6 such as the locations and arrangement of several minor capsid proteins, could be discerned at ~13 Å resolution. Analysis of these new features demonstrates that CIV differs from PRD1, Bam35, and adenovirus in that the minor capsid proteins of CIV that connect the MCPs are globular instead of extended polypeptides.
Results and Discussion
CryoEM and 3D image reconstruction of CIV at 13 Å resolution
Samples of CIV (~3mg/ml) were purified and prepared for cryoEM as described earlier.6 Images of vitrified virions were recorded on film and processed (Materials and Methods) (Fig. 1a). The resolution of the 3D map was estimated to be ~13 Å, based on the standard 0.5 Fourier Shell Correlation threshold.31 The CIV structure seen at 13 Å resolution has the same morphology and gross features as were observed in the earlier, 26 Å reconstruction.6 However, numerous details can be identified in the 13 Å resolution but not in the 26 Å resolution map. The improved resolution was primarily the result of the increased number of particle images (1800 vs. 460) included in the new reconstruction, but also due to the use of higher voltage (300 vs. 200 keV), a wider range of defocus settings (0.8-3.0 vs. 2-2.5 μm), and use of enhanced processing and refinement methods.32
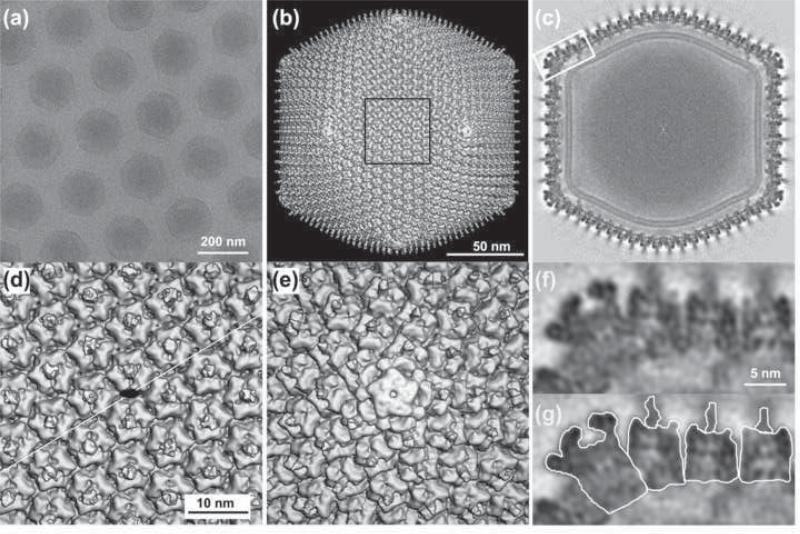
Fig. 1. 3D reconstruction of CIV at 13 Å resolution.
(a). Micrograph of vitrified CIV virions suspended over a hole in a carbon support film. The core contents (dsDNA genome and DNA binding proteins) do not uniformly fill the volume enclosed by the capsid. (b) Shaded-surface representation of CIV 3D density map viewed along an icosahedral 2-fold axis. (c) Central cross section, one pixel thick, viewed as in (b). A lipid bilayer, ~40 Å thick, follows the inner contour and icosahedral symmetry of the capsid shell. (d) Magnified view of the region outlined in (b). A black ellipse marks the location of an icosahedral 2-fold axis and the thin white line indicates the cleavage plane between two trisymmetrons. (e) Magnified view of the pentamer complex at the 5-fold vertex. This complex is easily distinguished from the trimeric capsomers in that it is larger, has a small axial hole and lacks a fiber. (f) Magnified view of boxed region in (c). (g) Same as (f) with the pentamer complex, three capsomers and their fibers individually outlined.
The most conspicuous features of the CIV structure are the 1460 hexameric capsomers and 12 pentameric vertex complexes on the viral surface (Fig. 1b-g) and their organization into tri- and pentasymmetrons (Fig. 2a). Each of the 1460 capsomers is ~76 Å thick, has a hexameric morphology, and a central fiber (~20 Å diameter) that extends outwards along the capsomer 3-fold rotational axis (Fig. 1f, g). All fibers, as measured in unprocessed micrographs, appear to project radially about 350 Å beyond the capsid surface. However, they only appear ~60 Å long in the cryoEM reconstruction, indicating that the fibers are flexible and their distal parts are positioned more randomly with respect to the capsid, which leads to smearing of the fiber density in the icosahedrally symmetrized reconstruction.
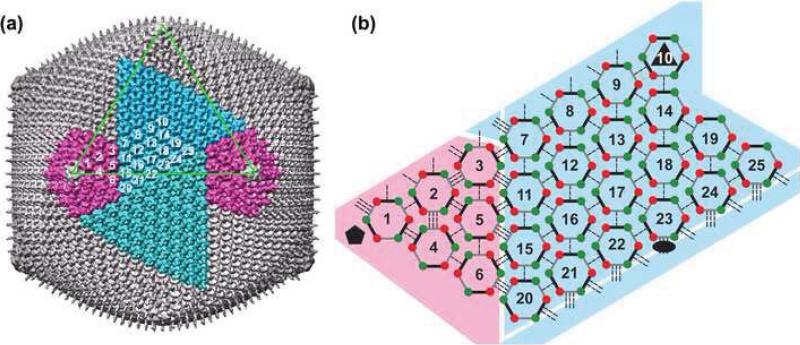
Fig. 2. Organization of trimeric capsomers in CIV.
(a) The CIV capsid has twenty trisymmetrons (one shown in light blue and another in dark blue) and twelve pentasymmetrons (two shown in magenta). One triangular face of the icosahedron is outlined in green. The asymmetric unit of the capsid includes 24 1/3 trimers (numbering scheme is arbitrary). Only one monomer of capsomer 10 (at an icosahedral 3-fold axis) is included in the icosahedral asymmetric unit. (b) Schematic diagram of inter-capsomer interactions within one asymmetric unit. Each capsomer is represented as a pseudo-hexamer (three double jelly-rolls, each depicted as a pair of red-green dots connected by a thick line). Capsomers form three classes (I, II, and III; Table 1) of interactions with neighboring capsomers as indicated by single, double, and triple dashed lines. The black ellipse, triangle, and pentagon symbols mark the icosahedral 2-, 3-, and 5-fold axes, respectively. Diagram does not depict curvature present in the arrangement of capsomers in the CIV structure.
The P50 capsomers
In CIV, as in PBCV-16 and PpV01,5 the capsomers are grouped into 20 trisymmetrons and 12 pentasymmetrons (Fig. 2a). Within a trisymmetron capsomers are packed with a mean separation of ~75 Å and all adopt a similar orientation. However, capsomers on either side of the boundary between two trisymmetrons are packed with inter-capsomer separations that alternate between slightly longer (~78 Å) and shorter (~73 Å)8 distances. A similar alternating pattern was also observed in the arrangement of capsomers in PBCV-1,7 where the corresponding inter-capsomer separations are ~78 Å and ~75 Å,8 respectively. This pattern is what creates the lines of cleavage, first observed in Sericesthis iridescent virus,9 between adjacent trisymmetrons and between trisymmetrons and pentasymmetrons (Table 1 and Fig. 2b). Although all trimers within a trisymmetron are similarly oriented, two of the six in each asymmetric unit of a pentasymmetron (capsomers 1 and 2 in Fig. 2) are rotated by ~60° relative to the others.
Table 1.
Interactions between capsomers.
| Contact type8 |
Number† |
Mean distance (Å) |
r.m.s. deviations of mean distance (Å) |
Mean rotation (°) |
r.m.s. deviations of mean rotation (°) |
|---|---|---|---|---|---|
| Similar orientation: Type I‡ | |||||
| In trisymmetron | 45 |
75 |
0.7 |
4.1 |
2.2 |
| In pentasymmetron |
8 |
75 |
0.5 |
7.8 |
4.7 |
| Opposite orientation: Type II‡ | |||||
| Between trisymmetrons | 6 |
78 |
0.2 |
57.5 |
1.9 |
| In pentasymmetron | 3 |
76 |
1.6 |
55.9 |
2.6 |
| Between penta- and trisymmetron |
3 |
77 |
0.8 |
58.5 |
0.4 |
| Opposite orientation: Type III‡ | |||||
| Between trisymmetrons | 6 |
73 |
0.1 |
57.9 |
1.5 |
| In pentasymmetron | 3 |
74 |
0.2 |
57.6 |
1.1 |
| Between penta-and trisymmetron | 3 | 73 | 0.4 | 58.9 | 0.4 |
Number of interactions in an asymmetric unit.
Type I contact: Similar rotational orientation between capsomers
Type II contact: Opposite orientation and long interaction (~76-78 Å) between capsomers
Type III contact: Opposite orientation and short interaction (~73 Å) between capsomers
A capsomer must sit at the center of each face of the icosahedron, coincident with the icosahedral 3-fold axes whenever the h and k integers that define the triangulation symmetry (T) are both odd, as is the case for CIV with T=147 (h=7, k=7). In contrast, in viruses like the T=25 (h=5, k=0) PRD1,14 the T=169d (h=7, k=8) PBCV-1,6 and the T=219 (h=7, k=10) PpV01,33 the icosahedral 3-fold axes pass between capsomers.
The crystal structures of the trimers of different MCPs, including Vp54 of PBCV-1,7 P3 of PRD1,14 and P2 of PM2,15 have a consistent structural motif that consists of six jelly-roll domains with a hollow, hydrophobic13 cavity in the middle of the trimer. However, in CIV, this cavity is plugged by the long, flexible fiber that, most likely, is the first component of the virion that comes into contact with and recognizes the surface of a potential host. The presence of 1460 of these fibers creates a halo around each virion. It seems unlikely that this halo is required merely to assure effective interaction with the host, and may include other roles such as protection against harsh environments. In PBCV-1 and PpV01 there is one special capsomer per icosahedral asymmetric unit that has a central fiber-like protrusion,5 whereas in CIV every capsomer has a fiber.
There are 18 1/3 capsomers in the asymmetric unit of the trisymmetron and six capsomers per asymmetric unit of the pentasymmetron in CIV. This contrasts with PBCV-1, which has 22 and six capsomers, respectively. The capsomer structure in CIV shows subtle differences in pentasymmetrons and trisymmetrons as evidenced by fitting the homology model of a CIV capsomer (Materials and Methods) into the 25 distinct capsomer densities in the CIV cryo-reconstruction. The fitting scores for capsomers in the trisymmetron (Averaged Sumf38 = 51.9) were about 6% better than those in the pentasymmetron (Averaged Sumf38 = 48.9). These differences might reflect conformational variations among the capsomers and might also signify that the pentasymmetrons are formed not from P50 but, instead, a different CIV coded protein that is homologous to P50.
Structure of the pentameric complex
Adenovirus capsids have fibers associated with their pentameric vertices12 but CIV has a complex, mushroom-like structure at its vertices (Figs. 1e-g and 3). The head of the complex has the appearance of a five-blade propeller, with a maximum outer diameter of ~110 Å. Each blade points downward and, looking at the vertex from outside of the virus, the tip of each propeller blade is twisted in an anti-clockwise direction to make contact with two peripentonal MCP monomers. A small, ~10 Å diameter, hole at the center and top of the pentameric complex leads into a 20 Å deep, flask-shaped cavity that is 25 Å wide at its base. The stalk of the complex, ~90 Å diameter, extends radially downward towards the viral membrane and makes additional contacts with the peripentonal capsomers. The size of the pentameric complex far exceeds that of a P50 capsomer. The diameter and maximum height of the CIV pentameric complex are 110 and 127 Å versus 73 and 76 Å for a capsomer, respectively.
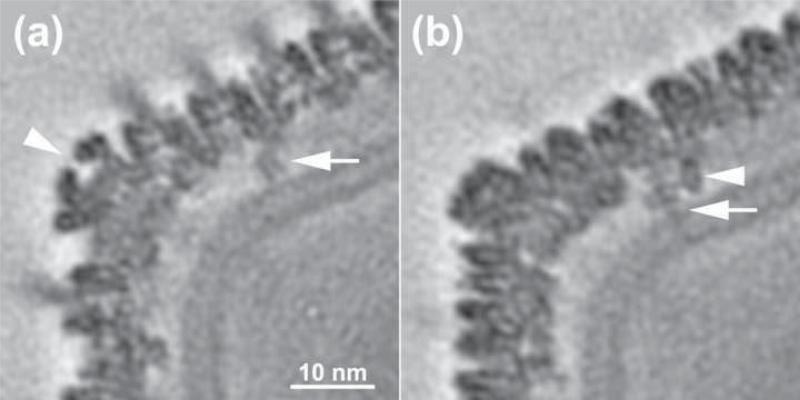
Fig. 3. Transmembrane anchor protein beneath CIV pentasymmetrons.
(a) An equatorial-section, one pixel thick, through the CIV density map showing the pentamer vertex complex (arrow head) and a portion of the anchor protein (arrow). The bilayer membrane sharply curves in this region. (b) Similar to (a), but showing a section (parallel to but displaced ~21 Å from the equatorial section) that reveals additional details of the anchor protein. Two stick-like densities are apparent. The longer of the two (arrow) crosses both leaflets of the bilayer, whereas the other stops at the outer leaflet.
Caspar and Klug10 had correctly predicted that the capsids of icosahedral viruses would generally be assembled of hexamers and of pentamers at 5-fold vertices with the monomers in the hexamers and pentamers having quasi-equivalent environments. However, larger viruses with their greater coding capacity have allowed divergent evolution from a primordial single jelly-roll structure to quasi-hexameric capsomers and specialized pentameric complexes to give greater freedom for these proteins to fit into their respective environments. The adenovirus33 pseudo-6-fold capsomers are assembled from three double jelly-roll monomers as also occurs in CIV, PBCV-1,7 Sulfolobus turreted icosahedral virus (STIV) 34 and PRD1.14 However, the special vertex proteins of adenovirus and PRD114 have a simpler, single jelly-roll structure.35 Another example is the special vertex protein, gene product 24 (gp24) of bacteriophage T4, which has an “HK97” fold33 and is homologous to gp23, the T4 MCP that forms hexameric capsomers. Considering the structure of the adenovirus vertex protein35 and assuming the vertex proteins of larger viruses like CIV have evolved similarly, the pentameric vertex complex of CIV likely consists of monomers with a single jelly-roll motif. However, the estimated molecular mass of about 40 kDa for the vertex subunit, based on a comparison of volumes with the P50 capsomers (Materials and Methods), suggests that there are large insertions (~12 kDa total) in the single jelly-roll.
Lipid membrane and virion core
The lipid membrane in the PRD136 virion is derived from the host's plasma membrane on budding, whereas the membrane in CIV originates from the host cell's cytoplasm.29 This difference might reflect their different assembly mechanisms. In CIV there is a layer of moderate density, with distinct features, attributed to several minor proteins (see below), that lies immediately beneath the 1460 trimeric capsomers and 12 pentameric vertex complexes. This density merges with a region of disordered, low density structure on the outer leaflet of the viral membrane.
The prominent, non-spherical CIV bilayer in the virion interior has a “railroad track” profile characteristic of the two lipid containing leaflets of a typical cellular membrane (Fig. 1c), reminiscent of the lipid membrane such as in flaviviruses.37 The distinctive structure of the membrane means that its morphology must be dictated at least in part by the symmetry of the capsid, perhaps owing to attractive electrostatic forces between the inner surface of the capsid and the outer surface of the membrane. These forces maintain a fairly constant distance between membrane and capsid. Most of the viral membrane is ~40 Å thick, which is typical of membranes in eukaryotic cells, although it is only ~30 Å thick in the regions just below the 5-fold vertices.
The inner layer of the membrane is ~30 Å from the nucleocapsid core. The outer membrane layer is ~70 Å from the capsomer densities near the 2- and 3-fold axes, and ~40 Å near the pentameric complex. The large volume of density in the virion core, representing the viral genome and other minor proteins, is relatively featureless. In many unprocessed CIV images, the core is partially separated from the lipid membrane, especially in the vicinity of the 5-fold vertices (Fig. 1a).
Removal of the MCP density from the cryoEM map
In the absence of a crystal structure for the CIV MCP P50, a homology model was built (Materials and Methods) of trimeric P50 based on its sequence similarity to Vp54, the MCP of PBCV-1.18 The programs Situs38 and EMfit39 were used to fit this preliminary homology model into each of the densities corresponding to the 25 CIV capsomers within the icosahedral asymmetric unit. The Situs procedure depends on density gradients and, thus, primarily fits surface features of the model to the density. The EMfit procedure aims at maximizing the fit of all model atoms to the density map. The rms difference in the position and orientation of each capsomer as determined by these distinct procedures was less than 2.0 Å and 2.0°, respectively. The accuracy of the fitting was further improved by using a polynomial function that computed the position of all 55 capsomers in a trisymmetron, assuming a smooth variation of the translational and rotational parameters across the surface of the virus. Each capsomer was assigned two indices to identify its position within the hexagonal array of capsomers (Materials and Methods). The rms difference between the observed and calculated (by means of the polynomial function) capsomer positions and orientations was about 1.5 Å and 1.2°, respectively, less than the difference between the Situs and EMfit results. Thus, the polynomial function seems to provide a reasonable means to minimize the error of the independently fitted capsomer model to each of the capsomer densities in the cryoEM map.
A difference “-(P50)” map of the complete CIV particle was computed by setting to zero the density within a 4 Å radius of every atom in the final homology model after it was translated and rotated to each of the 1460 capsomers in the original CIV cryoEM map. In addition, all density enclosed by the inner leaflet of the membrane was set to zero. 40 The resultant “-(P50, core)” difference map depicted all density inside and outside the membrane not accounted for by the homology model fitted into each of the capsomer densities. Aside from membrane-associated density remaining in the “-(P50,core)” map, a series of prominent, repetitive structures were observed along with some other, lower density features, presumably corresponding to the presence of icosahedrally ordered minor capsid proteins. The largest densities with repeating features were situated below the trisymmetrons (Fig. 4), whereas the density below the pentasymmetrons did not show any obvious repeating features.
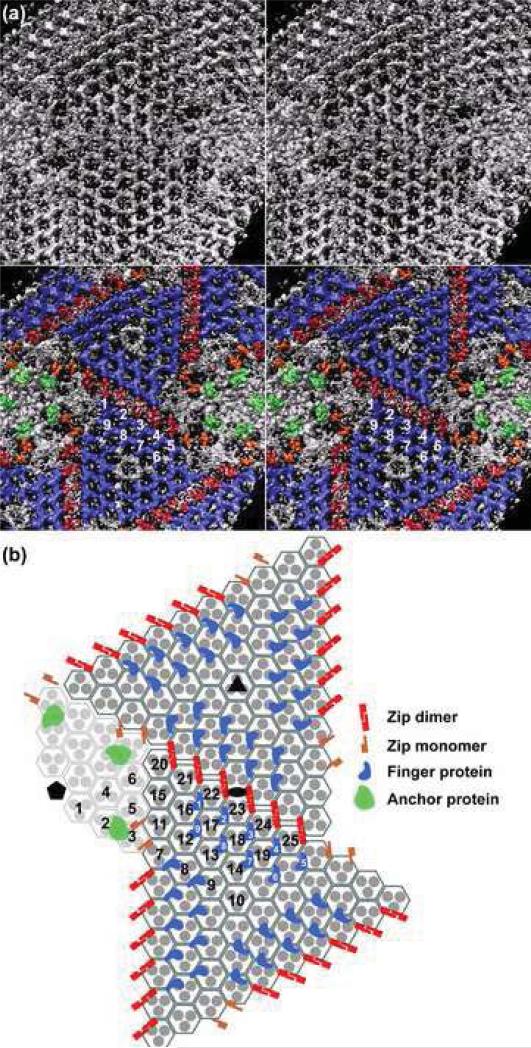
Fig. 4. Minor proteins in CIV.
(a). Upper panel: Inside view of “-(P50,core)” map with membrane density removed for clarity, in stereo and along an icosahedral 2-fold axis, shows a complex distribution of repeating density features. Some are arranged in a hexagonal array under the trisymmetrons whereas others are associated with the pentasymmetrons. Lower panel: Same as upper panel but colored to highlight similarly shaped features that were identified as putative, unique minor proteins. Twenty-seven finger proteins (colored blue, numbered 1 to 9 in the icosahedral asymmetric unit) form a tight network underneath each trisymmetron and their finger-like protrusions point towards the lipid membrane. Six zip dimers (red) and four zip monomers (orange) lie below the interface between adjacent trisymmetrons. Transmembrane anchor proteins (green) are the most readily identified features located underneath the pentasymmetrons. (b) Schematic, planar diagram of region depicted in (a), with the pentamer complex omitted for clarity. The ellipse, triangle, and pentagon symbols highlight the positions of 2-, 3-, and 5-fold icosahedral axes, respectively. All P50 trimers are depicted as three filled disks enclosed by a hexagon. The icosahedral asymmetric unit contains 24 1/3 of these capsomers (numbered in black). Each trisymmetron contains 55 capsomers, all oriented similarly and rotated by 60° relative to those in the adjacent trisymmetron. Twenty-seven finger proteins (blue) bind to each trisymmetron (nine within one asymmetric unit are numbered in white). Eighteen zip dimers (red) form the interface between one trisymmetron and its three adjacent trisymmetrons. Six zip monomers (orange) form a portion of the interface between a trisymmetron and its neighboring pentasymmetrons. The transmembrane anchor proteins (green) are associated with capsomers 2 and 3 beneath the pentasymmetron.
Minor capsid proteins: the finger protein
The largest uninterpreted densities in the difference map have a finger-like morphology that is repeated by translations, found immediately below the conjunction of sets of three neighboring capsomers within the trisymmetrons (Fig. 4) as would occur in a hexagonally close packed arrangement. Each finger protein has a globular base that interacts with the low radii portions of three neighboring capsomers, and has an ~55 Å-long, finger-like feature that points radially inwards towards the lipid membrane. However, there is no verification that this is a single molecular species. Seven such finger proteins, clearly visible in the “-(P50,core)” map (finger proteins 1-4 and 7-9 in Fig. 4), are arranged in two rows in each asymmetric unit of a trisymmetron. In addition, two similar, but less well-defined finger-like proteins appear at one end of the two rows (Fig. 4, finger proteins 5 and 6). The density for each of the seven unambiguous finger proteins was individually segmented based on density gradients and connectivity40 (Materials and Methods).
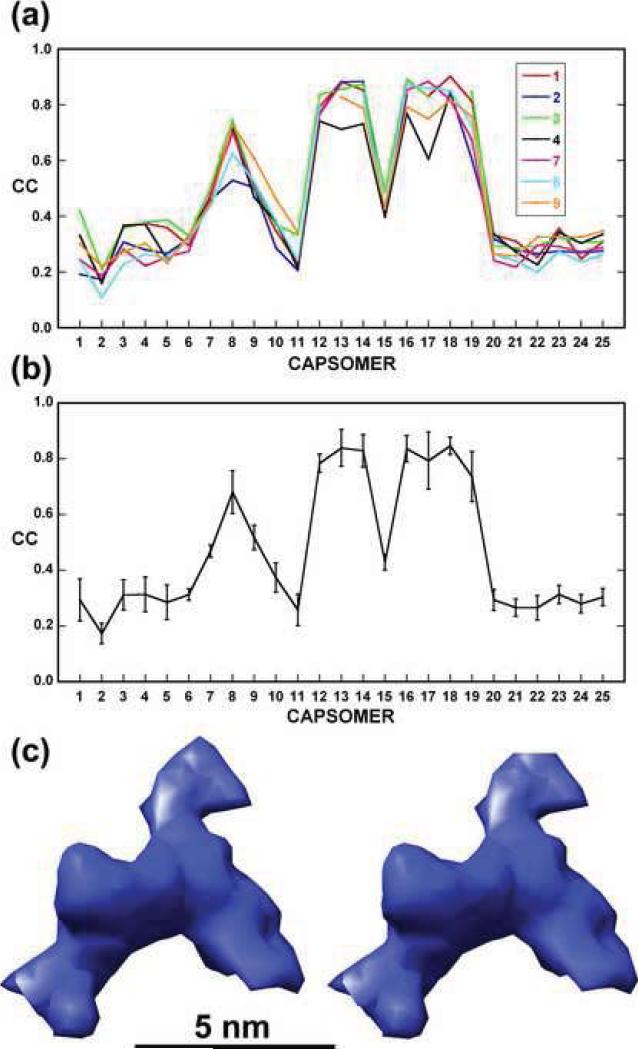
Assignment of density to a particular structure such as a finger protein was substantiated by computing correlation coefficients between pairs of corresponding, segmented densities. An average correlation of about 0.9 was found for comparisons among the seven, best-defined finger densities (Fig. 5a and b). The two less well-defined finger proteins had average correlations between 0.7 and 0.5 with the other seven (Fig. 5a and b). Furthermore, the correlation between any of these seven finger proteins and densities at other corresponding sites was about 0.4 or lower (Fig. 5a and b), suggesting no finger proteins occur at these other sites. The seven well-defined finger proteins were averaged based on the rotational and translational relationships of their associated capsomers (Fig. 5c).
Fig. 5. Correlation coefficient analysis and averaging of finger protein densities.
(a) Plots of correlation coefficients computed between each of the seven, most prominent finger densities (1-4 and 7-9 in Fig. 4) and the corresponding densities at each of the other 24 capsomer sites. Densities associated with capsomers 12-14 and 16-19 show the highest correlations (average = 0.81). This analysis also indicates a high correlation for capsomer 8 (CC = 0.68) and capsomer 9 has the next highest correlation (CC = 0.52). (b) Same as (a) but only plotting the average correlation and its standard deviation (vertical bar) at each capsomer site. (c) Stereo view of the average density for the seven prominent finger proteins.
In PRD1,41 PM2,42, and STIV,34 as in CIV, similar minor capsid proteins bridge the space between the capsomers and the outer leaflet of the lipid membrane. By comparison with PRD1,43 the CIV finger proteins might serve as “cement” or “glue” proteins required for capsid assembly.
Minor capsid proteins: the zip protein
A mask generated from the average finger density was used to identify and zero out density attributed to all 540 finger proteins in the “-(P50,core)” map. The resultant “-(P50,core,finger)” map showed density located primarily just below the boundaries between neighboring trisymmetrons and between trisymmetrons and pentasymmetrons. However, the density under the boundaries between neighboring trisymmetrons exhibited a translationally repeating pattern. Like the finger proteins, the higher density features of this pattern were centered under the holes created by the convergence of three capsomers to produce a hexagonally close packed assembly. Because the orientation of capsomers is different in neighboring trisymmetrons, the additional densities below the trisymmetron boundaries have a different environment than the finger proteins. Thus, these densities (“zip proteins”) probably represent a different minor capsid protein that helps bind neighboring trisymmetrons together.
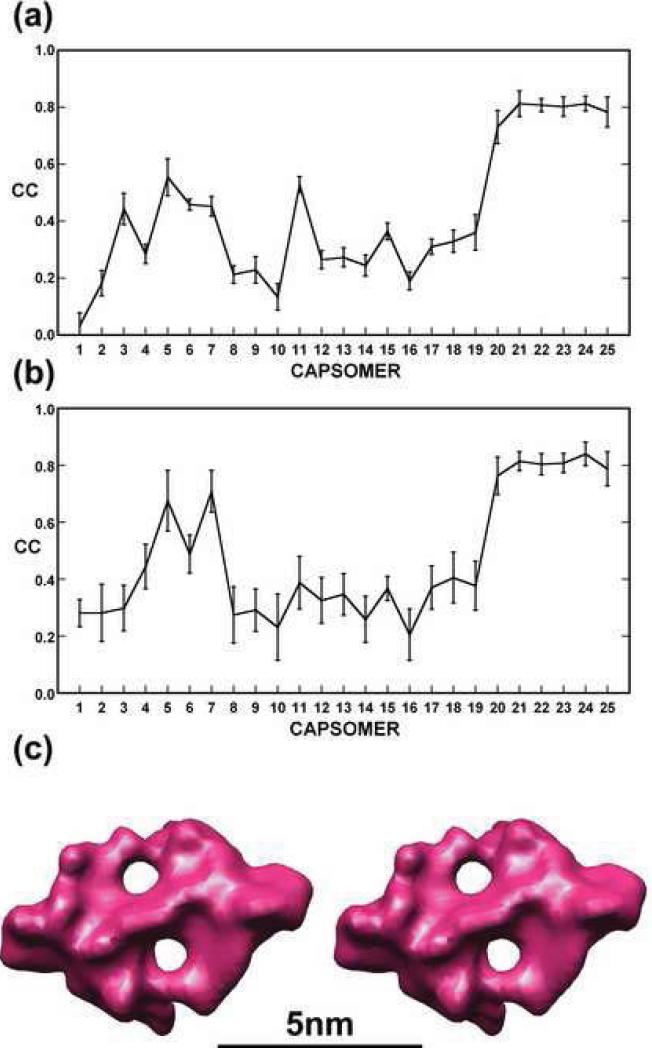
Six repeating units of zip protein densities were identified at successive quasi-2-fold axes that relate capsomers in adjacent trisymmetrons8 (associated with capsomers 20, 21, 22, 23, 24 and 25 in Fig. 4b). The densities of these six zip proteins had approximate 2-fold symmetry coincident with the quasi-2-fold axes between the overlaying capsomers.8 These zip proteins connect capsomers across trisymmetron boundaries separated by distances that are systematically longer than the distances between capsomers within trisymmetrons (Table 1). The correlation coefficient between each zip protein and its superimposed 2-fold related structure was 0.8 to 0.9 (Fig. 6a). However, the correlations with densities in the corresponding gaps between other capsomers were less than 0.6 (Fig. 6a). Therefore, the six zip dimers were averaged based on the rotational and translational relationships of the capsomers with which they interact (Fig. 6c).
Fig. 6. Correlation coefficient analysis and averaging of zip protein densities.
(a) Plot of average correlation coefficient computed between each of the six, prominent densities representing zip dimers and the corresponding densities at each of the other 24 capsomer sites. Densities associated with capsomers 20-25 show the highest correlations (average = 0.79). (b) Same as (a) for density of a zip monomer. Two zip monomers (associated with capsomers 5 and 7) have a correlation greater than 0.67. (c) Stereo view of the average density for the six prominent zip dimers. Note that no 2-fold symmetry was enforced during this averaging procedure.
The densities associated with capsomers 5 and 7 (Fig. 6b) correlated much better (0.8 and 0.9, respectively) with the density of a zip monomer than with the density of a zip dimer (Fig. 6a). This result might indicate that densities associated with capsomers 5 and 7 are monomeric, but not dimeric, zip proteins. Therefore, there are a total of eight zip protein monomers along one edge of a trisymmetron, six of which form dimers with the zip proteins in the neighboring trisymmetron and the other two are monomers at the interface between trisymmetron and pentasymmetron (Fig. 4).
Minor capsid proteins: the anchor protein
Additional density is situated below capsomer 2 (Fig. 4) and traverses the lipid membrane. This structure (“anchor protein”) appears to tether the lipid membrane to the pentameric vertex complex. It is the only detectable, icosahedrally-ordered transmembrane protein in CIV. Similar transmembrane tethering proteins near the 5-fold vertices have been reported to occur in PRD1,14 Bam35,16 and STIV.34 The CIV anchor protein may help stabilize membrane curvature in the vicinity of the pentasymmetrons.
The putative identification of finger, zip, and anchor proteins leaves significant unassigned regions of density in the CIV cryo-reconstruction outside the viral core (Fig. 4a). Indeed, extra densities occur under the pentasymmetrons, below the boundaries between trisymmetrons and pentasymmetrons, below the 20 capsomers at the icosahedral 3-fold axes, and between groups of finger proteins. However, density in these regions is either too weak, non-periodic, or not extensive enough to yield definitive interpretations in the 13 Å resolution cryoEM map.
Molecular mass estimates
The molecular masses of the minor capsid proteins were estimated using the known molecular mass of the MCP (51.4 kDa) as a standard. Thus, it was necessary to determine the averaged volume of the MCPs and compare it with the volume of each of the minor capsid proteins. As the spatial limits of the envelope were not known precisely, estimates of molecular masses were computed using a series of density contour levels (Supplementary Fig. 1). The molecular mass of each minor capsid protein (finger protein, zip protein, pentameric complex and anchor protein,) was given by
Where M represents mass and V represents volume.
The ratios of the Volumes should be independent of the contour level used to determine the volumes. However when choosing the contour level greater than the maximum density of that minor capsid protein then the mass of the minor capsid protein would calculate as zero. Thus the ratio of the volumes becomes constant only when the contour level truly encompasses the volume of the protein. Hence, at progressively lower contour levels the estimated molecular mass of the minor capsid protein increased until further lowering of the contour level did not increase the apparent volume of the minor capsid protein. The common contour level at which there was no further significant increase in the estimated mass of the minor capsid protein was used to define the envelope of each protein. Conversely, if the maximum density of the minor capsid protein is greater than that of the MCP (as is the case in the pentameric vertex), the estimated molecular mass will decrease until the density level has reached the limits of the envelope. In all cases, the contour level that yields reliable mass estimates was found to be ~30 arbitrary units of density, which corresponded to 0.55 Da/Å3 for the MCP. At this contour level, the estimated molecular mass for the finger, zip, pentameric complex monomer, and anchor proteins were found to be 19.7, 11.9, 39.3 and 32.4 kDa. The standard deviations of the molecular mass estimates for the finger and zip proteins were 1.5 and 1.4 kDa, which provides an approximate estimate of the error in the estimate of the mass. However, these estimates do not take into account error caused by variation in occupancy and order of the proteins. Resolution of the bands in SDS-PAGE gels of the whole virion (data not shown) was insufficient to identify these proteins with any degree of certainty when considering their position on the gel appropriate for their estimated mass and the intensity of the band that should be roughly proportional to the copy number.
CIV assembly
What roles, if any, do the minor proteins serve in directing the assembly of CIV? The finger and zip proteins both appear to stabilize the capsid by cross-linking sets of three adjacent capsomers. Finger proteins stabilize trisymmetrons, whereas zip proteins link trisymmetrons to neighboring trisymmetrons and pentasymmetrons. In phage PRD1, the elongated minor capsid protein, P30, is located between the outer icosahedral capsid and the lipid membrane and extends along the edge of each icosahedral face.14 Thus, P30 has been likened to a “tape measure protein” that regulates the size of each face and, hence, the size of the virus.14 P30 also likely contributes to the stability of PRD1 by linking capsomers together. In contrast, no tape measure proteins have been identified in adenovirus44 or in STIV.34 We show here that CIV has several different minor capsid proteins situated between the assembled outer capsomers and the viral membrane, but there is no clear evidence for an extended, tape measure protein. What then determines the final size of capsids in CIV, adenovirus or STIV? Since the number of minor capsid proteins increases with virus size, it may be that minor proteins affect assembly by regulating the sizes as well as interactions of sub-assembly units such as trisymmetrons and pentasymmetrons.
Materials and Methods
CryoEM and 3D image reconstruction of CIV
Samples of CIV were purified and prepared for cryoEM as described earlier.6 The final concentration of the virus sample vitrified was ~3mg/ml. Images were recorded on Kodak SO-163 film in an FEI/Philips CM300 FEG electron microscope at 33,000x nominal magnification with defocus levels ranging from 0.8 to 3.0 μm and with an electron dose of ~22 e-/Å2. Of the 210 micrographs recorded, 180 exhibiting minimal astigmatism and effects of specimen drift and charging were digitized at 7 μm intervals on a Zeiss Phodis microdensitometer and these data were bin-averaged to yield an effective pixel size of 4.24 Å. The program RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt) was used to extract 6925 individual CIV virion images from these micrographs. Model-based procedures45 were used to determine and refine particle origin and orientation parameters for each particle. The previously published CIV reconstruction6 served as the initial model. Challenges inherent in the analysis of the 4712 pixel dimension CIV images stimulated the development of the AUTO3DEM automated image reconstruction software.32 The 3D reconstruction of CIV reported here was computed from 1800 particle images to a resolution limit of 10 Å, with Fourier coefficients beyond 11 Å damped by a Gaussian fall-off. The effective resolution, defined as the point where the Fourier shell correlation31 fell below 0.5, was found to be 13 Å.
The P50 trimer homology model
CIV P50 and PBCV-1 Vp54 have 21% amino acid identity and 33% similarity. Thus, a homology model of P50 was constructed by building the CIV P50 amino acid sequence into the PBCV-1 Vp54 structure. The resultant model was refined using the program O46 to obtain idealized geometry. Insertions in the P50 sequence were not included in the initial model. The biggest omitted loop consisted of about 20 amino acids. This homology model of the CIV P50 monomer was then superimposed onto each of the three monomers of the crystallographically determined Vp54 trimer7 to create the CIV capsomer model. Manual adjustments of the model were made to relieve steric clashes between adjacent P50 monomers.
Refinement of the positional and orientational parameters for the capsomers
Translational and rotational transformations that relate the coordinates of the trimeric homology model to each capsomer change smoothly from one capsomer to the next on the viral surface, but are discontinuous at the trisymmetron and pentasymmetron boundaries. Therefore, it should be possible to improve the accuracy of the position and orientation with which the homology model was placed into each capsomer density, at least within one trisymmetron. This was accomplished by determining the coefficients of a third degree polynomial that gave the best agreement with observed values for the orientation and central position of each trimer. The polynomial function used to examine the positions of all capsomers in one trisymmetron was:
where the position of each capsomer is given by x1, x2, x3 relative to a coordinate system used to represent the cryoEM density. Similarly, the orientation relative to a standard orientation of the homology model is given by the Eulerian angles x4, x5, x6. The h and k indices identify each capsomer on the surface of a trisymmetron. The origin of this (h, k) hexagonal system is taken to be in the middle of a trisymmetron, co-incident with an icosahedral 3-fold axis. The h and k values that determine the triangulation number10 of the icosahedron are related to the h and k indices by an origin shift from a 5-fold to a 3-fold icosahedral axis.
Both Situs38 and EMfit39 were used to fit the capsomer model into each of the 18 capsomer densities within one asymmetric unit of a trisymmetron as well as the capsomer in the center of the trisymmetron on the icosahedral 3-fold axis. The polynomial coefficients, Dij, were determined using a least squares procedure that minimized the difference between the observed and calculated positional and rotational parameters for all 55 capsomers within one trisymmetron (Table 2).
Table 2.
Differences in position and orientation determined by the programs Situs (Situs), EMfit (EMfitobs) and from the refinement of the EMfitobs values using the polynomial functions (EMfitcalc).
| Capsomer number | Difference in position (Å)† | Difference in orientation (°)‡ | ||
|---|---|---|---|---|
| Situs vs. EMfitobs | EMfitobs vs. EMfitcal | Situs vs. EMfitobs | EMfitcal vs. EMfitobs | |
| 7 | 1.2 | 0.4 | 2.5 | 1.2 |
| 8 | 2.1 | 0.5 | 0.8 | 1.8 |
| 9 | 1.4 | 1.2 | 2.0 | 2.3 |
| 10 | 2.0 | 1.5 | 1.2 | 0.2 |
| 11 | 1.0 | 1.0 | 2.0 | 1.3 |
| 12 | 1.4 | 0.7 | 2.5 | 1.2 |
| 13 | 2.1 | 0.6 | 0.9 | 1.4 |
| 14 | 1.6 | 1.0 | 2.1 | 1.0 |
| 15 | 1.7 | 1.4 | 1.9 | 1.8 |
| 16 | 1.5 | 1.5 | 0.7 | 1.0 |
| 17 | 1.8 | 0.8 | 1.1 | 1.5 |
| 18 | 1.5 | 1.1 | 2.9 | 1.5 |
| 19 | 1.5 | 1.1 | 2.4 | 0.4 |
| 20 | 1.4 | 0.9 | 1.5 | 0.2 |
| 21 | 1.3 | 0.7 | 2.5 | 0.3 |
| 22 | 1.7 | 1.4 | 1.4 | 1.5 |
| 23 | 1.3 | 0.5 | 1.5 | 1.2 |
| 24 | 1.6 | 1.5 | 2.4 | 0.3 |
| 25 | 1.0 | 0.7 | 2.1 | 1.3 |
| Average | 1.7 | 0.9 | 1.8 | 1.1 |
Notes: Capsomer numbering same as in Fig. 2.
sqrt (x12+x22+x32)
kappa=2*acos(cos(x5/2)cos((x4+x6)/2))
Averaged density of the P50 capsomer
If [E(h, k)] represents the rotational and translational matrices that relate each capsomer, identified by the indices (h, k), to the homology model in the reference coordinate system (x), then u = [E(h, k)] x. If ρh,k(u) is the density at position u of capsomer (h, k), then ρh,k(u)≈ref(x). The [E] matrices will have been determined by the Situs or EMfit fitting procedures. The average density was calculated from Σ(ρh,k(u)) taken over all capsomers in the asymmetric unit of the trisymmetron. These calculations were applied to all positions within a mask that defines the boundaries of the capsomer. The envelope used to define this capsomer mask was generated by setting a 4 Å radius around each atom of the homology model. The exact boundary of this mask was not critical to the overall process.
The averaged CIV capsomer density was used to modify and improve the P50 homology model by manually adding the previously omitted loops into the vacant density. The modified capsomer model was then refitted into the capsomer densities in the original CIV cryoEM density map in order to refine the [E] matrices.
Density correlation to estimate similarities among different copies of finger and zip proteins
The envelope of each of the seven visually recognizable finger proteins was determined (“segmented”) with the AutoSeg routine40 in program VolRover47 and used to establish a separate density mask for each protein with the program MAMA48. Given the close positional match between the hexagonal arrays of capsomers and the hexagonal arrays of finger proteins, it was reasonable to apply the same translational and rotational operators to the finger proteins as had been used to average the P50 capsomers. The program IMP48 was used to compute the correlation between each of the seven obvious finger proteins with the corresponding regions associated with every capsomer in the icosahedral asymmetric unit, and taking into account three possible orientations for each capsomer owing to its local 3-fold rotational symmetry. Hence, a total of 70 correlations were computed for each finger protein but only the highest correlation value associated with each capsomer was recorded in the plot (Fig. 5a and b). The finger protein transformation matrices, [E], were used to compute the average finger protein density distribution (Fig. 5c). The averaging procedure enhanced the density of the finger protein, whereas the surrounding density became more diffuse further away from the center of the finger protein where the transformation matrices [E] no longer described accurately the relationship between the proteins. Similar operations were also performed on the six zip dimers and six zip monomers (Fig. 6).
Illustrations
RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt) was used to produce Figures 1 and 3; Chimera49 to produce Figures 2a, 4a, 5c, and 6c; Adobe Illustrator to produce Figures 2b and 4b; and KaleidaGraph to produce Figures 5a, b and 6a, b.
Supplementary Material
Acknowledgments
We thank W. Zhang, X. Zhang and R. Sinkovits for helpful discussions. We are grateful for support provided by the Keck foundation and Purdue University matching funds for the purchase of a CM300 FEI transmission electron microscope. The work was supported by NIH grants to MGR (AI 11219) and TSB (R37 GM-033050).
Footnotes
Depositions
The CIV cryo-reconstruction map has been deposited with the EBI (accession number XXXX).
References
- 1.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Etten JL, Graves MV, MÜller DG, Boland W, Delaroque N. Phycodnaviridae - large DNA algal viruses. Arch. Virology. 2002;147:1479–1516. doi: 10.1007/s00705-002-0822-6. [DOI] [PubMed] [Google Scholar]
- 3.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 4.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:1104–1114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan X, Chipman PR, Castberg T, Bratbak G, Baker TS. The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry. J. Virol. 2005;79:9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan X, Olson NH, Van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containing dsDNA viruses. Nature Struct. Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl Acad. Sci. USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr. sect. D. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 9.Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J. Gen. Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 10.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 11.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 12.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson SD, Bamford JKH, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 14.Abrescia NGA, Cockburn JJB, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, Bamford DH, Bamford JKH. Insights into assembly from structural analysis of bacteriophage PRD1. Nature (London) 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 15.Abrescia NGA, Grimes JM, Kivelä HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JKH, Bamford DH, Stuart DI. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Laurinmaki PA, Huiskonen JT, Bamford DH, Butcher SJ. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure. 2005;13:1819–1828. doi: 10.1016/j.str.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Lin T, Chen Z, Usha R, Stauffacher CV, Dai J-B, Schmidt T, Johnson JE. The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- 18.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Tsao J, Chapman MS, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith TJ, Rossman MG, Compans RW, Parrish CR. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 20.McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, Rossmann MG, Olson NH, Baker TS, Incardona NL. Atomic structure of single-stranded DNA bacteriophage phiX174 and its functional implications. Nature (London) 1992;355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devauchelle G, Attias J, Monnier C, Barray S, Cerutti M, Guerillon J, Orangebalange N. Chilo iridescent virus. Curr. Top. Microbiol. Immunol. 1985;116:37–48. doi: 10.1007/978-3-642-70280-8_3. [DOI] [PubMed] [Google Scholar]
- 22.Jakob NJ, Muller K, Bahr U, Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology. 2001;286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- 23.Goorha R, Murti KG. The genome of frog virus 3, an animal DNA virus, is circularly permuted and terminally redundant. Proc. Natl. Acad. Sci. USA. 1982;79:248–252. doi: 10.1073/pnas.79.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darai G, Anders K, Koch HG, Delius H, Gelderblom H, Samalecos C, Flugel RM. Analysis of the genome of fish lymphocystis disease virus isolated directly from epidermal tumours of pleuronectes. Virology. 1983;126:466–479. doi: 10.1016/s0042-6822(83)80005-1. [DOI] [PubMed] [Google Scholar]
- 25.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnitzler P, Soltau JB, Fischer M, Reisner H, Scholz J, Delius H, Darai G. Molecular cloning and physical mapping of the genome of insect iridescent virus type 6: further evidence for circular permutation of the viral genome. Virology. 1987;160:66–74. doi: 10.1016/0042-6822(87)90045-6. [DOI] [PubMed] [Google Scholar]
- 27.Cerutti M, Devauchelle G. Characterization and localization of CIV polypeptides. Virology. 1985;145:123–131. doi: 10.1016/0042-6822(85)90207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stohwasser R, Raab K, Schnitzler P, Janssen W, Darai G. Identification of the gene encoding the major capsid protein of insect iridescent virus type 6 by polymerase chain reaction. J. Gen. Virol. 1993;74:873–879. doi: 10.1099/0022-1317-74-5-873. [DOI] [PubMed] [Google Scholar]
- 29.Balange-Orange N, Devauchelle G. Lipid composition of an iridescent virus type 6 (CIV). Arch. Virol. 1982;73:363–367. doi: 10.1007/BF01318091. [DOI] [PubMed] [Google Scholar]
- 30.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 32.Yan X, Sinkovits RS, Baker TS. AUTO3DEM--an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. U S A. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cusack S. Adenovirus complex structures. Curr. Opin. Struct. Biol. 2005;15:237–243. doi: 10.1016/j.sbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Bamford DH, Caldentey J, Bamford JKH. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 1995;45:281–319. doi: 10.1016/s0065-3527(08)60064-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 40.Yu Z, Bajaj C. Automatic ultrastructure segmentation of reconstructed cryoEM maps of icosahedral viruses. IEEE Trans. Image Process. 2005;14:1324–1337. doi: 10.1109/tip.2005.852770. [DOI] [PubMed] [Google Scholar]
- 41.San Martin C, Huiskonen JT, Bamford JK, Butcher SJ, Fuller SD, Bamford DH, Burnett RM. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nature Struct. Biol. 2002;9:756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- 42.Huiskonen JT, Kivela HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 43.Martins MD, Cavalcanti de Araujo V, Raitz R, Soares de Araujo N. Expression of cytoskeletal proteins in developing human minor salivary glands. Eur. J. Oral. Sci. 2002;110:316–321. doi: 10.1034/j.1600-0722.2002.21360.x. [DOI] [PubMed] [Google Scholar]
- 44.Saban SD, Silvestry M, Nemerow GR, Stewart PL. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 2006;80:12049–12059. doi: 10.1128/JVI.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Yu Z, Bajaj C. Computational Approaches for Automatic Structural Analysis of Large Bio-molecular Complexes. IEEE/ACM Trans. Comput. Biol. Bioinfor. 2008 doi: 10.1109/TCBB.2007.70226. in press. [DOI] [PubMed] [Google Scholar]
- 48.Kleywegt GJ, Jones TA. Software for handling macromolecular envelopes. Acta Crystallogr. D Biol. Crystallogr. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.