SUMMARY
Crossovers between meiotic homologs are crucial for their proper segregation, and crossover number and position are carefully controlled. Crossover homeostasis in budding yeast maintains crossovers at the expense of non-crossovers when double-strand DNA break (DSB) frequency is reduced. The mechanism of maintaining constant crossover levels in other species has been unknown. Here we investigate in fission yeast a different aspect of crossover control – the near invariance of crossover frequency per kb of DNA despite large variations in DSB intensity across the genome. Crossover invariance involves the choice of sister chromatid vs. homolog for DSB repair. At strong DSB hotspots, intersister repair outnumbers interhomolog repair ~3:1, but our genetic and physical data indicate the converse in DSB-cold regions. This unanticipated mechanism of crossover control may operate in many species and explain, for example, the large excess of DSBs over crossovers and the repair of DSBs on unpaired chromosomes in diverse species.
INTRODUCTION
DNA double-strand breaks (DSBs) are introduced into the genome as part of the meiotic program to segregate homologs and form haploid gametes (Keeney, 2001). Repair of DSBs using the homolog but not the sister chromatid as a template is required to produce functional (interhomolog) crossovers, which are essential in most organisms for the proper segregation of homologs at the first meiotic division. Interhomolog, but not intersister, repair also promotes genetic diversification, important for the evolution of species. How DSB repair is controlled to occur productively between homologs rather than non-productively between sisters is a critical, unsolved problem in meiosis, one that we address in this study.
Although DSBs are concentrated at preferred sites (hotspots) on chromosomes, crossovers are nearly uniformly distributed along chromosomes in the fission yeast Schizosaccharomyces pombe studied here (Young et al., 2002; Cromie et al., 2007). Our studies on the requirements for intersister and interhomolog repair lead us to propose a novel mechanism for crossover control – the controlled repair of DSBs by differential interaction with the sister chromatid or with the homologous chromosome. This mechanism maintains a nearly constant level of crossovers, measured in centimorgans per physical unit of DNA, in the face of wide variations in the frequency of DSBs along the genome. The findings reported here are also relevant to meiotic recombination in other contexts and to mitotic cells, in which DSB repair with the sister chromatid appears to be preferred, precisely to avoid crossovers. (Here and below we use “crossover” to mean that between homologs, since only these produce genetic recombinants and the chiasmata that facilitate proper meiotic homolog disjunction.)
Since crossovers are crucial for proper homolog segregation in meiosis, their number and position are exquisitely controlled by various means. In most species crossovers interfere with each other, resulting in their being farther apart than randomness would predict. In the budding yeast Saccharomyces cerevisiae when the number of DSBs is modestly decreased, the number of crossovers is maintained at a nearly constant level at the expense of non-crossover outcomes of DSB repair (Martini et al., 2006; Chen et al., 2008). The molecular basis of this crossover homeostasis is not known, but Martini et al. (2006) suggested that it is related to that of crossover interference, whose molecular basis is also unclear. In support of this view mutations in certain genes affect both types of control (Chen et al., 2008). An additional level of control lies at the initiation of recombination, the formation of DSBs, which varies markedly across genomes, with some loci, called hotspots, having much more frequent DSBs than other regions. In S. pombe DSBs at hotspots, which are separated on average by ~65 kb, can be as much as 400-times more frequent than DSBs in other intervals (Hyppa et al., 2008). The frequency of crossing over in a chromosomal interval is determined by a complex interplay of each of these factors. Here we focus on the interplay of DSB formation and partner choice for DSB repair in meiotic recombination.
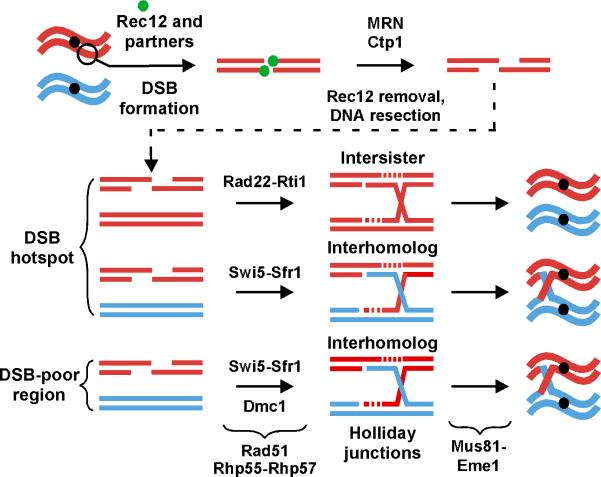
The central mechanics of meiotic recombination appear to be similar in the several species examined and perhaps in all sexually reproducing species (Keeney, 2001; Cromie and Smith, 2008). In S. pombe, meiotic DSBs are formed by the action of Rec12, the homolog of Spo11 in S. cerevisiae (Cervantes et al., 2000). Aided by other proteins, Rec12 breaks each DNA strand and becomes covalently linked to each 5’ end of the DNA at the DSB (Cromie et al., 2007; Hyppa et al., 2008; Milman et al., 2009; Rothenberg et al., 2009) (Figure 1). Rec12 is removed attached to an oligonucleotide (Milman et al., 2009; Rothenberg et al., 2009), and the DNA end is thought to be resected in the 5’ to 3’ direction (Farah et al., 2009). A free DNA end is thereby created and, assisted by multiple proteins studied here, invades homologous duplex DNA, either the sister chromatid or the homolog, and uses it as a template for DNA synthesis and DSB repair. Invasion of duplex DNA by single-stranded (ss) DNA is thought to create a displacement loop (D-loop), which in turn is cut and anneals to the second end of the initially broken DNA to establish a stable four-stranded DNA molecule – a Holliday junction (Cromie et al., 2006). Resolution of the Holliday junction into two duplex DNA molecules by the Mus81-Eme1 complex (Boddy et al., 2001; Cromie et al., 2006) can result in the reciprocal exchange of DNA flanking the DSB to produce a crossover. Interhomolog, but not intersister, exchange provides a physical connection important for meiotic homolog segregation and increased genetic diversification.
Figure 1. Model for Meiotic Recombination in S. pombe.
Meiotic replication (not shown) produces sister chromatids, each a DNA duplex (thick lines, red and blue distinguishing the homologs). 1. A DSB is made in one duplex by Rec12 (with assistance by other proteins), and Rec12 (green ball) remains covalently linked to the 5’ ends of each DNA strand (thinner lines). 2. The MRN complex (Rad32-Rad50-Nbs1) with Ctp1 clips off Rec12 and resects one DNA strand to form long ss DNA with a 3’-end. 3. This ss DNA forms a nucleoprotein filament with Rad51, and strand invasion, aided by Rhp55-Rhp57, is promoted in three possible ways to form single Holliday junctions (HJs). At strong DSB hotspots Rad22-Rti1 promotes intersister HJ formation, and Swi5-Sfr1 promotes interhomolog HJ formation; both reactions are independent of Dmc1. Rad22-Rti1 plays a minor role in interhomolog gene conversion, perhaps by SDSA (Octobre et al., 2008). In DSB-poor regions, Swi5-Sfr1 and Dmc1 promote interhomolog HJ formation. 4. The HJs are resolved by Mus81-Eme1 into crossovers as shown or non-crossovers (not shown). The crossovers aid chromosome segregation at the first meiotic division and promote genetic diversification. See Cromie and Smith (2008) and Milman et al. (2009) for references and further discussion.
Several aspects of crossover control appear to operate at a critical step of recombination, the formation of the initial joint DNA molecule by strand invasion (Figure 1, step 3). The choice of unbroken partner – sister chromatid or homolog – and the stability of the joint molecule determine whether this intermediate is further processed into a crossover or not. [In certain mutants of S. cerevisiae some DSBs appear to be repaired by interaction with both the sister chromatid and the homolog (Oh et al., 2007; Jessop and Lichten, 2008; Oh et al., 2008), but the relevance of these events to those in wild type is unclear.] Stand invasion requires multiple proteins. Rad51, also called Rhp51 in S. pombe (Muris et al., 1997), is a homolog of bacterial RecA protein and has robust strand-exchange activity resulting from its coating 3’ ss DNA ends and facilitating the invasion of duplex DNA (Aboussekhra et al., 1992; Shinohara et al., 1992; Sung, 1994; Haruta et al., 2006). As expected, in S. pombe rad51Δ mutants meiotic DSBs are made but not repaired, and meiotic recombination and spore viability are severely reduced, indicating the essential role of Rad51 in meiotic DSB repair (Muris et al., 1997; Grishchuk and Kohli, 2003; Young et al., 2004).
Like many other species, S. pombe has another, meiosis-specific RecA homolog, Dmc1 (Bishop et al., 1992; Fukushima et al., 2000; Haruta et al., 2006). In S. pombe dmc1Δ mutants, meiotic DSBs are formed and repaired as rapidly as in wild type and spore viability is high, but recombinant frequencies are reduced 3 – 6-fold in the several intervals reported (Fukushima et al., 2000; Grishchuk and Kohli, 2003; Ellermeier et al., 2004; Young et al., 2004). These data indicate that DSBs are repaired in S. pombe dmc1Δ mutants but less frequently with the homolog than in wild type. In S. cerevisiae, Dmc1 is needed for essentially all interhomolog recombination, and dmc1Δ mutants have a severe meiotic defect, at least in some strains, as there is an apparent barrier to redirecting DSB repair from the homolog to the sister (Bishop et al., 1992; Shinohara et al., 1997; Hayase et al., 2004; Tsubouchi and Roeder, 2004; Niu et al., 2009).
Rad51 requires two distinct accessory (mediator) complexes for efficient strand exchange; in S. pombe these are Rhp55-Rhp57 (homolog of Rad55-Rad57 in S. cerevisiae) (Khasanov et al., 1999; Tsutsui et al., 2000; Tsutsui et al., 2001) and Swi5-Sfr1 (homolog of Sae3-Mei5 in S. cerevisiae) (Akamatsu et al., 2003; Ellermeier et al., 2004; Hayase et al., 2004). Both purified complexes stimulate Rad51-promoted strand exchange reactions, and Swi5-Sfr1 additionally stimulates Dmc1-promoted reactions (Sung, 1997; Haruta et al., 2006; Ferrari et al., 2009). While mutants lacking either single complex have only a mild meiotic DSB repair defect (Young et al., 2004), mutants lacking both have a severe defect comparable to that of a rad51Δ mutant (Hyppa et al., 2008). From these and other observations it was proposed that the Rhp55-Rhp57 and Swi5-Sfr1 complexes act in different sub-pathways of Rad51-dependent recombination (Akamatsu et al., 2003; Grishchuk and Kohli, 2003; Ellermeier et al., 2004). When both sub-pathways are blocked, Rad51 cannot function in strand exchange.
To determine the roles of these gene products in the mechanics and control of meiotic DSB repair, we assayed Holliday junctions (HJs) at two unlinked, strong meiotic DSB hotspots, meiotic break site 1 (mbs1) and ade6-3049 (Steiner et al., 2002; Young et al., 2002; Cromie et al., 2005; Steiner and Smith, 2005). Diploids with heterozygous restriction-site markers flanking these hotspots allowed us to differentiate interhomolog and intersister HJs and thereby measure partner choice for DSB repair (Cromie et al., 2005, 2006); recombination between markers on homologs provided an additional measure of interhomolog repair.
Our analysis of the gene products required for HJ formation reveal a novel, differential requirement for Dmc1 at strong DSB hotspots vs. weaker DSB sites. To reconcile a major discrepancy between the nearly uniform distribution of crossovers but strikingly non-uniform distribution of meiotic DSBs (Young et al., 2002; Cromie et al., 2007), we show here that this aspect of crossover control, which we call crossover invariance, is effected by the repair of DSBs at strong hotspots predominantly by intersister HJ formation without Dmc1 and at weaker DSB sites by interhomolog HJ formation with Dmc1 (see Discussion and Figure 6). This mechanism of crossover control contrasts with the crossover homeostasis reported in S. cerevisiae (Martini et al., 2006; Chen et al., 2008). We discuss the choice of partner for meiotic DSB repair and the biological consequences of these two types of crossover control in these two markedly different yeasts and other species.
Figure 6. Model for Crossover Invariance by Differential Choice of Homolog vs. Sister Chromatid for DSB Repair.
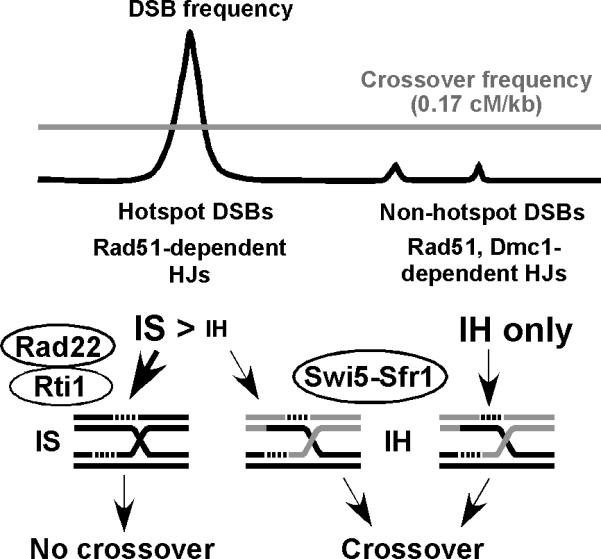
DSB repair at strong DSB hotspots is predominantly with the sister chromatid and therefore yields few crossovers per DSB. At weaker DSB sites repair is predominantly with the homolog and yields more crossovers per DSB. The result is a more uniform distribution of crossovers (nearly constant cM/kb; crossover invariance) than of DSBs, as observed (Young et al., 2002; Table 1B). The proteins required for DSB repair are also differential, as indicated (see Figure 1).
RESULTS
Formation of Meiotic Holliday Junctions at the DSB Hotspot mbs1 Is Rad51-dependent but Dmc1-independent
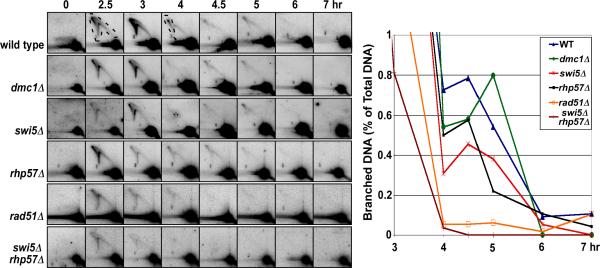
To determine the genetic requirements for the formation of HJs, an intermediate essential for crossover formation, we used a previously developed physical assay for HJs at the strong DSB hotspot mbs1 (Cromie et al., 2006). DNA was extracted from meiotically induced cells and analyzed via two-dimensional gel electrophoresis and subsequent Southern blot hybridization. Replication and recombination intermediates, both of which appear in this analysis, can be differentiated by the timing of the G1 to G2 transition and by Rec12-dependence: recombination intermediates occur after DNA replication and, unlike replication intermediates, are Rec12-dependent (Cromie et al., 2006). Replication intermediates were visible at 2.5 and 3 hr (Figure 2), which corresponded to the timing of replication measured by flow cytometry (Figure S1). Recombination intermediates (HJs, indicated by dashed lines in the 4 hr panel) were visible from 4 to 5 hr in a wild-type strain and accumulated to high levels in mus81Δ strains; rec12Δ blocks the appearance of HJs at these late times in both mus81+ and mus81Δ strains (Cromie et al., 2006), implying that these HJs are recombination intermediates. In a rad51Δ mutant, replication intermediates were formed with nearly wild-type kinetics and frequency, but essentially no recombination-related HJs were formed (Figures 2 and S2A). (Hereafter, “HJs” refers to recombination-related HJs.) Thus, most or all HJ-formation depends on Rad51 in S. pombe. S. cerevisiae rad51 mutants, however, retain a significant level of HJs (Schwacha and Kleckner, 1997).
Figure 2. Holliday Junction-formation at the DSB Hotspot mbs1 Is Dependent on Rad51 and Its Mediators But Is Independent of Dmc1.
DNA of meiotically induced cells with the indicated mutations was digested with PvuII, separated by two-dimensional gel electrophoresis, and Southern blot-hybridized with a ds DNA probe specific for mbs1 (see Figure 3, upper left panel for diagram). Images of Southern blots of DNA from HJ resolvase-proficient (mus81+) strains show the formation and repair of HJs from the start of meiotic induction (0 hr). The corresponding graph shows the quantification of branched DNA recombination intermediates indicated by the dashed lines (4 hr panel, top row); these recombination intermediates migrate above the linear DNA arc and are formed after 3 hr, when replication is complete. The quantification of replication intermediates (dashed lines in 2.5 hr panel, top row) is omitted here for clarity (see Figure S2A for the complete time-course). Replication and recombination intermediates are inferred from the timing of DNA replication (Figures S1), dependence on Rec12, and accumulation in mus81Δ mutants (Cromie et al. 2006). The half-hour delay in maximal HJ abundance in the dmc1Δ mutants is within our experimental error. Each measurement is the mean of two independent meiotic inductions, and nearly all values are within 20% of their respective mean; error bars are omitted for clarity. See also Figures S1, S2, and S3.
In marked contrast, dmc1+ and dmc1Δ strains showed the same levels of HJs in both mus81+ strains and mus81Δ mutants, in which HJs accumulate and in which a more precise determination is possible (Figures 2 and S2C). Dmc1-independence was unexpected based on the reduction of recombination in several intervals in dmc1Δ mutants (see Introduction), in particular the 6-fold crossover-reduction in the ura1 – rqh1 interval, which contains the mbs1 hotspot assayed here (see Table 1 and Figure 5C discussed below). A dmc1Δ mutant deficient in strand exchange would be expected to give fewer interhomolog HJs to account for the fewer crossovers observed. This seeming discrepancy will be addressed below and in the Discussion. These data show that all detectable HJ formation depends on Rad51 but is independent of Dmc1, at least at the mbs1 DSB hotspot.
Table 1.
Dmc1-dependence of Recombination Becomes Stronger at Lower DSB Levels
| (A) | ade6 alleles crossed | DSB level (%)2 | Ade+/103 viable spores1 |
dmc1 ratio3 | |
|---|---|---|---|---|---|
| dmc1+ | Dmc1Δ | ||||
| 3057 × M375 | <0.1 | 0.51 ± 0.17 (3) | 0.17 ± 0.02 (3) | 3.0 | |
| M26 × 52 | 0.74 | 5.6 ± 0.41 (7) | 2.5 ± 0.12 (7) | 2.5 | |
| 3074 × 52 | — 5 | 8.7 ± 0.47 (6) | 5.6 ± 0.25 (7) | 1.5 | |
| 3049 × M375 | 5.84 | 14 ± 3 (3) | 12 ± 4 (3) | 1.2 | |
| 3049 × M266 | 6.54 | 35 ± 2 (4) | 32 ± 3 (4) | 1.1 | |
| (B) | Intergenic interval tested | kb | Maximal DSB peak7 | wt cM/kb8 | Genetic distance (cM)9 |
dmc1 ratio3 | ||
|---|---|---|---|---|---|---|---|---|
| + | dmc1Δ | swi5Δ | ||||||
| lys3 – aur1 | 68 | 13 | 0.19 (0.16) | 12.5 | 1.9 | 0.3 | 6.6 | |
| lys3 – ura1 | 212 | 49 | 0.12 (0.11) | 24.4 | — | — | — | |
| lys3 – ura1 (mbs1Δ)10 | 200 | 85 | 0.13 | 25.2 | 4.3 | 2.6 | 5.9 | |
| ura1 – rqh1 | 91 | 149 | 0.19 (0.12) | 17.4 | 3.0 | — | 5.8 | |
| ura1 – rqh1 (mbs1Δ) | 79 | 20 | 0.14 | 11.0 | 1.0 | 0.5 | 11.0 | |
| ura2 – leu2 | 14 | 107 | 0.15 | 2.1 | 0.55 | 0.5 | 3.8 | |
Data are the mean ± SEM of the Ade+ recombinant frequency from the number of crosses in parentheses. See also Table S1B.
Percent of ade6 DNA broken in rad50S meiosis (Steiner et al., 2002).
Ade+ frequency or cM in dmc1+ divided by that in dmc1Δ.
Calculated as the mean of the DSB values in the homozygous diploids.
Not determined.
Ade+ frequency from ade6-3049 × ade6-M26 in swi5Δ was 5.8 ± 0.1 (4), giving a ratio of 6. In sfr1Δ it was 8.8 ± 0.6 (4), giving a ratio of 4. Thus, this recombination is Swi5-Sfr1-dependent.
Maximal value of median-normalized Rec12-DNA linkages, measured by ChIP-on-chip analysis, in the indicated interval in side-by-side assays of mbs1+ and mbs1Δ (Figures 5 and S6) (Hong et al., 2001; Cromie et al., 2007; G. Cromie, R.W.H., and G.R.S., unpublished data).
Data in parentheses are from Young et al. (2002).
Recombinant frequencies in Table S1B were converted to cM using Haldane's equation.
mbs1-19, a 12 kb deletion.
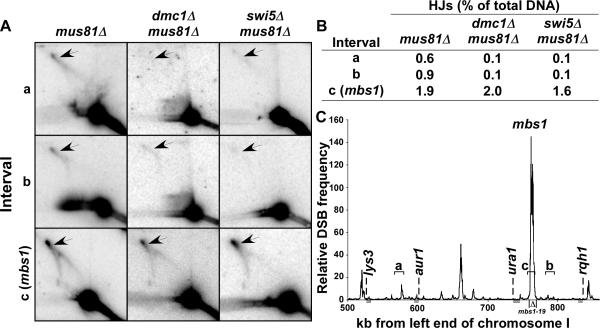
Figure 5. HJ Formation in DSB-poor Regions Requires Dmc1.
(A) DNA from the indicated mutants extracted 5 hr after meiotic induction was digested with PvuII, separated by two-dimensional gel electrophoresis, and probed for HJs at the positions shown in (C).
(B) The fraction (%) of total probed DNA in the position of HJs was determined for blots in (A) or similar blots. Data are the means of 2 – 3 determinations; SEM is <20% of the mean.
(C) Map of the left portion of NotI fragment J on chromosome I shows genes used for crosses in Table 1B, the positions (indicated by horizontal brackets labeled a, b, and c) of the restriction fragments analyzed in (A), and the level of Rec12-DNA covalent linkages (relative DSB frequency) in a rad50S strain 5 hr after meiotic induction, normalized to the genome median (Cromie et al., 2007; Hyppa et al., 2008). See also Figure S1.
Holliday Junction-formation Depends on a Combination of Swi5-Sfr1 and Rhp55-Rhp57 Mediator Complexes
We next determined the roles of the mediator complexes Swi5-Sfr1 and Rhp55-Rhp57 in HJ formation at mbs1. A swi5Δ mutant formed and repaired HJs with wild-type kinetics, but the frequency was reduced to ~60% of the wild-type level (Figure 2). Total (accumulated) HJs, measured in a swi5Δ mus81Δ double mutant, were also reduced to about ~60% (Figure S2C). swi5Δ and sfr1Δ single mutants and the double mutant had similar viable spore yields, as high as 40% of the wild-type yield (Table S1A), and the sfr1Δ mutation partially suppressed the viable spore yield defect of mus81Δ (Table S1A), as does swi5Δ (Ellermeier et al., 2004). This suppression likely reflects fewer HJs being formed, thereby alleviating the mus81Δ resolution defect. These data provide further evidence that Swi5 and Sfr1 act as a complex important, but not essential, for HJ formation (Akamatsu et al., 2003; Ellermeier et al., 2004; Haruta et al., 2006).
To test the role of the Rhp55-Rhp57 complex on HJ formation, we used the rhp57Δ mutant; rhp55Δ, rhp57Δ, and the double rhp55Δ rhp57Δ mutants have similar phenotypes in genetic assays (Khasanov et al., 1999; Tsutsui et al., 2000; Grishchuk and Kohli, 2003; Ellermeier et al., 2004). The rhp57Δ mutant formed and repaired HJs with wild-type kinetics but with a reduced frequency, about 75% of the wild-type level (Figure 2), slightly higher than that of swi5Δ. In an rhp57Δ mus81Δ double mutant, total (accumulated) HJs at mbs1 were reduced about 2- to 3-fold (Figure S2C). The low viable spore yield of mus81Δ was partially suppressed by rhp55Δ or rhp57Δ (Table S1A), as true for swi5Δ or sfr1Δ (Ellermeier et al., 2004). Thus, elimination of either the Swi5-Sfr1 or the Rhp55-Rhp57 complex reduced but did not eliminate HJ formation.
Removal of components of both mediator complexes, however, showed a much more dramatic effect. A swi5Δ rhp57Δ double mutant formed HJs at a severely reduced frequency, similar to that of a rad51Δ mutant; that is, essentially no HJs were formed at the DSB hotspot mbs1 (Figure 2). This result is consistent with DSBs remaining unrepaired in this double mutant (Hyppa et al., 2008). Collectively, these results agree with earlier studies indicating that the two complexes are part of two alternative pathways of Rad51-dependent recombination (Akamatsu et al., 2003; Ellermeier et al., 2004).
Swi5-Sfr1 Is Necessary for the Formation of Interhomolog, but not Intersister, HJs
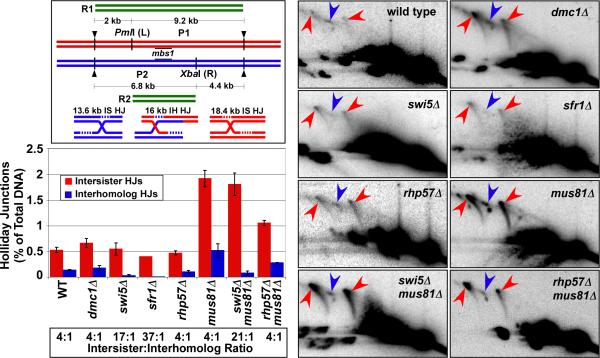
To investigate the roles of the two Rad51-dependent pathways of HJ formation, we determined the relationship between interhomolog (IH) and intersister (IS) HJs formed at the mbs1 hotspot. In particular, we tested whether the mediator complexes influenced the formation of one or both types of HJs. Heterozygous restriction sites flanking mbs1 were used to assay IH and IS HJs via two-dimensional gel electrophoresis (Cromie et al., 2006; Figure 3, upper left panel ). IH HJs were strongly reduced, by a factor of 5, in both swi5Δ and sfr1Δ mutants, but IS HJs were formed at nearly wild-type frequency (Figure 3). In a swi5Δ mus81Δ mutant, accumulated IH HJs were also reduced by a factor of 5, but IS HJs remained at the wild-type level, as observed in swi5Δ mus81+ strains. The IS:IH ratio in the swi5Δ mus81Δ double mutant was nearly the same as that in the swi5Δ mus81+ strain (17:1 and 21:1, respectively), much higher than the wild-type ratio of 4:1 (Figure 3). Thus, the specific reduction of IH HJ frequency was not an artifact of IH HJs being more rapidly resolved than IS HJs in mus81+ strains. In a dmc1Δ mutant both IS and IH HJs appeared at the wild-type level (Figure 3), consistent with the total HJ assays noted above (Figure 2).
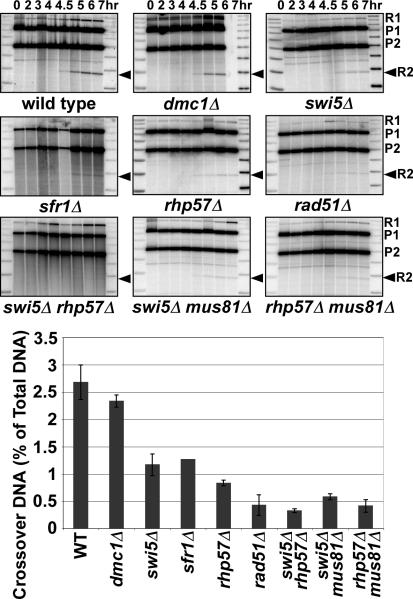
Figure 3. Swi5-Sfr1 Is Necessary for the Formation of Interhomolog, but Not Intersister, HJs at the DSB Hotspot mbs1.
The relative amounts of IH and IS HJs in the indicated mutants were determined as in Figure 2 using diploids with heterozygous restriction sites as indicated in the diagram in the upper left panel. The black bars at mbs1 indicate the ds DNA probe. IH and IS HJs were determined by differences in their masses, 18.4 and 13.6 kb for IS HJs, and an intermediate mass of 16 kb for IH HJs. Parental fragments are 9.2 kb (P1) and 6.8 kb (P2). Gel images from 4.5 or 5 hr (the time of maximal HJs) for the indicated mutants are shown. Red arrows indicate IS HJs; blue arrows, IH HJs. Quantification of HJs in 2 – 5 experiments (a single experiment for sfr1Δ) is displayed on the bar graph; data are the mean, and the error bars indicate the range or SEM. The ratio of IS:IH HJs is given below for comparison. See also Figures S1, S2, and S4.
To determine the function of the other mediator complex, Rhp55-Rhp57, we examined rhp57Δ and rhp57Δ mus81Δ mutants. Unlike the result with a swi5Δ mutant, both IH and IS HJs were reduced slightly in an rhp57Δ mutant, and the IS:IH ratio remained at the wild-type level (4:1; Figure 3). In the rhp57Δ mus81Δ double mutant both IH and IS HJs were reduced 2-fold compared to rhp57+ mus81Δ, and again the IS:IH ratio remained at the wild-type level (Figure 3). In summary, our data indicate that the Swi5-Sfr1 complex functions specifically in the formation of IH HJs, while Rhp55-Rhp57 functions non-specifically in all HJ formation.
Reduction of Crossover DNA at mbs1 Reflects the Observed HJ Levels in the Mutants
The flanking heterozygous markers at mbs1 allowed measuring interhomolog crossover DNA as diagnostic restriction fragments (Cromie et al., 2006; Figure 3, upper left panel). Of the mutants tested, rad51Δ and swi5Δ rhp57Δ mutants showed the lowest level of detected crossovers, reduced to 0.4 – 0.5% from the wild-type frequency of 2.7% (Figure 4), comparable to the reduction previously seen in mus81Δ (Cromie et al., 2006). The low levels of crossover DNA are consistent with the lack of DSB repair or HJ formation in the absence of either Rad51 or both the Swi5-Sfr1 and Rhp55-Rhp57 complexes (Young et al., 2004; Hyppa et al., 2008; Figure 2). The low, residual level of the restriction fragment assayed as crossover DNA in the rad51Δ and mus81Δ mutants likely reflects conversion of the right-hand restriction site rather than crossing over (Cromie et al., 2005, 2006).
Figure 4. Crossover DNA at the DSB Hotspot mbs1 Is Dependent on Rad51 and Its Mediators, but Not on Dmc1.
The level of crossover DNA at mbs1 was measured by the accumulation of the R2 recombinant DNA fragment (black arrowhead; see Figure 3, upper left panel for diagram). Crossover frequency is 2 × (R2 DNA)/total DNA. Each measurement is the average of the crossover DNA fragment at 6 or 7 hr in two independent meiotic inductions (one for sfr1Δ); the error bars indicate the range. Based on tetrad analyses, the residual level of crossover DNA in rad51Δ, swi5Δ rhp57Δ, and mus81Δ mutants can be accounted for by gene conversion of the right-hand marker (Cromie et al., 2005). See also Figures S1, S2, and S5.
The crossover reductions seen in swi5Δ, sfr1Δ, and rhp57Δ mutants (Figure 4) are consistent with their previously described phenotypes based on both genetic and physical assays (Akamatsu et al., 2003; Ellermeier et al., 2004; Khasanov et al., 2008) (Figures 2 and 3, Table 1). As expected, both crossover DNA (Figure 4) and IH HJs (Figure 3) were reduced in rhp57Δ, swi5Δ, and sfr1Δ mutants, although the precise degree of reduction is uncertain because of the low IH HJ levels in the mutants and the likely conversion product interfering with crossover determination. The residual crossovers in these mutants were Mus81-dependent, since crossover levels in the swi5Δ mus81Δ and rhp57Δ mus81Δ double mutants were lower than those in swi5Δ and rhp57Δ single mutants but comparable to those in themus81Δ single mutant (Figure 4) (Cromie et al., 2006).
In contrast, in the dmc1Δ mutant we observed a marked discrepancy between the genetic and physical data. As noted in the Introduction, in dmc1Δ there is a 3 – 6-fold reduction of intergenic recombination measured genetically in several intervals, including the 91 kb ura1 – rqh1 interval, which includes the mbs1 hotspot studied here (Table 1). We observed, however, no significant reduction of crossover DNA (Figure 4) or IH HJs (Figure 3) in the short (4.8 kb) interval encompassing the mbs1 hotspot. A possible explanation for this discrepancy is that recombination at strong DSB hotspots, such as mbs1, is Dmc1-independent but recombination elsewhere is Dmc1-dependent. To address this possibility, we investigated recombination and HJ formation at another hotspot, ade6-3049, amenable to both genetic and physical analyses.
Gene Conversion of the ade6-3049 DSB Hyper-hotspot, but Not of Other ade6 Alleles, Is Dmc1-independent
Intragenic recombination between alleles of the S. pombe ade6 gene results exclusively from gene conversion (Gutz, 1971). Previous measurements of ade6 intragenic recombination in a dmc1Δ mutant showed a consistent reduction by a factor of 2 – 3 relative to wild type (Fukushima et al., 2000; Grishchuk and Kohli, 2003; Ellermeier et al., 2004; Table 1A). However, ade6 intragenic recombination using the ade6-3049 allele, a very intense DSB and recombination hotspot (Steiner et al., 2002; Steiner and Smith, 2005), manifested no significant dependence on Dmc1 (Table 1A). In contrast, crosses with ade6-3057, a non-hotspot control for ade6-3049, showed a 3-fold reduction in the dmc1Δ mutant, similar to that reported for other intragenic intervals (Fukushima et al., 2000; Grishchuk and Kohli, 2003; Ellermeier et al., 2004). ade6-3074, whose hotspot activity is intermediate between those of the hyper-hotspot ade6-3049 and the weaker hotspot ade6-M26 (Steiner and Smith, 2005), gave an intermediate reduction (~1.5-fold). Thus, these data are consistent with Dmc1 becoming less important for recombination, and hence IH HJ formation, as the intensity of the DSB hotspot goes up.
In contrast, Swi5-Sfr1 is strongly required for ade6 intragenic recombination regardless of the alleles crossed. ade6-3049 × ade6-M26 recombination was reduced by a factor of 4 – 6 in both swi5Δ and sfr1Δ strains (Table 1A, footnote 6); recombination of other ade6 alleles is also reduced in each mutant by factors of 4 – 14 (Schmidt et al., 1987; DeVeaux et al., 1992; Ellermeier et al., 2004; Khasanov et al., 2008). These data show that recombination at the hotspot ade6-3049, like that at mbs1, requires Swi5-Sfr1 but not Dmc1. These results imply that the requirements for strand invasion and recombination differ for DSBs at hotspots and DSBs in other intervals (see Discussion).
HJ Formation and Crossing over at the ade6-3049 DSB Hyper-hotspot Parallel the Events at mbs1
To complement these genetic analyses and to determine the requirements for HJ formation at another DSB hotspot, we assayed the formation of HJs and crossover DNA at ade6-3049. The frequency of HJs formed at ade6-3049 was comparable to that at mbs1 (2.1% at ade6-3049 and 2.8% at mbs1, as measured in the mus81Δ mutant background) (Figure S3). As expected from the genetic data (Table 1A, footnote 6), HJs at ade6-3049 were reduced by a factor of ~3 in the swi5Δ strain, while only a slight decrease (25%) was seen in the dmc1Δ mutant (Figure S3), similar to the data at mbs1 (Figure 2).
At the ade6-3049 hotspot as at mbs1 (Figure 3), in both wild-type and mus81Δ backgrounds IS HJs were more frequent than IH HJs, by ratios of 2:1 and 3:1, respectively (Figure S4), only slightly lower than the 4:1 ratio seen at mbs1 (Figure 3). The frequencies of IS and IH HJs at ade6-3049 were also similar to those at mbs1 (Cromie et al., 2006; Figure 3). In a swi5Δ mutant (mus81+) IH HJs at ade6-3049 were reduced about 3-fold, but IS HJs were not significantly reduced. Therefore, at both mbs1 and ade6-3049, a swi5Δ mutation reduces IH HJs more than IS HJs, but both are independent of Dmc1 (Figure S4). These data agree with the genetic data for ade6-3049 (Table 1A) and the assays of total HJs at ade6-3049 (Figure S3). Thus, by these assays HJ formation is controlled similarly at the two strong DSB hotspots mbs1 and ade6-3049.
Crossover DNA at ade6-3049 was assayed with heterozygous restriction site mutations flanking ade6 (Figure S5, lower right panel), as was done at mbs1. Crossover DNA was reduced 2-fold in swi5Δ, but no significant reduction was seen in dmc1Δ. Thus, these physical data are in accord with the genetic data (Table 1A): the Swi5-Sfr1 complex has an important role at ade6-3049 similar to that at mbs1, while Dmc1 does not influence any of the events of recombination – the frequency of HJ formation, crossover DNA formation, or gene conversion – at either DSB hotspot.
Holliday Junction Formation and Crossing Over in Low-level DSB Regions Strongly Require Dmc1
HJ formation, crossover DNA formation, and gene conversion at the mbs1 and ade6-3049 hotspots do not require Dmc1 (Figures 2, 3, 4, S3, S4, and S5; Table 1A). Paradoxically, Dmc1 is required for wild-type levels of crossing over in multiple genetic intervals (Fukushima et al., 2000; Grishchuk and Kohli, 2003), suggesting that HJ formation in these chromosomal regions does require Dmc1. We therefore tested the possibility that HJ formation in intervals lacking strong DSB hotspots requires Dmc1. We examined two such low-level DSB intervals (i.e., hotspot-poor regions), each about 15 kb long; one is ~20 kb to the right of mbs1, and the other ~200 kb to the left of mbs1 (Figure 5). In both intervals total (IS plus IH) HJs were readily detectable in dmc1+ strains but were strongly reduced in dmc1Δ mutants (Figure 5A, B). In the mus81Δ background HJs accumulated to 0.6 – 0.9% in dmc1+ strains but to only ~0.1% in dmc1Δ strains. In contrast, there was no significant reduction at the mbs1 hotspot, as noted above (Figures 5 and S2C). These data are consistent with the genetic data: Dmc1 is required for HJ formation and recombination in some genetic intervals but not in others. In both classes of intervals, HJ formation is Swi5-dependent (Figures 5 and S2C), in accord with genetic recombination in all tested intervals being Swi5-dependent (Schmidt et al., 1987; Ellermeier et al., 2004). As only IH HJs are Swi5-dependent (Figures 3 and S4), this result suggests that the HJs in DSB hotspot-poor intervals are predominantly IH HJs.
To complement these physical assays, we measured crossing over between markers that bracket DSB hotspot-poor and hotspot-rich intervals. In the 68 kb DSB hotspot-poor interval between lys3 and aur1, about 200 kb to the left of mbs1, the strongest DSB site is <1/10 as intense as mbs1 in the ura1 – rqh1 interval (Young et al., 2002; Cromie et al., 2007) (Figure 5C; Table 1B). Crossovers in the lys3 – aur1 interval were strongly reduced, by a factor of 6.6, in the dmc1Δ mutant (Table 1B), in accord with the strong reduction of HJs in the 14 kb sub-interval of lys3 – aur1 in the dmc1Δ mutant (Figure 5A and B). Thus, both HJ formation and crossing over are strongly dependent on Dmc1 in this DSB hotspot-poor region, in stark contrast to the Dmc1-independence at strong DSB hotspots. As expected, crossing over in the small (14 kb) ura2 – leu2 interval, which contains a strong hotspot (Figure S6A) (Cromie et al., 2007; Hyppa et al., 2008), was reduced by a factor of only 3.8 in the dmc1Δ mutant (Table 1B).
We directly tested the Dmc1 requirement for crossing over in an interval (ura1 – rqh1) with and without a strong hotspot (mbs1+ and mbs1-19, respectively). The 12 kb mbs1-19 deletion strongly reduced the intensity of the DSB hotspot (by a factor of ~7; Figure S6B; Table 1B) but only modestly reduced crossing over (by ~35%; Table 1B). The dmc1Δ mutation reduced crossing over in the ura1 – rqh1 interval by a greater factor (11.0) in mbs1Δ than in mbs1+ (5.8). These data are in accord with the physical assay for HJs (Figures 2, 3, and 5) and the increased Dmc1-dependence of gene conversion and crossing over as the level of DSBs decreases (Table 1). In summary, both the physical and genetic data show that Dmc1 is more strongly required for recombination in DSB hotspot-poor regions than in DSB hotspot-rich regions. Below, we discuss the implications of these observations.
DISCUSSION
The data reported here bear on two questions – how is the intact DNA partner (homolog vs. sister chromatid) chosen for DSB repair, and how is crossing over maintained at constant level, in spite of highly focused DSBs (hotspots)? We present below a basis for this novel aspect of crossover control, which we call crossover invariance, stemming from the genetic control of partner choice for DSB repair demonstrated by the data reported here.
Distinct Genetic Requirements for Interhomolog vs. Intersister HJ formation
Our results show that the S. pombe Swi5-Sfr1 complex is necessary for IH but not IS HJ formation at the two DSB hotspots examined on chromosomes 1 and 3 (Figures 3 and S4), and genetic recombination data suggest that this function extends to DSBs across the genome (Schmidt et al., 1987; Ellermeier et al., 2004; Khasanov et al., 2008; Table 1). To our knowledge, this is the first report of a differential genetic requirement for IH vs. IS HJ formation in any species other than S. cerevisiae, in which several proteins function specifically in meiotic interhomolog HJ formation. Lack of Dmc1, Red1, Hop1, Rad51, Rad55, or Rad57 reduces the frequency of IH HJs but not of IS HJs (Schwacha and Kleckner, 1994, 1997), and Mnd1-Hop2 and Hop1-Red1-Mek1 appear to promote specifically interhomolog recombination (Tsubouchi and Roeder, 2002; Niu et al., 2005, 2007). Latypov et al. (2010) have inferred from genetic experiments that S. pombe Hop1 and Mek1 are similarly involved in partner choice for meiotic recombination. In S. pombe swi5Δ and sfr1Δ mutants, most meiotic DSBs at hotspots are repaired, albeit with delayed kinetics (Young et al., 2004; unpublished data). These DSBs are repaired almost exclusively with the sister chromatid; DSBs that are repaired in wild type with the homolog may be repaired in these mutants via synthesis-dependent strand annealing (SDSA), as no increase of IS HJs was seen in these mutants (Figures 3 and S4), resulting in only modestly reduced viable spore yields but strongly reduced recombinant frequencies (Ellermeier et al., 2004) .
IS HJ formation may be mediated by Rad22 and Rti1, S. pombe homologs of S. cerevisiae Rad52 (van den Bosch et al., 2001). In an assay measuring intrachromosomal recombination, the rad22Δ mutant shows a 6-fold reduction compared to rad22+, and a double mutant with rti1Δ has a 100-fold reduction (Octobre et al., 2008). In contrast, interhomolog crossing over is not detectably reduced in rad22Δ or rti1Δ mutants; gene conversion at two loci is reduced about 2-fold and may occur by an HJ-independent mechanism, such as SDSA, in these mutants. These results support an IS-specific role for Rad22-Rti1 to complement the IH-specific Swi5-Sfr1. Consistent with this view, Rad22 inhibits loading of Dmc1 but stimulates loading of Rad51 onto RPA-coated ss DNA in vitro (Kurokawa et al., 2008; Y. Murayama and H. Iwasaki, pers. comm.). These results suggest that Rad22 and Rti1 mediate IS HJ formation and that the Swi5-Sfr1 complex mediates IH HJ formation by controlling the access of Rad51 and Dmc1 to the ss DNA end of resected DSBs.
Dmc1 Acts Primarily in DSB Hotspot-poor Regions in S. pombe
Our genetic and physical data indicate that the requirement for Dmc1 in meiotic recombination is inversely related to the strength of DSB hotspots in the chromosomal interval tested. For both intragenic recombination (gene conversion) and intergenic recombination (crossing over) the dependence on Dmc1 is strong in intervals with few DSB hotspots but weaker or absent in intervals with strong DSB hotspots (Table 1). Our physical analyses confirm that Dmc1 is not required for HJ formation, either IS or IH, or for crossover DNA at two strong hotspots (Figures 2, 3, 4, S3, S4, S5). In two intervals without strong hotspots, total (IS plus IH) HJ formation depends on Dmc1 and on Swi5-Sfr1, indicating that these HJs are primarily IH (Figure 5). Thus, the genetic and physical data indicate that Dmc1 is required only for IH HJ formation and only in DSB hotspot-poor intervals. To our knowledge, this is the first report of a locus-dependent requirement for Dmc1 in HJ formation.
What might be the basis of Dmc1's differential action at DSB hotspots vs. other intervals? Once a DSB is made, the double-strand (ds) DNA end would seem to be the same, whether at a hotspot or not. As noted above, the recombination mediators Rad22-Rti1 and Swi5-Sfr1 influence the choice of partner for DSB repair. In addition, we propose that some aspect of chromatin structure, broadly defined as DNA and closely associated proteins, distinguishes DSBs at hotspots from DSBs elsewhere and makes repair of the latter Dmc1-dependent. This feature of chromatin may in turn dictate strong (frequent) vs. weak (rare) DSB formation. Thus, chromatin structure may differentiate chromosomal intervals into two types – DSB hotspots with Dmc1-independent recombination and non-hotspot intervals with Dmc1-dependent recombination.
DSBs at hotspots in dmc1Δ mutants are repaired with wild-type frequency and efficiency (Young et al., 2004; unpublished data). Spore viability is also high in dmc1Δ mutants (Grishchuk and Kohli, 2003; Ellermeier et al., 2004), indicating that the non-hotspot DSBs are also repaired efficiently. We conclude that these DSBs, which are repaired primarily with the homolog in wild-type cells, are repaired primarily with the sister in dmc1Δ mutants, thereby accounting for the high spore viability but low recombinant frequency in these intervals. This repair may be via SDSA, as very few HJs are formed at non-hotspots in dmc1Δ mutants. Thus, there seems to be a flexibility of DSB repair in S. pombe not seen in S. cerevisiae, in which failure to repair DSBs via IH interaction is not compensated by increased IS repair (Schwacha and Kleckner, 1997), except when the IS-preventing activity of the Hop1-Red1-Mek1 complex is disabled (Niu et al., 2007, 2009). In S. pombe, there are abundant crossovers on each chromosome – about 10, 15, and 20, on chromosomes 3, 2, and 1, respectively (Munz, 1994). Prevention of IS repair may not be needed in S. pombe to ensure a crossover on each homolog pair for their efficient disjunction.
There is additional evidence that partner choice differs in DSB hotspot-rich and -poor intervals. Regions of the S. pombe genome with few detectable DSBs have about as many crossovers per unit physical distance as regions with hotspots (Young et al., 2002; Cromie et al., 2007) (Figures 5C and S6; Table 1). This uniformity of crossover density, arising from a strikingly non-uniform pattern of DSB density, could result either from control of the crossover:non-crossover ratio or from control of partner choice for DSB repair. Since in S. pombe the frequency of crossovers associated with gene conversion, 65 – 80%, is similar at hotspots and in hotspot-poor intervals (Cromie et al., 2005), we favor the second possibility, partner choice.
A Mechanism for Crossover Invariance Stemming from Partner Choice for DSB Repair: Implications for the Global Control of Meiotic DSB Repair and Crossing over
Our data indicate that low-level DSBs are repaired preferentially with the homolog and that high level DSBs, at strong hotspots such as mbs1 and ade6-3049, are repaired preferentially with the sister. This differential use of partner for DSB repair would result in crossover invariance across the genome, i.e., a nearly constant level of crossing over per unit physical distance across the genome (Figure 6). Recent analysis by ChIP-on-chip of genome-wide Rec12-DNA covalent linkages (DSBs) has revealed low-level DSBs between the strong DSB hotspots (Cromie et al., 2007; Hyppa et al., 2008). Collectively, these low-level DSBs may contribute as much meiotic recombination in S. pombe as the strong hotspots do. Strong DSB hotspots are widely spread across the chromosomes of S. pombe, spaced on average about 65 kb apart with weaker DSB sites between them (e.g., see Figures 5C and S6), but there is a nearly uniform frequency of crossovers per kb across the genome. (Young et al., 2002) observed a nearly constant crossover density, ~0.12 cM per kb, in >20 intervals surrounding mbs1, similar to the 0.17 cM/kb genome-wide and 0.12 – 0.19 cM/kb for the six intervals tested here (Table 1B). In the short (4.8 kb) interval centered on the DSB hyper-hotspot mbs1 the density is about six-fold higher (~1.0 cM/kb) (Cromie et al., 2005, 2006), but these crossovers are only about half of the total crossovers in the ura1 – rqh1 interval even though nearly all of the DSBs are in this small interval (Figure 5C). The partner choice we describe here accounts for a more uniform crossover density (crossover invariance) than the strikingly non-uniform pattern of DSBs would predict.
A related but distinct concept of crossover control was introduced by Martini et al., (2006) after they observed in S. cerevisiae that, when DSBs were reduced by certain spo11 non-null mutations, crossovers were not reduced in parallel but rather were maintained near the wild-type level at the expense of non-crossovers. They designated this control “crossover homeostasis” and suggested that it occurs by the same mechanism as crossover interference. Genome-wide analysis of crossovers and gene conversions (a measure of DSBs) confirmed crossover homeostasis in wild-type S. cerevisiae (Chen et al., 2008); zip2 and zip4 mutants are deficient in both crossover homeostasis and crossover interference, suggesting that the two mechanisms are closely related. Since there is no crossover interference in S. pombe (Munz, 1994), Martini et al. (2006) suggested that there would be no crossover homeostasis in S. pombe.
We find that a different type of crossover control – crossover invariance – exists in S. pombe. The two different ways of maintaining a constant level of crossovers may reflect similar biological requirements for meiotic recombination but different patterns of meiotic DSB formation. In both yeasts there is genetically controlled, differential repair of DSBs – with or without crossing over in S. cerevisiae (Martini et al., 2006) and with the sister or with the homolog in S. pombe (this study). The primary role of crossover control in S. cerevisiae may be to help ensure at least one crossover per homolog pair for meiotic homolog disjunction, whereas in S. pombe the primary role may be to enhance recombination and the generation of genetic diversity across the entire genome. The abundant crossing over on each S. pombe chromosome obviates the need for interference, and the high density of strong DSB hotspots on each S. cerevisiae chromosome allows frequent recombination between all genes (Buhler et al., 2007). Thus, these two distantly related yeasts may have adopted different strategies to ensure that meiotic recombination provides both of its two vital functions – aiding homolog disjunction and promoting genetic diversity. These two yeasts may represent the ends of a spectrum of crossover control. Other species may use a mixture of the two types of crossover control to achieve chromosome segregation and genetic diversification during meiosis.
DSB Repair by Interaction with the Sister Chromatid in Other Contexts
Mitotic recombination, perhaps a reflection of DSB repair, is predominantly with the sister chromatid in S. cerevisiae (Kadyk and Hartwell, 1992) and perhaps in other species. IS repair avoids crossovers between homologs, which can have deleterious effects for at least two reasons. First, if the repair is between repeated sequences on different chromosomes, translocations can be produced. Second, if the repair is between allelic positions on the same chromosome, the part of the chromosome centromere-distal to the exchange becomes homozygous in about half of the subsequent cell divisions. Both of these events can uncover recessive phenotypes, leading for example to cell inviability or cancer. Thus, choosing the sister as partner for mitotic DSB repair is important for health. The mechanisms that govern partner choice in S. pombe meiosis described here may apply to mitotic DSB repair as well.
Repair of meiotic DSBs with the sister seems to be rarely considered in discussions of meiotic crossover control, apparently because at the two DSB hotspots tested in S. cerevisiae IH HJs outnumber IS HJs by ~5:1 (Schwacha and Kleckner, 1994, 1997; Allers and Lichten, 2001; Oh et al., 2007; Jessop and Lichten, 2008). As assayed by light microscopy, IH exchanges also appear to outnumber IS exchanges in grasshopper (Tease and Jones, 1979). In S. pombe, however, IS HJs clearly outnumber IH HJs, by ~3:1, at both DSB hotspots tested (Cromie et al., 2006) (Figures 3 and S4). Furthermore, recent evidence indicates that in S. cerevisiae about 1/3 of meiotic DSBs are repaired with the sister chromatid, apparently by a mechanism, such as SDSA, that does not involve detectable HJs (T. Goldfarb and M. Lichten, pers. comm.); DSBs very near centromeres may also be repaired with the sister (Chen et al., 2008). In animals one gender generally has distinct sex chromosomes, e.g., the X and Y chromosomes of mammalian males. Pairing and exchange between these chromosomes is limited to a small pseudoautosomal region, yet DSBs (measured as Rad51 foci) are abundant across these chromosomes (e.g., Ashley et al., 1995). In Caenorhabditis elegans, XO individuals also repair DSBs that arise on the unpaired X (Jaramillo-Lambert and Engebrecht, 2010). Presumably, the DSBs in these cases are repaired with the sister chromatid.
Given the wide diversity of mechanisms of meiotic chromosome behavior in the several species examined (Moens, 1987; Egel, 2008a, b) and even at different loci in the same species (Martini et al., 2006; Table 1; Figures 2 and 5), it is important to consider the possibility that IS repair is common. For example, Rad51 microscopic foci, interpreted as DSBs, outnumber genetic crossovers by ~10:1 in mice (Baudat and de Massy, 2007), ~15:1 in Arabidopsis (Mercier et al., 2005), and ~20:1 in maize (Franklin et al., 1999). This result is usually interpreted to mean that DSBs are repaired by interaction with the homolog but resulting in a non-crossover (e.g., Baudat and de Massy, 2007). An alternative explanation is that most DSBs are repaired by interaction with the sister chromatid, as is the case at DSB hotspots in S. pombe. In Drosophila melanogaster crossing over between sister chromatids, measured as non-disjunction of a heterozygous ring chromosome, occurs in approximately 30% of meioses and even more frequently in certain mutants deficient in sister chromatid cohesion (Webber et al., 2004). Thus, repair of meiotic DSBs with the sister appears to be wide-spread.
Our results encourage further investigations, which may reveal additional, unexpected aspects of the control of meiotic DSB repair and recombination.
EXPERIMENTAL PROCEDURES
S. pombe Culture and Meiosis Conditions
Diploid pat1-114 strains were thermally induced for meiosis and analyzed for DNA content by flow cytometry as described by Cervantes et al. (2000). Meiotic crosses were conducted and analyzed as described by Young et al. (2002).
Gel Electrophoresis and Hybridization Quantification
Cells imbedded in agarose plugs were lysed with enzymes and treated with Proteinase K as described by Cervantes et al. (2000). The DNA was digested with appropriate restriction enzymes and analyzed by gel electrophoresis and Southern blot hybridization as described by Young et al. (2002) and Cromie et al. (2006). The DNA probe for mbs1 is described by Cromie et al. (2006). The DNA probe for ade6 corresponds to bp 31550 – 32778 of cosmid SPCC1322 (GenBank accession no. AL035259.1). The DNA probes to detect HJs in the lys3 – aur1 interval (Figure 5, interval a) and 20 kb to the right of mbs1 (Figure 5, interval b) correspond to bp 573,115 – 574,115 and 787,609 – 788,599 of chromosome I, respectively (GenBank accession no. NC_003424.3)
Branched DNA structures were quantified using a Phosphorimager (GE Healthcare) and ImageQuant TL (Amersham) software. A line was drawn around the replication or recombination DNA structures, as shown in Figure 2 (top row, 2.5 and 4 hr panels), and around the linear (parental) DNA fragments. This represented the signal (“volume”) contributed by each structure. Each outline was copied and placed to the side of the structures in an area of the gel representative of the background, and this value was subtracted from the branched or linear DNA value. The signal of branched DNA (minus background) was then divided by the signal of total DNA (branched plus linears minus their background) to give the fraction of branched DNA at each time-point during meiosis.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Hiroshi Iwasaki for strains; Chad Ellermeier, Joseph Farah, and Emily Higuchi for unpublished recombination data; Michael Lichten and Hiroshi Iwasaki for unpublished information; Gareth Cromie for critical discussions and unpublished DSB data from the mbs1-19 mutant; and Sue Amundsen, Michael Lichten, and Naina Phadnis for helpful comments on the manuscript. This work was supported by research grant GM031693 from the National Institutes of Health to G.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
The Supplemental Data include six figures, one table, and experimental procedures.
REFERENCES
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HE, Farah JA, Grewal SHIS, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genetics. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Smith GR. Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. In: Egel R, Lankenau D-H, editors. Recombination and meiosis: Models, means, and evolution. Springer-Verlag; Berlin: 2008. pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. Recombination and Meiosis: Crossing-over and disjunction. Springer-Verlag; Berlin: 2008a. [Google Scholar]
- Egel R. Recombination and meiosis: Models, means, and evolution. Springer-Verlag; Berlin: 2008b. [Google Scholar]
- Ellermeier C, Schmidt H, Smith GR. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah JA, Cromie GA, Smith GR. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic DNA repair and recombination. Proc Natl Acad Sci USA. 2009;106:9356–9361. doi: 10.1073/pnas.0902793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K, Yoshimi T, Nabeshima K, Yoneki T, Tougan T, Tanaka S, Nojima H. Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 2000;28:2709–2716. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk AL, Kohli J. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics. 2003;165:1031–1043. doi: 10.1093/genetics/165.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51 and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- Hyppa RW, Cromie GA, Smith GR. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 2008;4:e1000267. doi: 10.1371/journal.pgen.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Engebrecht J. A single unpaired and transcriptionally silenced X chromosome locally precludes checkpoint signaling in the Caenorhabditis elegans germ line. Genetics. 2010;184:613–628. doi: 10.1534/genetics.109.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Khasanov FK, Salakhova AF, Khasanova OS, Grishchuk AL, Chepurnaja OV, Korolev VG, Kohli J, Bashkirov VI. Genetic analysis reveals different roles of Schizosaccharomyces pombe sfr1/dds20 in meiotic and mitotic DNA recombination and repair. Curr Genet. 2008;54:197–211. doi: 10.1007/s00294-008-0212-z. [DOI] [PubMed] [Google Scholar]
- Khasanov FK, Savchenko GV, Bashkirova EV, Korolev VG, Heyer W-D, Bashkirov VI. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics. 1999;152:1557–1572. doi: 10.1093/genetics/152.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol Cell Biol. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogue F, Doutriaux MP, Horlow C, Grelon M, et al. Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr Biol. 2005;15:692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. Meiosis. Academic Press; New York: 1987. [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DFR, Vreeken K, Schmidt H, Ostermann K, Clever B, Lohman PHM, Pastink A. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+, and rad22+ genes. Current Genetics. 1997;31:248–254. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octobre G, Lorenz A, Loidl J, Kohli J. The Rad52 homologs Rad22 and Rti1 of Schizosaccharomyces pombe are not essential for meiotic interhomolog recombination, but are required for meiotic intrachromosomal recombination and mating-type-related DNA repair. Genetics. 2008;178:2399–2412. doi: 10.1534/genetics.107.085696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY-H, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing the formation of multi-chromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Kapitza P, Gutz H. Switching genes in Schizosaccharomyces pombe: their influence on cell viability and recombination. Curr Genet. 1987;11:303–308. [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa AH, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hotspot M26. Mol Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Tease C, Jones GH. Analysis of exchanges in differentially stained meiotic chromosomes of Locusta migratoria after BrdU-substitution and FPG staining. II. Sister chromatid exchanges. Chromosoma. 1979;73:75–84. [Google Scholar]
- Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The budding yeast Mei5 and Sae3 proteins act together with Dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y, Khasanov FK, Shinagawa H, Iwasaki H, Bashkirov VI. Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics. 2001;159:91–105. doi: 10.1093/genetics/159.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y, Morishita T, Iwasaki H, Toh H, Shinagawa H. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics. 2000;154:1451–1461. doi: 10.1093/genetics/154.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M, Vreeken K, Zonneveld J, Brandsma J, Lombaerts M, Murray J, Lohman PH, Pastink A. Charcterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mut Res. 2001;461:311–323. doi: 10.1016/s0921-8777(00)00060-4. [DOI] [PubMed] [Google Scholar]
- Webber HA, Howard L, Bickel SE. The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol. 2004;164:819–829. doi: 10.1083/jcb.200310077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.