Abstract
Despite numerous reports about the acute and sub-chronic toxicities caused by MDMA (3,4-methylenedioxymethamphetamine, ecstasy), the underlying mechanism of organ damage is poorly understood. The aim of this review is to present an update of the mechanistic studies on MDMA-mediated organ damage partly caused by increased oxidative/nitrosative stress. Because of the extensive reviews on MDMA-mediated oxidative stress and tissue damage, we specifically focus on the mechanisms and consequences of oxidative-modifications of mitochondrial proteins, leading to mitochondrial dysfunction. We briefly describe a method to systematically identify oxidatively-modified mitochondrial proteins in control and MDMA-exposed rats by using biotin-N-maleimide (biotin-NM) as a sensitive probe for oxidized proteins. We also describe various applications and advantages of this Cys-targeted proteomics method and alternative approaches to overcome potential limitations of this method in studying oxidized proteins from MDMA-exposed tissues. Finally we discuss the mechanism of synergistic drug-interaction between MDMA and other abused substances including alcohol (ethanol) as well as application of this redox-based proteomics method in translational studies for developing effective preventive and therapeutic agents against MDMA-induced organ damage.
Keywords: MDMA, metabolism, oxidative/nitrosative stress, protein oxidation, functional redox proteomics, mitochondrial dysfunction, organ damage
1. INTRODUCTION
Recent epidemiological studies indicate that 3,4-methylenedioxymethamphetamine (MDMA, Ecstasy)1, a synthetic derivative of amphetamine with a range of psychotropic actions, is widely abused by mostly young people during rave parties (large social gatherings characterized by dancing and loud music). According to a United Nations report, the use of MDMA has reached epidemic proportions in the United States as well as in Europe [1–3]. Repeated usages of MDMA pose significant public health and social problems in the United States and other countries [1–3]. In fact, acute or sub-chronic exposure to MDMA alone or in combination with other abused substances (e.g., alcohol, cocaine, cannabis) can damage several organs such as the heart, liver, kidney, and brain, potentially leading to death, as evidenced by multiple deaths related to MDMA abuse [4–7]. Acute MDMA toxicity can lead to myocardial infarction and arrhythmia often accompanied with tachycardia, hypertension, and hyperthermia, all of which usually precede disseminated intravascular coagulation, rhabdomyolysis, and multiple organ failure or death [8, 9]. The life threatening clinical manifestations of MDMA toxicity also include acute hepatic damage [10–12], hyponatremia, and rhabdomyolysis-induced renal failure [13–15]. In addition, MDMA is known to interfere with endocrine, gonadal, and immune functions [16–18]. In the brain, MDMA was shown to deplete serotonergic neurotransmitter and cause neurodegeneration through serotonin transporter action, nitric oxide and peroxynitrite, and formation of neurotoxic MDMA metabolites [19–21].
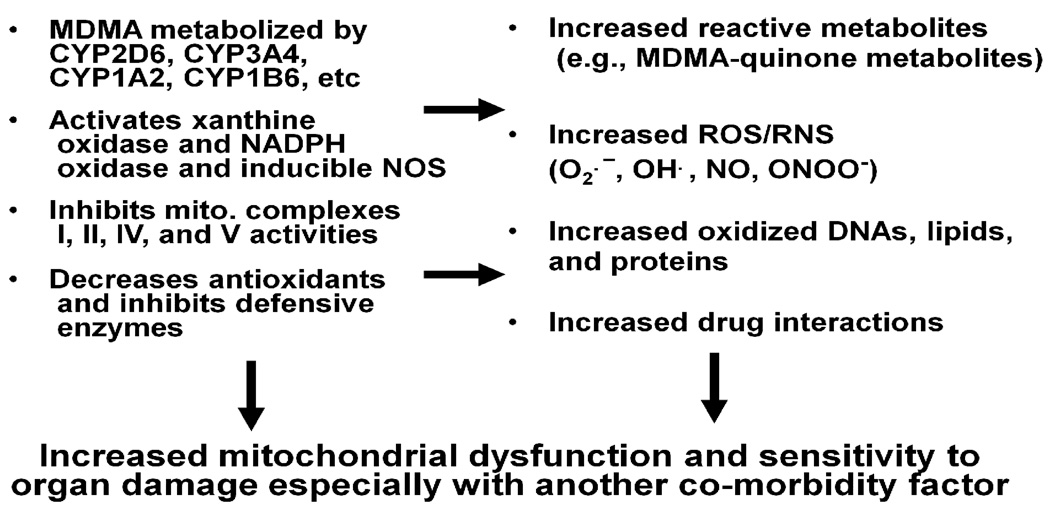
Although MDMA-mediated toxicities are well-established, the general public does not seem to fully appreciate the adverse consequences associated with MDMA usage. In fact, MDMA is a major cause of liver injury in people under the age of 25 years [12]. All these points suggest an urgent need to educate the public about the toxicities of MDMA. Although many reports have demonstrated MDMA-induced organ damage [11–13], the underlying mechanism accounting for tissue toxicity is poorly understood. Various factors contribute to the extent of MDMA-induced injury to different tissues. For instance, MDMA metabolism [22], the increased efflux of neurotransmitters [22, 23], hyperthermia [24], increased oxidative/nitrosative stress, and the oxidation of catecholamines and various proteins [25, 26] are suggested to be involved in MDMA-related neuronal injury. Several reports indicate that reactive quinone metabolites of MDMA contribute to MDMA-mediated toxicities [22, 27]. The metabolism of MDMA involves N-demethylation to 3,4-methylenedioxyamphetamine while both MDMA and 3,4-methylenedioxyamphetamine are demethylated to catecholamines [N-methyl-α-methyldopamine and α-methyldopamine, respectively] that can undergo oxidation to corresponding quinones. Quinones are highly redox-active molecules that can undergo the following pathways: (a) a redox cycle producing semiquinones radicals, leading to the generation of reactive oxygen species (ROS) [28, 29]; (b) irreversible 1,4-intramolecular cyclization with subsequent formation of aminochromes [25]; (c) conjugation with reduced glutathione (GSH) to form a glutathionyl adduct that can further react with GSH and protein thiols, leading to GSH depletion [27, 30]; and d) formation of MDMA-protein adducts, leading to inactivation of the target proteins [31]. Taken together, these results indicate that MDMA metabolism with increased production of ROS and/or toxic oxidation products coincident with GSH depletion may be responsible for liver damage. It is also believed that other tissues including brain, heart and kidney would be affected in a similar mechanism following the metabolism of MDMA [32, 33]. Since antioxidant defense systems become severely impaired upon MDMA exposure, administration of small molecule antioxidants such as N-acetylcysteine and ascorbic acid or over-expression of antioxidant enzymes like superoxide dismutase, can potentially neutralize the toxic effects of MDMA [27, 34]. Depletion of GSH levels following MDMA exposure correlates with increased oxidative stress accompanied with lipid peroxidation, DNA damage, protein oxidation and cell damage [27, 29, 35, 36]. Furthermore, it was shown that reactive nitrogen species (RNS) including nitric oxide (NO) and peroxynitrite (ONOO−) are also involved in MDMA-mediated toxicity [19, 37–40]. The overall processes for increased production of MDMA-induced oxidative/nitrosative stress and functional consequences are summarized in Fig. (1).
Fig. 1.
MDMA-induced oxidative/nitrosative stress and functional consequences. Hepatic metabolism of MDMA and activation of other enzymes contribute to increased ROS/RNS production and reduced levels of anti-oxidants. These changes promote oxidative modifications of many mitochondrial proteins, resulting in their inactivation followed by mitochondrial dysfunction and tissue damage after MDMA exposure.
2. MITOCHONDRIAL DYSFUNCTION BY OXIDATIVE PROTEIN MODIFICATIONS IN MDMA-EXPOSED RATS
It is well-established that increased oxidative/nitrosative stress and mitochondrial dysfunction serve as common factors in many disease states, although the etiological cause for each disease may be different. These disease states include: Alzheimer’s disease, alcoholism-related tissue injury, cancer, cardiovascular diseases, chronic inflammation, diabetes, eye diseases, kidney diseases, non-alcoholic liver diseases, neurological disorders including Huntington’s disease and Parkinson disease, sepsis, stroke, etc [41, 42, 43]. Exposure to potentially toxic and environmental contaminants such as heavy metals, smoking and UV/irradiation or abusive drugs such as MDMA and methamphetamine can also produce increased levels of ROS/RNS [41, 42, 44].
In addition to the well-established role of increased oxidative/nitrosative stress in MDMA-induced tissue damage [27–29, 37, 38], oxidation and deletion of DNA especially in the mitochondria may also contribute to tissue damage [17, 35, 36]. Since there are many excellent review papers about the mechanisms and consequences of MDMA-mediated oxidative/nitrosative stress [19, 20, 28, 39, 40], in this review, we specifically describe MDMA-induced oxidative stress and oxidative modifications of mitochondrial proteins, leading to inhibition of mitochondrial function (i.e., mitochondrial dysfunction) and ultimately organ damage. Mitochondria, which play a critical role in energy supply, fat oxidation, ammonia-urea detoxification, anti-oxidant defense, and apoptosis, is known to be a major site of ROS production while mitochondrial proteins are known to be major targets of increased oxidative/nitrosative stress upon exposure to toxic compounds and/or in pathophysiological conditions [41, 42, 43]. It is possible that MDMA and/or its reactive metabolites (including N-methyl-α-methyldopamine, orthoquinone, and quinone thioesters) [27–29, 31] suppress the mitochondrial function by directly inhibiting the activities of mitochondrial complexes I, II, IV, and V [36, 44] or interacting with various proteins in the mitochondria [31]. Interruption of the mitochondrial electron transport system would lead to increased leakage of ROS (including hydrogen peroxide) from the mitochondrial respiratory chain, as demonstrated in neurodegenerative diseases [43] or after exposure to toxic agents [41, 42, 44]. In addition to ROS production from mitochondria, other enzymes are also known to produce ROS/RNS following MDMA exposure. These enzymes may include: cytochrome P450 2D6 (CYP2D6) and CYP3A (that are known to be involved in MDMA metabolism), NADPH-oxidase, xanthine oxidase, and inducible nitric oxide synthase [45]. Moreover, MDMA exposure increases nitrosative stress [37, 38, 44]. Increased production of ROS and RNS usually result in production of more potent peroxynitrite (ONOO−), which interacts with cellular macromolecules such as DNA, proteins, and lipids to produce oxidized DNA, proteins, and lipid peroxides, respectively [46]. The modification of these macromolecules has deleterious effects on their physiological functions. Peroxynitrite is known to nitrate Tyr residues [46] as well as S-nitrosylate Cys residues in many proteins [47]. Production of peroxynitrite after MDMA exposure likely leads to the oxidative modifications of some mitochondrial complex proteins, resulting in their inhibition, as described [48, 49]. The inactivation of certain mitochondrial proteins likely contributes to mitochondrial dysfunction and organ damage especially in the presence of another co-morbidity factor such as alcohol (ethanol) and cocaine.
Despite many reports about MDMA-mediated oxidative stress and lipid peroxidation [36, 44, 50], the identities of the oxidatively-modified proteins in MDMA-exposed tissues have not been reported. To our knowledge, few systematic proteomic analyses have been performed to investigate the changes in protein expression or identify oxidatively-modified proteins in MDMA-exposed tissues. Instead of studying the overall pattern changes in protein and/or mRNA expression following MDMA treatment, we specifically focused on the oxidative-modifications of mitochondrial proteins to evaluate their functional changes and roles in MDMA-mediated organ damage. Based on the increased oxidative stress following MDMA exposure, we hypothesized that oxidative modifications of certain mitochondrial proteins lead to inhibition of their activities, resulting in mitochondrial dysfunction and ultimately contributing to MDMA-mediated organ damage. To test this hypothesis and to elucidate the mechanism of mitochondrial dysfunction, we systematically analyzed the oxidatively-modified mitochondrial proteins in MDMA-exposed tissues. Characterization of the oxidatively-modified proteins accompanied with biochemical analysis would aid our understanding of the molecular mechanisms of mitochondrial dysfunction and tissue injury associated with MDMA. In this review, we briefly describe our systematic identification of oxidatively-modified mitochondrial proteins using the redox-based Cys-targeted proteomic method that was recently developed in this laboratory and the implications of our findings in the understanding the mechanism of MDMA- induced liver damage [51–54]. We also discuss the application of this redox-based proteomics method for studying oxidized proteins in other sub-cellular organelles of various tissues as well as the advantages and limitations of this method. Although our studies have focused on hepatic proteins, our approach should be amenable for characterizing the mechanisms of organ damage in other tissues.
It is known that many amino acids such as Cys, Met, His, Arg, Lys, Pro, Thr, Tyr, and Trp can undergo oxidative modifications [55]. We specifically studied the oxidative modifications of Cys residues in proteins because of the availability of Cys-specific reagents such as N-ethylmaleimide and biotin-N-maleimide (biotin-NM). As described in our reports [44,51], the oxidatively-modified protein thiols in MDMA-exposed rat livers (versus controls) were labeled with biotin-NM and then detected by immunoblot analysis with the monoclonal antibody against biotin. To further determine the identity of each protein, biotin-NM-labeled oxidized proteins from controls and MDMA-exposed tissues were purified by streptavidin-agarose beads. The oxidized proteins bound to streptavidin-agarose beads were washed three times with the buffer containing 1% CHAPS to remove any contaminating proteins and then analyzed by 2-D gel electrophoresis followed by silver staining. Densities of both gels representing the oxidized proteins in control animals or MDMA-exposed rats were adjusted for comparable exposure levels of an internal control protein in 2 different gels. Under these comparable conditions, we observed that the intensity and number of oxidized mitochondrial proteins were increased in MDMA-exposed tissues than those of control [44]. Mass spectral analysis of the protein spots excised from the 2-D gels revealed that many mitochondrial proteins were oxidatively modified in MDMA-exposed rat livers. The list of the oxidized mitochondrial proteins is summarized in Fig. (2). These proteins are involved in: protein folding (chaperone proteins), oxidative phosphorylation, energy production, anti-oxidant defense, fat metabolism, electron transport, etc. In addition, the activities of certain enzymes such as NADH-ubiquinone oxidoreductase (complex I), ATP synthase (complex V), aldehyde dehydrogenase 2 (ALDH2), and 3-ketoacyl-CoA thiolase were significantly inhibited in MDMA-exposed tissues. Inactivation of these enzymes correlated with increased hydrogen peroxide, decreased ATP level, accumulation of lipid peroxides such as malondialdehyde (MDAL), and triglyceride fat accumulation, respectively [44]. Moreover, mitochondrial dysfunction with decreased activities of mitochondrial proteins correlated with biochemical measurements of plasma transaminase activities and histological examinations of the liver specimens [44]. Although we did not measure the activities of all of the proteins listed in Fig. (2) and [44], it is likely that many of their activities including ALDH6 (methylmalonate semi-aldehyde dehyrogenase) and ALDH7 (alpha-aminoadipic semialdehyde dehydrogenase) is inhibited by oxidative modifications, as demonstrated with mitochondrial ALDH2 [56] and cytosolic ALDH1 [57].
Fig. 2.
Summary of oxidized mitochondrial proteins in MDMA-exposed rat livers. Male rats were treated with MDMA (10 mg/kg each) orally twice for 2 days. Treated rats were euthanized 12 h after the last dose of MDMA and oxidized mitochondrial proteins were analyzed, as described [44]. Oxidized mitochondrial proteins are summarized under different functions.
Although we showed that many mitochondrial proteins were oxidatively-modified in MDMA-exposed tissues, we believe that the actual number of oxidized proteins could be much higher since some of the mitochondrial proteins, expressed in small quantities, might not be detected by the current method. In addition, some of the oxidized or nitrated proteins could be rapidly degraded [55, 58, 59] and thus could not be detected by our analysis. In fact, many smaller fragments of their full-length parent proteins such as α-ATP synthase and glutamate dehydrogenase were identified in our study [44]. These data indicate that oxidatively-modified parent proteins were spontaneously broken into smaller fragments [55] or subjected to proteolytic degradation [58, 59]. However, no detection by this Cys-targeted proteomics approach does not necessarily mean no or little oxidative modification of a certain protein. In fact, oxidation of a certain protein, especially expressed in small quantity and thus not detected by this technique, can be demonstated by immunoblot analysis with an antibody against S-NO-Cys or glutathione (for detection of mixed disulfides) after the target protein is immunoprecipitated with a specific antibody against it, as discussed below.
2.1 APPLICATIONS OF THE REDOX-BASED PROTEOMICS APPROACH IN MDMA-EXPOSED TISSUES
The redox-based, Cys-targeted approach using biotin-NM, as we originally described [51–54], exhibited a major advantage in detecting subtle increments in the amounts of oxidatively-modified proteins over the previously existing methods of using biotin-conjugated iodoacetamide (BIAM) [60], 4-iodobutyl-triphenylphosphonium [61], or isotope-coded affinity tag (ICAT) reagent [62], mainly because the levels of oxidized proteins detected with these reagents (e.g., BIAM, 4-iodobutyl-triphenylphosphonium, or ICAT) are inversely related to the increased levels of oxidative stress. Furthermore, the biotin-switch method using biotin-NM as a probe does not need special reagents as stated [51]. This is in contrast to the requirement of special reagents such as a specific antibody to 4-iodobutyltriphenyl-phosphonium [61] or a cysteine-specific ICAT reagent [62]. All reagents used in this method are available commercially and easy to obtain.
Many proteomics approaches including 2-D Fluorescence Difference Gel Electrophoresis (2-D-DIGE) have been recently developed to efficiently determine different levels of protein expression in a variety of samples (e.g., treated versus untreated controls) [63, 64]. Although this newly-developed 2-D DIGE method may allow us to evaluate different levels of protein expression following MDMA exposure, it does not always predict functional alterations of certain proteins due to their post-translational modifications. In fact, many proteins are known to be functionally altered (i.e., inhibited) through post-translational modifications without changes in their protein contents, as described for mitochondrial ALDH2 in hepatic cancer patients [65, 66]. In contrast, the Cys-targeted redox proteomics approach allows us to identify oxidized proteins/enzymes and further predict their functional alterations (e.g., inhibition) even in the absence of any changes in protein contents by simply searching the literature. For instance, PubMed search revealed that both mitochondrial and peroxisomal 3-keto-acyl-CoA thiolases contain 2 Cys residues and 1 His residue in its active site [67], while these two isozymes share 37% structural identity [68]. Based on pre-existing literature and our data [summarized in Fig. (2)], we predicted inhibition of mitochondrial 3-keto-acyl-CoA thiolase activity. Inhibition of this enzyme was experimentally confirmed by comparing the activities in MDMA-exposed tissues relative to control [44]. We would also predict the inhibition of ALDH6 and ALDH7 isozymes by oxidative modification. Our prediction is based on the fact that all ALDH isozymes contain a highly conserved Cys residue in their active sites and can be S-nitrosylated, as demonstrated with mitochondrial ALDH2 [56] and cytosolic ALDH1 [57].
By comparing the patterns of oxidized proteins in different sub-organelles and/or tissues, we can also predict the functional alterations of specific proteins in each organ. In fact, this redox-related proteomics method can be used to systematically identify oxidatively-modified proteins in different sub-cellular organelles [e.g., cytoplasm, mitochondria, endoplasmic reticulum (ER), and nuclear fractions] and in different organs/tissues of interest (e.g., liver, brain, kidney, heart, intestine, etc) after exposure to potentially toxic agents [51–53] or under different pathophysiological conditions [54, 69]. Based on the MDMA-mediated oxidative stress, we also expected that an increased number of oxidized cytosolic proteins would be identified in MDMA-exposed tissues. Consistent with this prediction, our unpublished, preliminary results showed that the number and intensity of oxidized proteins in the cytoplasm are significantly increased in MDMA-exposed liver compared to those of controls. Moreover, cytosolic peroxiredoxin, involved in anti-oxidant defense, was oxidized in MDMA-exposed rat tissues, as it was in ethanol-exposed hepatoma cells and mouse livers [52]. Since oxidative inactivation of peroxiredoxin was shown to be correlated with activation of c-Jun N-terminal protein kinase (JNK) and/or p38 kinase [70, 71], we expect the increased activation of JNK and p38 kinase in MDMA-exposed tissues. These stress-activated protein kinases may contribute to apoptosis with concomitant release of mitochondrial cytochrome c into cytoplasm and caspase activation [72]. Phosphorylated JNK and/or p38 kinase activated after MDMA exposure can phosphorylate pro-apoptotic Bax, which is then translocated to mitochondria to promote mitochondrial permeability change and apoptosis, as demonstrated [73].
This redox-based method can also be used for future translational research in treating MDMA-related behavioral and pathological damage. Understanding of the molecular mechanisms of MDMA-induced toxicities would be very important in developing efficient preventive or therapeutic agents for MDMA-abused people. The benefits of various anti-oxidants such as N-acetylcysteine, ascorbic acid, and glutathione analogs were demonstrated in the models of MDMA-exposed cell/tissue damage [27, 36]. Usage of protease inhibitors was also suggested in treating MDMA-related neuronal damage, since the MDMA-induced neuronal damage is mediated through the time-dependent degradation of structural proteins such as cytoskeletal αII-spectrin and microtubule-associated tau, leading to cytoskeletal damage [74]. The redox-based Cys-targeted approach described here can be also used to evaluate or monitor the efficacy of certain beneficial agents against MDMA-exposed tissue injury by studying the pattern of oxidatively-modified proteins before and after treatment with the beneficial agent. Based on our recent data [75], we expect that the levels of oxidatively-modified proteins in MDMA-treated samples would be significantly decreased when the animals are pretreated with certain anti-oxidants or other cell protective agents. However, our expectations need to be experimentally supported in the future.
2.2 POTENTIAL LIMITATIONS AND ALTERNATIVE APPROACHES IN MDMA-EXPOSED TISSUES
Despite many advantages and application potential, this Cys-targeted proteomics approach also possesses some limitations. Common to all proteomics methods including 2-D DIGE method [63, 64], the Cys-targeted redox proteomics approach only allows us to detect oxidatively-modified proteins expressed in large quantities, as we originally described [51]. For instance, it is unlikely that we can detect oxidative modifications of certain enzymes or transcription factors that are expressed in very low quantities, although these proteins may contain Cys residues that can be oxidatively-modified following MDMA exposure. The failure to detect these proteins could be simply due to their low levels of expression relative to other proteins in the specific tissues of interest. However, despite this limitation, it is possible to successfully demonstrate oxidative modifications of these proteins by immunoprecipitation with a specific antibody against each target protein and then immunoblot analysis with a specific antibody against S-NO-Cys or glutathione (for detecting mixed disulfides). This alternative approach should be accompanied by the measurements of the specific enzyme activity or protein levels to further confirm functional implications of their oxidative modifications. For instance, it was shown that MDMA inhibits the activity of tryptophan hydroxylase (TPH), the initial and rate limiting enzyme in the biosynthesis of the neurotransmitter serotonin, through elevated peroxynitrite, which can nitrate Tyr residues and S-nitrosylate Cys residues in many proteins [46, 47]. However, Kuhn and coworkers carefully demonstrated that the inhibition of TPH was mediated through nitrosylation of Cys residues in TPH [76]. They also showed that nitration of Tyr residues of TPH has minimal impact on the inhibition of TPH catalytic activity. This mechanism provides one potential cause of serotonin depletion and neuropsychiatric behavioral deficits observed after MDMA usage, although many other mechanisms probably exist. Tyrosine hydroxylase, the first and rate-limiting enzyme in the biosynthesis of the neurotransmitter dopamine, was also inhibited by a similar mechanism of peroxynitrite-mediated S-nitrosylation of Cys residues [77], although this enzyme can be inhibited through nitration of critical Tyr residues [78]. In case of transcription factors, one can measure the mRNA and protein levels of their down-stream targets. It is expected that these critical enzymes or many transcription factors may not be detected by the Cys-targeted analysis due to their low levels of expression compared to other proteins expressed in large amounts.
Another limitation of the redox-related Cys-targeted proteomics approach is that Cys residues of various proteins can be modified by many different reactions, as discussed [79]. For instance, protein thiols can be modified through: S-nitrosylation, S-glutathionylation, S-succinylation, ADP-ribosylation, disulfide (including mixed disulfide bonds with glutathione), sulfenic acid, sulfinic acid, sulfonic acid, and Cys-adducts (with electrophilic MDMA metabolites), which cannot be reduced by DTT/Fe+2 ferrous iron) [80]. Furthermore, Cys residues can undergo adduct formation with lipid peroxides such as MDAL, which can be elevated under oxidative stress [81, 82]. In fact, MDMA exposure increased the levels of lipid peroxides [44, 83]. In many cases, the activities of certain proteins are inhibited although their levels are not altered, suggesting that the catalytic and other critical Cys residues of these proteins are likely oxidatively-modified or conjugated with reactive MDMA metabolites, as reviewed [84]. The covalent modifications of Cys residues in the latter cases can be demonstrated by measuring the enzyme activity after incubation with DTT, a strong reducing agent. If the suppressed enzyme activities are recovered by the addition of DTT [51–54], the critical Cys residue(s) of target proteins are likely to be oxidatively modified to sulfenic acid, S-nitrosylation, or disulfides (including mixed disulfides with glutathione). If the catalytic activities are not restored even after incubation with DTT, this likely represents the irreversible, covalent modifications (e.g., conjugation adducts with MDMA-quinone adducts) or hyperoxidation of Cys residues to sulfinic/sulfonic acids. Alternatively, other amino acid residues (e.g., Met, His, Trp, Lys, Tyr, Ser, etc) of the target proteins may also be covalently modified (e.g., oxidation or phosphorylation) and thus contribute to irreversible inactivation of the target protein [79, 84, 85]. Therefore, it is advised to consider many possible routes of oxidative modifications in analyzing the data from MDMA-exposed tissues.
3. MDMA-MEDIATED ORGAN DAMAGE IN VARIOUS TISSUES
In addition to damage to neuronal tissues which were studied extensively, MDMA can cause many other peripheral tissues such as heart, liver, kidney, testes, etc. Similar to liver damage, MDMA and its metabolites are also known to damage kidney during their excretion through the kidney. MDMA-induced rhabdomyolysis (necrosis of myocytes from a rapid rise in cellular calcium), which can lead to myoglobin deposition in the kidney, along with extreme dehydration and electrolyte imbalances may also contribute to acute and chronic renal failure [13–15, 86]. MDMA also interferes with endocrine function, altering the levels of corticotrophin (i.e., ACTH), cortisol and prolactin [16] while it inhibits gonadal function [17]. Although the toxic mechanisms for MDMA-induced peripheral organs are not clearly elucidated, it is likely that increased levels of oxidative/nitrosative stress and MDMA metabolites also contribute to tissue damage in the peripheral organs. Since the extra-hepatic tissues contain very low levels of MDMA-metabolizing P450 isozymes (compared to the liver), MDMA metabolites produced in the liver are likely to be responsible for the toxic effects of MDMA in the brain, heart and other peripheral tissues. This notion was supported by the fact that direct administration of MDMA into the brain or heart did not cause tissue damage [32, 33]. Therefore, it would be of interest to determine which proteins are oxidatively-modified and how the oxidized proteins correlate with the degree of tissue damage in these extra-hepatic tissues following MDMA exposure.
4. POTENTIATION OF MDMA TOXICITY WITH OTHER ABUSED SUBSTANCES: HARMFUL DRUG-DRUG INTERACTION
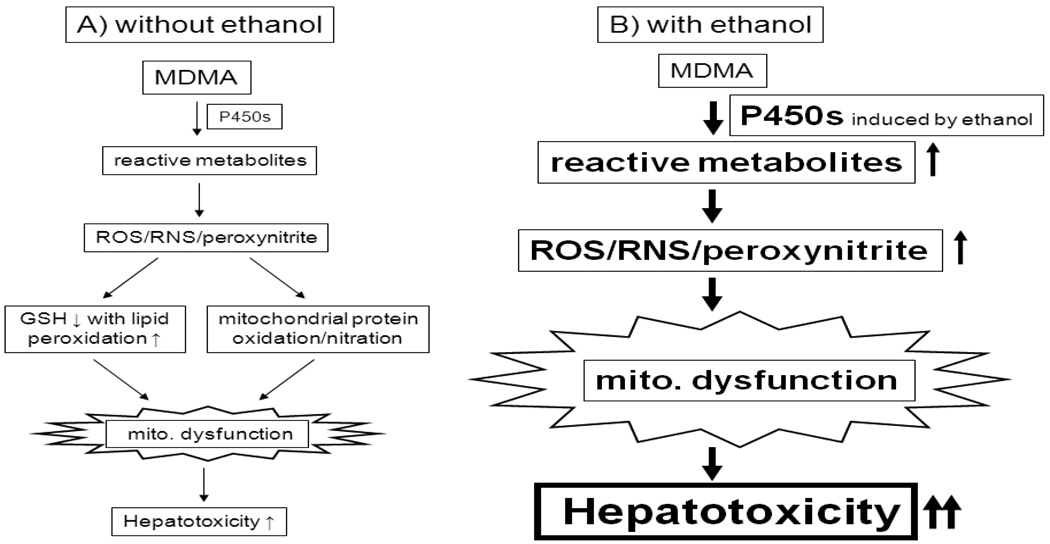
Recent epidemiological studies revealed that MDMA is frequently co-abused with other substances such as alcohol (ethanol), cannabis (marijuana), cocaine, opioids, and amphetamines [87–91]. Up to 70% of MDMA users co-abuse alcohol at dangerous levels. Evidence indicates that the combination of MDMA and ethanol further increases the risk of cell and organ damage (i.e., harmful drug-drug interactions) since acute or sub-chronic exposure to each abused substance, MDMA or alcohol, can itself cause organ damage. For instance, acute and binge exposure to alcohol, a widely-abused substance and legally available, is known to cause oxidative/nitrosative stress and tissue injury in many organs including liver, brain, pancreas, and heart [92–95]. As described earlier in this review, MDMA alone promotes hyperthermia and tissue damage in many organs. Combination of these 2 abused substances (e.g., MDMA and alcohol) can accelerate the pathological processes as demonstrated in cultured hepatocytes, animal models, and humans [96–101]. Moreover, ethanol or its metabolite, acetaldehyde, enhanced MDMA-mediated long term serotonergic neurotoxicity [96], while it increased MDMA concentration in the brain [102], most likely through disruption of the blood brain barrier [103]. It is believed that one of the main causes of the potentiating effects of MDMA and alcohol is increased oxidative/nitrosative stress accompanied by mitochondrial dysfunction. For instance, alcohol-mediated inactivation of mitochondrial enzymes such as ALDH2 [53] and its isozymes [57], involved in cellular defense against toxic acetaldehyde, acrolein, MDAL, and other reactive carbonyl compounds, may increase sensitivity toward tissue damage by MDMA or vice versa [44, 96–101]. We recently showed that concomitant administration of MDMA with ethanol resulted in markedly suppressed ALDH2 and ALDH1 activities with elevated blood levels of acetaldehyde (the quintessentially toxic primary metabolite of ethanol) and enhanced hepatotoxicity in rats [101]. In addition, some of these abused substances such as ethanol and cocaine may increase the activities of P450 enzymes that are involved in the metabolism of MDMA, causing increased production of MDMA-mediated metabolites. For instance, ethanol intake is known to activate CYP3A, CYP1A2 and CYP2B in the liver (and the extra-hepatic tissues) [104], probably resulting in increased production of reactive MDMA metabolites [Fig. (1)]. However, ethanol is not supposed to compete with MDMA metabolism by these P450 enzymes. In fact, the majority of ethanol is being metabolized by cytosolic alcohol dehydrogenase while acetaldehyde produced from ethanol metabolism is catalyzed by mitochondrial ALDH2. The latter 2 enzymes are not known to be involved in MDMA metabolism, thus causing no or little competition between MDMA and ethanol. However, ethanol and acetaldehyde are known to decrease the levels of various anti-oxidants such as glutathione and vitamin C, likely contributing to increased oxidative stress in the presence of reactive MDMA metabolites. All these events eventually contribute to greater tissue damage initiated by MDMA, as exemplified in Fig. (3). Although potentiation of MDMA-mediated liver damage by ethanol is illustrated in Fig. (3), the mechanism of organ damage in other tissues such as brain and kidney could be similar to that in the liver.
Fig. 3.
Synergistic interaction between MDMA and alcohol (or other abused drugs). MDMA, alcohol, or cocaine is known to decrease GSH levels, facilitating increased oxidative stress. Ethanol (or cocaine) increases some P450 isozymes involved in MDMA metabolism, leading to greater production of MDMA metabolites with decreased levels of cellular antioxidants, contributing to increased oxidative/nitrosative stress, mitochondrial dysfunction and sensitivity toward MDMA-mediated tissue damage, as exemplified with hepatotoxicity.
5. CONCLUSION AND FUTURE STUDIES
Despite numerous reports about MDMA-mediated tissue injuries, the molecular mechanisms of its pathological effects are still poorly understood. Acute and sub-chronic usage of MDMA increases oxidative/nitrosative stress, which contributes to tissue damage in many tissues and deaths in some cases. Various metabolites of MDMA including potentially toxic quinone and thioester compounds may directly or indirectly interfere with the mitochondrial electron transport system, leading to increased leakage of ROS from the mitochondria. ROS produced from mitochondria and the catalytic cycles of P450-mediated MDMA metabolism are likely to oxidatively modify cellular macromolecules such as lipids, DNA, and proteins. Oxidation of lipids likely causes accumulation of lipid peroxides such as MDAL and 4-hydroxynonenal, which are toxic to the cells [81, 82, 105]. Oxidation of DNA or adduct formations would lead to oxidized DNA bases and/or DNA strand breaks, contributing to increased chances of mutations (and carcinogenesis). Oxidative modifications of various proteins in many different sub-cellular organelles (e.g., mitochondria, ER, etc) would promote their inactivation, resulting in mitochondrial dysfunction and ER stress. Our Cys-targeted proteomics analysis revealed that many mitochondrial proteins were oxidatively-modified in MDMA-exposed rat livers, contributing to mitochondrial dysfunction [Fig. (2)]. The oxidized proteins in other sub-cellular organelles and extra-hepatic tissues should be also characterized to further understand the roles of specific proteins in MDMA-mediated damage in a given tissue of interest. However, this Cys-targeted proteomics analysis should be complemented by the enzymatic activity measurement of an individual target protein and immunoprecipitation followed by immunoblot analysis with S-NO-Cys, glutathione, or 3-nitro-tyrosine. In addition, MDMA-mediated signaling pathways should be studied since increased ROS/RNS may directly or indirectly activate the number of cell signaling pathways including many different protein kinases. For instance, elevated ROS may oxidize the highly-conserved, active site Cys residue of mitogen-activated protein kinase phosphatases [106, 107], resulting in persistent activation (phosphorylation) of mitogen-activated protein kinases such as JNK and p38 kinase, contributing to MDMA-induced apoptosis/necrosis. Furthermore, some of the potential targets (including Bcl-2 family proteins involved in apoptosis) of these stress-activated protein kinases need to be experimentally demonstrated in the future. Clear understanding of the toxicity mechanisms would lead to future translational research including development of efficient therapeutic agents against MDMA-mediated organ damage or deaths. Finally, the biomedical research data should be used to educate the public including MDMA users about the potential harms of MDMA use especially in conjunction with other abused substances including alcohol.
ACKNOWLEDGMENT
This research was supported by the Intramural Program Fund at the National Institute on Alcohol Abuse and Alcoholism. We are grateful to Drs. Timothy D. Veenstra, Li-Rong Yu, and Xiaoying Ye at the Laboratory of Proteomics and Analytical Technologies, Advanced Technology Program, SAIC-Frederick, Inc. for determining the protein sequences of oxidatively-modified mitochondrial proteins, as described [44]. The authors do not have any conflict of interest.
Footnotes
The abbreviations used are: MDMA, 3,4-methylenedioxymethamphetamine; ROS, reactive oxygen species; RNS, reactive nitrogen species; GSH, reduced glutathione; Complex I, NADH-ubiquinone oxidoreductase; Biotin-NM, biotin-N-maleimide; ALDH2, mitochondrial aldehyde dehydrogenase 2; MDAL, malondialdehyde; S-NO-Cys, S-nitrosylated Cys; BIAM, biotin-conjugated iodoacetamide; ICAT, isotope coded affinity tag; 2-D DIGE, 2-D fluorescence difference gel electrophoresis; ER, endoplasmic reticulum; JNK, c-Jun N-terminal protein kinase; TPH, tryptophan hydroxylase.
REFERENCES
- 1.Landry MJ. MDMA: a review of epidemiologic data. J. Psychoactive Drugs. 2002;34(2):163–169. doi: 10.1080/02791072.2002.10399950. [DOI] [PubMed] [Google Scholar]
- 2.Schifano F, Corkery J, Deluca P, Oyefeso A, Ghodse AH. Ecstasy (MDMA, MDA, MDEA, MBDB) consumption, seizures, related offences, prices, dosage levels and deaths in the UK (1994–2003) J. Psychopharmacol. 2006;20(3):456–463. doi: 10.1177/0269881106060147. [DOI] [PubMed] [Google Scholar]
- 3.Reid LW, Elifson KW, Sterk CE. Ecstasy and gateway drugs: initiating the use of ecstasy and other drugs. Ann. Epidemiol. 2007;17(1):74–80. doi: 10.1016/j.annepidem.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry JA, Hill IR. Fatal interaction between ritonavir and MDMA. Lancet. 1998;352(9142):1751–1752. doi: 10.1016/s0140-6736(05)79824-x. [DOI] [PubMed] [Google Scholar]
- 5.Harrington RD, Woodward JA, Hooton TM, Horn JR. Life-threatening interactions between HIV-1 protease inhibitors and the illicit drugs MDMA and gamma-hydroxybutyrate. Arch. Intern. Med. 1999;159(18):2221–2224. doi: 10.1001/archinte.159.18.2221. [DOI] [PubMed] [Google Scholar]
- 6.Kuypers KP, Samyn N, Ramaekers JG. MDMA and alcohol effects, combined and alone, on objective and subjective measures of actual driving performance and psychomotor function. Psychopharmacology. 2006;187(4):467–475. doi: 10.1007/s00213-006-0434-z. [DOI] [PubMed] [Google Scholar]
- 7.Fineschi V, Centini F, Mazzeo E, Turillazzi E. Adam (MDMA) and Eve (MDEA) misuse: an immunohistochemical study on three fatal cases. Forensic Sci. Int. 1999;104(1):65–74. doi: 10.1016/s0379-0738(99)00095-x. [DOI] [PubMed] [Google Scholar]
- 8.Lai TI, Hwang JJ, Fang CC, Chen WJ. Methylene 3, 4 dioxymethamphetamine-induced acute myocardial infarction. Ann. Emerg. Med. 2003;42(6):759–762. doi: 10.1016/s0196-0644(03)00511-0. [DOI] [PubMed] [Google Scholar]
- 9.Cerretani D, Riezzo I, Fiaschi AI, Centini F, Giorgi G, D’Errico S, Fiore C, Karch SB, Neri M, Pomara C, Turillazzi E, Fineschi V. Cardiac oxidative stress determination and myocardial morphology after a single ecstasy (MDMA) administration in a rat model. Int. J. Legal Med. 2008;122(6):461–469. doi: 10.1007/s00414-008-0262-2. [DOI] [PubMed] [Google Scholar]
- 10.Ellis AJ, Wendon JA, Portmann B, Williams R. Acute liver damage and ecstasy ingestion. Gut. 1996;38(3):454–458. doi: 10.1136/gut.38.3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreu V, Mas A, Bruguera M, Salmeron JM, Moreno V, Nogué S, Rodés J. Ecstasy: a common cause of severe acute hepatotoxicity. J. Hepatol. 1998;29(3):394–397. doi: 10.1016/s0168-8278(98)80056-1. [DOI] [PubMed] [Google Scholar]
- 12.Beitia G, Cobreros A, Sainz L, Cenarruzabeitia E. Ecstasy-induced toxicity in rat liver. Liver. 2000;20(1):8–15. doi: 10.1034/j.1600-0676.2000.020001008.x. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho M, Hawksworth G, Milhazes N, Borges F, Monks TJ, Fernandes E, Carvalho F, Bastos ML. Role of metabolites in MDMA (ecstasy)-induced nephrotoxicity: an in vitro study using rat and human renal proximal tubular cells. Arch. Toxicol. 2002;76(10):581–588. doi: 10.1007/s00204-002-0381-3. [DOI] [PubMed] [Google Scholar]
- 14.Ninkovic M, Selakovic V, Dukic M, Milosavljevic P, Vasiljevic I, Jovanovic M, Malicevic Z. Oxidative stress in rat kidneys due to 3,4-methylenedioxymethamphetamine (ecstasy) toxicity. Nephrology. 2007;13(1):33–37. doi: 10.1111/j.1440-1797.2007.00886.x. [DOI] [PubMed] [Google Scholar]
- 15.Campbell GA, Rosner MH. The agony of ecstasy: MDMA (3,4-methylenedioxymethamphetamine) and the kidney. Clin. J. Am. Soc. Nephrol. 2008;3(6):1852–1860. doi: 10.2215/CJN.02080508. [DOI] [PubMed] [Google Scholar]
- 16.Mas M, Farré M, de la Torre R, Roset PN, Ortuño J, Segura J, Camí J. Cardiovascular and neuroendocrine effects and pharmacokinetics of 3, 4-methylenedioxy methamphetamine in humans. J. Pharmacol. Exp. Ther. 1999;290(1):136–145. [PubMed] [Google Scholar]
- 17.Barenys M, Macia N, Camps L, de Lapuente J, Gomez-Catalan J, Gonzalez-Linares J, Borras M, Rodamilans M, Llobet JM. Chronic exposure to MDMA (ecstasy) increases DNA damage in sperm and alters testes histopathology in male rats. Toxicol. Lett. 2009;191(1):40–46. doi: 10.1016/j.toxlet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Connor TJ, McNamara MG, Finn D, Currid A, O'Malley M, Redmond AM, Kelly JP, Leonard BE. Acute 3,4-methylenedioxymethamphetamine (MDMA) administration produces a rapid and sustained suppression of immune function in the rat. Immunopharmacology. 1998;38(3):253–260. doi: 10.1016/s0162-3109(97)00084-2. [DOI] [PubMed] [Google Scholar]
- 19.Quinton MS, Yamamoto BK. Causes and consequences of methamphetamine and MDMA toxicity. AAPS J. 2006;8(2):E337–E347. doi: 10.1007/BF02854904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capela JP, Carmo H, Remião F, Bastos ML, Meisel A, Carvalho F. Molecular and cellular mechanisms of ecstasy-induced neurotoxicity: an overview. Mol. Neurobiol. 2009;39(3):210–271. doi: 10.1007/s12035-009-8064-1. [DOI] [PubMed] [Google Scholar]
- 21.Bai F, Jones DC, Lau SS, Monks TJ. Serotonergic neurotoxicity of 3,4-(+/−)-methylenedioxyamphetamine and 3,4−(+/−)-methylendioxymethamphetamine (ecstasy) is potentiated by inhibition of gamma-glutamyl transpeptidase. Chem. Res. Toxicol. 2001;14(7):863–870. doi: 10.1021/tx010011l. [DOI] [PubMed] [Google Scholar]
- 22.de la Torre R, Farre M, Roset PN, Pizarro N, Abanades S, Segura M, Segura J, Camí J. Human pharmacology of MDMA: pharmacokinetics, metabolism, and disposition. Ther. Drug Monit. 2004;26(2):137–144. doi: 10.1097/00007691-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 23.O'Shea E, Escobedo I, Orio L, Sanchez V, Navarro M, Green AR, Colado MI. Elevation of ambient room temperature has differential effects on MDMA-induced 5-HT and dopamine release in striatum and nucleus accumbens of rats. Neuropsychopharmacology. 2005;30(7):1312–1323. doi: 10.1038/sj.npp.1300673. [DOI] [PubMed] [Google Scholar]
- 24.Green AR, O'Shea E, Colado MI. A review of the mechanisms involved in the acute MDMA (ecstasy)-induced hyperthermic response. Eur. J. Pharmacol. 2004;500(1–3):3–13. doi: 10.1016/j.ejphar.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Bindoli A, Rigobello MP, Deeble DJ. Biochemical and toxicological properties of the oxidation products of catecholamines. Free Radic. Biol. Med. 1992;13(4):391–405. doi: 10.1016/0891-5849(92)90182-g. [DOI] [PubMed] [Google Scholar]
- 26.Milhazes N, Cunha-Oliveira T, Martins P, Garrido J, Oliveira C, Rego AC, Borges F. Synthesis and cytotoxic profile of 3,4-methylenedioxymethamphetamine ("ecstasy") and its metabolites on undifferentiated PC12 cells: A putative structure-toxicity relationship. Chem. Res. Toxicol. 2006;19(10):1294–1304. doi: 10.1021/tx060123i. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho M, Remião F, Milhazes N, Borges F, Fernandes E, Carvalho F, Bastos ML. The toxicity of N-methyl-alpha-methyldopamine to freshly isolated rat hepatocytes is prevented by ascorbic acid and N-acetylcysteine. Toxicology. 2004;200(2–3):193–203. doi: 10.1016/j.tox.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem. Res. Toxicol. 2000;13(3):135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 29.Montiel-Duarte C, Ansorena E, Lopez-Zabalza MJ, Cenarruzabeitia E, Iraburu MJ, Cenarruzabeitia E, Iraburu MJ. Role of reactive oxygen species, glutathione and NF-kappaB in apoptosis induced by 3,4-methylenedioxy methamphetamine ("Ecstasy") on hepatic stellate cells. Biochem. Pharmacol. 2004;67(6):1025–1033. doi: 10.1016/j.bcp.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Miller RT, Lau SS, Monks TJ. 2,5-Bis-(glutathion-S-yl)-alpha-methyldopamine, a putative metabolite of (+/−)-3,4-methylenedioxyamphetamine, decreases brain serotonin concentrations. Eur. J. Pharmacol. 1997;323(2–3):173–180. doi: 10.1016/s0014-2999(97)00044-7. [DOI] [PubMed] [Google Scholar]
- 31.Fisher AA, Labenski MT, Malladi S, Gokhale V, Bowen ME, Milleron RS, Bratton SB, Monks TJ, Lau SS. Quinone electrophiles selectively adduct "electrophile binding motifs" within cytochrome c. Biochemistry. 2007;46(39):11090–11100. doi: 10.1021/bi700613w. [DOI] [PubMed] [Google Scholar]
- 32.Paris JM, Cunningham KA. Lack of serotonin neurotoxicity after intraraphe microinjection of (+)-3,4-methylenedioxymethamphetamine (MDMA) Brain Res. Bull. 1992;28(1):115–119. doi: 10.1016/0361-9230(92)90237-r. [DOI] [PubMed] [Google Scholar]
- 33.Carvalho M, Remião F, Milhazes N, Borges F, Fernandes E, Monteiro Mdo C, Gonçalves MJ, Seabra V, Amado F, Carvalho F, Bastos ML. Metabolism is required for the expression of ecstasy-induced cardiotoxicity in vitro. Chem. Res. Toxicol. 2004;17(5):623–632. doi: 10.1021/tx049960f. [DOI] [PubMed] [Google Scholar]
- 34.Jayanthi S, Ladenheim B, Andrews AM, Cadet JL. Overexpression of human copper/zinc superoxide dismutase in transgenic mice attenuates oxidative stress caused by methylenedioxymethamphetamine (Ecstasy) Neuroscience. 1999;91(4):1379–1387. doi: 10.1016/s0306-4522(98)00698-8. [DOI] [PubMed] [Google Scholar]
- 35.Alvarenga TA, Andersen ML, Ribeiro DA, Araujo P, Hirotsu C, Costa JL, Battisti MC, Tufik S. Single exposure to cocaine or ecstasy induces DNA damage in brain and other organs of mice. Addict. Biol. 2010;15(1):96–99. doi: 10.1111/j.1369-1600.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 36.Alves E, Binienda Z, Carvalho F, Alves CJ, Fernandes E, de Lourdes Bastos M, Tavares MA, Summavielle T. Acetyl-L-carnitine provides effective in vivo neuroprotection over 3,4-methylenedioximethamphetamine-induced mitochondrial neurotoxicity in the adolescent rat brain. Neuroscience. 2009;158(2):514–523. doi: 10.1016/j.neuroscience.2008.10.041. [DOI] [PubMed] [Google Scholar]
- 37.Darvesh AS, Yamamoto BK, Gudelsky GA. Evidence for the involvement of nitric oxide in 3,4-methylenedioxymethamphetamine-induced serotonin depletion in the rat brain. J. Pharmacol. Exp. Ther. 2005;312(2):694–701. doi: 10.1124/jpet.104.074849. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Y, Laverty R. Role of brain nitric oxide in (+/−)3,4-methylenedioxy methamphetamine (MDMA)-induced neurotoxicity in rats. Brain Res. 1998;795(1–2):257–263. doi: 10.1016/s0006-8993(98)00313-8. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto BK, Bankson MG. Amphetamine neurotoxicity: Cause and consequence of oxidative stress. Crit. Rev. Neurol. 2005;17(2):87–117. doi: 10.1615/critrevneurobiol.v17.i2.30. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto BK, Raudensky J. The role of oxidative stress, metabolic compromise, and inflammation in neuronal injury produced by amphetamine-related drugs of abuse. J. Neuroimmune Pharmacol. 2008;3(4):203–217. doi: 10.1007/s11481-008-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amacher DE. Drug-associated mitochondrial toxicity and its detection. Curr. Med. Chem. 2005;12(16):1829–1839. doi: 10.2174/0929867054546663. [DOI] [PubMed] [Google Scholar]
- 42.Begriche K, Igoudjil A, Pessayre D, Fromenty B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion. 2006;6(1):1–28. doi: 10.1016/j.mito.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 44.Moon KH, Upreti VV, Yu LR, Lee IJ, Ye X, Eddington ND, Veenstra TD, Song BJ. Mechanism of 3,4-methylenedioxymethamphetamine (MDMA, ecstasy)-mediated mitochondrial dysfunction in rat liver. Proteomics. 2008;8(18):3906–3918. doi: 10.1002/pmic.200800215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;287(2):L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- 46.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls: The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 1991;266(7):4244–4250. [PubMed] [Google Scholar]
- 48.Cardoso SM, Pereira C, Oliveira R. Mitochondrial function is differentially affected upon oxidative stress. Free Radic. Biol. Med. 1999;26(1–2):3–13. doi: 10.1016/s0891-5849(98)00205-6. [DOI] [PubMed] [Google Scholar]
- 49.Murray J, Taylor SW, Zhang B, Ghosh SS, Capaldi RA. Oxidative damage to mitochondrial complex I due to peroxynitrite. Identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003;278(39):37223–37230. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]
- 50.Sprague JE, Nichols DE. The monoamine oxidase-B inhibitor L-deprenyl protects against 3,4-methylenedioxymethamphetamine-induced lipid peroxidation and long-term serotonergic deficits. J. Pharmacol. Exp. Ther. 1995;273(2):667–673. [PubMed] [Google Scholar]
- 51.Suh SK, Hood BL, Kim BJ, Conrads TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics. 2004;4(11):3401–3412. doi: 10.1002/pmic.200400971. [DOI] [PubMed] [Google Scholar]
- 52.Kim BJ, Hood BL, Aragon RA, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Increased oxidation and degradation of cytosolic proteins in alcohol-exposed mouse liver and hepatoma cells. Proteomics. 2006;6(4):1250–1260. doi: 10.1002/pmic.200500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon KH, Hood BL, Kim BJ, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology. 2006;44(5):1218–1230. doi: 10.1002/hep.21372. [DOI] [PubMed] [Google Scholar]
- 54.Moon KH, Hood BL, Mukhopadhyay P, Rajesh M, Abdelmegeed MA, Kwon YI, Conrads TP, Veenstra TD, Song BJ, Pacher P. Oxidative inactivation of key mitochondrial proteins leads to dysfunction and injury in hepatic ischemia reperfusion. Gastroenterology. 2008;135(4):1344–1357. doi: 10.1053/j.gastro.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272(33):20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 56.Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579(27):6115–6120. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moon KH, Abdelmegeed MA, Song BJ. Inactivation of cytosolic aldehyde dehydrogenase via S-nitrosylation in ethanol-exposed rat liver. FEBS Lett. 2007;581(21):3967–3972. doi: 10.1016/j.febslet.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 1996;385(1–2):63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 59.Widmer R, Kaiser B, Engels M, Jung T, Grune T. Hyperammonemia causes protein oxidation and enhanced proteasomal activity in response to mitochondria-mediated oxidative stress in rat primary astrocytes. Arch. Biochem. Biophys. 2007;464(1):1–11. doi: 10.1016/j.abb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 60.Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000;283(2):214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 61.Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, Darley-Usmar V, Bailey SM. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286(4):G521–G527. doi: 10.1152/ajpgi.00399.2003. [DOI] [PubMed] [Google Scholar]
- 62.Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell Proteomics. 2004;3(3):273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 64.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1(3):377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 65.Park KS, Cho SY, Kim H, Paik YK. Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int. J. Cancer. 2002;97(2):261–265. doi: 10.1002/ijc.1585. [DOI] [PubMed] [Google Scholar]
- 66.Kim J, Kim SH, Lee SU, Ha GH, Kang DG, Ha NY, Ahn JS, Cho HY, Kang SJ, Lee YJ, Hong SC, Ha WS, Bae JM, Lee CW, Kim JW. Proteome analysis of human liver tumor tissue by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of disease-related proteins. Electrophoresis. 2002;23(24):4142–4156. doi: 10.1002/elps.200290032. [DOI] [PubMed] [Google Scholar]
- 67.Zeng J, Li D. Expression and purification of His-tagged rat mitochondrial 3-ketoacyl-CoA thiolase wild-type and His352 mutant proteins. Protein Expr. Purif. 2004;35(2):320–326. doi: 10.1016/j.pep.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 68.Arakawa H, Takiguchi M, Amaya Y, Nagata S, Hayashi H, Mori M. cDNA-derived amino acid sequence of rat mitochondrial 3-oxoacyl-CoA thiolase with no transient presequence: structural relationship with peroxisomal isozyme. EMBO J. 1987;6(5):1361–1366. doi: 10.1002/j.1460-2075.1987.tb02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdelmegeed MA, Moon KH, Hardwick JP, Gonzalez FJ, Song BJ. Role of peroxisome proliferator-activated receptor-alpha in fasting-mediated oxidative stress. Free Radic. Biol. Med. 2009;47(6):767–778. doi: 10.1016/j.freeradbiomed.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang SW, Chang TS, Lee TH, Kim ES, Yu DY, Rhee SG. Cytosolic peroxiredoxin attenuates the activation of JNK and p38 but potentiates that of ERK in Hela cells stimulated with tumor necrosis factor-alpha. J. Biol. Chem. 2004;279(4):2535–2543. doi: 10.1074/jbc.M307698200. [DOI] [PubMed] [Google Scholar]
- 71.Lee YM, Park SH, Shin DI, Hwang JY, Park B, Park YJ, Lee TH, Chae HZ, Jin BK, Oh TH, Oh YJ. Oxidative modification of peroxiredoxin is associated with drug-induced apoptotic signaling in experimental models of Parkinson disease. J. Biol. Chem. 2008;283(15):9986–9998. doi: 10.1074/jbc.M800426200. [DOI] [PubMed] [Google Scholar]
- 72.Montiel-Duarte C, Varela-Rey M, Osés-Prieto JA, López-Zabalza MJ, Beitia G, Cenarruzabeitia E, Iraburu MJ. 3,4-Methylenedioxymethamphetamine ("Ecstasy") induces apoptosis of cultured rat liver cells. Biochim. Biophys. Acta. 2002;1588(1):26–32. doi: 10.1016/s0925-4439(02)00112-6. [DOI] [PubMed] [Google Scholar]
- 73.Kim BJ, Ryu SW, Song BJ. JNK- and p38 kinase-mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J. Biol. Chem. 2006;281(30):21256–21265. doi: 10.1074/jbc.M510644200. [DOI] [PubMed] [Google Scholar]
- 74.Warren MW, Kobeissy FH, Liu MC, Svetlov SI, Hayes RL, Gold MS, Wang KK. Ecstasy toxicity: A comparison to methamphetamine and traumatic brain injury. J. Alz. Dis. 2006;25(4):115–123. doi: 10.1300/J069v25n04_11. [DOI] [PubMed] [Google Scholar]
- 75.Song BJ, Moon KH, Olsson NU, Salem N., Jr Prevention of alcoholic fatty liver and mitochondrial dysfunction in the rat by long-chain polyunsaturated fatty acids. J. Hepatol. 2008;49(2):262–273. doi: 10.1016/j.jhep.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuhn DM, Geddes TJ. Peroxynitrite inactivation of tryptophan hydroxylase via sulfhydryl oxidation: Coincident nitration of enzyme tyrosyl residues has minimal impact on catalytic activity. J. Biol. Chem. 1999;274(42):29726–29732. doi: 10.1074/jbc.274.42.29726. [DOI] [PubMed] [Google Scholar]
- 77.Kuhn DM, Aretha CW, Geddes TJ. Peroxynitrite inactivation of tyrosine hydroxylase: mediation by sulfhydryl oxidation, not tyrosine nitration. J. Neurosci. 1999;19(23):10289–10294. doi: 10.1523/JNEUROSCI.19-23-10289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ara J, Przedborski S, Naini AB, Jackson-Lewis V, Trifiletti RR, Horwitz J, Ischiropoulos H. Inactivation of tyrosine hydrozylase by niration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Proc. Natl. Acad. Sci. USA. 1998;95(13):7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moon KH, Lee YM, Song BJ. Inhibition of hepatic mitochondrial aldehyde dehydrogenase by carbon tetrachloride through JNK-mediated phosphorylation. Free Radic. Biol. Med. 2010;48:391–398. doi: 10.1016/j.freeradbiomed.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wrona MZ, Dryhurst G. A putative metabolite of serotonin, trypamine-4,5-dione, is an irreversible inhibitor of tryptophan hydroxylase: Possible relevance to the serotonergic neurotoxicity of methamphetamine. Chem. Res. Toxicol. 2001;14(9):1184–1192. doi: 10.1021/tx010037c. [DOI] [PubMed] [Google Scholar]
- 81.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 82.Catalá A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys. Lipids. 2009;157(1):1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 83.Zhou JF, Chen P, Zhou YH, Zhang L, Chen HH. 3,4-Methylenedioxymethamphetamine (MDMA) abuse may cause oxidative stress and potential free radical damage. Free Radic. Res. 2003;37(5):491–497. doi: 10.1080/1071576031000076286. [DOI] [PubMed] [Google Scholar]
- 84.Monks TJ, Lau SS. The pharmacology and toxicology of polyphenolic-glutathione conjugates. Annu Rev Pharmacol Toxicol. 1998;38:229–255. doi: 10.1146/annurev.pharmtox.38.1.229. [DOI] [PubMed] [Google Scholar]
- 85.Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signal. 2003;5(5):577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- 86.Holt SG, Moore KP. Pathogenesis and treatment of renal dysfunction in rhabdomyolysis. Intensive Care Med. 2001;27(5):803–811. doi: 10.1007/s001340100878. [DOI] [PubMed] [Google Scholar]
- 87.Topp L, Hando J, Dillon P, Roche A, Solowij N. Ecstasy use in Australia: patterns of use and associated harm. Drug Alcohol Depend. 1999;55(1–2):105–115. doi: 10.1016/s0376-8716(99)00002-2. [DOI] [PubMed] [Google Scholar]
- 88.Winstock AR, Griffiths P, Stewart D. Drugs and the dance music scene: a survey of current drug use patterns among a sample of dance music enthusiasts in the UK. Drug Alcohol Depend. 2001;64(1):9–17. doi: 10.1016/s0376-8716(00)00215-5. [DOI] [PubMed] [Google Scholar]
- 89.Boyd CJ, McCabe SE, d’Arcy H. Ecstasy use among college undergraduates: gender, race and sexual identity. J. Subst. Abuse Treat. 2003;24(3):209–215. doi: 10.1016/s0740-5472(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 90.Barrett SP, Darredeau C, Pihl RO. Patterns of simultaneous polysubstance use in drug using university students. Hum. Psychopharmacol. 2006;21(4):255–263. doi: 10.1002/hup.766. [DOI] [PubMed] [Google Scholar]
- 91.Wu LT, Parrott AC, Ringwalt CL, Yang C, Blazer DG. The variety of ecstasy/MDMA users: results from the National Epidemiologic Survey on alcohol and related conditions. Am. J. Addict. 2009;18(6):452–461. doi: 10.3109/10550490903206049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34(1):9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 93.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2008;44(5):723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haorah J, Ramirez SH, Floreani N, Gorantla S, Morsey B, Persidsky Y. Mechanism of alcohol-induced oxidative stress and neuronal injury. Free Radic. Biol. Med. 2008;45(11):1542–1550. doi: 10.1016/j.freeradbiomed.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Keefe JH, Bybee KA, Lavie CJ. Alcohol and cardiovascular health: the razor-sharp double-edged sword. J. Am. Coll. Cardiol. 2007;50(11):1009–1014. doi: 10.1016/j.jacc.2007.04.089. Review. [DOI] [PubMed] [Google Scholar]
- 96.Izco M, Orio L, O’Shea E, Colado MI. Binge ethanol administration enhances the MDMA-induced long-term 5-HT neurotoxicity in rat brain. Psychopharmacology (Berl) 2007;189(4):459–470. doi: 10.1007/s00213-006-0602-1. [DOI] [PubMed] [Google Scholar]
- 97.Nutt DJ. A tale of two Es. J. Psychopharmacol. 2006;20(3):315–317. doi: 10.1177/0269881106064592. [DOI] [PubMed] [Google Scholar]
- 98.Pontes H, Sousa C, Silva R, Fernandes E, Carmo H, Remião F, Carvalho F, Bastos ML. Synergistic toxicity of ethanol and MDMA towards primary cultured rat hepatocytes. Toxicology. 2008;254(1–2):42–50. doi: 10.1016/j.tox.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 99.Pontes H, Santos-Marques MJ, Fernandes E, Duarte JA, Remião F, Carvalho F, Bastos ML. Effect of chronic ethanol exposure on the hepatotoxicity of ecstasy in mice: an ex vivo study. Toxicol. In Vitro. 2008;22(4):910–920. doi: 10.1016/j.tiv.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 100.Pontes H, Duarte JA, Guedes de, Pinho P, Soares ME, Fernandes E, Dinis-Oliveira RJ, Sousa C, Silva R, Carmo H, Casal S, Remião F, Carvalho F, Bastos ML. Chronic exposure to ethanol exacerbates MDMA-induced hyperthermia and exposes liver to severe MDMA-induced toxicity in CD1 mice. Toxicology. 2008;252(1–3):64–71. doi: 10.1016/j.tox.2008.07.064. [DOI] [PubMed] [Google Scholar]
- 101.Upreti VV, Eddington ND, Moon KH, Song BJ, Lee IJ. Drug interaction between ethanol and 3,4-methylenedioxymethamphetamine ("ecstasy") Toxicol. Lett. 2009;188(2):167–172. doi: 10.1016/j.toxlet.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hamida SB, Tracqui A, de Vasconcelos AP, Szwarc E, Lazarus C, Kelche C, Jones BC, Cassel JC. Ethanol increases the distribution of MDMA to the rat brain: possible implications in the ethanol-induced potentiation of the psychostimulant effects of MDMA. Int. J. Neuropsychopharmacol. 2009;12(6):749–759. doi: 10.1017/S1461145708009693. [DOI] [PubMed] [Google Scholar]
- 103.Haorah J, Knipe B, Gorantla S, Zheng J, Persidsky Y. Alcohol-induced blood-brain barrier dysfunction is mediated via inositol 1,4,5-triphosphate receptor (IP3R)-gated intracellular calcium release. J. Neurochem. 2007;100(2):324–336. doi: 10.1111/j.1471-4159.2006.04245.x. [DOI] [PubMed] [Google Scholar]
- 104.Sinclair JF, McCaffrey J, Sinclair PR, Bement WJ, Lambrecht LK, Wood SG, Smith EL, Schenkman JB, Guzelian PS, Park SS. Ethanol increases cytochromes P450IIE, IIB1/2, and IIIA in cultured rat hepatocytes. Arch. Biochem. Biophys. 1991;284(2):360–365. doi: 10.1016/0003-9861(91)90308-6. [DOI] [PubMed] [Google Scholar]
- 105.Soh Y, Jeong KS, Lee IJ, Bae MA, Kim YC, Song BJ. Selective activation of the c-Jun N-terminal protein kinase pathway during 4-hydroxynonenal-induced apoptosis of PC12 cells. Mol. Pharmacol. 2000;58(3):535–541. doi: 10.1124/mol.58.3.535. [DOI] [PubMed] [Google Scholar]
- 106.Kim HS, Song MC, Kwak IH, Park TJ, Lim IK. Constitutive induction of p-Erk1/2 accompanied by reduced activities of protein phosphatases 1 and 2A and MKP3 due to reactive oxygen species during cellular senescence. J. Biol. Chem. 2003;278(39):37497–37510. doi: 10.1074/jbc.M211739200. [DOI] [PubMed] [Google Scholar]
- 107.Heneberg P, Dráber P. Regulation of cys-based protein tyrosine phosphatases via reactive oxygen and nitrogen species in mast cells and basophils. Curr. Med. Chem. 2005;12(16):1859–1871. doi: 10.2174/0929867054546636. [DOI] [PubMed] [Google Scholar]