Abstract
Streptococcus pneumoniae is a common bacterial pathogen that is well-known for its ability to cause acute respiratory disease (pneumonia), ear infections, and other serious illnesses. This Gram-positive bacterium relies on its carbohydrate metabolizing capabilities for full virulence in its host; however, the range of glycan targets that it can attack is presently not fully appreciated. S. pneumoniae is known to have a fucose utilization operon that in the TIGR4 strain plays a role in its virulence. Here we identify a second type of fucose utilization operon that is present in a subset of S. pneumoniae strains, including the serotype 3 strain SP3-BS71. This operon contains a transporter with a solute-binding protein, FcsSBP (Fucose Solute Binding Protein), that interacts tightly (Ka ~1 × 106 M−1) and specifically with soluble A- and B-antigen trisaccharides but displays no selectivity between these two sugars. The structure of the FcsSBP in complex with the A-trisaccharide antigen, determined to 2.35 Å, reveals its mode of binding to the reducing end of this sugar thus highlighting this protein’s requirement for soluble blood-group antigen ligands. Overall, this report exposes a heretofore unknown capability of certain S. pneumoniae strains to transport and potentially metabolize the histo-blood group antigen carbohydrates of its host.
Keywords: Streptococcus pneumoniae, crystal structure, solute-binding protein, blood group antigen, carbohydrate transport
INTRODUCTION
Streptococcus pneumoniae is a capsule forming, Gram-positive bacterium that colonizes the throat and upper respiratory tract of humans. Although S. pneumoniae inhabits 10–40% of humans as a commensal, this opportunistic pathogen frequently takes on a more aggressive role and is the etiological agent of several serious infections, including pneumonia, meningitis, septicaemia and otitis media 1. Recent large-scale signature-tagged mutagenesis studies to identity genes necessary for full virulence of S. pneumoniae in pneumoniae and otitis media models have revealed that full pneumococcal virulence results from the complicated interplay of numerous factors 2; 3; 4. An interesting outcome of these studies is that carbohydrate metabolism pathways, particularly their extracellular components, are critical to pneumococcal virulence (see 5 for a review). Once such carbohydrate utilization pathway that has emerged in importance is dedicated to the harvesting and processing of the sugar fucose 6.
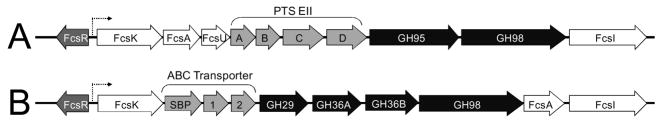
The fucose utilization operon of Streptococcus pnuemoniae was initially described in the TIGR4 strain (serotype 4) 7. This operon comprises eleven open reading frames whose products can be broadly classified as having 4 general functions: operon regulation (fcsR), glycan hydrolysis (gh95 and gh98), carbohydrate transport (eIIA-D), and fucose processing (fcsU, I, K, and A) (Figure 1A). fcsR encodes a repressor protein that controls the expression of the operon. Based on similarity to E. coli genes, fcsU, I, K, and A encode putative mutarotase, isomerase, kinase and aldolase enzymes, respectively. The products of these genes are proposed to act sequentially to process fucose into lactaldehyde and dihyroxyacetone phosphate. Transport of carbohydrates across the membrane is performed by the sugar specific components of a phosphoenolpyruvate-dependent transporter system (PTS) of which EIIA and B represent the phosphorelay components and EIIC and D represent the membrane spanning permease components. The gene products, GH95 and GH98, the latter sometimes referred to as “fuco-lectin related protein”, show substantial amino acid sequence identity to family 95 and family 98 glycoside hydrolases. None of these enzymes have been biochemically characterized; however, we previously demonstrated that non-catalytic carbohydrate-binding modules (CBM) present in the GH98, the only predicted extracellular component of this operon, specifically recognized the LewisY carbohydrate antigen [Fucα1–2Galβ1–4(Fucα1–3)GlcNAc]8.
Figure 1.
The fucose utilization operons of Streptococcus pneumoniae. A) The type 2 fucose utilization operon of Streptococcus pneumoniae TIGR4 comprises eleven open reading frames including a divergently transcribed gene encoding the regulator. Boxed arrows represent genes encoding the regulator (dark grey), a phosphoenolpyruvate-dependent phosphotransferase system (light grey), fucose processing enzymes (white) and hydrolytic glycan processing enzymes (black). B) The organization of the fucose utilization operon in Streptococcus pneumoniae SP3-BS71. The color coding is the same as that used in panel A except that a putative ABC transporter is coloured light grey. In particular, the putative ABC transporter comprises a solute-binding component (SBP) and two permease components (1 and 2).
Expression of the fucose utilization operon in the TIGR4 strain is induced by the presence of fucose yet S. pneumoniae is unable to grow on fucose as a sole carbon source 6; 7. Despite this inability to use fucose as an energy source, in the TIGR4 strain individual disruption of fcsK, eIIA, eIIC, or gh98, or deletion of the entire operon, comprised the bacterium’s ability to cause acute respiratory disease in a mouse model 2; 6. Rather at odds with the apparent importance of this operon to pneumococcal virulence, the comparative genomic hybridization of several S. pneumoniae genomes from serotypes 6 and 10 indicated that the complete fucose utilization operon of S. pneumoniae was not conserved across all of the tested isolates 9. However, a subset of the operon components were present in all of the studied isolates. Considered together, these observations create somewhat of a mystery regarding the structure and composition of this operon, and its role in pathogenesis.
The recent availability of several new pneumococcal genomes, whose sources span a variety of strains and serotypes, has allowed a more thorough examination of the fucose utilization operon. Here we describe the presence of an ABC transporter, which replaces the PTS transporter, in the fucose utilization operon of a serotype 3 strain of S. pneumoniae, SP3-BS71. Biophyscial analysis of the solute-binding protein (SBP), here called FcsSBP, from this transporter provides evidence that S. pneumoniae SP3-BS71 is able to specifically bind and potentially transport blood-group A- and B- antigens. To the best of our knowledge, this represents the first structural and mechanistic insight into how a SBP binds a complex human glycan antigen.
RESULTS AND DISCUSSION
Identification of an ABC transporter in a fucose utilization operon
BLAST 10 searches reveal that the fcsR, I, K, and A genes, which encode the fucose processing enzymes, are present with no less than 98% DNA sequence identity in all of the 23 sequenced strains of S. pneumoniae, indicating that all of these pneumococcal strains have a core component of a fucose utilization operon. Closer examination of this locus revealed that only 19 of the strains posses the same operon, that which is identical in gene composition and organisation to the type described in the S. pneumoniae TIGR4 strain. Inspection of the remaining four strains, including S. pneumoniae SP3-BS71, showed that these strains contain a complete fucose utilization operon that comprises eleven open reading frames (Figure 1B). The organization and content of this undescribed fucose utilization operon, however, are somewhat different from the TIGR4 operon (compare Figure 1A and 1B). One of the most striking differences in this second type of operon is the presence of components of an ABC transporter rather than a PTS transporter.
The fucose utilization operon of the TIGR4 strain employs a PTS to transport extracellular carbohydrates across the membrane and into the cytoplasm 11. Through its phospho-relay system, a phosphate is transferred from phosphoenolpyruvate sequentially via the sugar-independent components to EIIA, then to EIIB and ultimately to the translocated sugar to yield an intracellular phospho-sugar. In contrast, the SP3-BS71 strain has three genes encoding components of a putative ABC transporter (ATP-dependent Binding Cassette), instead of a PTS, is downstream and adjacent to the gene encoding the fuculose kinase (FcsK) (not shown). ABC transporters couple the hydrolysis of ATP to the accumulation of metabolites within the cell and adopt a common ultrastructure consisting of three main components: two membrane spanning permeases that form a translation pore and a cytoplasmic ATPase 12; 13. A SBP is often positioned on the extra-cytosplasmic surface and functions as the specificity determinant of the transporter by sequestering target ligands and delivering them to the mouth of the translation pore 14; 15. In Gram-negative bacteria, the SBP is free floating within the periplasmic compartment, a fundamental difference from Gram-positive bacteria, such as the pneumococcus, where the SBP is tethered to the extracellular membrane by a short N-terminal lipidated anchor or covalently linked to the transporter 16. Within the SP3-BS71 fucose utilization operon, the fcssbp gene is immediately down stream of the fuculose kinase gene (Fig. 1B). Downstream of fcssbp are two contiguous genes (Locus tags: CGSSp3BS71_10428 CGSSp3BS71_10423) (Fig. 1B) that are distantly related to each other (18% identity at the DNA level) and encode proteins with predicted topologies of six and five transmembrane segments, respectively. These components likely comprise the transmembrane permease channel. The third component of the transporter, the ATPase, is traditionally positioned on the cytoplasmic side of the membrane and operates as a functional homodimer. This domain interacts with the transmembrane segments and provides the mechanical energy required for solute transport and accumulation. Interestingly, we were unable to detect the presence of an ATPase within this operon. This is similar to the operon containing the chitobiose transporter of Streptomyces coelicolor, which also appears to lack the ATPase component yet has been demonstrated to transport chitobiose 17. Likewise, the Bifidobacterium longum operon that is dedicated to the transport and metabolism of lacto-N-biose has a carbohydrate transporter with permease and solute-binding protein components but lacks an ATPase 18; 19. These operons, and the fucose utilization operon in S. pneumoniae SP3-BS71, may fulfill the energy requirement of carbohydrate transport by utilizing an ATPase that is encoded within another region of the genome. Support for this concept was recently provided by gene-deletion studies of two carbohydrate-specific ABC transporters from Streptococcus mutans, which demonstrated that other ATPase components within the genome complement the mutated genes and enable the bacterium to fulfill the ATPase requirement of these compromised transporters 20.
Given the placement of the gene encoding FcsSBP in a putative fucose utilization operon we hypothesized that this protein may bind fucose or a fucosylated glycan, a binding specificity not yet observed for a SBP. Furthermore, understanding its specificity may provide insight into the dedicated ligand(s) for the ABC transporter in this pneumococcal fucose utilization operon. To approach this hypothesis we undertook structural and functional studies of the FcsSBP using a cloned and heterologously overproduced soluble form of the protein that lacked its secretion signal peptide and lipid anchoring motif.
The pneumococcal FcsSBP binds blood group antigens
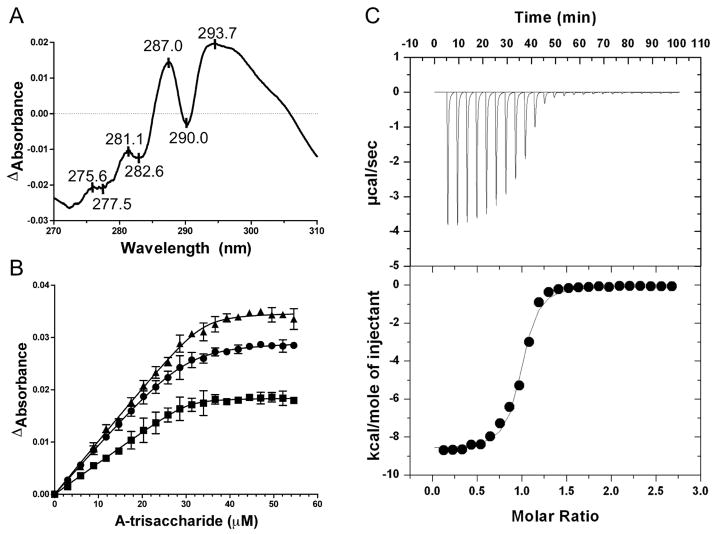
Initially, the binding specificity of FcsSBP was qualitatively investigated by UV difference studies. UV difference scans using recombinant FcsSBP were used to screen a variety of fucose containing glycans (fucose, LewisA, LewisX, LewisB, LewisY, A-antigen, B-antigen, and type 2 H-trisaccharide) that on the basis of their fucose content and presence in human tissues were deemed to be possible substrates for the fucose utilization operon. Only the A-trisaccharide [GalNAcα1–3(Fucα1–2)Gal] and the B-trisaccharide [Galα1–3(Fucα1–2)Gal] revealed a strong UV difference signals that indicated binding to the sugars (Figure 2A). The complex UV difference profile comprising at least four peaks is consistent with the involvement of both tryptophan residues and tyrosine residues in carbohydrate recognition by this protein 21. Synthetic versions of the A-tetrasaccharide [GalNAcα1–3(Fucα1–2)Galβ1–4GlcNAcβ-CH2-CH2-N3] and the B-tetrasaccharide [Galα1–3(Fucα1–2)Galβ1–4GlcNAcβ-CH2-CH2-N3] with blocked reducing ends failed to give a UV difference signal showing a lack of binding to these sugars. Quantitative analysis by UV difference titrations gave an association constant (Ka) of 1.2 (±0.3) × 106 M−1 and a stoichiometry (n) of 1.2 (±0.1) for the A-trisaccharide indicating relatively tight binding and the formation of a 1:1 complex (Figure 2B). Further experiments using this method failed to identify any significant binding to disaccharide fragments of the blood group antigens: GalNAcα1–3Gal (A-determinant), Galα1–3Gal (B-determinant), and Fucα1–2Gal (H-disaccharide).
Figure 2.
The interaction FcsSBP with carbohydrates. A) The UV difference spectrum induced by the binding of the A-trisaccharide to FcsSBP. Peaks and troughs in the spectrum are labelled. B) An isotherm of FcsSBP binding to the A-trisaccharide produced by a UV difference titration. Each curve corresponds to a different peak-to-trough wavelength pair used to measure the change in UV absorbance upon ligand binding: triangles, 287.0-282.6 nm pair; circles, 293.7-290.0 nm pair; and squares, 287.0–290.0 nm pair. Error bars show the standard deviation of triplicate measurements. Solid lines show the fits to a one-site binding module accounting for ligand depletion. C) Representative isothermal titration calorimetry titration for the A-trisaccharide. The upper panel shows the unintegrated data corrected for baseline. The lower panel shows the integrated heats fit to a one-site binding model.
Analysis by isothermal titration calorimetry yielded n values of 1.0 (±0.1) for the A-trisaccharide and 1.2 (±0.1) for the B-trisaccharide. The Kas of 1.01 (±0.16) × 106 M−1 and 1.15 (±0.27) × 106 M−1 for the A- and B-trisaccharides, respectively, were in good agreement with the Ka determined by UV difference and within the range of affinities determined for other SBPs 18; 22; 23; 24. The similar Kas for the distinct ligands also demonstrated that FcsSBP lacks the ability to discriminate between the two carbohydrates (Figure 2C). The ΔH and ΔS values were determined to be −9210 (±540) cal/mole and −3.31 (±1.85) cal/mole/K, respectively, for the A-trisaccharide and −8400 (±460) cal/mole and −0.46 (±0.49) cal/mole/K, respectively, for the B-trisaccharide. The thermodynamic values for the two sugars are similar and reveal a signature of binding that is consistent with the vast majority of protein-carbohydrate interactions 25. Overall, these results reveal the specificity of FcsSBP for the blood group A- and B-antigens. However, the protein displays a surprising requirement for the unadorned trisaccharide antigens and no capacity to bind their disaccharide components. Thus the binding specificity of FcsSBP appears to be limited the intact and soluble trisaccharides.
The structural basis of blood group antigen recognition
The recognition of blood group antigens is unprecedented among solute binding proteins. To investigate the molecular basis of FcsSBP’s specificity the X-ray crystal structure of the protein in complex with the A-trisaccharide was determined to 2.35 Å by optimized single-anomalous dispersion using selenomethionine derivatized protein.
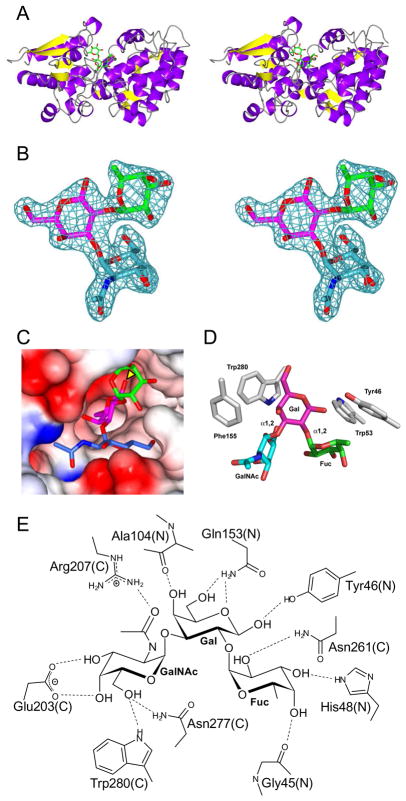
All currently described SBP structures display a similar overall (α/β) fold and operate by a similar mechanism 26; 27. FcsSBP adopts a bimodular conformation, consisting of a smaller N-terminal α/β/α sandwich domain and a larger C-terminal domain possessing a more complicated organization of α-helical and β-strand content, which is typical of all SBPs (Figure 3A). These two globular domains are connected by a flexible hinge region that lacks secondary structure. The general SBP binding mechanism has been described as “Venus-flytrap”-like because the protein exists in two major conformations, an open conformation and a closed conformation with the closed conformation being stabilized by the binding of solute 14. FcsSBP was crystallized in the presence of its A-trisaccharide ligand and adopts an overall conformation that is consistent with the closed form observed for solute binding proteins 22. The FcsSBP did not crystallize in the absence of its ligand suggesting an unliganded conformation that is substantially different from the closed form and likely consistent with the “Venus-flytrap”-like model of binding for other solute binding proteins.
Figure 3.
X-ray crystal structure of the FcsSBP from Streptococcus pneumoniae SP3-BS71. A) The divergent stereo diagram of the FcsSBP structure shows the bilobed structure of the protein and the general location of the ligand binding site. The blood group A-trisaccharide ligand is shown in green stick representation. B) The high quality of the electron density of the bound A-trisaccharide ligand is shown in the divergent stereo maximum-likelihood/σa-weighted Fobs-Fcalc maps 47, 50 (contoured at 3σ = 0.21 e−/Å2) produced by refinement of the FcsSBP structure with the A-trisaccharide coordinates omitted. The electron density is shown as blue mesh while the ligand is shown in stick representation (green, fucose; pink, galactose; and blue/cyan, N-acetylgalactosamine). C) The binding site of the FcsSBP is a deep pocket formed upon the convergence of the N- and C-terminal domains. The reducing end of the sugar (indicated with a yellow arrow) pointing into the binding site. The sugar is color coded as in panel B with the protein shown as a solvent accessible surface colored by electrostatic potential (red, negative; blue, positive). D) The aromatic “cradle” of the active site comprises three sidechains, which are shown in grey stick representation and labelled. The ligand is shown as in panel C. E) Hydrogen bonding schematic of the binding site. The contributions of amino acids from the N- and C-terminal domains are indicated with a N and C respectively.
FcsSBP was crystallized in the presence of the A-antigen trisaccharide and indeed the complete sugar could be easily modelled into unambiguous electron density in the binding site (Figure 3B). The carbohydrate-binding site of FcsSBP is deep pocket at the interface of both the N- and C-terminal domains that almost fully envelopes the ligand (Figure 3C). The reducing end of the trisaccharide ligand is tightly fitted into the base of the binding site with the equatorial O1 of galactose involved in a determinant interaction, clearly indicating that extensions at the reducing end of the central galactose residue would preclude binding. This is consistent with our observation that FcsSBP is unable to bind the synthetic blood group tetrasaccharides, which have an extended and azido-ethyl modified reducing end. Significantly, the determination that FcsSBP must access the reducing end of its target ligands, thereby interacting exclusively with soluble blood group antigen trisaccharides, indicates that the protein functions in a foraging role rather than aiding in the adherence of the bacterium to host tissues.
Three aromatic residues in the FcsSBP active site, Tyr46, Trp53, and Trp280 make classic pyranose ring-aromatic sidechain hydrophobic interactions with both the fucose and galactose residues and provide a contoured foundation for the binding site (Figure 3D). Phe155 is positioned close to Trp280 forms a hydrophobic pocket with Trp280. These four aromatic residues are contributed by both the N-terminus (Try46 and Trp53) and the C-terminus (Phe155 and Trp280), which come together to form a hydrophobic ring that encircles the bound ligand. The involvement of both tryptophan and tyrosine residues is in good agreement with the observed UV difference spectrum induced upon carbohydrate binding, which showed a complicated profile suggestive of overlapping tyrosine and tryptophan signals. A number of putative hydrogen bonds are formed between the binding site and all three residues of the oligosaccharide (Figure 3E). Of particular interest, the determinant galacto-configured axial O4 of the N-acetylglucosamine is in hydrogen bond contact with the carboxylate functional group of Glu203 and the axial O4 of fucose forms a hydrogen bond with the backbone carbonyl of Gly45. In addition to these direct hydrogen bonds an extensive network of water mediated hydrogen bonds is also present (not shown). This constellation of interactions provides to molecular signature for recognizing the stereochemical chemistries of each of the three carbohydrate residues and the overall three-dimensional shape of the branched carbohydrate. In this regard the binding site of the solute binding protein family is emerging as a plastic scaffold for the accommodation of diverse ligands.
The blood group A- and B-antigens are distinguished by their terminal N-acetylgalactosamine (GalNAc) and galactose residues, respectively, which are in turn differentiated by the 2-acetamido group of the GalNAc. Thus, the acetamido group is a potentially important chemical group in the recognition of these antigens. The acetamido group of the A-determinant N-acetylgalactosamine (GalNAc) sits in a water lined pocket where O7 makes a direct hydrogen bond with a Nη2 of Arg207 and a water mediated hydrogen bond with Nη1. N2 of the GalNAc makes a water mediated hydrogen bond with the backbone carbonyl of A104 and Oδ of Asn106. This latter interaction is the only one that is probably conserved with the O2 of the B-determinant galactose. Despite the additional hydrogen bonding and van der Waals interactions afforded by the GalNAc acetamido group, FcsSBP did not display any preference for this sugar over the B-antigen trisaccharide indicating that these interactions make no net contribution to the free energy of binding.
There are four major classes of non-catalytic proteins that can bind carbohydrates: antibodies, lectins, solute-binding proteins (i.e. the periplasmic-binding protein superfamily), and carbohydrate-binding modules (CBMs). Specific members of three of these classes, antibodies 28, lectins 29; 30, and CBMs 31, are able to bind A- and/or B-blood group antigens. We have now demonstrated that the fourth major class of carbohydrate-binding protein also has at least one member that can bind these carbohydrates. However, FcsSBP binds the A- and B-blood group antigens through a different mode than antibodies, lectins, and CBMs. The latter classes of protein, including viral adhesins 32, bind these sugars via their non-reducing end and thus are capable of interacting with antigens displayed on proteins or lipids. Furthermore, only the blood group antigen binding CBM from Clostridium perfringens utilizes all three sugar residues of the A- and B-trisaccharides as recognition determinants 31. In contrast, FcsSBP not only makes extensive interactions will all of the sugar residues in the A- and B-trisaccharides but it specifically recognizes the reducing end of the sugar (Figures 3C and 3E). Thus, FcsSBP can only bind soluble blood group antigens setting it apart from all other examples of blood group antigen binding proteins.
Blood group specific SPBs in other bacteria
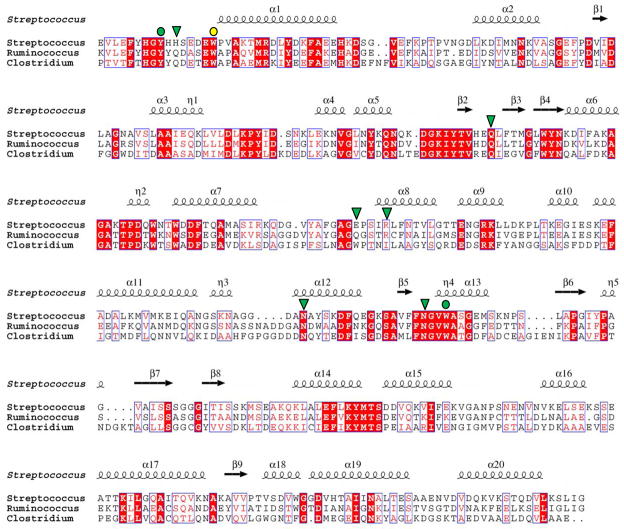
The protein with the highest primary structure identity and whose structure and function is known, RafE from S. pneumoniae, displays only ~20% amino acid sequence identity with FcsSBP. Structural comparison RafE, which is implicated in raffinose transport 33, has a root-mean-squared-deviation with FcsSBP of 3.0 Å over 307 matched Cαs for the ligand bound form of RafE (PDB code 2i58). Thus, though FcsSBP displays a general fold that is shared among SBPs, it appears to be quite distantly related to currently characterized SBPs. Indeed, blood group antigen specificity in a solute binding protein is highly unusual prompting us to ask the question if proteins with similar specificity can be predicted in other organisms. The closest relatives of FcsSBP are the uncharacterized products of open reading frames found in Clostridium scindens (ATCC 35704) and Ruminococcus gnavus (ATCC 29149), which are both found as commensals in the human gut. The putative SBPs from R. gnavus (referred to by its locus tag RUMGNA_03829) and C. scindens (locus tag CLOSCI_02090) have 50% and 33% amino acid sequence identity, respectively, with FcsSBP. The amino acids involved in blood group antigen recognition by FcsSBP are generally well conserved in both RUMGNA_03829 and CLOSCI_02090 (Figure 4). In all but one case, Glu203 in FcsSBP, which is a tryptophan in CLOSCI_02090, non-identical residues are substituted with functionally equivalent amino acids (Figure 4). Importantly, the aromatic residues that form the hydrophobic binding platform are absolutely conserved (Figure 4). Neither SPB from R. gnavus or C. scindens resides in an obvious fucose utilization operon; however, CLOSCI_02090 is proximal in the C. scindens genome to genes annotated as encoding an α-L-fucosidase and a fucose isomerase, while RUMGNA_03829 is near a gene annotated as encoding an α-L-fucosidase. The proximity of RUMGNA_03829 and CLOSCI_02090 to genes encoding fucose active proteins and the similarities in primary structure between these putative SBPs and FcsSBP suggest that these two human gut symbionts, R. gnavus and C. scindens, may transport and process the A/B-blood group antigen of their hosts, such as would be found in the muco-oligosaccharides of the gut. This is in keeping with recent studies of Bacteroides thetaiotaomicron, another gut symbiont, which indicate that upon limiting the host’s intake of dietary polysaccharides that the bacterium then metabolizes the muco-oligosaccharides of the gut lining 34.
Figure 4.
Alignment of FcsSBP with its most closely related proteins in the database. Streptococcus denotes FcsSBP from Streptococcus pneumoniae SP3-BS71, Ruminococcus denotes RUMGNA_03829 from Ruminococcus gnavus (ATCC 29149), Clostridium denotes CLOSCI_02090 from Clostridium scindens (ATCC 35704). The secondary structure of FcsSBP is indicated above the sequence. Residues in FcsSBP that are involved in hydrogen bonding with substrate are indicated above the alignment by green shapes (green triangle, hydrogen bonding only; green circle, apolar and hydrogen bonding interaction). Aromatic residues involved in apolar interactions are indicated by circles (yellow circle, apolar interaction only; green circle, apolar and hydrogen bonding interaction).
Conclusion
Carbohydrate-specific solute binding components of ABC transporters are quite well studied. However, the specificities of solute-binding proteins studied to date have primarily been limited to simple sugars or homo-oligosaccharides originating from storage polysaccharides, plant cell walls or bacterial cell walls. An interesting stand-out example is the solute-binding protein from Bifidobacterium longum, which binds the non-reducing ends of lacto-N-biose and galacto-N-biose linear components of O-linked glycans 18. Unlike the B. longum solute-binding protein, FcsSBP can only recognize soluble ligand thus precluding its participation in tissue adherence. The FcsSBP from S. pneumoniae SP3-BS71 is therefore unique among solute-binding proteins, and indeed all carbohydrate-binding proteins, in its ability to specifically bind the reducing end of soluble blood-group A- and B-antigen trisaccharides.
The specificity of the FcsSBP and its possible role in metabolism provides new evidence of this bacterium’s ability to utilize specific complex host glycans. In particular, this is the first study that demonstrates that the carbohydrate transport machinery of S. pneumoniae is capable of recognizing distinctive carbohydrate antigens, namely the A- and B-blood group antigens, and suggests that this operon in S. pneumoniae SP3-BS71 serves to harvest the fucose from these specific carbohydrates. FcsSBP is only identifiable in 4 of the 23 available S. pneumoniae genomes indicating that the function of this particular operon is not a general feature of the species. Indeed, we suggest that the different transporter and glycoside hydrolases of the fucose utilization operon that is found in the other 19 S. pneumoniae genomes, may enable these strains to target a different, but as yet unknown, fucosylated glycan. Given the link between the pneumococcal fucose utilization operon and the virulence of the bacterium 2; 4; 6, it raises the possibility that there may be a strain-specific relationship between the virulence or colonizing capacity of the pneumococcus and the specific carbohydrate antigens presented by the host.
MATERIAL AND METHODS
Cloning, recombinant SPB production, and FcsSBP purification
FcsSBP is a predicted cell-membrane attached lipoprotein, therefore, to produce it in a soluble form, the FcsSBP gene was engineered to lack the secretion signal peptide and lipoprotein attachment motif. The FcsSBP gene fragment encoding amino acids 24–430 of SPB (CGSSp3BS71_10433) was amplified by PCR from S. pneumoniae SP3-BS71 genomic DNA using the forward primer SBPF1 (5′-TTTGGAGCTAGCGGAACATCTAAGGATGCAAG-3′) and reverse primer SBPR1 (5′-CTTTGTCTCGAGTTAACCAATCAATGATTTC-3′). The DNA fragments were inserted into pET 28 via engineered 5′ NheI and 3′ XhoI restriction sites to generate pSBP. This expression plasmid encodes the module of interest fused to an N-terminal His6-tag via a thrombin protease cleavage site.
pSBP was transformed into BL21 DE3 for protein production. Cultures were grown at 37°C for 5–6 hours then expression was induced by the addition of IPTG to 0.5 mM. Growth was continued at 16 °C overnight. Cells were harvested by centrifugation then disrupted by chemical lysis. Polypeptides were purified from the cell lysate, previously cleared by centrifugation, by immobilized metal affinity chromatography using 2 ml of Ni2+ affinity resin (GE Healthcare). Purified protein was concentrated and buffer exchanged into 20 mM Tris HCl, pH 8.0, in a stirred cell ultrafiltration device with a 5000 MWCO membrane (Millipore). The concentrations of pure FcsSBP preparations were determined by UV absorbance at 280 nm using a calculated extinction coefficient of 49390 M−1cm−1 35.
Binding analysis
Qualitative and quantitative UV difference binding studies were performed and analyzed as described previously using a bimolecular binding model that accounted for ligand depletion and included a term for stoichiometry (n) 21; 36. UV difference titrations were performed in 50 mM Tris HCl, pH 7.5. The protein concentration used, 28.5 μM, was well in excess of the Kd (~1 μM) thus giving C-values 37 approaching 30 and allowing the experimental determination of n in addition to the association constant (Ka).
Isothermal titration calorimetry (ITC) was performed as described previously using a VP-ITC (MicroCal, Northampton, MA) 22; 38. Briefly, protein samples were extensively dialyzed against buffer (50 mM Tris HCl, pH 7.5) then concentrated in a stirred ultrafiltration cell as above. Sugar solutions were prepared by mass in buffer saved from the ultrafiltration step. Both protein and sugar solutions were filtered and degassed immediately prior to use. The protein concentration used was ~50 μM giving C-values in excess of 50. Titrations were performed in triplicate at 25 °C. The data were fit with a single binding site model to determine Ka, n, and ΔH (change in enthalpy). ΔS (change in entropy) was calculated using ΔG=ΔH−TΔS. Errors represent the standard deviations of the triplicate determinations.
Crystallization, data collection and structure solution
Selenomethionine labeled SPB (SeMet- FcsSBP) was produced using previously described procedures 39. Prior to crystallization, SeMet-FcsSBP was further purified by gel filtration chromatography using Sephacryl-200 resin and 20 mM Tris HCl, pH 8.0, as a buffer. Crystals of SeMet- FcsSBP (15 mg/ml) were grown using the hanging-drop vapor diffusion method at 18 °C in 25% PEG 1500, 0.1 M Bis-Tris, pH 6.5, and 0.1 M sodium iodide. Crystals of SeMet- FcsSBP were cryoprotected by a short soak in the crystallization solution supplemented with 25% ethylene glycol and flash cooled in liquid nitrogen. An X-ray diffraction data set was collected at the selenium edge (0.97884 Å) on beamline 9.2 at the SSRL (Stanford, CA) and processed with CrystalClear/d*trek 40. Using anomalous signal and data extending to 3.5 Å ShelxC/D41 was able to find the positions of 18 selenium atoms corresponding to the 9 selenomethionine residues in each of the two SeMet-FcsSBP in the asymmetric unit. Refinement of heavy atom parameters and initial phasing was performed with SHARP 42. PROFESS was then used to determine the NCS operators describing the relative orientations of the two SeMet-FcsSBP molecules in the AU 43. Solvent flattening and NCS averaging with phase extension to 2.35 Å using DM yielded phases that when submitted to ARP/wARP resulted in a model with ~30% of the backbone 44; 45. Two round of successive manual building with COOT 46, refinement with REFMAC 47, solvent flattening with NCS averaging, and resubmission to ARP/wARP ultimately produced phases sufficient for ARP/wARP to build an 80% complete model with docked sidechains. This model was corrected and completed manually using COOT and refined with REFMAC. Final refinement with REFMAC incorporated NCS restraints and inclusion of 10 TLS groups (determined with TLSMD web server 48) per FcsSBP monomer in the AU. Water molecules were added using the REFMAC implementation of ARP/wARP and inspected visually prior to deposition. Five percent of the observations were flagged as “free” and used to monitor refinement procedures 49. All data collection and model statistics are given in Table 1. Coordinates and structure factors have been deposited with the PDB code of 2w7y.
Table 1.
Data collection and structure statistics.
| SeMet-FcsSBP + A-trisaccharide | |
|---|---|
| Data collection | |
| Space Group | P21 |
| Resolution (Å) | 30.00–2.35 (2.48–2.35)* |
| Cell Dimensions | |
| (a, b, c) | 36.9, 104.9, 97.7 |
| (α, β, γ) | 90, 89.95, 90 |
| Rmerge | 0.097 (0.390) |
| I/σI | 22.5 (5.5) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 10.4 (10.4) |
| # Reflections | 323208 |
| # unique reflections | 31126 |
| Refinement | |
| Rwork/Rfree | 0.21/0.26 |
| No. atoms | |
| Protein | 2983 (A) 2983 (B) |
| Ligand/ion atoms | 72 (sugar) 2 (IOD) |
| Water molecules | 410 |
| B-factors | |
| Protein | 23.3 (A) 23.3 (B) |
| Ligand/ion | 17.5 (sugar) 41.2 (IOD) |
| Water | 27.6 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.212 |
| Ramachandran statistics (%) | |
| Preferred | 94.3 |
| allowed | 5.5 |
| outliers | 0.1 |
Highest resolution shell is shown in parenthesis.
Acknowledgments
The authors are very thankful to Dr. Garth Erlich for his kind gift of the SP3-BS71 genomic DNA. We are grateful to Core D of the Consortium for Functional Glycomics for providing the blood group A- and B-tetrasaccharides used in the binding studies. The resources and collaborative efforts provided by The Consortium for Functional Glycomics were funded by NIGMS - GM62116. We are also thankful for the assistance of the beamline 9.2 staff at the SSRL. This work was supported by Canadian Institutes of Health Research (CIHR) grant. MAH is supported by doctoral fellowships from the Natural Sciences and Engineering Research Council of Canada and the Michael Smith Foundation for Health Research (MSFHR). MJB holds a CIHR New Investigator award and is a MSFHR Career Scholar. ABB is a Canada Research Chair in Molecular Interactions and a MSFHR Career Scholar.
References
- 1.Kadioglu A, Andrew PW. The innate immune response to pneumococcal lung infection: the untold story. Trends Immunol. 2004;25:143–9. doi: 10.1016/j.it.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–406. [PMC free article] [PubMed] [Google Scholar]
- 3.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–9. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Ma Y, Yang J, O’Brien CJ, Lee SL, Mazurkiewicz JE, Haataja S, Yan JH, Gao GF, Zhang JR. Genetic requirement for pneumococcal ear infection. PLoS ONE. 2007;3:e2950. doi: 10.1371/journal.pone.0002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008;16:318–25. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embry A, Hinojosa E, Orihuela CJ. Regions of Diversity 8, 9 and 13 contribute to Streptococcus pneumoniae virulence. BMC Microbiol. 2007;7:80. doi: 10.1186/1471-2180-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan PF, O’Dwyer KM, Palmer LM, Ambrad JD, Ingraham KA, So C, Lonetto MA, Biswas S, Rosenberg M, Holmes DJ, Zalacain M. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J Bacteriol. 2003;185:2051–8. doi: 10.1128/JB.185.6.2051-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006 doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
- 9.Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–77. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotrba P, Inui M, Yukawa H. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J Biosci Bioeng. 2001;92:502–17. doi: 10.1263/jbb.92.502. [DOI] [PubMed] [Google Scholar]
- 12.Locher KP. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2004;14:426–31. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Davidson AL, Maloney PC. ABC transporters: how small machines do a big job. Trends Microbiol. 2007;15:448–55. doi: 10.1016/j.tim.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 15.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–21. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 16.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–64. doi: 10.1128/MMBR.00031-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito A, Shinya T, Miyamoto K, Yokoyama T, Kaku H, Minami E, Shibuya N, Tsujibo H, Nagata Y, Ando A, Fujii T, Miyashita K. The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N′-diacetylchitobiose in Streptomyces coelicolor A3(2) Appl Environ Microbiol. 2007;73:3000–8. doi: 10.1128/AEM.02612-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki R, Wada J, Katayama T, Fushinobu S, Wakagi T, Shoun H, Sugimoto H, Tanaka A, Kumagai H, Ashida H, Kitaoka M, Yamamoto K. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283:13165–73. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 19.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99:14422–7. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb AJ, Homer KA, Hosie AH. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J Bacteriol. 2008;190:168–78. doi: 10.1128/JB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boraston AB, Warren RA, Kilburn DG. beta-1,3-Glucan binding by a thermostable carbohydrate-binding module from Thermotoga maritima. Biochemistry. 2001 Dec 4;40(48):14679–85. doi: 10.1021/bi015760g. [DOI] [PubMed] [Google Scholar]
- 22.Abbott DW, Boraston AB. Specific recognition of saturated and 4,5-unsaturated hexuronate sugars by a periplasmic binding protein involved in pectin catabolism. J Mol Biol. 2007;369:759–70. doi: 10.1016/j.jmb.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 23.Nataf Y, Yaron S, Stahl F, Lamed R, Bayer EA, Scheper TH, Sonenshein AL, Shoham Y. Cellodextrin and laminaribiose ABC transporters in Clostridium thermocellum. J Bacteriol. 2009;191:203–9. doi: 10.1128/JB.01190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quiocho FA, Spurlino JC, Rodseth LE. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 25.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine tuning polysaccharide recognition. Biochem J. 2004;382:769–782. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer MA, Hellinga HW. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr Opin Struct Biol. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Medintz IL, Deschamps JR. Maltose-binding protein: a versatile platform for prototyping biosensing. Curr Opin Biotechnol. 2006;17:17–27. doi: 10.1016/j.copbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Patenaude SI, MacKenzie CR, Bilous D, To RJ, Ryan SE, Young NM, Evans SV. Production, crystallization and diffraction to atomic resolution of an antibody Fv specific for the blood-group A oligosaccharide antigen. Acta Crystallogr D Biol Crystallogr. 1998;54:1456–9. doi: 10.1107/s0907444998005824. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni KA, Katiyar S, Surolia A, Vijayan M, Suguna K. Generation of blood group specificity: new insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin. Proteins. 2007;68:762–9. doi: 10.1002/prot.21428. [DOI] [PubMed] [Google Scholar]
- 30.Walser PJ, Haebel PW, Kunzler M, Sargent D, Kues U, Aebi M, Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure. 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Gregg KJ, Finn R, Abbott DW, Boraston AB. Divergent modes of glycan recognition by a new family of carbohydrate-binding modules. J Biol Chem. 2008;283:12604–13. doi: 10.1074/jbc.M709865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81:5949–57. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson NG, Riboldi-Tunnicliffe A, Mitchell TJ, Isaacs NW. Purification, crystallization and preliminary X-ray diffraction analysis of RafE, a sugar-binding lipoprotein from Streptococcus pneumoniae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:676–9. doi: 10.1107/S1744309106021695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–9. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 35.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boraston AB, Ghaffari M, Warren RA, Kilburn DG. Identification and glucan-binding properties of a new carbohydrate-binding module family. Biochem J. 2002;361:35–40. doi: 10.1042/0264-6021:3610035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 38.Ficko-Blean E, Boraston AB. The interaction of carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor. J Biol Chem. 2006 doi: 10.1074/jbc.M606126200. [DOI] [PubMed] [Google Scholar]
- 39.van Bueren AL, Higgins M, Wang D, Burke RD, Boraston AB. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol. 2007;14:76–84. doi: 10.1038/nsmb1187. [DOI] [PubMed] [Google Scholar]
- 40.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55 (Pt 10):1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 41.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 42.Evans G, Bricogne G. Triiodide derivatization and combinatorial counter-ion replacement: two methods for enhancing phasing signal using laboratory Cu Kalpha X-ray equipment. Acta Crystallogr D Biol Crystallogr. 2002;58:976–91. doi: 10.1107/s0907444902005486. [DOI] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 45.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 49.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 50.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst A. 1986;42:140–149. [Google Scholar]