Abstract
Protein structure refinement is an important but unsolved problem; it must be solved if we are to predict biological function that is very sensitive to structural details. Specifically, Critical Assessment of Techniques for Protein Structure Prediction (CASP) shows that the accuracy of predictions in the comparative modeling category is often worse than that of the template on which the homology model is based. Here we describe a refinement protocol that is able to consistently refine submitted predictions for all categories at CASP7. The protocol uses direct energy minimization of the knowledge-based potential of mean force that is based on the interaction statistics of 167 atom types (Summa and Levitt, Proc Natl Acad Sci USA 2007; 104:3177–3182). Our protocol is thus computationally very efficient; it only takes a few minutes of CPU time to run typical protein models (300 residues). We observe an average structural improvement of 1% in GDT_TS, for predictions that have low and medium homology to known PDB structures (Global Distance Test score or GDT_TS between 50 and 80%). We also observe a marked improvement in the stereochemistry of the models. The level of improvement varies amongst the various participants at CASP, but we see large improvements (>10% increase in GDT_TS) even for models predicted by the best performing groups at CASP7. In addition, our protocol consistently improved the best predicted models in the refinement category at CASP7 and CASP8. These improvements in structure and stereochemistry prove the usefulness of our computationally inexpensive, powerful and automatic refinement protocol.
Keywords: Refinement, Comparative Modeling, CASP7, ENCAD, MESHI, Knowledge-based, Stereochemistry
INTRODUCTION
Comparative modeling is a widely established approach that provides reliable structural information on naturally occurring protein sequences1,2. Even with the best possible template and sequence alignment, comparative models have coordinate errors compared to the true native structures. Refinement protocols that aim at detecting and correcting these errors are an indispensable part of the process. Model refinement has received a good deal of attention and the previous studies can be broadly classified by the magnitude of change they cause. On the one end of the scale are methods that explore significant changes in the model3,4,5 (i.e. replacing secondary structure elements, reorienting helices, etc.). These methods have the potential of adding significant structural information to the model6, but are computationally intensive and perform inconsistently across models. On the other end of the scale are methods that make small, local changes to the model. These methods usually fix obvious errors in the models such as gaps or clashes7,8,9, or change side-chains conformations10,11. Refinement of this kind is computationally much less expensive, shows consistent performance on the problem it is designed to solve, but generally does not move the structure significantly closer to the native coordinates.
Although refinement generally has a small effect on the coordinates of structures, the effect on the energy can be much larger. Thus, local refinement could aid the selection of the best final models out of large sets of decoys using quality assessment programs, such as ProQ12 and SELECTpro13. Ever since the first energy calculations on protein macromolecules14,15 showed that local refinement is very useful in combination with experimental data, this has been an active area. Most recently, low-resolution data such as electron density maps from electron microscopy have been used to deform a model to better fit experiment16,17.
All local refinement methods sample the conformational space around the initial model so as to optimize energy or more general scoring function, the final value of which is then used to select the best refined model. Two general procedures are common to ensure local changes in the initial model. The first, uses direct minimization or short simulated annealing molecular dynamics runs; sampling is restricted and never goes outside the energy landscape basin that surrounds the initial model which greatly speeds the calculation. The second ensures locality by adding restraining terms that are based on the initial model. Such terms remold the energy landscape to have a deeper basin around the initial model, making it very difficult for any sampling strategy to move too far. The details of the restraining terms vary, but they usually involve the initial coordinates14 or distances between atoms8,16,18.
In this work we present a refinement protocol that is based on minimization of the model with our knowledge-based potential, KB01. Previously we showed this potential to be effective in refining near-native decoy structures19,20. Here, we test our protocol on a more realistic protein modeling task, applying it to thousands of model submitted by participants in recent CASP events21. The CASP benchmark is highly diverse in the difficulty of the prediction targets, the variety the prediction methods used and the success of participants. Our protocol shows consistent average improvement of all models across prediction groups, target difficulty, and CASP category. Thus, we expect the protocol to be a useful addition to current local refinement tools, and discuss its possible inclusion in more general optimization pipelines.
METHODS
PREPARATION OF CASP7 MODELS AND REFINEMENT TARGETS
The seventh Critical Assessment of Techniques for Protein Structure Prediction (CASP7) experiment posted 114 targets from various experimental sources21. Submitted models from all human and server expert groups provide the dataset used for this work. We discarded models with more than 3% missing residues. We also discarded models that did not have side-chain represented with atomic detail. This resulted in a total of 36,802 models from 178 groups (11,537 models were discarded). We also used a total of 21 refinement targets from both CASP7 (9 models) and CASP8 (12 models) refinement categories (CASPR) for this study. These refinement targets are selected by the CASP organizers as the best predicted models from all groups for the target. For each target, the native structures were downloaded from the PDB and pre-processed using the domain definitions for each target mentioned on the CASP website.
THE KB01 REFINEMENT PROTOCOL
Our KB01 refinement protocol is composed of two steps: (a) structure refinement by energy minimization using a statistical knowledge-based potential of mean force derived for a bin width of 0.1Å (KB01)19 and (b) stereochemistry correction using MESHI22.
We initially parameterized the KB01 term19 more accurately in the ENCAD force field23. The physics based and knowledge-based hybrid force field in ENCAD is given by the following energy terms:
| (1) |
where Ebonded-terms are the published ENCAD bonded energy terms and EKB01 is the KB01 potential. We ran a parameter search on the KB01 weight (w) and tracked the refinement as improvement in weighted root mean square deviation20 of the near-native decoys (Fig. S3). The near-native decoys used were those used by Summa and Levitt19. We see that the optimal weight range for the KB01 energy term is from 0.1 and 2.0 (Fig. S3), confirming that the original work19 done at w=1.0 was near-optimally weighted. We used the optimum weight of w=0.381 for all KB01 minimization in this work.
KB01 minimization of each model consisted of 200,000 steps of energy minimization or until convergence to machine precision. The limited memory Broyden-Fletcher-Goldfarb-Shannon (L-BFGS) algorithm24 was used as the minimizer. We also ran a parameter search on part of the CASP7 models and got the same result as the near-native decoys for the optimal parameter range for the KB01 term in ENCAD (Fig. S3). We see this as a validation of our decoy testing methods in that the best conditions for the near-native decoys used earlier19,20 are also optimal for the CASP7 homology models.
MESHI used in the second step of the protocol restores stereochemistry spoilt by the KB01 minimization in the first step. MESHI consists of two stages. In the first MESHI stage, short fragments with disallowed {ϕ, Ψ} torsion angles are identified by scanning for high-energy residues with a Ramachandran energy term25. These fragments were replaced by fragments of corresponding lengths from native structures. The new fragments were selected according to the lowest RMSD fit to the Cαand Cβatoms of the starting fragments. The energy threshold for replacing fragment was set at a level that mark high-energy residues within the PROCHECK disallowed regions in the Ramachandran map26,27. These fragments were usually 1 to 3 residues in length.
In the second MESHI stage the model was subjected to 20,000 steps of energy minimization with the following energy:
| (2) |
where Ebonded-terms are the bonded terms of the MESHI force field22, ELJ is a Lennard-Jones term with united atom radii28, Ehydrogen-bonds is the count of all the hydrogen bond forming atom pairs that are closer than 3.3Å, ERamach is a knowledge-based formulation of the Ramachandran plot25 and Etether is a tethering term of the Cαand Cβatoms of the model to their initial positions with spring constants of 1 EnergyUnit/Å. The side-chains of the model were restrained to their initial rotamers through a quadratic term on every side-chain torsion angle (last term of Eq. 2).
METRICS USED FOR STRUCTURAL ASSESSMENT
We used the Cαroot mean square deviation (CαRMSD) and Global Distance Test (GDT) scores29 to measure the accuracy of any model with respect to its native structure. The GDT method first computes the maximum percentage of residues (not necessarily contiguous) that can be superimposed within a certain cutoff from their corresponding native structure residues. The GDT_TS score is an average of such percentages at cutoffs of 1Å, 2Å, 4Å and 8Å, and the GDT_HA score (high accuracy) is an average at cutoffs of 0.5Å, 1Å, 2Å and 4Å.
The GDT calculation produces a list of the superimposing residues at different cutoffs and thus enables a breakdown of the score according to different secondary structure components. Residues of the native structure were classified as “Helix”, “Sheet” or “Coil” according to DSSP30. The GDT contribution of a specific secondary structure class was then counted as the number of superimposing residues on the list that belong to a particular class, normalized by the total number of residues in that class.
A stereochemical index change (ΔSCI) is the average measure of the improvement in stereochemistry for models treated with our KB01 refinement protocol. This was calculated by averaging the stereochemical features, obtained using PROCHECK26,27, after normalization with their characteristic magnitudes.
We represent, σfeature, the ‘characteristic change magnitude’ for each feature as:
| (3) |
We create the stereochemistry index change (ΔSCI) for each group by averaging the features after normalization with their characteristic change magnitudes to give Z-scores:
| (4) |
where, Δ(Ramachandran outliers) is the percentage reduction of residues in Ramachandran generously allowed and disallowed regions, Δ(Chi 1,2 outliers) is the percentage reduction in number of rotamer outliers, Δ(Angle and bonds outliers per residue) is the reduction in number of bad bond angles and bond lengths per residue and Δ(Clashes per residue) is the reduction in number of van der Waals clashes per residue.
RESULTS
We first describe the overall results obtained by using the KB01 refinement protocol on all the models submitted to CASP7 from both human and server groups. We then present in more detail the improvements for the models predicted by the top performing groups at CASP7. We also test our protocol on targets in the refinement subcategory. Lastly, we assess the stereochemical improvements that the refinement protocol adds to the models of various groups at CASP7.
CONSISTENT REFINEMENT OF CASP7 MODELS
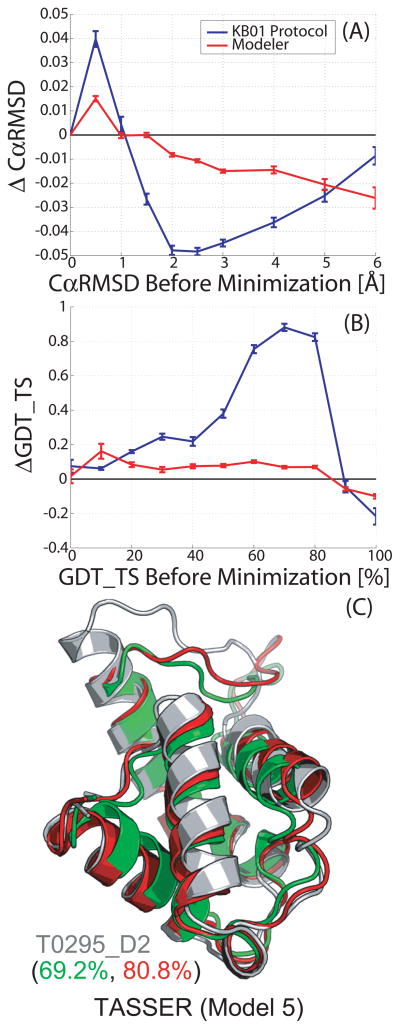
We run our KB01 refinement protocol on all the models submitted to CASP7, a task that was possible because of its low computation requirements. It takes only a few minutes (less than 5 minutes) to refine models of typical chain length (Fig. S4). This allows us to assess its performance on a diverse set of proteins and prediction techniques. Figure 1 shows the average accuracy changes following refinement, across all models, as a function of the initial model accuracy. The overall contribution of refinement is small but favorable, as indicated by both the decrease in the CαRMSD and the increase in GDT_TS score. A total of 22,336 models (out of 36,802) refined with an average ΔGDT_TS of 1.2%. The protocol is most successful for GDT_TS in the range of 50 to 80% (CαRMSD range of 1.5 to 3.5Å), which coincides with the range of values encountered in homology or comparative modeling. Outside this range the results degrade significantly. At the lower GDT_TS range, which corresponds to the most difficult comparative modeling and ab-initio approaches, the protocol still achieves minor but consistent improvement over the initial models. At the higher end of the GDT_TS range, which corresponds to situation with high sequence identity between target and template and a starting model that is highly accurate, the protocol slightly degrades the initial models. In Figure 1, we overlay the average results obtained by a single run of MODELLER31 (Ver 9.2), which is a widely used end-step in comparative modeling. Our protocol outperforms MODELLER in accuracy improvement by a significant amount and for all levels of target difficulty.
FIGURE 1.
Comparing improvements over all CASP7 submitted models caused by our KB01 refinement protocol (blue line) and by MODELLER (red line) for the CαRMSD (in A) and GDT_TS (in B). The changes are averaged in bins of width 0.5 Å for CαRMSD and 10% for GDT_TS, respectively, and are plotted as a function of the initial CαRMSD or GDT_TS scores of the models. The standard deviation of the mean is shown as error bars for both CαRMS and GDT_TS. Successful refinement that brings the structure closer to the native structure is associated with a negative change in CαRMSD and positive change in the GDT_TS. Our KB01 refinement protocol (blue line) improves the models more than does MODELLER, which is a commonly used method (red line). Very accurate initial modes (CαRMSD < 1 Å or GDT_TS > 90%) are not improved by either method. For models with initial GDT_TS score in the range 50% to 80% our KB01 refinement protocol leads to an average improvement of 1%. (C) Showing model 5 for target T0295_D2 submitted by TASSER, a good case with KB01 protocol in the 50 to 80% initial GDT_TS range. The improvement in GDT_TS is 11.6%, from initial GDT_TS of 69.2% to final GDT_TS of 80.8%. The refinement is clearly seen in the movement of the alpha-helices from the initial model (green) to the refined model (red), which moves them towards their positions in the native structure (gray).
The changes caused to the models by the protocol are generally local and moderate as expected from energy minimization. The small CαRMSD change (Fig. 1A) indicates that the global folds of the models do not change. The overall secondary structure composition for all CASP7 models is 36.85% helix, 19.97% sheet and 43.18% coil. Table II shows the contributions from different secondary structures to the average GDT change induced by the KB01 refinement protocol at various CαRMSD movements (0.5Å, 1Å, 2Å, 4Å and 8Å) for residues in the “Helix”, “Sheet” and “Coil” state of particular models in the comparative modeling category. We observe that most improvement in GDT_TS score are for the helical residues of the protein models, followed by some improvement in coil residues and least improvement in sheet residues. In general, both helical and coil residues are refined for nearly the entire range of modeling difficulty whereas sheet residues are not refined for the initial GDT_TS range from 20 to 60% (Fig. S5). This could be a limitation of the KB01 protocol or due to bad models submitted by many groups. One of the best refinement cases (Fig. 1C) exemplifies the changes in the model following refinement. It shows an 11.6% improvement in the GDT_TS score of a model submitted by the TASSER group to CASP target T0295_D2. This significant accuracy improvement stems from better spacing and orientation of model parts that are near the correct conformation. As expected, energy minimization with its limited sampling is not able to improve the parts of the models that were initially wrong.
Table II.
The contributions of different secondary structures towards the average GDT change for the KB01 refinement protocol. The GDT change is averaged across models from all CASP7 groups with initial GDT_TS scores of 50%–80%. The contributions are normalized by the abundance of the secondary structures in each target. Positive values indicate successful refinement.
| Secondary Structure | Cutoffs | ||||
|---|---|---|---|---|---|
| 0.5Å | 1Å | 2Å | 4Å | 8Å | |
| Helix | 3.1% | 3.1% | 1.7% | 0.5% | 0% |
| Sheet | −1.4% | 0.1% | 0.1% | 0% | 0% |
| Coil | 0.1% | 0.5% | 0.8% | 0.3% | 0.1% |
The structural improvements for all models are almost entirely due to ENCAD energy minimization with the KB01 potential; the affect of the MESHI force field on KB01 minimized models is negligible for the CαRMSD, GDT_TS and GDT_HA scores for the entire range of target difficulty (Fig. S1). Clearly, KB01 minimization consistently improves structures by moving them towards the native structure. This indicates that the KB01 potential of mean force is a good energy function for refinement with energy minima near the native structure.
IMPROVEMENTS FOR BEST PERFORMING GROUPS AT CASP7
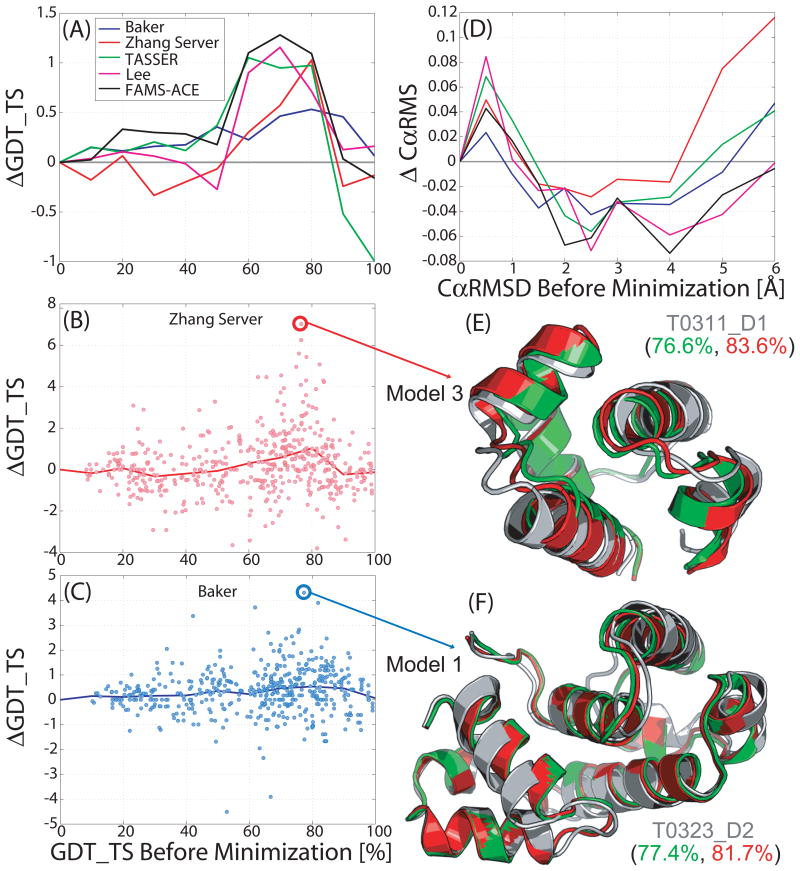
How does our KB01 refinement protocol work on predictions from the top performing groups at CASP732? We focus on five groups: Baker, Zhang Server, TASSER, Lee and FAMS-ACE. The average improvement profiles for each group (Figures 2A and 2D) vary both in the maximal extent of improvement and the initial model accuracy for which it occurs. The Baker group benefits the least from the protocol with a peak improvement in GDT_TS of just 0.5% compared to 1.0% in GDT_TS for Zhang Server, TASSER and Lee and 1.25% for FAMS-ACE group. Nevertheless, we are heartened to see that all the profiles are positive in the GDT_TS of 50 to 80%, corresponding to comparative modeling. This suggests that the protocol can provide a consistent improvement irrespective of the methodology producing the initial model.
FIGURE 2.
Showing the average KB01 refinement protocol improvement for the five best performing groups at CASP7. (A) Showing the average improvement in GDT_TS for all submitted models for each of these groups. Most improvement is again seen for the initial GDT_TS between 50 and 80%. Average improvement over this GDT_TS range is 0.42% for Baker, 0.42% for Zhang Server, 0.65% for TASSER, 0.71% for Lee and 1.05% for FAMS-ACE. (B) Showing the spread of improvement in GDT_TS for all models of Zhang Server (red points) and the average improvement in GDT_TS (red line). There are 13 models with more than 4% improvement in GDT_TS. (C) Showing the spread of improvement in GDT_TS for all models of Baker (blue points) and the average improvement in GDT_TS (blue line). There is only one model with more than 4% improvement in GDT_TS. (D) Showing consistent improvement in CαRMSD for all five top-performing groups at CASP7. As seen in Figure 1, the KB01 refinement protocol is not able to improve models very close (< 1 Å CαRMSD), i.e. for high homology cases (GDT_TS > 90%). (E) Showing the most refined submitted model for Zhang Server. An improvement of 7.0% in GDT_TS is evident for model 3 of target T0311_D1 as the KB01 refined structure (red) is closer to the native structure (grey), compared to the starting model (green). (F) Showing best improvement with Baker submitted model 1 of target T0323_D2 with an improvement of 4.3% in GDT_TS. Local refinements all over the protein structure are shown with no significant shift in secondary structure units that was seen for the Zhang Server result in (E).
For greater detail, we focused on the impact of our refinement on individual models from two groups: Baker and the Zhang Server. Figures 2B and 2C show the scatter of GDT_TS score changes following refinement for all the models submitted by these two groups. The magnitude of the scatter is about 1% for the Baker group and 2% for the Zhang Server group. We see that the scatter is about twice as large as the peak average improvements. Still, cases of significant degradation are rare; less than one percent of the models are degraded by more than 2% in GDT_TS score for both groups. Thus, we can consider the protocol as a ‘safe’ refinement strategy. We show the best refinement cases from the two groups with improvements of 4.3% and 7% in GDT_TS. As seen from the example presented in the previous section, refinement causes local changes to the model that appear to mainly influence the relative spacing of segments of polypeptide chain.
We also compared GDT_HA scores, which are more sensitive to small changes in structure than GDT_TS, for the three groups, Baker, the Zhang Server and TASSER, over all CASP7 targets (Fig. S2). The average improvement in these individual groups (solid lines in Fig. S2 for Baker, Zhang Server and TASSER) follow the same trend of average improvement in GDT_HA for the entire CASP7 corpus for KB01 protocol with the maximum improvement in the comparative modeling range; also observed for GDT_TS scores (Fig. 1). The affect of the KB01 refinement protocol on secondary structure for these five groups is consistent with the result for all the CASP7 modes. There are seven models with more than 4% improvement in GDT_HA for Baker, six models with more than 6% improvement for Zhang Server and three models with more than 10% improvement in GDT_HA for TASSER.
REFINEMENT OF MODELS FROM THE CASP REFINEMENT CATEGORY
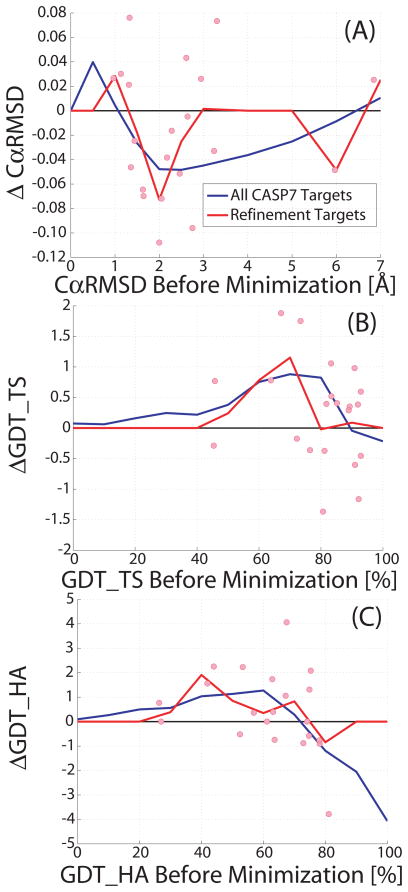
The consistency of KB01 refinement protocol was further tested on the refinement targets in the refinement category (CASPR) of CASP7 and CASP8. These comprise nine individual models from CASP7 and twelve from CASP8 that were chosen by the CASP organizers from the best submitted models for each respective target. These models are a mix of comparative modeling targets with small inaccuracies in alignment compared to the experimental structure21. The goal of this category is to test if further refinement of the predicted models can be achieved using the information about problematic areas which are given by the CASP organizers for many of the CASPR structures.
We ran our KB01 refinement protocol on all twenty one refinement CASPR targets but choose not to use the information provided by the organizers about the problematic regions in the model. Also, our protocol was applied on the entire target structure with no sub-division into individual domains, even when the target was known to have two or more domains. These choices were made to provide a true consistency check of our protocol on the best predictions at CASP. Figure 3 details the changes to the accuracy scores of the twenty refinement models following our protocol. Although the sample size is small, the improvement obtained for the refinement targets is consistent with the average trend seen above for all the CASP7 models. More specifically, the maximum average improvement is 1% in GDT_TS for both the refinement targets and all of CASP7 corpus (Fig. 3B). The affect of the KB01 refinement protocol on secondary structure for the refinement targets is consistent with the result on all the CASP7 models. The two best results for GDT_TS (Fig. 3(B)) are for the comparative modeling (50 to 80% GDT_TS) category for the targets TR368 and TR462. Most residues in the native structure of TR368 are alpha-helical and TR462 contains more alpha-helical and coil residues than sheet residues. Large improvements in GDT_TS for the high homology category also have similar secondary structure characteristics in the native structure. For all the targets that were degraded, there was a mix of sheet and coil residues. The best improvement in GDT_TS was around 2% and the best improvement for GDT_HA around 4% for refinement targets in the comparative modeling category. MacCallum et al.33 reasoned that metrics like high-accuracy metrics like GDT_HA would be more important for the refinement targets, as most of the starting models would already be close to the native structure.
FIGURE 3.
Showing improvements of CASP7 and CASP8 refinement targets (red circles) with our KB01 refinement protocol as measured by the (A) CαRMSD, (B) GDT_TS and (C) GDT_HA scores. Improvement caused by refinement is evident from a negative change in CαRMSD and positive changes in GDT_TS and GDT_HA. Most improvement is again seen in the 50 to 80% GDT_TS range. The average improvement for all twenty one refinement targets (red lines) shows a similar trend to that seen for all CASP7 models (blue lines) with improvement over nearly the entire range modeling difficulties. This shows the consistency of our KB01 refinement protocol. A sharp decline in GDT_HA for all CASP7 models with high initial GDT_HA values shown in (C) is due to the few high homology targets that KB01 was not able to improve.
We tested our KB01 refinement protocol on the twelve refinement targets of CASP8 refinement category to assess the performance of the current protocol with the actual data from the CASP8 refinement category experiment. Had we submitted our refined structure as Model 1, we would have obtained an average improvement of 0.36% in GDT_TS and 0.15% in GDT_HA score for all twelve targets. In the actual CASP8 experiment, the greatest average improvement for Model 1 was 0.09% in GDT_TS and 0.27% in GDT_HA (see LEE in Table II of MacCallum et al.33). This shows that our current protocol can be safely used as a final refinement step.
STEREOCHEMICAL CORRECTION
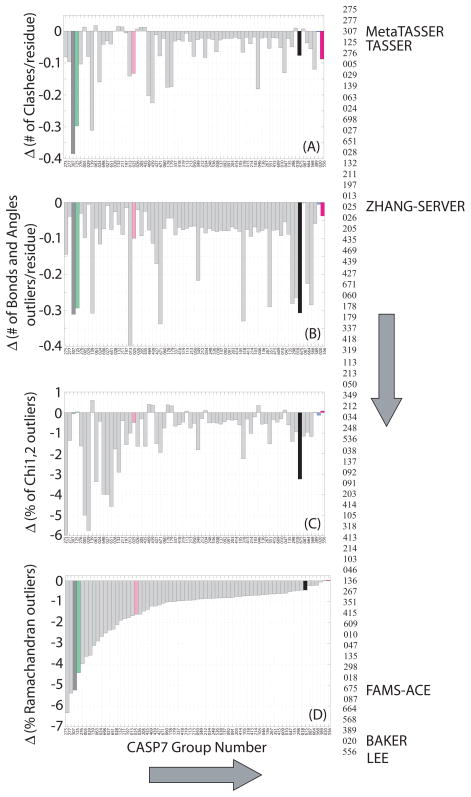
We tested the effect of our KB01 refinement protocol on stereochemistry by comparing the PROCHECK26,27 reports of the original and refined models. We focused on four PROCHECK criteria used in two recent assessment studies33,34, namely: (i) number of clashes, indicating van der Waals violations, (ii) number of angles and bonds outliers, (iii) number of side-chain chi1 and chi2 outliers, and (iv) number of backbone ({ϕ, Ψ}) angles that are outliers on the Ramachandran map. We also restricted our analysis to models with a GDT_TS in the comparative modeling range of 50 to 80%. Figure 4 shows the variation of these different stereochemical criteria for different groups of predictors. Such variation is expected as different groups used a wide range of methodologies for prediction. However, average improvement is seen on all the criteria and for nearly all the groups.
FIGURE 4.
Showing the effect of KB01 protocol on the stereochemical components of all CASP7 models submitted by all groups with initial GDT_TS from 50% to 80%. A negative change for any feature indicates stereochemical improvement. Almost all groups show consistent improvement in all stereochemical features. The Baker models submissions were stereochemically correct models and were improved least over all features of stereochemistry (blue bar). The Zhang Server models (red bar) and FAMS-ACE (black bar) were corrected for all features of stereochemistry. A large number of clashes (number of bad contacts) and number of bond or angle outliers were reduced in Lee models (magenta bar) with slight degradation in side chain chi 1 and 2 angles. The number of clashes, number of bond and angle outliers and percentage of backbone dihedral outliers on the Ramachandran map were greatly reduced for submitted models from both TASSER (green bar) and MetaTASSER (dark grey bar) with the KB01 protocol. The vertical string of 3 digit numbers are the group IDs for various groups at CASP7, appearing in the same order for the respective bar plots for every feature.
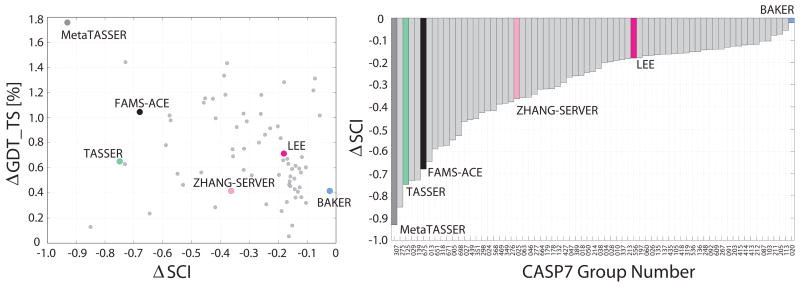
The value of stereochemical index change (ΔSCI) allows us to compare the average improvement our protocol causes to the stereochemistry of models belonging to each prediction group (Fig. 5, right). It is encouraging that all groups have negative ΔSCI values, indicating improvement. Also, the ordering of the groups is easy to explain. For example, the Baker group emphasizes the submission of stereochemical plausible models3, thus it is not surprising that it gets a ΔSCI of nearly 0. On the other hand, TASSER models use a coarse-grained lattice grid for greater speed35 and consequently lack good stereochemistry. We find that the ΔSCI and ΔGDT_TS values of a particular group are uncorrelated (Fig. 5, left). This shows that our protocol does not improve GDT_TS through correction of stereochemistry.
FIGURE 5.
Showing correlation between average improvements in GDT_TS and average change in Stereochemical Index (ΔSCI) for all submitted models by all groups at CASP7 that have an initial GDT_TS score between 50% and 80%. The Stereochemical Index is a score that equally weights the four components in Figure 4. A negative change in ΔSCI indicates stereochemical improvement and a positive change in ΔGDT_TS indicates structural improvement. The improvement in GDT_TS is not due to the improvement in stereochemistry as no significant correlation exists (CC=−0.29 left plot); there is more correlation between ΔGDT_TS and ΔSCI for the six named groups (CC=−0.77), which is mainly due to MetaTASSER results. A varying degree of improvement in stereochemistry is seen for all CASP7 groups (right plot), with the most improvement in stereochemistry as well as GDT_TS for the models submitted by MetaTASSER.
DISCUSSION AND CONCLUSION
This work presents a refinement protocol that utilizes the KB01 knowledge-based potential. The protocol is based on direct energy minimization with that potential and thus requires a few minutes (typically less than 5 minutes) of CPU time for proteins of varying chain length in CASP7. This low computational cost allowed us to run it on the thousands of models submitted to CASP7, an unparalleled model repository of protein types and prediction techniques. We find that the protocol improves the accuracy improvements to models that had an initial GDT_TS accuracy of 50 to 80%. We are very heartened to see an average improvement in coordinate accuracy for all groups on all the models they submitted in that GDT_TS range (Table I).
Table I.
Average change in the Stereochemical Index (ΔSCI) and GDT_TS score for all groups across submitted models in the 50 to 80% GDT_TS range. A negative value of ΔSCI and a positive value of ΔGDT_TS show improvement with our KB01 refinement protocol. Groups that are mentioned in Figure 5 are named. Entries are sorted by decreasing ΔGDT_TS value.
| Group Name | Group ID | ΔSCI | ΔGDT_TS | Group Name | Group ID | ΔSCI | ΔGDT_TS | Group Name | Group ID | ΔSCI | ΔGDT_TS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MetaTASSER | 307 | −0.93 | 1.76 | 50 | −0.25 | 0.86 | 427 | −0.29 | 0.54 | ||
| 139 | −0.73 | 1.44 | 10 | −0.19 | 0.84 | 319 | −0.16 | 0.53 | |||
| 276 | −0.38 | 1.43 | 47 | −0.27 | 0.80 | 248 | −0.14 | 0.51 | |||
| 469 | −0.39 | 1.33 | 651 | −0.59 | 0.78 | 105 | −0.16 | 0.49 | |||
| 205 | −0.07 | 1.31 | 63 | −0.36 | 0.75 | 435 | −0.16 | 0.47 | |||
| 197 | −0.18 | 1.28 | 178 | −0.32 | 0.75 | 698 | −0.53 | 0.46 | |||
| 211 | −0.08 | 1.22 | Lee | 556 | −0.18 | 0.71 | 418 | −0.16 | 0.45 | ||
| 18 | −0.26 | 1.18 | 46 | −0.36 | 0.69 | 415 | −0.13 | 0.44 | |||
| 349 | −0.38 | 1.18 | 212 | −0.12 | 0.69 | 92 | −0.14 | 0.43 | |||
| 298 | −0.43 | 1.15 | 26 | −0.17 | 0.67 | Baker | 20 | −0.02 | 0.42 | ||
| 351 | −0.45 | 1.15 | 213 | −0.18 | 0.66 | Zhang Server | 25 | −0.36 | 0.42 | ||
| 439 | −0.46 | 1.12 | TASSER | 125 | −0.75 | 0.65 | 91 | −0.13 | 0.41 | ||
| FAMS-ACE | 675 | −0.68 | 1.05 | 29 | −0.73 | 0.63 | 413 | −0.12 | 0.37 | ||
| 389 | −0.26 | 1.03 | 135 | −0.17 | 0.63 | 609 | −0.14 | 0.36 | |||
| 664 | −0.32 | 1.03 | 267 | −0.14 | 0.62 | 60 | −0.17 | 0.33 | |||
| 113 | −0.06 | 1.02 | 87 | −0.10 | 0.60 | 103 | −0.10 | 0.32 | |||
| 318 | −0.58 | 1.02 | 203 | −0.13 | 0.59 | 214 | −0.24 | 0.31 | |||
| 568 | −0.42 | 1.00 | 536 | −0.15 | 0.59 | 24 | −0.42 | 0.29 | |||
| 671 | −0.57 | 0.98 | 179 | −0.32 | 0.58 | 337 | −0.19 | 0.26 | |||
| 132 | −0.31 | 0.97 | 27 | −0.47 | 0.56 | 13 | −0.65 | 0.24 | |||
| 34 | −0.20 | 0.93 | 414 | −0.12 | 0.56 | 136 | −0.15 | 0.14 | |||
| 38 | −0.23 | 0.92 | 5 | −0.55 | 0.55 | 275 | −0.85 | 0.13 | |||
| 277 | −0.34 | 0.92 | 28 | −0.20 | 0.54 | 137 | −0.16 | 0.05 |
The general success of the protocol in improving accuracy makes it a safe and beneficial end-step strategy for any prediction method. The only exception is application to very high homology models (i.e. with initial GDT_TS>80%) for which the protocol is not effective. Fortunately, the high homology cases are usually the result of high sequence similarity, which is easily identified. Moreover, most improvement in GDT_TS and GDT_HA scores is for the helical residues of the protein models for nearly the entire range of modeling difficulty, followed by some improvement in coil residues and least improvement in sheet residues (Fig. S5). Thus, a consistent improvement in helical residues for nearly the entire range of modeling difficulty suggests that KB01 potential works by preserving pairwise distances, in that the spacing between the alpha-helical subunits in the proteins is corrected as shown in the cartoons of Figures 1 and 2. We therefore recommend that the protocol is only run on models that result from templates with less than 25% sequence identity as well as on models from nearly the entire range of modeling difficulty, which contain a high percentage of helical and coil residues to get the maximum benefit from the KB01 refinement protocol. In our experience, this still leaves the majority of CASP models amenable to the protocol.
Table I shows that predictions of every group are improved in terms of fit to the native structure (GDT_TS) and stereochemistry (SCI). Predictions by the MetaTASSER server were most improved with our protocol. It is well-known that MetaTASSER predictions can be significantly improved by small manual intervention, such as done in TASSER36. Even top performing groups like the Zhang Server can benefit from our KB01 refinement protocol with an improvement in stereochemistry and GDT_TS. There is consistent improvement in stereochemistry averaged over all models and groups at CASP7 for models in the 50 to 80% GDT_TS range. Specifically, percentage reduction in the number of outliers on the Ramachandran map is 1.40%, in the number of rotamer outliers is 1.09%, the reduction in number of van der Waals clashes per residue is 0.06 and in the number of bond length and angle outliers is 0.11. Overall the ΔSCI value, a Z-score, is reduced by 0.31.
The success of our KB01 refinement protocol on unknown homology models is a validation of our decoy testing methods19,20 in that we did as well as we had expected to do. For the refinement category at CASP8, improvements in the models were achieved by simply refining structures provided by the CASP8 organizers with our protocol. Many groups did not refine from the given initial structure and re-ran their prediction methodology. Many also used the information provided by the organizers to refine certain parts of the protein. Our refinement protocol did not use any information provided by the organizers and refined the entire model in a completely automated and computationally inexpensive procedure.
The protocol is a combination of two minimization steps: KB01 and MESHI. We compared the contributions of both steps to the accuracy improvement by running them separately on the initial models (Fig. S1). The comparison indicates that the improvement in model accuracy is chiefly caused by KB01 minimization and not by the MESHI stereochemical correction step. We introduced the MESHI step after preliminary trials of the project showed that the strong weight given to the KB01 potential distorted of bonded stereochemistry. This led to the use of MESHI additional minimization step to restore proper stereochemistry.
We find it intriguing that the KB01 refinement is qualitatively similar across models from so many groups (Fig. 2A), including the top ranking ones. This suggests that the KB01 potential has a local minimum closer to the native structure than do many of the other force-fields; it is consistent with previous studies of the KB01 energy surface19,20. The KB01 pairwise potential encodes information about favored pairwise interatomic distances (see local minima of Fig. 2 in Summa and Levitt19), which seems to be essential for structural improvement. The lower resolution potentials like KB0219, KB0519 and the physics-based non-bonded potentials do not contain such information and also do not refine. Co-localization of the native structure and minimum is a very desirable property for a potential, but it must also be accompanied by a suitable sampling. In this work sampling was limited by direct minimization and this resulted in limited atomic movement. The consistency of our KB01 refinement protocol indicates that the KB01 potential energy surface fits native proteins structures well. This suggests that further refinement could be achieved by using better sampling methodologies. This is beyond the scope of the current work, but we are currently testing various stochastic optimization techniques to better sample the KB01 potential energy surface.
We conclude that our KB01 refinement protocol performs better than MODELLER, in that it improves both the structure as well as stereochemistry of the predicted models consistently. A refined model with improved GDT score and better stereochemistry makes our refinement protocol a natural “end” step for any modeling scenario.
Supplementary Material
Acknowledgments
We thank João Rodrigues for his help during the revision of the paper and the reviewers for a very careful review. This work was supported by National Institutes of Health Grant GM63817 (to G.C., N.K. and M.L.). Simulations were done on the Bio-X2 Dell supercluster and supported by National Science Foundation Grant CNS-0619926.
References
- 1.Chandonia JM, Brenner SE. The impact of Structural Genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 2.Levitt M. Growth of novel protein structural data. Proc Natl Acad Sci USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misura KM, Chivian D, Rohl CA, Kim DE, Baker D. Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc Natl Acad Sci USA. 2006;103:5361–5366. doi: 10.1073/pnas.0509355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joo K, Lee J, Lee S, Seo J, Lee S, Lee J. High accuracy template based modeling by global optimization. Proteins. 2007;69(Suppl 8):83–89. doi: 10.1002/prot.21628. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 6.Qian B, Raman VS, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 8.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 9.Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics. 2006;Suppl 15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Canutescu AA, Dunbrack RL. SCWRL and MolIDE: programs for protein side-chain prediction and homology modeling. Nat Protoc. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77(4):778–95. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12(5):1073–86. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randall A, Baldi P. SELECTpro: effective protein model selection using a structure-based energy function resistant to BLUNDERs. BMC Struct Biol. 2008;8:52. doi: 10.1186/1472-6807-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levitt M, Lifson S. Refinement of protein conformations using a macromolecular energy minimization procedure. J Mol Biol. 1969;46:269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 15.Jack A, Levitt M. Refinement of Large Structures by Simultaneous Minimization of Energy and R Factor. Acta Crystallogr. 1978;A34:931–935. [Google Scholar]
- 16.Schröder GF, Brunger AT, Levitt M. Combining Efficient Conformational Sampling with a Deformable Elastic Network Model Facilitates Structure Refinement at Low Resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of Protein Structures into Low-Resolution Density Maps Using Rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summa C, Levitt M. Near-native structure refinement using in vacuo energy minimization. Proc Natl Acad Sci USA. 2007;104:3177–3182. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopra G, Summa CM, Levitt M. Solvent dramatically affects protein structure refinement. Proc Natl Acad Sci USA. 2008;105:20239–20244. doi: 10.1073/pnas.0810818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalisman N, Levi A, Maximova T, Reshef D, Zafriri-Lynn S, Gleyzer Y, Keasar C. MESHI: a new library of Java classes for molecular modeling. Bioinformatics. 2005;21:3931–3932. doi: 10.1093/bioinformatics/bti630. [DOI] [PubMed] [Google Scholar]
- 23.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput Phys Commun. 1995;91:215–231. [Google Scholar]
- 24.Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Math Programming. 1989;45:503–528. [Google Scholar]
- 25.Amir ED, Kalisman N, Keasar C. Differentiable multi-dimensional, knowledge-based energy terms for torsion angle probabilities and propensities. Proteins. 2008;72(1):62–73. doi: 10.1002/prot.21896. [DOI] [PubMed] [Google Scholar]
- 26.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: Standard radii and volumes. J Mol Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 29.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A, Sali A. Modeller: generation and refinement of homologybased protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 32.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 33.MacCallum JL, Hua L, Schnieders MJ, Pande VS, Jacobson MP, Dill KA. Assessment of the protein-structure refinement category in CASP8. Proteins. 2009;77(Suppl 9):66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Arakaki AK, Skolnick JR. TASSER: an automated method for the prediction of protein tertiary structures in CASP6. Prot Struct Funct Bioinform. 2005;61(Suppl 7):91–98. doi: 10.1002/prot.20724. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H, Pandit SB, Lee SY, Borreguero J, Chen H, Wroblewska L, Skolnick J. Analysis of TASSER-based CASP7 protein structure prediction results. Proteins. 2007;69(Suppl 8):90–97. doi: 10.1002/prot.21649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.