Introductory Paragraph
Soft tissue sarcomas, which encompass approximately 10,700 diagnoses and 3800 deaths per year in the US1, exhibit remarkable histologic diversity, with more than 50 recognized subtypes2. However, knowledge of their genomic alterations is limited. We describe an integrative analysis of DNA sequence, copy number, and mRNA expression in 207 samples encompassing seven major subtypes. Frequently mutated genes included TP53 (17% of pleomorphic liposarcomas), NF1 (10.5% of myxofibrosarcomas and 8% of pleomorphic liposarcomas), and PIK3CA (18% of myxoid/round-cell liposarcomas). PIK3CA mutations in myxoid/round-cell liposarcomas were associated with AKT activation and poor clinical outcomes. In myxofibrosarcomas and pleomorphic liposarcomas, we found both point mutations and genomic deletions affecting the tumor suppressor NF1. Finally, we found that shRNA-based knockdown of several genes amplified in dedifferentiated liposarcoma, including CDK4 and YEATS4, decreased cell proliferation. Our study yields a detailed map of molecular alterations across diverse sarcoma subtypes and provides potential subtype-specific targets for therapy.
Current knowledge of the key genomic aberrations in soft tissue sarcoma is limited to the most recurrent alterations or translocations. Subtypes with simple, near-diploid karyotypes bear few chromosomal rearrangements but have pathognomonic alterations: translocations in myxoid/round-cell liposarcoma (MRC) [t(12;16)(q13;p11), t(12;22)(q13;q12)] and synovial sarcomas (SS) [t(X;18)(p11;q11)]; activating mutations in KIT or PDGFRA in gastrointestinal stromal tumors (GIST)3,4. The discovery of the latter mutations led to the clinical deployment of imatinib for the treatment of GIST5, providing a model for genotype-directed therapies in molecularly defined sarcoma subtypes. Conversely, sarcomas with complex karyotypes, including dedifferentiated and pleomorphic liposarcoma, leiomyosarcoma, and myxofibrosarcoma, have no known characteristic mutations or fusion genes, although abnormalities are frequently observed in the Rb, p53, and specific growth-factor signaling pathways6.
Recent large-scale analyses7–10 have established a standard for cancer genome studies, but soft tissue sarcomas have not yet been a focus of this type of effort. Given the urgent need for new treatments for the ~4000 patients who die each year in the US of soft tissue sarcoma1, we sought to identify novel genomic alterations that could serve as therapeutic targets. Here, we describe complementary genome and functional genetic analyses of seven subtypes of high-grade soft tissue sarcoma (Table 1 and Supplementary Table 1) to discover subtype-specific events. Several of our findings, detailed below, could have nearly immediate therapeutic implications.
Table 1.
Summary of clinical and pathologic information for 207 soft-tissue sarcoma patients
| Characteristic | Value |
|---|---|
| No. of patients | 207 |
| Age [mean±SD (range)] | 56±16 (7–84) |
| Gender (%) † | |
| Female | 102 (50.2) |
| Male | 101 (49.8) |
| Tumor size § | |
| 0–5 cm | 35 (17.4) |
| 5–10 cm | 65 (32.3) |
| 10–15 cm | 43 (21.4) |
| >15 cm | 58 (28.9) |
| Primary site (%) † | |
| Retro-intrabdominal | 60 (29.6) |
| Visceral | |
| Gastrointestinal | 23 (11.3) |
| Genitourinary | 4 (2) |
| Gynecological | 1 (0.5) |
| Thoracic | 12 (5.9) |
| Extremity | 93 (45.8) |
| Trunk | 8 (3.9) |
| Head and Neck | 2 (1) |
| Stage at time of sample procurment ‡ | |
| Primary | 139 (68.8) |
| Local recurrence | 29 (14.4) |
| Distant recurrence | 34 (16.8) |
| Histology | |
| Dedifferentiated liposarcoma | 50 (24.2) |
| Myxoid/round cell liposarcoma | 21 (10.1) |
| Pleomorphic liposarcoma | 24 (11.6) |
| Leiomyosarcoma | 27 (13) |
| Gastrointestinal stromal tumor | |
| Epithelioid | 4 (1.9) |
| Spindle | 11 (5.3) |
| Mixed or unspecified | 7 (3.4) |
| Myxofibrosarcoma | |
| Myxofibrosarcoma | 35 (16.9) |
| Pleomorphic MFH | 3 (1.5) |
| Synovial sarcoma ‖ | |
| Monophasic | 19 (9.2) |
| Biphasic | 4 (1.9) |
| Median follow-up (months) | 35.65 |
| Time to distant recurrence (months) | 15.7 |
| Co-morbidities | 57 (27.5) |
One synovial sarcoma not specified
Data available for §201, †203, and ‡202 patients respectively
To study the genomic alterations in sarcomas, we initially analyzed 47 tumor/normal DNA pairs encompassing six soft tissue sarcoma subtypes by sequencing 722 protein-coding and microRNA genes, followed by verifying discovered mutations with mass spectrometry-based genotyping (see Methods, Supplementary Figure 1A, and Supplementary Table 2). The results revealed 28 somatic non-synonymous coding point mutations and 9 somatic insertions/deletions (indels) involving 21 genes in total (Table 2 and Supplementary Figure 1B). No mutations were detected in microRNAs genes. We extended the analysis to an additional 160 tumors, where we genotyped each of the mutations found above and re-sequenced exons of NF1 and ERBB4 in pleomorphic liposarcoma and myxofibrosarcoma, PIK3CA and KIT in myxoid/round cell liposarcoma, and CDH1 in dedifferentiated liposarcoma; this revealed nine additional mutations (Table 2 and Supplementary Table 3).
Table 2.
Mutations identified in soft tissue sarcoma
| Gene | No. of mut.a |
Subtype | Tumor ID | Cases affected (%)b |
mRNA | Protein |
|---|---|---|---|---|---|---|
| CDH1 | 2 | DDLPS | PT7DD | 2 | 712A>AG | N238D |
| GIST | PT61GT | 4.5 | 1849G>AG | A617Te | ||
| CTNNB1 | 2 | DDLPS | PT18DD | 2 | 122C>CT | T41Id |
| Synovial | PT195SYN | 4 | 95A>AT | D32Vd | ||
| EPHA1 | 1 | DDLPS | PT10DD | 2 | 634G>GA | A212T |
| EPHA5 | 1 | Pleomorphic | PT182PL | 4.2 | 2386A>AG | Y796H |
| EPHA7 | 1 | MYXF | PT106MF | 2.6 | 1649C>CT | S550N |
| ERBB4 | 2 | MYXF | PT130MF | 2.6 | 3437A>AT | D1146V |
| Pleomorphic | PT167PL | 4.2 | 1558A>AT | C520S | ||
| FBXW7 | 2 | DDLPS | PT38DD | 2 | 338_342delTCATC>TC | E113fs |
| GIST | PT58GT | 4.5 | 563G>GT | C188F | ||
| IRS1 | 1 | GIST | PT61GT | 4.5 | 3406C>CT | E1136K |
| KIT | 6 | GIST | PT57GT | 23 | 1727T>CT | L576Pd |
| GIST | PT63GT | 1961T>CT | V654Ad | |||
| GIST | PT61GT | 1667_1674delAGTGGAAG>AG | Q556fs | |||
| GIST | PT60GT | 1667_1687delc | Q556_I563>Q | |||
| GIST | PT59GT | 1670_1675delGGAAGG | W557_V559>Fe | |||
| MRC | PT149MRC | 4.8 | 2334G>CG | K778N | ||
| LTK | 1 | Synovial | PT190SYN | 4 | 2243_2244delTT>T | C748fs |
| MOS | 1 | GIST | PT61GT | 4.5 | 898A>AG | S300P |
| MST1R | 1 | GIST | PT60GT | 4.5 | 1229G>AG | P410L |
| NF1 | 7 | MYXF | PT104MF | 10.5 | 7972C>CT | H2658Y |
| MYXF | PT104MF | 7790C>CT | S2597L | |||
| MYXF | PT127MF | 910C>T | R304*d | |||
| MYXF | PT134MF | 910C>T | R304*d | |||
| MYXF | PT102MF | 7010T>TG | L2337R | |||
| Pleomorphic | PT176PL | 8.3 | 1105C>CT | Q369*d | ||
| Pleomorphic | PT179PL | 4006C>CT | Q1336* | |||
| NTRK1 | 1 | MYXF | PT101MF | 2.6 | 2338C>CT | R780W |
| PI4KA | 2 | MYXF | PT137MF | 2.6 | 4081_4088delTCTTATCT>TCT | 1361fs |
| Synovial | PT203SYN | 4 | 4081_4088delTCTTATCT>TCT | 1361fs | ||
| PIK3CA | 6 | MRC | PT143MRC | 18 | 1633G>AG | E545Ke |
| MRC | PT149MRC | 1633G>AG | E545Ke | |||
| MRC | PT138MRC | 3140A>AG | H1047Re | |||
| MRC | PT158MRC | 3140A>AG | H1047Re | |||
| Pleomorphic | PT173PL | 4.2 | 1660delC | H554fs | ||
| Synovial | PT195SYN | 4 | 1659delT | S553fs | ||
| PTEN | 2 | MYXF | PT100MF | 2.6 | G>CG | Splice site |
| Synovial | PT206SYN | 4 | 106G>AA | G36Re | ||
| PTK2B | 1 | Pleomorphic | PT163PL | 4.2 | G>AG | Splice site |
| RB1 | 1 | Pleomorphic | PT167PL | 4.2 | 1818T>TA | Y606*e |
| SYK | 1 | Pleomorphic | PT163PL | 4.2 | 52G>AA | G18S |
| TP53 | 4 | Pleomorphic | PT163PL | 16.7 | 404C>AA | C135Fe |
| Pleomorphic | PT169PL | 464G>AA | T155I | |||
| Pleomorphic | PT173PL | C>CT | Splice site | |||
| Pleomorphic | PT164PL | C>TT | Splice site |
DDLPS, dedifferentiated liposarcoma; GIST, gastroinstestinal stromal tumor; MRC, myxoid/round-cell liposarcoma; MYXF, myxofibrosarcoma.
Number of nonsynonymous or splice site mutations detected in either primary sequencing or extended genotyping.
Percentage of cases by subtype.
Reference allele: GTGGAAGGTTGTTGAGGAGAT.
Mutations previously identified in d soft-tissue sarcoma or in any e cancer type (COSMIC; http://www.sanger.ac.uk/genetics/CGP/cosmic/).
KIT was frequently mutated in GISTs and unexpectedly, in one myxoid/round cell liposarcoma sample (Supplementary Note). The next most frequently mutated genes observed within specific sarcoma subtypes were PIK3CA, in 18% of myxoid/round cell liposarcomas, TP53 in 17% of pleomorphic liposarcomas (interestingly, the only subtype in which mutations of this gene were found), and NF1 in 10.5% of myxofibrosarcomas and 8% of pleomorphic liposarcomas (Table 2 and Figure 1). Additional genes, including protein and lipid kinases, as well as known or candidate tumor suppressor genes, were found mutated in just one sample for each sarcoma subtype (Table 2, Figure 1, and Supplementary Note). Further studies will be needed to establish the functional impact of these mutations in sarcoma.
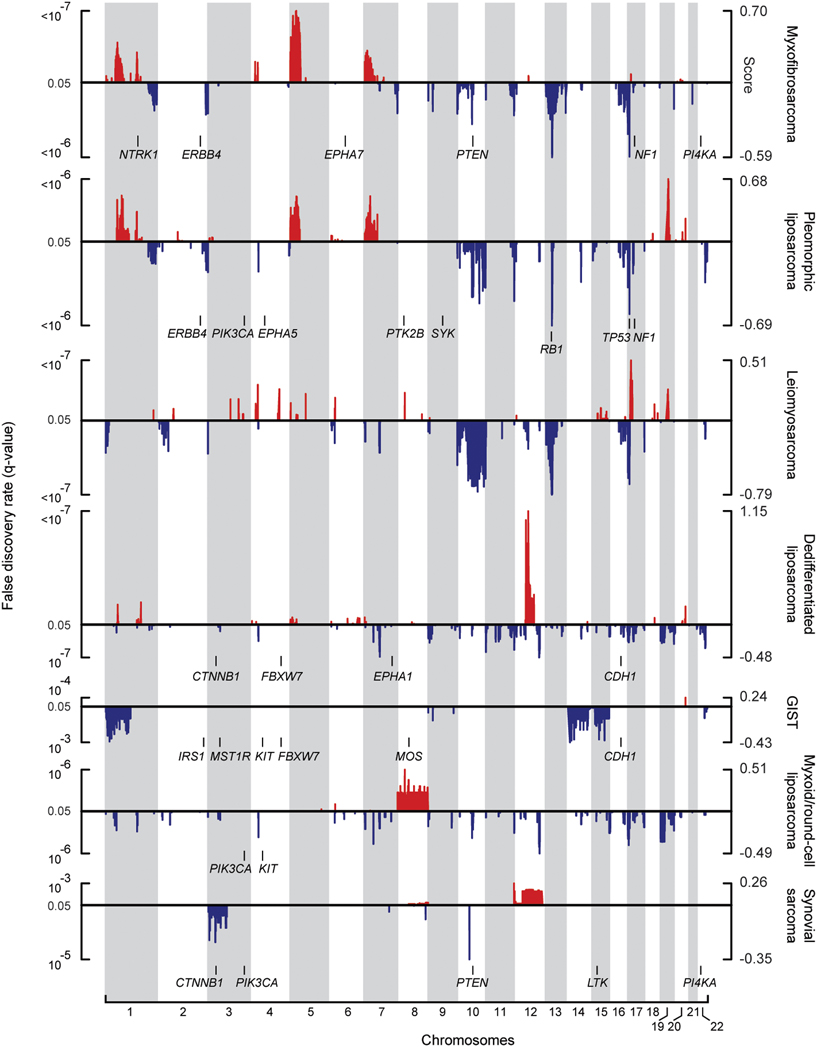
Figure 1. Nucleotide and copy number alterations in soft-tissue sarcoma subtypes.
The statistical significance of genomic aberrations for each subtype is shown. RAE q-values [left axis; for visualization, q-values ≤ 0.05 are considered significant, corresponding false discovery rate (FDR) ≤ 5%] and scores (right axis) for gains and amplifications (red) and losses and deletions (blue) are plotted across the genome (chromosomes indicated at bottom). Genes harboring somatic nucleotide alterations in this study are indicated in each subtype in which they were discovered (Table 2).
Below, we focus on three major specific genomic findings with therapeutic implications: point mutation and deletion of NF1 in a subset of soft tissue sarcomas, point mutation of PIK3CA in myxoid/round cell liposarcoma, and the complex pattern of amplification of chromosome 12q in dedifferentiated liposarcoma.
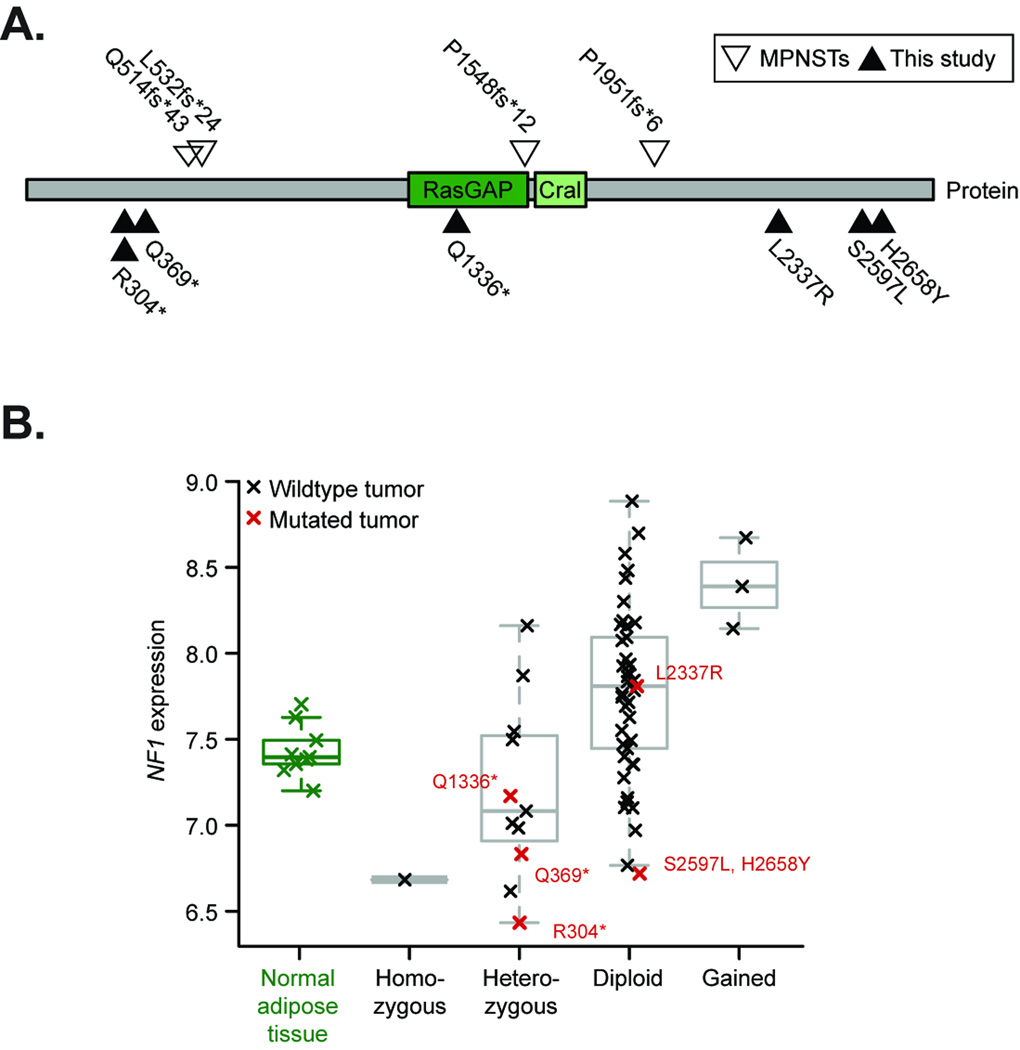
Integrated analysis of DNA copy number, expression, and mutation data uncovered diverse alterations of the Neurofibromatosis type 1 gene (NF1) in several sarcoma subtypes. While germline and somatic inactivation of NF1 is associated with malignant peripheral nerve sheath tumors11 and GISTs in Neurofibromatosis type 1 patients12, no somatic NF1 alterations have been reported in other sarcomas. We detected six point mutations and twelve genomic deletions encompassing the NF1 locus, occurring in both myxofibrosarcoma and pleomorphic liposarcoma (Table 2 and Figure 1, 2A–B; copy number analysis discussed further below). Two of the mutations, R304* and Q369*, were previously reported as germline mutations in patients with Neurofibromatosis type 113,14, while the other four mutations (three missense and one nonsense) have not been previously reported. In some tumors, biallelic inactivation was evident, with heterozygous point mutations accompanied by deletion of the wild-type allele and correspondingly reduced gene expression compared to normal adipose tissue15 in most cases (Figure 2B). Together, these data indicate a diverse pattern of NF1 aberrations in myxofibrosarcomas and pleomorphic liposarcomas. These results complement recent reports of NF1 alterations in lung cancers and glioblastomas7,8.
Figure 2. NF1 alterations in karyotypically complex sarcomas.
A. Somatic mutations in the NF1 protein in myxofibrosarcoma and pleomorphic liposarcoma (black triangles) and the position of the RasGAP and Cral domains (dark and light green respectively) are juxtaposed to known mutations in malignant peripheral nerve sheath tumors (MPNSTs; open triangles). B. Transcript expression according to copy number and sequence status in myxofibrosarcoma and pleomorphic liposarcoma compared to normal adipose tissue samples (black/red and green respectively, log2 expression from Affymetrix array profiling data; p-value=1.94×10−5, ANOVA; mutated tumors are indicated). One of the two R304* mutant tumors lacked expression data.
PIK3CA, encoding the catalytic subunit of phosphatidylinositol 3-kinase (PI3K), had one of the highest somatic mutation frequencies among the genes in this analysis (Table 2). Nucleotide substitutions in PIK3CA were initially detected in 4 of 21 myxoid/round-cell liposarcomas (MRCs). We measured the frequency of point mutations in PIK3CA in this subtype by genotyping an independent cohort of 50 MRCs16 for 13 common sites of PIK3CA mutation, including those discovered in our initial sequencing; mutations were detected in 9 additional patients (in total, 13 of 71). The mutations were clustered in two domains, the helical domain (E542K and E545K) and the kinase domain (H1047L and H1047R) (Table 2); both these domains are also mutated in epithelial tumors17.
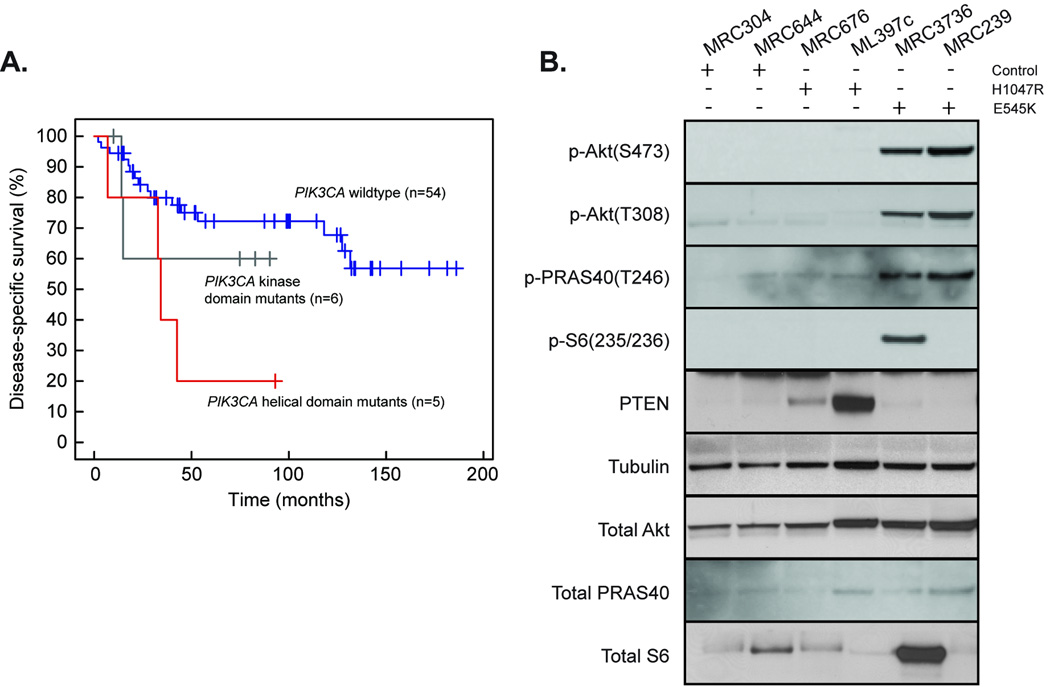
MRC patients whose tumors harbored mutations in PIK3CA had a shorter duration of disease–specific survival than did those with wildtype PIK3CA (p=0.036, log-rank test). Similar to observations in breast cancers18, patients with helical-domain PIK3CA mutations had worse outcomes than those with kinase-domain mutations (Figure 3A). However, this difference was not statistically significant given the small number of cases in our study.
Figure 3. Different effect of helical and kinase domain PIK3CA mutations on PI3K pathway activation and survival in myxoid/round-cell liposarcoma.
A. Survival for patients with tumors that harbor helical-domain mutations (red) versus kinase-domain mutations (grey), and wildtype PIK3CA (blue). The analysis includes the 65 patients for whom outcome information was available. Patients with mutations in either the helical or the kinase domain had a shorter disease–specific survival compared to those with wildtype PIK3CA (p-value = 0.0363, log-rank test). The difference in disease-specific survival between patients with helical-domain mutant tumors and those with wildtype PIK3CA tumors was significant (p-value=0.013, log-rank test). B. Western blots of myxoid/round-cell liposarcoma tumor lysates comparing the phosphorylation levels of Akt, PRAS40, and S6 kinase, as well as their protein levels, in patients with wild-type PIK3CA or with mutations in PIK3CA helical or kinase domains.
As both helical- and kinase-domain PIK3CA mutants are believed to activate Akt, although through different mechanisms19–21, we assessed Akt activation in MRC tumors harboring wildtype and mutated PIK3CA. Of note, only E545K helical-domain mutations were associated with increased Akt phosphorylation relative to wildtype, both at serine-473 and threonine-308 (TORC2 and PDK1 phosphorylation sites, respectively), and with increased phosphorylation of Akt substrates PRAS40 and S6 kinase (Figure 3B). Surprisingly, tumors with H1047R kinase-domain mutations did not have similar increases in Akt phosphorylation or activation (Figure 3B). However, H1047R-mutant tumors exhibited variably higher levels of PTEN, a negative regulator of PI3K activity, which may partly explain lower Akt activity. In addition, we detected a single MRC tumor with homozygous PTEN deletion and high Akt phosphorylation levels (data not shown). Further studies are needed to determine the relationship between activated PI3K signaling (resulting from PIK3CA mutations) and the pathognomonic t(12;16)(q13;p11) translocation in this subtype.
In addition to sequencing, we characterized the spectrum of genomic aberrations in soft tissue sarcoma with 250K single nucleotide polymorphism (SNP) arrays for somatic copy number alterations (SCNAs: n=207; Figure 1 and Supplementary Figure 2A) and loss-of-heterozygosity (LOH) (n=200; Supplementary Figure 2B) and with oligonucleotide gene expression arrays (n=149) (see Methods). The patterns of statistically significant SCNAs22,23 (Figure 1) revealed substantial differences between subtypes with simple and complex karyotypes (Figure 1). Myxoid/round-cell liposarcoma, synovial sarcoma, and GIST had relatively normal karyotypes compared to dedifferentiated and pleomorphic liposarcoma, leiomyosarcoma, and myxofibrosarcoma. In addition, only the four complex subtypes harbored significant copy-neutral LOH (Supplementary Figure 2B and Supplementary Table 4). These types exhibit varied levels of complexity: both dedifferentiated liposarcoma and leiomyosarcoma are less complex than pleomorphic liposarcoma and myxofibrosarcoma (Figure 1). The latter two subtypes were strikingly similar (Figure 1 and Supplementary Figure 2A), indicating they might appropriately be considered a single entity in a molecular classification, as previously suggested24.
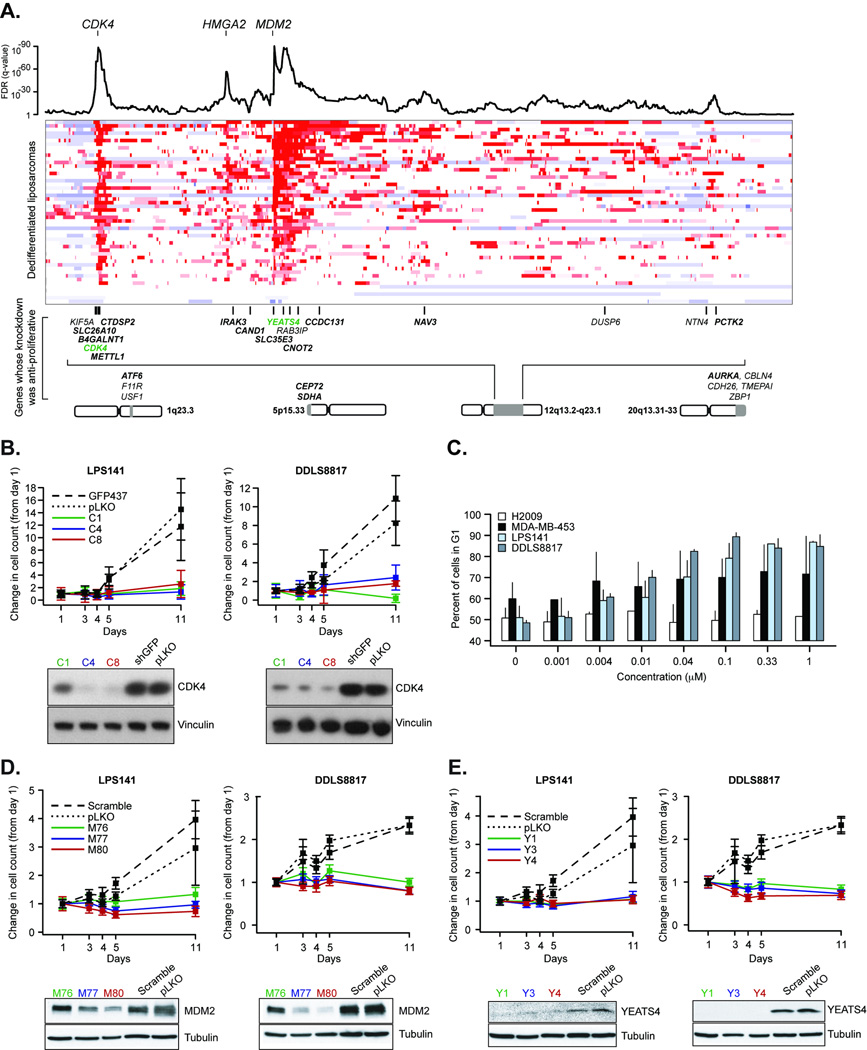
Our copy number profiling revealed both focal and broad regions of recurrent amplification (Supplementary Table 5). The alteration with the highest prevalence in any subtype was chromosome 12q amplification in dedifferentiated liposarcoma (~90%; Figure 1 and Figure 4A). As amplification is a common mechanism of oncogenic activation, we designed an RNA interference (RNAi) screen to help identify genes in amplified regions that are necessary for cancer cell proliferation in this subtype. We performed knockdown with short hairpin RNAs (shRNA) on 385 genes (Supplementary Table 2) in three dedifferentiated liposarcoma cell lines (LPS141, DDLS8817, and FU-DDLS-1) with copy number profiles similar to those observed in primary tumors of this subtype. A total of 2,007 shRNA lentiviruses, a median of five per gene, were tested for their effects on cell proliferation after 5 days (see Methods).
Figure 4. Genes whose knockdown is anti-proliferative in dedifferentiated liposarcoma and the consequences of CDK4, MDM2 and YEATS4 knockdown in dedifferentiated liposarcoma.
(A) Integrated profile of statistically significant genomic gains/amplifications as assessed by both RAE and GISTIC (combined as described in Methods; FDR, false-discovery rate) is followed by a heatmap of copy number segmentation on 12q13.2-q32.1 in 50 patient samples of dedifferentiated liposarcomas (red is amplification, blue is deletion, each row indicates one tumor sample). Below is the position of genes from our screen encoded by this region of 12q whose knockdown is anti-proliferative in dedifferentiated liposarcoma. Bold gene symbols indicate those whose amplification produced over-expression of its transcript or those over-expressed in tumor relative to normal adipose tissue. Genes in green are highlighted in panels B–C and E. Alternative genomic regions encoding genes not on 12q whose knockdown is anti-proliferative are also included. (B) Effect of three validated shRNAs targeting CDK4 on the proliferation of two cell lines, LPS141 and DDLS8817, at various time points (x-axis) with negative controls (pLKO empty vector and GFP473). Below are western blots showing the effect of shRNAs on levels of CDK4 protein (as indicated). (C) G1 arrest induced in LPS141 and DDLS8817 cell lines by treatment with the CDK4/CDK6 inhibitor PD0332991. MDA-MB-435 (Rb-positive) and H2009 (Rb-negative) were included as sensitive and insensitive controls. Error bars are s.d. of replicate measurements. (D–E) As in panel (B), effect on proliferation of three shRNAs targeting MDM2 (panel D) and YEATS4 (panel E) (negative controls: pLKO empty vector and scrambled shRNA) where each targeting shRNA resulted in reduced protein levels (at bottom). Error bars are propagated error from the ratio of mean and s.d. of measurements/replicates to time 0.
Using a statistical method, RSA (see Methods, Supplementary Note, and ref. 25), we identified 99 genes whose knockdown significantly decreased cell growth in at least one cell line (nominal p<0.05; Supplementary Table 6). For 91 of the 99 genes, two or more independent shRNAs had anti-proliferative activity, reducing the likelihood that our results are due to off-target effects. To determine whether the effect of gene knockdown on cell proliferation was specific for dedifferentiated liposarcoma, we compared our results to a pooled shRNA screen of ~9500 genes in 12 cancer cell lines of different types26 which included 58 of the 99 genes whose knock-down reduced proliferation. Only one of the 58 genes, PSMB4, was identified as a common essential gene, for which depletion reduced cell proliferation in ≥8 of 12 cancer cell lines in the prior study26.
27 of the 99 genes whose knockdown reduced proliferation were amplified in at least one of the three dedifferentiated liposarcoma cell lines used in our study (Supplementary Figure 3). Among these 27 genes, the most strongly overexpressed in dedifferentiated liposarcoma compared to normal fat15 was CDK4, a cell-cycle regulator and a known oncogene27. We confirmed that sustained knockdown of CDK4 (>10 days) inhibited proliferation when we assayed two of the three cell lines we screened (see Methods, Figure 4B). Furthermore, pharmacological inhibition of CDK4 in dedifferentiated liposarcoma cells with PD0332991, a selective CDK4/CDK6 inhibitor currently in clinical trials28, induced G1 arrest in the same two cell lines (Figure 4C).
For MDM2, another oncogene found in focal 12q amplifications, knockdown did not significantly impair proliferation in our arrayed screen in any of the three cell lines tested. Nevertheless, proliferation was impaired by subsequent knockdown lasting more than a week when we assayed two of those three cell lines (Figure 4D). Interestingly, another gene whose knockdown reduced proliferation of cells in which it was amplified was YEATS4 (GAS41), encoding a putative transcription factor that represses the p53 tumor suppressor network during normal cell proliferation29. YEATS4, frequently co-amplified with MDM2 (Figure 4A), was transcriptionally upregulated both in tumors relative to normal adipose tissue and in tumors with amplification compared to those copy-neutral for the locus (Supplementary Figure 3). Repeat shRNA experiments confirmed the effect of YEATS4 knockdown seen in the arrayed screen (Figure 4E), consistent with the hypothesis that YEATS4 and MDM2 amplification may cooperatively repress the p53 network in dedifferentiated liposarcoma, as recently suggested30. This finding may have consequences for Nutlin-based antagonism of the p53-MDM2 interaction15,31 in dedifferentiated liposarcomas. Our findings lend additional support for YEATS4 serving as a likely key amplified gene in cancer, as recently suggested through a weight-of-evidence classification scheme proposed for identifying such amplified cancer genes32.
This dataset provides the most comprehensive database of sarcoma genome alterations to date, revealing genes and signaling pathways not previously associated with this group of diseases. The study results are available as a community resource that might further the biological understanding of sarcomas and, eventually, shed light on additional strategies to improve patient care. Some of our findings already have potential therapeutic implications. For instance, the PIK3CA mutations found in MRC constitute the first report of such mutations in a mesenchymal cancer. These mutations identify a subset of tumors that might respond to treatment with PI3K inhibitors currently in clinical trials33. Our results also provide further rationale for use of CDK4 inhibitors in dedifferentiated liposarcoma and suggest the use of mTOR inhibitors in NF1-deficient sarcomas, since loss of NF1 function appears to cause mTOR pathway activation34. Finally, these data lend support for the clinical evaluation of agents targeting the p53/MDM2 interaction in dedifferentiated liposarcoma.
This work argues for the therapeutic importance of genomic alterations in sarcoma and encourages us to pursue next-generation sequencing strategies that will continue to define the landscape of genomic aberrations in these deadly diseases.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgements
For advice and discussion, we thank W. Lin, J. Boehm, C. Johannessen, A. Bass, M. Garber, S. Finn, J. Fletcher, W.C. Hahn, T. Golub, and all the members of the Spanish Group for Research on Sarcomas (GEIS). We are grateful for the technical assistance and support of B. Blumenstail, L. Ziaugra and S. Gabriel of the Broad Genetic Analysis Platform; J. Baldwin of the Broad Sequencing Platform; J. Franklin, S. Mahan and K. Ardlie of the Broad Biological Samples Platform; and H. Le, P. Lizotte, B. Wong, A. Allen, A. Derr, C. Nguyen and J. Grenier of the Broad RNAi Platform. We thank L. Borsu for assistance with Sequenom assays at MSKCC. The MSKCC Sequenom facility is supported by the Anbinder Fund. We also thank the members of the MSKCC Genomics Core Laboratory and N.H. Moraco for clinical data support. J. Fletcher and J. Nishio provided the LPS141 and FU-DDLS-1 cell lines respectively. J.B. is a Beatriu de Pinos fellow of the Departament d’Universitats, Recerca i Societat de la Informacio de la Generalitat de Catalunya. B.S.T is a fellow of the Geoffrey Beene Cancer Research Center at MSKCC. This work was supported in part by The Soft Tissue Sarcoma Program Project (P01 CA047179, S.S., M.L. and C.S.), The Kristen Ann Carr Fund, the Starr Foundation Cancer Consortium, and by a generous donation from Mr. Mortimer B. Zuckerman.
Footnotes
URLs
Sarcoma Genome Project (SGP) data portals, http://www.broadinstitute.org/sarcoma/ and http://cbio.mskcc.org/cancergenomics/sgp; The RNAi Consortium shRNA library, http://www.broadinstitute.org/rnai/trc/lib/; UCSC Genome Browser, http://genome.ucsc.edu/; Database of Genomic Variants (DGV), http://projects.tcag.ca/variation/; GenePattern, http://www.broadinstitute.org/genepattern/; Integrative Genomics Viewer (IGV), http://www.broadinstitute.org/igv/
Accession numbers
Study data is deposited in NCBI GEO under accession number GSE21124.
Author Contributions
Project conception: E.S.L, H.E.V., W.R.S., M.M., S. Singer. Study design and oversight by J.B., B.S.T, A.L., R.G.M., L.A.G., G.K.S., E.S.L, H.E.V., W.R.S., C.R.A., M.L., C. Sander, M.M., S. Singer. Sample selection and analyte processing was carried out by P.L.D, A.V., C.R.A, M.L., S. Singer. Sequencing and genotyping experiments were performed by J.B., A.H.R., K.S., C.H., R.N., M.H., T. Sharpe., T.F., K.C., R.C.O., C. Sougnez. W.W., H.G., T. Saito, N.S., C.L. RNA interference screen was performed by J.B., K.S., S. Silver, D.R. Validation experiments performed by S.B., M.L.Q., A.H., G.K.S. Statistical and bioinformatics analyses were performed by B.S.T, A.H.R, N.D.S, B.A.W, D.Y.C, B.R., C.M. G.G., Y.A., R.B., S.N., J.E.M. Analysis and interpretation of the results was carried out by J.B. and B.S.T. J.B., B.S.T., S.B., A.H.R., M.L., C.S., M.M., S. Singer drafted the manuscript. All authors contributed to critical review of the paper.
Author Information: The authors declare no competing financial interests.
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher C, Unni K, Mertens F, editors. Pathology and Genetics of Tumors of Soft Tissue and Bone. Lyon: International Agency for Research on Cancer Press; 2002. p. 427. [Google Scholar]
- 3.Heinrich MC, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 4.Hirota S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 5.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 6.van de Rijn M, Fletcher JA. Genetics of soft tissue tumors. Annu Rev Pathol. 2006;1:435–466. doi: 10.1146/annurev.pathol.1.110304.100052. [DOI] [PubMed] [Google Scholar]
- 7.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King AA, Debaun MR, Riccardi VM, Gutmann DH. Malignant peripheral nerve sheath tumors in neurofibromatosis 1. Am J Med Genet. 2000;93:388–392. [PubMed] [Google Scholar]
- 12.Maertens O, et al. Molecular pathogenesis of multiple gastrointestinal stromal tumors in NF1 patients. Hum Mol Genet. 2006;15:1015–1023. doi: 10.1093/hmg/ddl016. [DOI] [PubMed] [Google Scholar]
- 13.Maertens O, et al. Comprehensive NF1 screening on cultured Schwann cells from neurofibromas. Hum Mutat. 2006;27:1030–1040. doi: 10.1002/humu.20389. [DOI] [PubMed] [Google Scholar]
- 14.Upadhyaya M, et al. Characterization of the somatic mutational spectrum of the neurofibromatosis type 1 (NF1) gene in neurofibromatosis patients with benign and malignant tumors. Hum Mutat. 2004;23:134–146. doi: 10.1002/humu.10305. [DOI] [PubMed] [Google Scholar]
- 15.Singer S, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 16.Antonescu CR, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clin Cancer Res. 2001;7:3977–3987. [PubMed] [Google Scholar]
- 17.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 18.Barbareschi M, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–6069. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 19.Huang CH, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 20.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 21.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor BS, et al. Functional copy-number alterations in cancer. PLoS ONE. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idbaih A, et al. Myxoid malignant fibrous histiocytoma and pleomorphic liposarcoma share very similar genomic imbalances. Lab Invest. 2005;85:176–181. doi: 10.1038/labinvest.3700202. [DOI] [PubMed] [Google Scholar]
- 25.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 26.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 29.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol Cell Biol. 2006;26:4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Italiano A, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer. 2008;122:2233–2241. doi: 10.1002/ijc.23380. [DOI] [PubMed] [Google Scholar]
- 31.Muller CR, et al. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. Int J Cancer. 2007;121:199–205. doi: 10.1002/ijc.22643. [DOI] [PubMed] [Google Scholar]
- 32.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- 34.Johannessen CM, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutt A, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 37.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Major JE. Genomic mutation consequence calculator. Bioinformatics. 2007;23:3091–3092. doi: 10.1093/bioinformatics/btm339. [DOI] [PubMed] [Google Scholar]
- 40.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 41.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 42.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 43.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 44.Lin M, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 45.Ambrosini G, et al. Sorafenib inhibits growth and mitogen-activated protein kinase signaling in malignant peripheral nerve sheath cells. Mol Cancer Ther. 2008;7:890–896. doi: 10.1158/1535-7163.MCT-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrosini G, et al. Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene. 2007;26:3473–3481. doi: 10.1038/sj.onc.1210136. [DOI] [PubMed] [Google Scholar]
- 47.Nishio J, et al. Establishment of a novel human dedifferentiated liposarcoma cell line, FU-DDLS-1: conventional and molecular cytogenetic characterization. Int J Oncol. 2003;22:535–542. [PubMed] [Google Scholar]
- 48.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 49.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 50.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc, Ser B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.