Abstract
Platelets play a critical role in the maintenance of hemostasis as well as in thrombosis and vessel occlusion that underlie stroke and acute coronary syndromes. Anucleate platelets contain messenger RNAs (mRNAs) and are capable of protein synthesis, raising the issue of how these mRNAs are regulated. Here we show that human platelets harbor an abundant and diverse array of microRNAs (miRNAs), which are known as key regulators of mRNA translation. Further analyses revealed that platelets contain Dicer and Argonaute 2 (Ago2) complexes functional in exogenously supplied miRNA precursor (pre-miRNA) processing and the control of specific reporter transcripts, respectively. Detection of the receptor P2Y12 mRNA in Ago2 immunoprecipitates suggests that P2Y12 expression may be subjected to miRNA control in human platelets. Our study lends an additional level of complexity to the control of gene expression in these anucleate elements of the cardiovascular system.
Key regulators of gene expression, miRNAs are short 21- to 24-nucleotide (nt) RNA species expressed in the vast majority of eukaryotes, including humans. Encoded by the genome of nucleated cells, miRNA genes are transcribed into primary miRNAs (pri-miRNAs), which are trimmed into miRNA precursors (pre-miRNAs) by the nuclear ribonuclease (RNase) III Drosha1, acting in concert with the DiGeorge syndrome critical region 8 (DGCR8) protein within the microprocessor complex2–4. After export to the cytoplasm, the resulting ~60- to 70-nt pre-miRNAs are processed by the RNase III Dicer5–7. Assisted by TAR RNA-binding protein 2 (TRBP2)8, Dicer cleaves the stem of pre-miRNA substrates at the base of the loop to generate miRNA:miRNA* duplexes. The mature miRNAs are subsequently incorporated into effector ribonucleoprotein (RNP) complexes containing Argonaute 2 (Ago2)9 and Fragile X mental retardation protein (FMRP)10, guiding the miRNPs for the regulation of specific mRNAs, as reviewed previously11,12. miRNAs regulate mRNA translation through recognition of binding sites of imperfect complementarity, in which pairing of the miRNA nt 2 to 8, or seed region, is critical. Predicted to regulate between 30% to 92% of the genes in human13,14, miRNAs have been shown to control numerous biological processes15, including the megakaryocytic differentiation of CD34+ hematopoietic progenitor cells16.
Released into the blood stream from bone marrow megakaryocytes, circulating blood platelets are central players involved in a variety of pathophysiological conditions, such as cardiovascular diseases (proliferative, thrombotic and occlusive), inflammation and possibly cancer, causing substantial morbidity and mortality. Devoid of a nucleus and lacking genomic DNA, platelets are nevertheless capable of protein synthesis. They were shown to contain rough endoplasmic reticulum and ribosomes17, to incorporate 14C-labeled leucine into proteins18 and to retain a small amount of poly(A)+ RNA from their megakaryocyte progenitor cells19 sufficient to support de novo Bcl-3 (ref. 20) and TxA2 (ref. 21) protein synthesis. In fact, between 15% and 32% of the protein-coding genes are represented in the form of mRNAs in platelets22–24. A strong correlation between transcript abundance and protein expression was observed23,24, supporting the functionality of these platelet transcripts.
Important insights have emerged recently on the regulatory control of gene expression in human platelets, as the maturation of interleukin-1β25 and tissue factor26 mRNAs has been reported to occur through mRNA splicing. Raising important issues on the translational control of the mature mRNAs present in human platelets, these observations prompted us to ask whether circulating platelets harbor a gene regulatory pathway based on miRNAs. Primarily using purified human platelets, we were able to demonstrate the existence of a competent miRNA pathway in these anucleate elements of the cardiovascular system.
RESULTS
Platelets contain an abundant array of miRNAs
Since platelet preparations are often contaminated by leukocytes, and that a single platelet contains ~12,500-fold less mRNA than a nucleated cell27, we first established a procedure that consistently yielded highly purified human platelets. Analysis of our platelet preparations by reverse transcriptase-polymerase chain reaction (RT-PCR) amplification of the leukocyte marker CD45 mRNA, in parallel with that of the platelet-specific gene product glycoprotein IIb (GPIIb), indicated a marked depletion of leukocytes from the starting PRP (Fig. 1a), which was confirmed by hemocytometer counting (Fig. 1b). The level of leukocyte RNA contamination was estimated to <0.4%, yielding a degree of purity sufficient to permit a reliable interpretation of our platelet data and ~30-fold higher than that found not to interfere with platelet RNA profiling analyses28.
Figure 1. Human platelets contain an abundant and diverse array of miRNAs.
(a,b) Characterization of the purified platelet preparations from starting platelet-rich plasma (PRP) by reverse transcriptase-polymerase chain reaction (RT-PCR) (a) and hemocytometer counting (b). (c) Bioanalyzer assessment of RNA samples prepared from purified platelets (left) or megakaryocytes (right). (d) Platelet miRNA profiling analysis. MiRNA probes with signals above the detection threshold are shown in order of increasing relative fluorescence unit (RFI). (e) Northern blot validation of 4 selected miRNAs shown in order of decreasing micro-array RFI. A 10-nt RNA ladder was used as a size marker. cDNA, complementary DNA; GP, glycoprotein; Leuk., leukocyte; nt, nucleotide.
Initial characterization of the total RNA content of platelets suggested a relatively low abundance of mRNA transcripts as compared to megakaryocytes (Fig. 1c) or neutrophils (data not shown). On the other hand, like megakaryocytes, platelets seem to contain a well defined population of small RNAs (Fig. 1c, left panel). Locked nucleic acid (LNA)-based micro-array profiling positively identified more than 170 different miRNAs, or 42.7% of the 398 miRNA probes that could be analyzed, in human platelets (Fig. 1d), covering a range of ~2.5 log in expression levels (Supplementary Table 1). An additional 49 miRNAs, unpublished at the time of analysis, were also positively detected (Supplementary Fig. 1), bringing the number of known platelet miRNAs to 219. Comparative analysis unveiled a subset of miRNAs that were differentially expressed among platelets, megakaryocytes and neutrophils, the majority of which were found to be more abundant in platelets (Supplementary Fig. 2). As for the differential miRNA profile of platelets compared to neutrophils (Supplementary Fig. 3), it supported further the lack of leukocyte contribution to the platelet miRNA signals.
The micro-array profiling data of 4 selected miRNAs, present at various levels in platelets, were confirmed by Northern blot (Fig. 1e). The Northern signals were proportional to the relative micro-array relative fluorescence intensity (RFI), thereby establishing a good correlation between the two hybridization methods. By Northern blotting, we could also exclude the possibility that the signal may originate from the miRNA sequence embedded within pre-miRNA or pri-miRNA precursor molecules. Relatively strong signals corresponding to ~22- to 24-nt mature miRNAs were detected in platelets, as compared to human embryonic kidney 293 (HEK293), HeLa and megakaryocytic cells (Fig. 1e, lower panels), for the 4 selected miRNAs. As inferred by the variety and relative abundance of its miRNA content, platelets may represent the richest source of human miRNAs reported to date.
In the case of the three most abundant miRNAs (miR-223, let-7c and miR-19a), the presence of bands in the ~70-nt range corresponding to pre-miRNA species was more readily detectable, in cells other than platelets, upon a prolonged exposure of the blots (Fig. 1e, upper panels). As for the band slightly above that corresponding to the pre-miRNA species of miR-223 and let-7c, it may be due to modifications, such as uridylation29, though this requires further analysis. Regarding the ~32 to 34-nt miR-223 and let-7c RNA species, it is possible that they represent intermediates containing loop sequences resulting from asymmetrical processing of the pre-miRNA substrate30, although this remains to be tested. This miRNA profile supports a scenario in which pre-miRNAs would serve as a template for platelet miRNA synthesis.
Pre-miRNA conversion into miRNA in platelet extracts
In support to this assertion, we were able to detect, in human platelets, the known protein components of the pre-miRNA processing complex, i.e. Dicer and TRBP2, as well as Ago2, the core component of miRNA effector complexes (Fig. 2a). The intraplatelet localization of Dicer and Ago2 was confirmed by confocal immunofluorescence microscopy (Supplementary Fig. 4). In contrast, the nuclear microprocessor components Drosha (Fig. 2a) and DGCR8 (Fig. 2a and Supplementary Fig. 4) could not be detected in platelets, which is consistent with their anucleate nature.
Figure 2. Platelets can synthesize miRNAs from pre-miRNAs.
(a) Detection of the protein components of the miRNA-guided RNA silencing machinery in megakaryocytes (left) or platelets (right) by Western blot10. β-Actin is used as a control. (b) Assessment of Dicer processing activity in protein extracts and Dicer immunoprecipitates (IP) prepared from purified platelets upon incubation with 32P-labeled human let-7a-3 pre-miRNA30. (c) Extracts from purified platelets were separated by gel filtration on a Superose 6 column and the fractions analyzed by Western blot using anti-Dicer antibody, as described6,10. Selected (odd) fractions were tested for Dicer activity as in b. (d) Immunoblot analysis of TRBP2 IP derived from platelet extracts by using an anti-Dicer antibody. (e) Dicer processing activity assays on IP derived from platelet extracts using anti-TRBP2, control IgG or anti-Dicer antibodies. S10, 10,000 g supernatant fraction.
Next, we assessed whether platelet Dicer is functional and is capable of synthesizing miRNAs from pre-miRNAs in RNase activity assays using 32P-labeled pre-let-7a-3. Incubated in the presence of platelet S10 extracts or Dicer immunoprecipitates (IP), this typical Dicer substrate was converted into miRNA-sized RNA species (Fig. 2b).
To characterize further the miRNA biosynthetic complex of platelets, we analyzed platelet extracts by gel filtration chromatography. These analyses revealed a pre-miRNA processing activity cofractionating with a peak of Dicer corresponding to a protein complex of ~440 kDa (Fig. 2c). Coimmunoprecipitated from human platelets (Fig. 2d), Dicer and TRBP2 formed a complex catalytically active in pre-miRNA processing into miRNA (Fig. 2e). Similar to those obtained in megakaryocytes (Supplementary Fig. 5), these data support the presence of Dicer·TRBP2 complexes in platelets that are capable of miRNA biogenesis.
Platelet miRNAs can mediate RNA silencing
The ability of human platelet miRNAs to mediate sequence-specific gene silencing was next examined in RNA-induced silencing complex (RISC) activity assays. Incubation of platelet extracts in the presence of a 32P-labeled miR-223 sensor transcript, bearing a sequence perfectly complementary to endogenous miR-223, led to the accumulation of a 5′ end labeled 39-nt RNA species (Fig. 3a, left panel). Similar results were obtained when using extracts from megakaryocytes (Supplementary Fig. 6). These data are in agreement with Ago2-mediated cleavage of the sensor sequence 10 nt from the 5′ end of the small RNA guide31. This activity was optimal at 30°C (Fig. 3a, center-left panel), and sensitive to proteinase K (Fig. 3a, center-right panel) and ethylenediaminetetraacetic acid (EDTA) (Fig. 3a, right panel) treatment, further supporting the idea that Mg2+-dependent Ago2 enzyme is the RISC nuclease acting in platelets. Exhibiting an activity proportional to their levels in platelets, the gene silencing properties of 3 additional miRNAs were validated using specific 32P-labeled sensor transcripts in platelet extracts (Fig. 3b).
Figure 3. Platelets harbor Ago2·miRNA effector complexes functionally competent in gene silencing.
(a, b) In vitro RNA-induced silencing complex (RISC) activity assays. (a) Protein extracts were incubated in the presence of a 32P-labeled RNA sensor bearing a binding site complementary to miR-223 without or with proteinase K (Prot. K; 1 mg ml−1) (center-right panel) or EDTA (5 mM) (right panel). (b) Same as in a (left panel), but using sensor RNAs bearing a binding site complementary to let-7c (left panel), miR-19a (center panel) or miR-199a-3p (right panel). (c) Same as in Figure 2c, except using anti-Ago2 antibody for immunoblotting. (d) Pooled fractions were probed for miR-223 (upper panel) and let-7c (lower panel) content by Northern blot. (e) Selected (even) fractions were tested for RISC activity as in a. (f) Ago2 immunoprecipitates (IP) were analyzed by Northern blot for the presence of miR-223. −, indicates IP using control IgG. (g) Ago2 IP were analyzed for RISC activity. (h,i) Same as in a, except in the presence of an antisense RNA to miR-223 (+) or Rluc (−) (h), or upon disrupting pairing of either the miR-223 cleavage site (lane 2) or seed region (lane 3) on the sensor transcript (i). * Indicates the expected cleavage products (38–40 nt). Temp., temperature.
Characterization of the endogenous miRNA effector complexes by gel filtration chromatography unveiled enrichment of relatively low molecular weight fractions in Ago2 and miR-223 (Figs. 3c and 3d), concomitant with a platelet-derived miR-223 sensor cleavage activity (Fig. 3e). Similar findings were obtained in megakaryocytes (Supplementary Fig. 7). Let-7c exhibited a fractionation profile similar to that of miR-223 (Fig. 3c). The composition of platelet miRNA effector complexes may be distinct from human Ago2 complexes of higher molecular weight detected in HeLa cells (Supplementary Fig. 8), which may also contain Dicer and TRBP2 (ref. 8).
Platelets harbor functional Ago2·miRNA complexes
The association between Ago2 and the mature form of endogenous miR-223 was verified by Northern blot analysis of platelet Ago2 IP (Fig. 3f). The implication of platelet Ago2 in gene silencing was confirmed upon detection of RISC activity in Ago2 IP (Fig. 3g), whereas the role of miR-223 in guiding the effector complex was inferred by the inhibitory effects of an antisense RNA to miR-223 in platelet extracts (Fig. 3h) and by disrupting pairing at the cleavage site (Fig. 3i, lane 2) or of the miR-223 seed region (Fig. 3i, lane 3). Together, these data suggest that miRNA-associated Ago2 proteins, in a complex reminiscent of the recombinant RISC32, may form the endogenous miRNA effector complex in platelets.
Ago2·miR-223 complexes may regulate P2Y12 expression
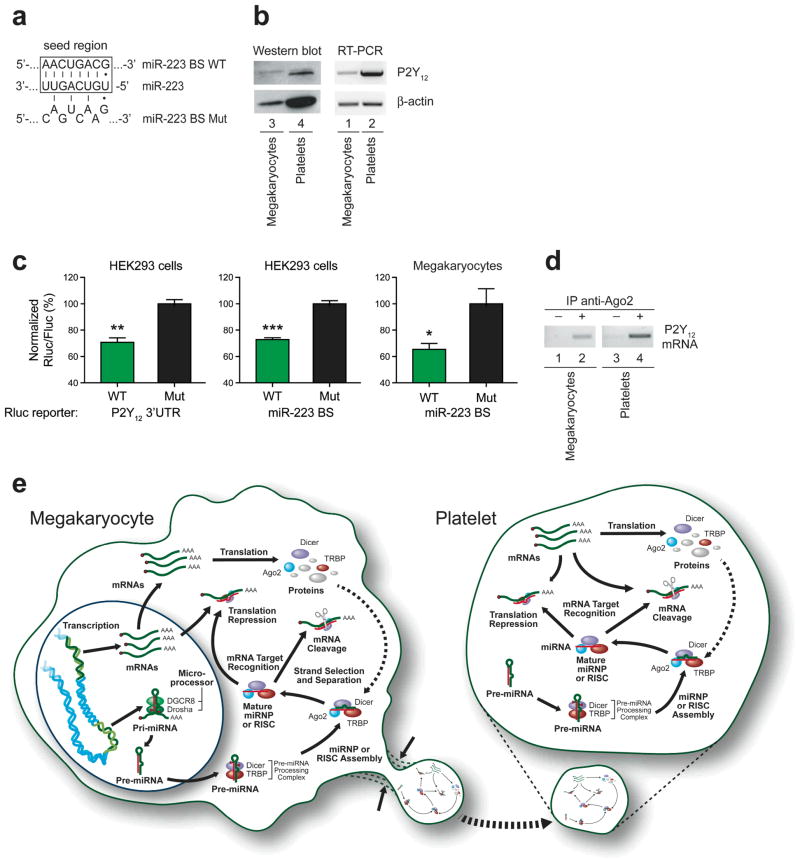
Browsing of the miRBase Target Database identified a putative binding site for miR-223 in the 3′ untranslated region (3′UTR) of the mRNA encoding for P2Y12 (Fig. 4a). This purinergic receptor is known to amplify aggregation induced by all known platelet agonists. With P2Y12 present in platelets at both the protein and mRNA level (Fig. 4b), our next aim was to validate the functionality of the predicted miR-223 binding site in the context of the full-length P2Y12 mRNA 3′UTR. For that purpose, we employed a reporter gene activity assay that involved coexpression of pre-miR-223 with a reporter construct containing P2Y12 3′UTR, either wild-type or mutated in the miR-223 binding site seed region, as depicted in Figure 4a, in the HEK293 cell line which does not express miR-223 (see Figure 1e, left panel). In these experiments, disruption of miR-223 pairing to the P2Y12 3′UTR impaired its ability to repress gene expression (Fig. 4c, left panel), implying a role for miR-223 in regulating P2Y12 expression. Similar results were obtained through the use of reporter constructs in which the Rluc gene is placed under the control of the isolated miR-223 binding site, either wild-type or mutated in its seed region, in HEK293 cells (Fig. 4c, center panel) as well as in megakaryocytes (Fig. 4c, right panel). These latter findings ascertained the ability of miR-223 to regulate expression of mRNAs harboring a miR-223 regulatory element in platelet precursor cells.
Figure 4. Ago2·miR-223 complexes may regulate P2Y12 mRNA expression in platelets.
(a) Base pairing of the miR-223 seed region to its predicted binding site, wild-type (WT) or mutated (Mut), in the 3′ untranslated region (3′UTR) of P2Y12 messenger RNA (mRNA). (b) Detection of P2Y12 protein (left) and mRNA (right) in extracts of platelets and megakaryocytes by Western blot and RT-PCR, respectively. (c) Functional validation of the predicted binding site for miR-223 present in P2Y12 mRNA 3′UTR by reporter gene assays performed in HEK293 cells (left and center panels) or megakaryocytes (right panel). Results are expressed as mean ± standard error of the mean (s.e.m.) (n = 5 to 7 experiments, in duplicate). * p<0.05; **p<0.001; *** p<0.0001 (two-tailed, unpaired Student’s t-test). (d) Detection of P2Y12 mRNA in Ago2 immunoprecipitates (IP) derived from either megakaryocyte or platelet extracts by RT-PCR. (e) Proposed model for platelet inheritance of a functional miRNA pathway, devoid of its initiation step, from megakaryocyte precursor cells. BS, binding site; miRNP, miRNA-containing ribonucleoprotein complex; RISC, RNA-induced silencing complex.
A role for miRNAs, such as miR-223, in regulating P2Y12 expression is further supported by the presence of platelet P2Y12 mRNA in Ago2 IP (Fig. 4d), which constitutes an established biochemical approach for the identification of miRNA targets in human cells33. P2Y12 mRNA may thus serve as a template for de novo P2Y12 protein synthesis in a process that may be regulated by Ago2·miR-223 complexes within human platelets.
DISCUSSION
In the present study, we have established the existence and functionality of a gene regulatory process based on miRNAs in human platelets. Harboring a small, but diverse transcriptome that represents up to a third of all protein-coding genes in human24, these anucleate elements of the cardiovascular system contain abundant quantities of miRNAs. Our findings confirm previous studies reporting the presence of miRNAs in PRP samples, which may be prone to leukocyte contamination, prepared either from healthy donors34 or patients with polycythemia vera35. This translates into a markedly increased miRNA/mRNA ratio versus nucleated cells of the hematopoietic system. Considering that the regulatory effect of miRNAs is dependent on the number of miRNA binding sites occupied in a given mRNA target, it is tempting to speculate that the few platelet mRNA transcripts are decorated with numerous Ago2·miRNA complexes, and may thus be repressed more strongly than in other cells.
In support of this scheme, SAGE analyses revealed that the average length of platelet mRNA 3′UTRs is much longer (1,047 nt) than nucleated cells (492 nt), with a large margin to the second-ranking fibroblasts (681 nt)36. An attractive explanation would be that a system that progressively loses its ability for transcriptional regulation, such as that occurring during platelet maturation from megakaryocytic precursor cells, could compensate this deficiency by enhancing its capacity for translational control and mRNA stability36. As such, it is relevant to note that the regulation of the bearded (Brd) box (consensus sequence: AGCUUUA)37, which likely involves the formation of RNA-RNA duplexes with complementary sequences found at the 5′ end of certain miRNAs37, has been found to be enriched in platelet transcripts36.
Regarding the origin of platelet miRNAs, the possibility that they derive from pri-miRNA transcripts within platelets is highly improbable, considering the absence of a nucleus and that of the nuclear components Drosha and DGCR8 required for their maturation. Encoded by the genome of its megakaryocytic precursor cells, pri-miRNAs are more likely to be converted into pre-miRNAs prior to platelet formation. Whether pre-miRNAs are actively processed into miRNAs in megakaryocytes and/or platelets remains unclear. The presence of detectable levels of pre-miRNA species and that of a Dicer·TRBP2 complex functionally competent in miRNA biosynthesis in platelets support the latter possibility, although its significance remains to be determined. On the other hand, the mature miRNA species are much more abundant than their pre-miRNA counterparts, suggesting that a large proportion of the mature miRNA content of platelets may have been inherited directly from megakaryocytes. Nevertheless, both Drosha and Dicer complexes are believed to be involved in the generation of platelet miRNAs, as implied by the overrepresentation of a U (48%) at the 5′ extremity of these miRNA sequences. This is in accordance with the preference of these RNase III enzymes to cleave on the 5′ side of a U.
It has been reported previously that mature miRNAs are bound by Ago proteins38 and that Ago2 is a crucial component of RNA silencing effector complexes in human cells9. The presence of miRNAs in relatively low molecular weight complexes, cofractionating and coimmunoprecipitating with Ago2 and sequence-specific RISC activity, attest to their likely functionality within miRNP complexes in platelets. While smaller than effector miRNPs of nucleated HeLa cells, the estimated molecular mass of platelet miRNP complexes could not be attributed to the dissociation of a higher molecular weight complex, and is instead compatible with a complex formed of Ago2·miRNA. The sequence- and site-specific processing of target RNAs in our in vitro cleavage assays imply that the endonucleotidic activity of Ago2 is guided by an associated miRNA. Since other groups have established the relevance of using Ago2 IP as a biochemical approach for the identification of miRNA-targeted mRNAs33,39,40, we opted for this methodology in order to circumvent the intrinsic limitations associated with working with primary human platelets, including their refractoriness to transfection that prevents the use and introduction of inhibitory (eg, 2′O-Me antisense oligonucleotides, short hairpin RNA, antagomirs) or stimulatory/mimicking (eg, vector expressing miRNA precursors, synthetic miRNA precursors) molecules traditionally used to identify miRNA targets in more amenable, tractable cellular models.
Using this approach, we identified P2Y12 mRNA in association with Ago2·miRNA complexes in platelets. Recognized by adenosine diphosphate (ADP), an important platelet agonist in vivo, P2Y12 is a 7-transmembrane domain receptor coupled to Gi2 protein that mediates a number of biological processes, such as platelet aggregation, granule secretion, and thrombus growth and stability41. The experimental validation of the predicted binding site for miR-223 in its natural 3′UTR context in P2Y12 mRNA, which is the most abundant platelet receptor for extracellular nucleotides (P2 receptors) expressed at the mRNA level42, and the demonstrated ability of miR-223 to regulate gene expression in the context of platelet precursor cells support further the concept that P2Y12 expression could be regulated by miRNAs, like miR-223, in human platelets. According to miRBase (http://microrna.sanger.ac.uk/targets/v5/), at least four additional miRNAs are predicted to target the 3′UTR of P2Y12 mRNA: let-7i, miR-21, miR-221 and let-7g. It is therefore possible that miRNAs act coordinately and/or in synergy to regulate P2Y12 mRNA translation via its 3′UTR, in which case they could play an important role in modulating platelet function.
In view of our data, we propose a model where platelets inherit of a partial, yet functional, miRNA pathway devoid of its nuclear initiation steps through entrapment of the cytoplasmic protein and RNA components of the miRNA machinery during their formation and release from megakaryocyte progenitor cells (Fig. 4e). The similarities that we observed between the pre-miRNA processing and effector complexes present in megakaryocytes and platelets are supportive of that possibility. In addition, we hypothesize that the miRNA-mediated repression of platelet mRNAs may be lifted upon activation, leading to mRNA translation and protein synthesis, thereby conferring to platelets the ability to respond to physiological stimuli and/or conditions. Considering that activated platelets have been shown to release microvesicles that may exert an extracellular function43, it is also tempting to speculate that platelet-derived microparticles (or exosomes) may act as a delivery system for mRNAs44 as well as miRNAs within the cardiovascular system.
Representing a new class of potential regulatory molecules in platelets, miRNAs lend an additional level of complexity to the control of gene expression in these anucleate elements. Finely tuning expression of specific gene products that are likely involved in governing platelet reactivity, a dysfunctional miRNA-based regulatory system could lead to the development of serious platelet-related cardiovascular diseases. Our study offers a new perspective to the etiology and therapeutic modulation of platelet function in human diseases.
METHODS
Platelet purification
We obtained venous blood from healthy volunteers, using sodium citrate (100 mM) as anticoagulant, and centrifuged (250 g, 20 min, at room temperature) to obtain the PRP. The PRP was then subjected to several rounds of leukocyte filtration, followed by negative selection using CD45+ beads (EasySep, StemCell Technologies), as previously described25. We characterized the purified platelet preparations by RT-PCR amplification of GPIIb and CD45 mRNAs, and by hemocytometer counting.
Cell culture
We maintained the megakaryocytic cell line MEG-01 (ATCC, CRL-2021) in Roswell Park Memorial Institute (RPMI) 1640 medium adjusted to contain 10% (v/v) fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units ml−1 penicillin and 100 μg ml−1 streptomycin in a humidified incubator under 5% (v/v) CO2 at 37°C. HEK293 and HeLa cells were grown under the same conditions, but in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units ml−1 penicillin, and 100 μg ml−1 streptomycin.
Protein analysis
We analyzed protein extracts by 7% (w/v) SDS-polyacrylamide gel elecrophoresis (PAGE) and immunoblotting followed by enhanced chemiluminescence (ECL) detection6,10, using anti-Drosha (Millipore), anti-DGCR8 (Protein Tech Group, Inc.), anti-Dicer6, anti-TRBP2 (Supplementary Fig. 9a), anti-Ago2 (Abnova) and anti-β-actin AC-40 (Sigma) antibodies. The anti-P2Y12 antibody was from Sigma. IP were prepared by using anti-Dicer6 (Supplementary Fig. 9b), anti-TRBP2, anti-Ago2 antibodies or control IgG (Santa Cruz Biotechnology, Inc.) and protein G agarose beads (Roche).
RNA extraction and analysis
Total RNA was extracted using Trizol (Invitrogen). We performed RT-PCR using gene-specific oligonucleotides (designed with GeneFisher available at: http://bibiserv.techfak.uni-bielefeld.de/genefisher2/), followed by agarose gel electrophoresis and ethidium bromide staining. For the detection of miRNA species, RNA was separated on a 12% (w/v) polyacrylamide gel containing 7 M urea, transferred to a nylon membrane followed by detection using randomly, 32P-labeled RNA probes complementary to the mature miRNA species of miR-223, let-7c, miR-19a or miR-199a-3p following a Northern blot procedure improved for the detection of small RNAs45.
miRNA profiling analysis
Total RNA was prepared using Trizol, subjected to RNeasy purification (Qiagen) and analyzed qualitatively using a Bioanalyzer 2100 (Agilent). The RNA samples were labeled using the miRCURY Hy3/Hy5 labeling kit and hybridized on the miRCURY LNA Array v.8.1 (Exiqon). Spike-in controls were added at various concentrations in both Hy3 and Hy5 labeling reactions to evaluate the labeling reaction, hybridization and performance of the array experiment. We obtained strong correlations (0.993 to 0.9995) for both Hy3 and Hy5 channels among the different slides. For each miRNA capture probe, the mean signal from 4 replicate spots were averaged. MiRNA detection was considered as positive when the calculated RFI for a given miRNA capture probe was above the detection threshold, which was conservatively set at 2 times the highest mean background signal observed in the array experiment.
Gel filtration chromatography
Cleared protein extracts (100,000 g × 45 min supernatant; S100) of human platelets resuspended in lysis buffer [50 mM Tris–HCl, 137 mM NaCl, 1% (v/v) Triton X-100, 1 mM phenylmethanesulfonyl fluoride, complete protease inhibitor cocktail (Roche), pH 8.0] and filtered through a 0.22 μm filter (Millipore) were separated by gel filtration on a Superose 6 column (10/300 GL) using an ÄKTA FPLC system (GE Healthcare). We monitored protein fractionation during elution by detection at 280 nm and analyzed the protein content of selected fractions by Western blot6,10.
Dicer activity assays
We assessed Dicer processing activity of cleared protein extracts (10,000 g × 15 min supernatant; S10), IP or FPLC fractions upon incubation with randomly 32P-labeled human let-7a-3 pre-miRNA (5′-UGAGGUAGUAGGUUGUAUAGUUUGGGGCUCUGCCCUGCUAUGGGAUAACUAUACAAUCUACUGUCUUUCC-3′; miRBase accession number MI0000062) and analysis by denaturing PAGE and autoradiography, as described30.
RISC activity assays
We prepared sensor RNA transcripts harboring binding sites complementary to endogenous miRNAs hsa-miR-223, hsa-let-7c, hsa-miR-19a and hsa-miR-199a-3p by in vitro transcription using T7 RNA promoter/polymerase (T7 MEGA shortscript kit, Ambion). We generated DNA templates by PCR filling of the T7 promoter oligonucleotide (T7p Fwd) annealed to miRNA binding site-specific oligonucleotides containing the reverse (Rev) sequence for T7p placed at their 3′ end. After gel purification, the RNA sensors were dephosphorylated with calf intestine alkaline phosphatase (Roche), 5′ end labeled with [γ-32P] ATP (Perkin-Elmer) using opti-kinase (USB) and purified by denaturing PAGE. The Open/Open sensor RNA transcript was detailed previously31.
We performed RISC activity assays essentially as reported31,46. Briefly, we prepared protein extracts from human platelets or megakaryocytes resuspended in lysis buffer (40 mM HEPES, 100 mM potassium acetate, 5 mM MgCl2, 2 mM dithiothreitol (DTT), 0.35% (v/v) Triton X-100, pH 7.6). Fifty μg of S100 protein extracts, 50-μl aliquots of FPLC fractions or Ago2 IP were incubated with the 32P-labeled RNA sensor (10,000 cpm) in assay buffer containing 20 mM HEPES, 50 mM potassium acetate, 2.5 mM MgCl2, 1 mM ATP, 0.2 mM GTP, 1 mM DTT, 2.5% (v/v) Superase·In, 0.18% (v/v) Triton X-100, pH 7.6 at 30°C for 90 min. Some experiments were performed at 4°C or in the presence of proteinase K (1 mg ml−1) or EDTA (5 mM), or involved a 30-min pre-incubation in the presence of an antisense RNA to miR-223. The reaction was stopped by adding 0.5 mg ml−1 proteinase K and incubating at 50°C for 30 min. After a phenol/chloroform extraction step, the RNA products were precipitated, resuspended in water, separated by 8% PAGE/7 M urea and analyzed by autoradiography.
RNA silencing assays
We plated HEK293 cells or MEG-01 megakaryocytes (2 × 105) in 24-well plates for cotransfection with a pre-miR-223 expression vector (500 ng DNA) and a reporter construct containing the P2Y12 3′UTR or isolated miR-223 binding site, either wild-type (WT) or mutated (Mut) in its seed region, fused downstream of the Rluc reporter gene (50 or 100 ng DNA) using Lipofectamine 2000 (Invitrogen). The psiSTRIKE and psiCHECK vectors used to prepare the pre-miR-223 and reporter gene constructs were obtained from Promega. psiNeg vector10 encodes a short hairpin RNA (shRNA) directed against a deleted region in Rluc mRNA and was used as control. Twenty-four h later, cells were harvested, lysates were prepared, and Renilla luciferase (Rluc) and Firefly luciferase (Fluc) activities were measured essentially as described10.
Supplementary Material
Acknowledgments
We thank Serge Picard (Université Laval) for providing the PRP, Keith Gull (University of Oxford) for the gift of anti-TAT1 antibody and the CHUL Research Center Computer Graphics Department for the illustrations. M.P.P. was supported by a doctoral studentship from Natural Sciences and Engineering Research Council of Canada. G.R. and P.P. are Senior Scholars from the Fonds de la Recherche en Santé du Québec (FRSQ). This work was supported by a Cardiovascular Research Award from Pfizer Canada Inc. and a CIHR/Rx&D Collaborative Research Grant (IRO-86239) to P.P.
Footnotes
Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
P.P. conceived and coordinated the study; P.L., G.R. and P.P. designed and planned the experiments; P.L. led the project; P.L., I.P., D.L.O. and M.P.P. performed the experiments; all authors analyzed the data and edited/commented on the manuscript; P.P. wrote the manuscript.
AUTHOR INFORMATION
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
References
- 1.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 2.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 4.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Provost P, et al. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, et al. Argonaute 2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 10.Plante I, et al. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 12.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 13.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Ouellet DL, Perron MP, Gobeil L-A, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotech. 2006 doi: 10.1155/JBB/2006/69616. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–83. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ts’ao CH. Rough endoplasmic reticulum and ribosomes in blood platelets. Scand J Haematol. 1971;8:134–40. doi: 10.1111/j.1600-0609.1971.tb01964.x. [DOI] [PubMed] [Google Scholar]
- 18.Warshaw AL, Laster L, Shulman NR. Protein synthesis by human platelets. J Biol Chem. 1967;242:2094–7. [PubMed] [Google Scholar]
- 19.Roth GJ, Hickey MJ, Chung DW, Hickstein DD. Circulating human blood platelets retain appreciable amounts of poly (A)+ RNA. Biochem Biophys Res Commun. 1989;160:705–10. doi: 10.1016/0006-291x(89)92490-x. [DOI] [PubMed] [Google Scholar]
- 20.Weyrich AS, et al. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc Natl Acad Sci U S A. 1998;95:5556–61. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evangelista V, et al. De novo synthesis of cyclooxygenase-1 counteracts the suppression of platelet thromboxane biosynthesis by aspirin. Circ Res. 2006;98:593–5. doi: 10.1161/01.RES.0000214553.37930.3e. [DOI] [PubMed] [Google Scholar]
- 22.Bugert P, Dugrillon A, Gunaydin A, Eichler H, Kluter H. Messenger RNA profiling of human platelets by microarray hybridization. Thromb Haemost. 2003;90:738–48. [PubMed] [Google Scholar]
- 23.Gnatenko DV, et al. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–93. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 24.McRedmond JP, et al. Integration of proteomics and genomics in platelets: a profile of platelet proteins and platelet-specific genes. Mol Cell Proteomics. 2004;3:133–44. doi: 10.1074/mcp.M300063-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Denis MM, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–91. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwertz H, et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med. 2006;203:2433–40. doi: 10.1084/jem.20061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink L, et al. Characterization of platelet-specific mRNA by real-time PCR after laser-assisted microdissection. Thromb Haemost. 2003;90:749–56. doi: 10.1160/TH03-02-0095. [DOI] [PubMed] [Google Scholar]
- 28.Rolf N, Knoefler R, Suttorp M, Kluter H, Bugert P. Optimized procedure for platelet RNA profiling from blood samples with limited platelet numbers. Clin Chem. 2005;51:1078–80. doi: 10.1373/clinchem.2005.049486. [DOI] [PubMed] [Google Scholar]
- 29.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Ouellet DL, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–12. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 32.Rivas FV, et al. Purified Argonaute 2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 33.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 34.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–10. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 35.Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93:1009–16. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- 36.Dittrich M, et al. Analysis of SAGE data in human platelets: features of the transcriptome in an anucleate cell. Thromb Haemost. 2006;95:643–51. [PubMed] [Google Scholar]
- 37.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 38.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 39.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. Rna. 2007;13:1198–204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–6. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahner BN, Shankar H, Murugappan S, Prasad GL, Kunapuli SP. Nucleotide receptor signaling in platelets. J Thromb Haemost. 2006;4:2317–26. doi: 10.1111/j.1538-7836.2006.02192.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, et al. Quantification of ADP and ATP receptor expression in human platelets. J Thromb Haemost. 2003;1:330–6. doi: 10.1046/j.1538-7836.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 43.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–9. [PubMed] [Google Scholar]
- 44.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 45.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–7. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 47.Provost P, et al. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J. 2001;359:255–63. doi: 10.1042/0264-6021:3590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woods A, et al. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93 (Pt 3):491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.