Abstract
If any proof were needed of DNA’s versatile roles and use, it is certainly provided by the numerous depositions of new three-dimensional (3D) structures to the coordinate databanks (PDB, NDB) over the last two years. Quadruplex motifs involving G-repeats, adducted sequences and oligo-2′-deoxynucleotides (ODNs) with bound ligands are particularly well represented. In addition, structures of chemically modified DNAs (CNAs) and artificial analogs are yielding insight into stability, pairing properties and dynamics, including those of the native nucleic acids. Besides being of significance for establishing diagnostic tools and in the analysis of protein-DNA interactions, chemical modification in conjunction with investigations of the structural consequences may yield novel nucleic acid-based therapeutics. DNA’s predictable and highly specific pairing behavior makes it the material of choice for constructing 3D-nanostructures of defined architecture. Recently the first examples of DNA nanoparticle and self-assembled 3D-crystals were reported. Although the structures discussed in this review are all based either on X-ray crystallography or solution NMR, small angle X-ray scattering (SAXS) and cryoEM are proving to be useful approaches for the characterization of nanoscale DNA architecture.
Introduction
With single-crystal structure determination of DNA entering its fourth decade, coordinate databanks now boast well above a thousand entries of ODNs (Nucleic Acid Database, NDB, http://ndbserver.rutgers.edu [1]). These structures have provided atomic-resolution snapshots of the duplex families and insights on the relationship between sequence and geometry, and revealed water structure, ion coordination and the binding modes of small molecule ligands and drugs. Despite all the progress, it cannot be overlooked that the sequence space has not been sampled very well. A handful of sequences make up a relatively large percentage of the entries. The Dickerson-Drew Dodecamer (DDD) of sequence d(CGCGAATTCGCG) is arguably the best example as this B-form duplex accounts for some 10% of all deposited structures.
Recently, the most frequent short sequences in non-coding DNA from several genomes including Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens were reported [2•]. This analysis led to the surprising conclusion that virtually none of the abundant DNA sequences have had their structure determined crystallographically. On the other hand, the DDD sequence occurs very rarely, in fact only once or not at all in the genomes studied (J.A. Subirana, pers. comm.). Nobody will argue that the structural dissection of the DDD has not helped our understanding of DNA geometry (narrow A-tract minor groove), bending (asymmetric kink), ion coordination (Mg2+ binding at the GpC and alkali metal ion binding at the ApT step in the major and minor grooves, respectively) and water structure (spine of hydration) [3–7]. In fact, we know now that narrowing of the minor groove and the resulting strongly negative electrostatic potential, first observed in the structure of the DDD, are widely used by proteins for DNA recognition (indirect readout) [8•]. Nevertheless, the fact that almost all of the most common DNA sequences remain structurally unexplored comes as somewhat of a shock. Because it is possible that such abundant sequences in non-coding regions of eukaryotic genomes serve specific purposes (i.e. the formation of nucleosome structures), their structural properties, potentials for drug binding and other features should be studied in more detail [2•].
Bioinformatics surveys have also mapped previously known sequence motifs such as G-rich stretches that were typically associated with the telomeric ends of eukaryotic chromosomes to other regions, such as promoters, especially those of oncogenes, and 5′-UTRs [9,10]. A better understanding of G-quadruplexes in promoters and the factors associated with their formation and breakdown is of interest in drug discovery and could lead to new cancer therapies. For example, the quadruplex of the myc oncogene that is extensively and intrusively involved in tumor biology could be targeted with small molecules in order to repress its transcriptional activation [11••]. Two drugs targeting G-quadruplexes, albeit not the one from myc, are currently in Phase II clinical trials. Both appear to exert their effects through the protein nucleolin that was shown to also bind to the myc G-quadruplex, thereby suppressing gene expression ([11••] and cited refs.). The structures of G-quadruplexes exhibit a high degree of conformational heterogeneity (strand polarities and loop arrangement) [12••], and thus offer opportunities for selective binding by small-molecule agents in principle [9]. For example, the structures of ODNs featuring human telomeric repeats determined in solution and in the crystal revealed significant differences. Conformational changes were also observed as a result of different conditions in solution (Na+ vs. K+ and between different potassium forms) [13–19]. New structures of G-quadruplexes and their complexes with small molecule binding agents have appeared at an enhanced rate in recent years [12••].
Here we highlight advances in the field of ODN structure determination during the last 24 months. Beyond the structural investigations focusing on native DNA we also review studies directed at CNAs and nucleic acid analogs of interest in the context of the pairing properties and function of native DNA and RNA. Along with the relevant citation(s) for the structures discussed, the coordinate entry codes in the Protein Data Bank (www.rscb.org [20]) are also provided.
G-quadruplex topology and recognition
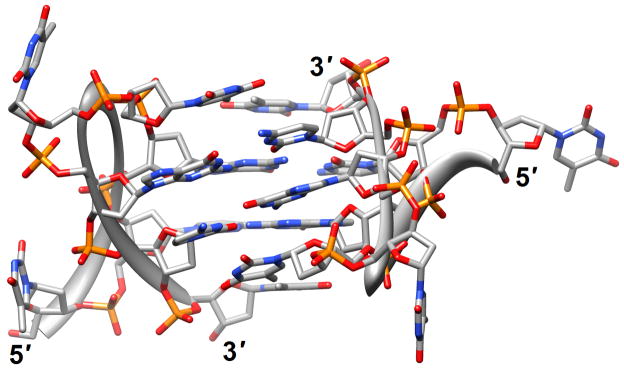
Structure determinations based on crystallography or solution NMR for natural human telomere sequences demonstrated significant deviations in conformation and loop geometry [13,14]. Subsequent investigations revealed that the symmetrical propeller-like structure in the crystal with parallel G-quartets and three double-chain reversal loops was not a major form in solution [15]. However, NMR structures of intramolecular G-quadruplexes formed by human telomere ODNs also exhibit significant differences as a result of the nature of the monovalent metal ions present (Na+ vs. K+) [13,17,18] (PDB entry codes 2jsk, 2jsl, 2jsm and 2jsq). Even under K+ conditions different conformations persist; a recently described third potassium form adopted by a human telomeric sequence with four G3 repeats folded into a basket-type quadruplex with just two layers of G-tetrads and two medium grooves and one narrow and one wide groove [19] (2kf7 and 2kf8; Figure 1a). This form appears to be more stable thermodynamically than the previously identified K+-forms and the increased stability was attributed to extensive pairing and stacking interactions in the loops, whereby each strand in the core featured both a parallel and an antiparallel adjacent strand. Overall, these findings provide further support for the heterogeneity of strand topology and loop conformation in G-quadruplexes. Loops of different orientation were also observed in structures of bimolecular antiparallel DNA quadruplexes [d(G3ACGTAGTG3)]2 and [d(G3T4G3)]2, whereby the former contains an autonomously stable mini-hairpin motif inserted within the diagonal loop [21] (2kaz). Thus, the nature of the loop sequence and the formation of interactions between loops and the G-quadruplex core are important elements in controlling quadruplex topology and stability.
Figure 1.
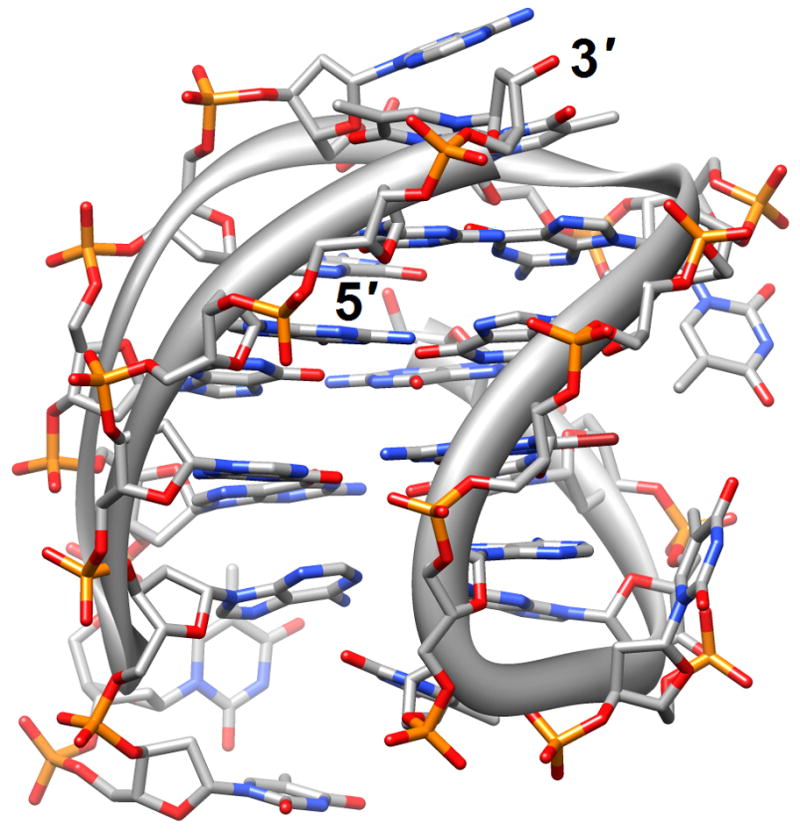
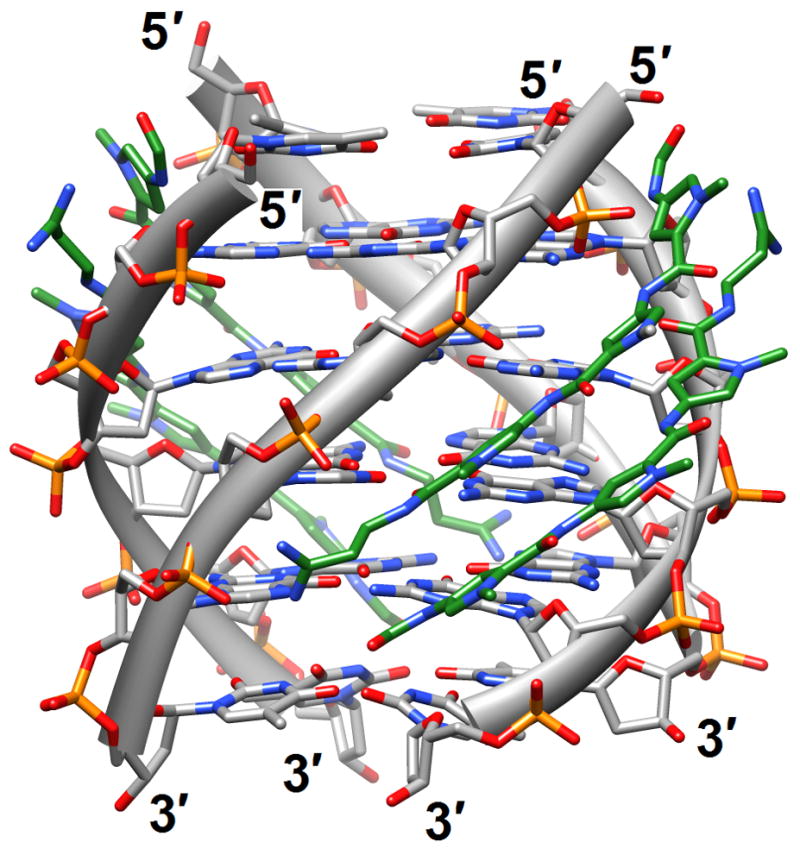
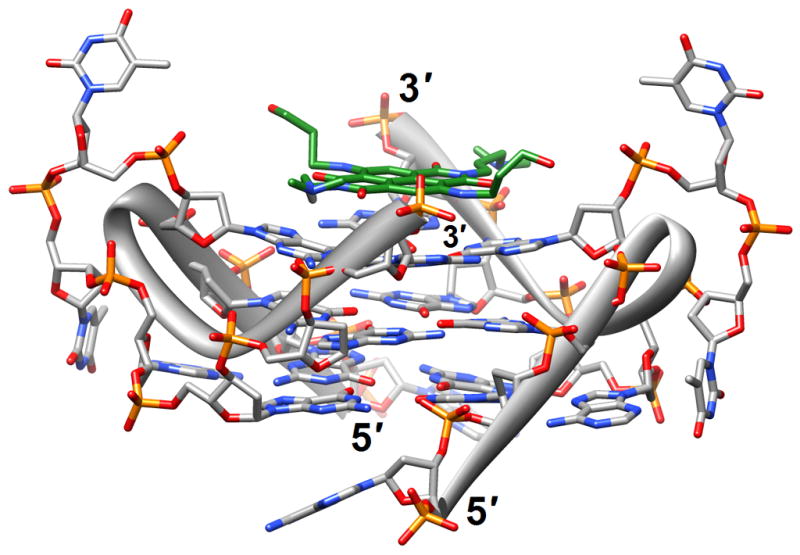
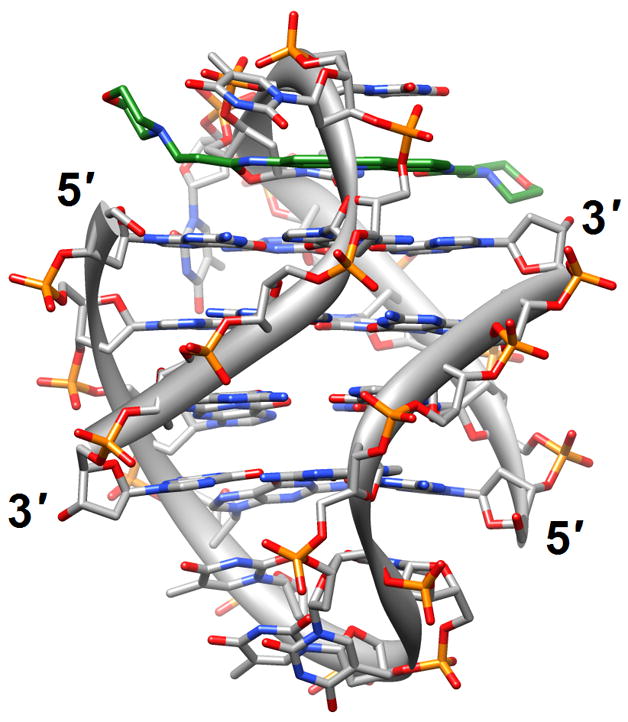
G-quadruplex topology and recognition. (a) The potassium-form structure of d(G3T2A)3G3T [19] (2kf7). (b) Groove interactions between distamycin A and the parallel-stranded quadruplex [d(TG4T)]4 [22] (2jt7). (c) Stacking of tetra-substituted naphthalene diimides on the terminal G-tetrads of the parallel-stranded quadruplex [d(TAG3T2AG3T)]2 [24] (3cco). (d) Acridine ligand interactions with the antiparallel-stranded G-quadruplex [d(G4T4G4)]2 [26] (3em2). DNA atoms are colored silver, red, blue and orange for carbon, oxygen, nitrogen and phosphorus, respectively, and carbon atoms of drug molecules are green.
Four-stranded motifs formed by G-rich strands present various surfaces for interactions with ligands. The lateral grooves are not unlike those present in DNA duplexes, although there are now four grooves instead of the well-known major and minor grooves in the B- and A-type duplex forms. But depending on the loop conformations and sequences, these grooves also exhibit different widths and depths. The terminal guanine tetrads of the quadruplex core constitute planar surfaces for favorable interactions with aromatic compounds of various dimensions. In principle, one should also expect such aromatic compounds to be able to insert themselves between neighboring layers of G-tetrads similar to the way intercalating agents penetrate base-pair stacks in DNA duplexes. Distamycin A is a familiar minor groove binding drug that can associate with duplexes in a 1:1 or a 2 (drug molecules):1 stoichiometry. The NMR solution structure of the complex between distamycin A and the parallel [d(TG4T)]4 quadruplex showed a groove interaction with a 4:1 stoichiometry, whereby two antiparallel distamycin dimers bind to two opposite grooves of the quadruplex [22] (2jt7; Figure 1b). Whether this interaction mode is sufficiently different from that seen with DNA duplexes with minor grooves of various widths to allow specific targeting of quadruplexes remains to be seen.
The binding of quinacridine-based ligands to a G-quaduplex was characterized by FRET-melting, microdialysis and 2D-NMR [23] (2jwq). The experimental approaches confirmed an interaction mode with extensive π-stacking between the outer surfaces of G-tetrads from the parallel-stranded quadruplex [d(T2AG3T)]4 and two ligands, with the cationic side chains of the latter directed toward the negatively polarized grooves. A similar binding mode between the outer G-tetrad surfaces of either an intramolecular quadruplex d[TAG3(TTAG3)3] or a bimolecular species [d(TAG3TTAG3T)]2 and a tetra-substituted naphthalene diimide ligand was observed in the respective crystal structures [24] (3cco and 3cdm; Figure 1c). The interaction with the ligands on either side of the G-stack doesn’t alter the (parallel-stranded) topology of these G-quadruplexes based on the human telomeric sequence, but contacts by the ligand to loop bases alter the conformations of the loops. This provides an indication that loop sequence and flexibility are likely acting as modulators of the binding affinity and specificity of aromatic ligands that interact with G-quadruplexes. Unlike in the case of the naphthalene diimide ligand, the crystal structure of the above bimolecular quadruplex in complex with an experimental acridine anticancer drug revealed a ligand sandwiched between a G-tetrad and a TATA tetrad from a neighboring quadruplex [25] (3ce5). X-ray crystal structures revealed that the large cavity established between the terminal G-tetrad and diagonal loops in the bimolecular quadruplex of antiparallel strand orientation formed by the sequence d(G4T4G4) from Oxytricha nova provides enough room for accommodation of the cyclic amino end-groups of bound disubstituted acridine ligands [26•] (3em2, 3eqw, 3eru, 3es0, 3et8, 3eui and 3eum; Figure 1d). In summary, it is clear that the loop geometry can affect binding to G-quadruplexes substantially and that the surfaces of G-tetrads at the ends of quadruplexes are the preferred sites of interaction for planar aromatic ligands. Conversely, intercalation between layers of G-tetrads does not seem to occur; perhaps stacked G-tetrads do not allow for sufficient breathing to enable an aromatic compound, especially one with bulky substituents, to insert itself between layers of guanines. The identification of ligands with sufficiently high affinity for G-quadruplexes and relatively low affinity for duplex DNA remains an important goal on the way to the development of drugs for regulating telomeric stability and G-rich regions of promoters as well as viral integration and recombination that have also been linked to G-rich sequences. Recently, guanidinium-modified phtalocyanines with very high affinity for G-quadruplexes were reported [27]. Interestingly, the fluorescence properties of these ligands also permit direct binding assays in vitro and imaging in vivo.
Minor groove binding agents
Among the three major interaction modes between small molecules and DNA, electrostatic, intercalation and groove binding, the latter affords the highest sequence specificity. Classic groove binders such as berenil, Hoechst 33258, netropsin and pentamidine prefer AT-rich regions with narrower, negatively polarized minor grooves that permit favorable interactions with their cationic moieties. The shapes of these molecules typically match the curvature of the groove, thus allowing a snugly fit under formation of van der Waals contacts and hydrogen bonds. Minor groove binding agents have been extensively studied as a class of potential therapeutics with antibacterial, antiprotozoal, antitumor and antiviral activities, and new classes of compounds have been investigated in recent years [28–32].
In the structure of a complex of pentamdine with the DNA duplex [d(ATATATATAT)]2 determined by X-ray crystallography, the drug was found to stabilize a coiled coil arrangement through formation of cross-links between neighboring duplexes [33•] (3ey0). The DNA duplex exhibits a mixture of Watson-Crick and Hoogsteen base pairs and, unlike in previous complexes with other DNA sequences, the drug is not entirely bound within the minor groove. Instead, only the central portion of pentamidine resides inside the groove, leaving the positively charged ends detached from the DNA and free to interact with phosphate groups from adjacent duplexes in the crystal. The potential implications of this binding mode for the antiprotozoal and antibacterial activities of pentamidine were discussed [33•]. To shed light on the different specificities of the antitrypanosomal agents 4,4′-bis(imidazolinylamino)- (CD27; prefers 5′-AATT over 5′-ATAT) and 4,4′-bis(guanidino)diphenylamine (CD25; binds more weakly and equally well to 5′-AATT and 5′-ATAT), the structure of CD27 bound to the duplex [d(CTTAATTCGAATTAAG)]2 in complex with a reverse transcriptase domain was determined by X-ray crystallography [34•] (3fsi). Rather than the expected highly twisted conformation, the ligand adopted a more planar crescent-like shape in the minor groove, with formation of bifurcated hydrogen bonds to the AATT portion. This binding mode results in widening of the minor groove and allows for formation of favorable van der Waals contacts. The observed induced fit with conformational changes in both the ligand and the DNA provided a rationalization for the different affinities and selectivities of CD27 and CD25. A more drastic perturbation of the DNA was observed in the crystal structure of the complex between an 8-ring pyrrole-imidazole polyamide and [d(CCAGGCCTGG]2 [35•] (3i5l; Figure 2). The ligand contacts the central six base pairs of the decamer duplex, prying open the minor groove by 4 Å and bending the helix axis toward the major groove by more than 18°. The distortions induced by this minor groove binder provide a molecular basis for the disruption of transcription factor-DNA interfaces by small molecules and the selective chemical regulation of expression.
Figure 2.
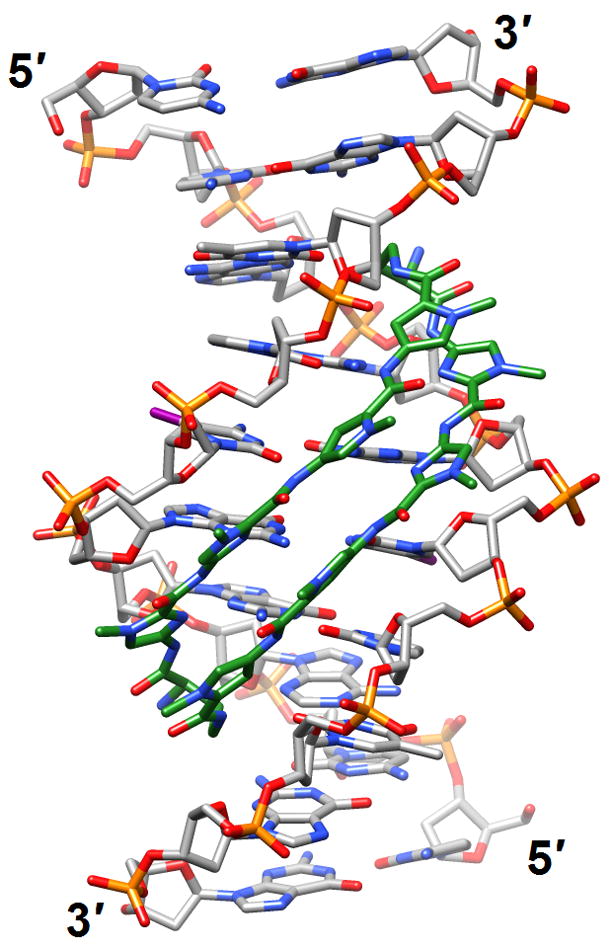
DNA bending and minor groove expansion in the crystal structure of a DNA:pyrrole-imidazole polyamide complex [35•] (3i5l). Carbon atoms of the ligand are highlighted in green and iodine atoms of 5-iodo-2′-deoxyuridine are magenta.
Conformational and dynamic heterogeneity of duplex DNA
Multiple studies were concerned with the sequence-dependent conformation and dynamics of double-helical DNA. NMR spectroscopy and molecular dynamics simulations provided insights into the intrinsic flexibility of a DNA sequence preferentially cleaved by topoisomerase II [36] (2jyk). The 21 base-pair ODN was found to exhibit opening of base pairs in an AT-rich stretch located immediately 5′ to one of the cleavage sites. The opening can be attributed to the particular sequence context as a similar dynamic behavior of bases was not observed with other AT pairs. This finding supports the notion that the enzyme may exploit the intrinsic dynamics of certain regions in the DNA duplex in its choice of cleavage sites. The sequence-dependent flexibility of certain regions could facilitate formation of a cleavage-competent complex and, therefore, the selection of such sites by topo II may be determined as much by conformational dynamics as by binding affinity. Sequence-dependent conformational alterations were also observed with a 16 base-pair DNA duplex studied by X-ray crystallography [37] (3bse). This enhancer element from Drosophila melanogaster is of importance in the regulation of a genetic switch between male and female patterns of gene expression and related sites are also conserved in the human genome. The ODN contains regulatory sequences related to recognition by several classes of eukaryotic transcription factors (DM motif, homeodomain, high mobility group box etc.) and the structure can serve as model for the conformational attributes of the DNA in the unbound state. In addition to particular patterns of groove widths and depths as well as metal ion coordination and hydration that were revealed by the 1.6-Å resolution crystal structure, the duplex also exhibited a sequence-dependent distribution of isotropic thermal B factors that expose variations in the local flexibility. The observed fluctuations in conformation and local dynamics allow the conclusion that intrinsic DNA flexibility is of crucial importance in a control element’s protein-directed rearrangement in a transcription preinitiation complex [37]. Whereas base pairs in the above duplexes exhibited the standard Watson-Crick pairing mode, A and T bases in the crystal structure of the duplex [d(ATATATCT)]2 all form Hoogsteen pairs [38] (2qs6; Figure 3a). In the asymmetric unit no fewer than seven antiparallel duplexes were found to stack in an end-to-end fashion, the formation of long helical stacks in crystals being a tendency previously seen with other of AT-rich ODNs. The 3′-terminal CT dimers are folded back such that T enters the minor groove and pairs with a T from the complementary strand, whereas Cs are extra-helical and stabilize the lattice under formation of a CAA triplet. The authors speculated that such interactions may provide a way to stabilize Hoogsteen-type duplexes in AT-rich regions in vivo.
Figure 3.
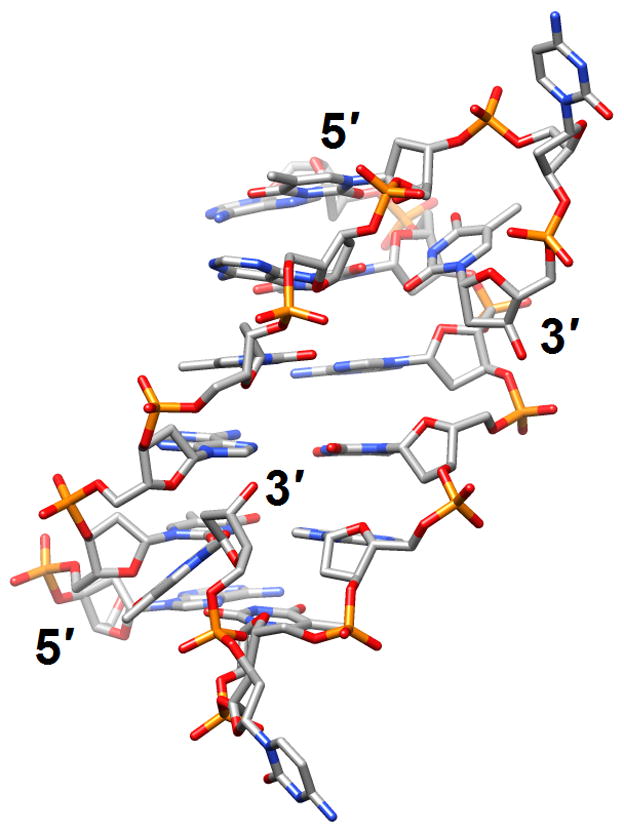
The versatile nature of DNA duplexes. (a) The Hoogsteen duplex [d(ATATATCT)]2 [38] (2qs6). (b) The symmetrical four-stranded motif formed by two octamers d(TCGTTGCT) and held together by minor groove C:G:G:C tetrads [42•] (2k8z).
Two studies, one examining an A-form DNA and relying on crystallography, the other a B-form DNA and employing solution NMR, explored the conformational distortions in DNA as a result of elevated pressure. In the first, the structure of the duplex [d(GGTATACC)]2 was determined at 295K at ambient pressure as well as at 0.55, 1.09 and 1.39 GPa to ca. 1.6 Å resolution [39] (2pkv, 2pl4, 2pl8 and 2plb). The only notable change was a continuous and smooth shortening of the duplex that could be monitored even in the 1.5 to 2 GPa range where the crystalline order was eventually lost. The average helical rise was lowered from 3.34 Å at ambient pressure to 3.07 Å at 2 GPa. Conversely, the base-pair geometry was virtually unaffected by exposing the crystal to increasing pressure. A slightly different outcome was observed in the high-resolution NMR experiment of a B-form DNA using data acquired at pressures of up to 0.2 GPa [40]. The structure of the DNA showed minimal compression along the axis but exhibited a widening of the minor groove with concomitant compression of Watson-Crick hydrogen bonds. The compression was more pronounced for AT pairs than GC pairs and the groove changes likely resulted in a compression of the water structure in the minor groove. Clearly, the different environments, crystal lattice versus solution, will account for the deviating outcomes of the two experiments. However, the overall relatively minor consequences provide an indication of the capability of nucleic acid duplexes to withstand even high-level pressures. Although we have no precise knowledge of the conditions that RNA duplexes were exposed to at the prebiotic stage, the picture of RNA as a molecular spring that is able to adapt to harsh conditions is an attractive one [39].
The short DNA duplex with a G1A2A3A4 tetraloop formed by the octamer d(GCGAAAGC) (loop residues are underlined) was used for assessing the quality of the structural models based on a combination of the NMR/DYANA and Biopolymer Chain Elasticity (BCE)/AMBER approaches compared to each method applied separately [41] (2k71). The combined approach yielded a more accurate molecular conformation that is compatible with all the NMR data and appears to require only minimal deformation of the standard backbone geometry. Specifically, the hairpin motif is stabilized by base-stacking interactions and a sheared G:A pair between G1 and A4 in the loop, thus creating a sharp turn between A3 and A4. The only significant deviation from the standard backbone geometry concerns the β and γ torsion angles of residue A3. A radically different arrangement of harpin DNAs was observed in the NMR solution structures of the sequences d(TCGTTGCT) and d(TGCTTCGT). Both octamers self-associate to form bimolecular complexes that feature minor groove C:G:G:C tetrads [42•] (2k8z, 2k90 and 2k97; Figure 3b). This four-stranded motif clearly differs from the G-tetrads discussed before and had previously been observed only with cyclic ODNs. Yet another unusual pairing mode of nucleobases was detected in the crystal structure of an asymmetric DNA junction that resembles an X with two sets of B-form arms which form semi-continuous coaxial stacks [43] (3igt). The wobble shape of an A:T pair located at the end of one of the stacked duplex arms and adjacent to the junction is indicative of a rare enol tautomeric form of thymine. Rather than ascribing it to the presence of the junction, the authors suggested that the occurrence of the unusual form of the A:T pair and the particular sequence context could serve as a model for transition mutations during DNA replication. Finally, a study of the nature of DNA conformation and the mechanism of homologous pairing (HP) in recombination demonstrated that diverse HP proteins use a non-canonical, extended and single-stranded DNA structural form as a common intermediate for HP, thereby providing support for a convergent mechanism of HP [44] (2rpd, 2rpe, 2prf and 2rph). Interestingly, RNA cannot adopt this single-stranded structure due to steric hindrance by 2′-OH groups. This may explain the absence of homologous recombination in organisms with RNA genomes. The switch to DNA genomes could have conferred a critical evolutionary advantage over RNA because DNA is particularly suited for both genomic stability and genomic diversity.
Adducted DNAs
Investigations of adducted DNAs have been productive and include single-base modifications (with G being particularly well represented), and intra- and inter-strand crosslinks. The majority of structures are based on solution NMR. The effects of methylation at N6 of adenine in the sequence 5′-GATC-3′ on stability and structure as well as the ability of factors interacting with GATC sites to differentiate between the fully and hemi-methylated forms were assessed by NMR [45] (2kal). Thus, a dodecamer duplex containing the hemi-methylated site displayed a unique major groove conformation that potentially allows the enzyme SeqA to discriminate against unmethylated or fully methylated forms. Moreover, hemi-methylation resulted in a higher base-pair opening rate for the flanking G:C pairs relative to the duplex with fully methylated As. The origins of the unusually high increases in stability (ΔTm ca. 16°C) afforded by the C2-(2-naphthyl)-substituted pyrrolo[2,1-c][1,4]benzodiazepine (PBD) 2:1 DNA adduct were investigated in the duplex [d(AATCTTTAAAGATT)]2 (PBDs preferentially interact with purine-guanine-purine sites) [46] (2k4l; Figure 4a). In contrast to favorable hydrogen-bonding interactions that occur with adducts of natural PBDs such as sibiromycin and anthramycin, the C2-naphtyl ring of the above compound was inserted into the minor groove, thus resulting in optimal hydrophobic contacts with the DNA. This observation could serve as the starting point for future design of synthetic PBDs with extended planar hydrophobic groups at the C2 position for maximization of the binding affinity.
Figure 4.
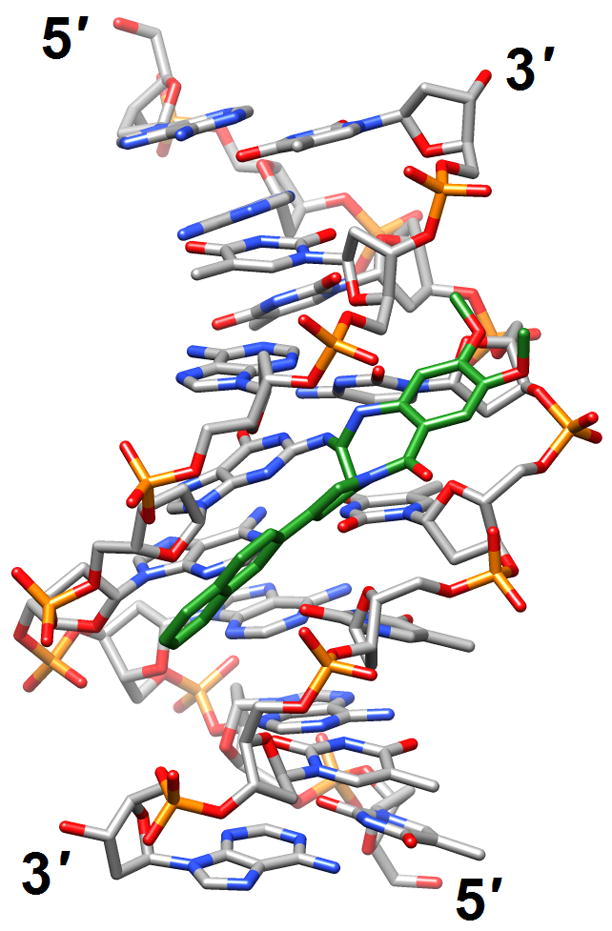
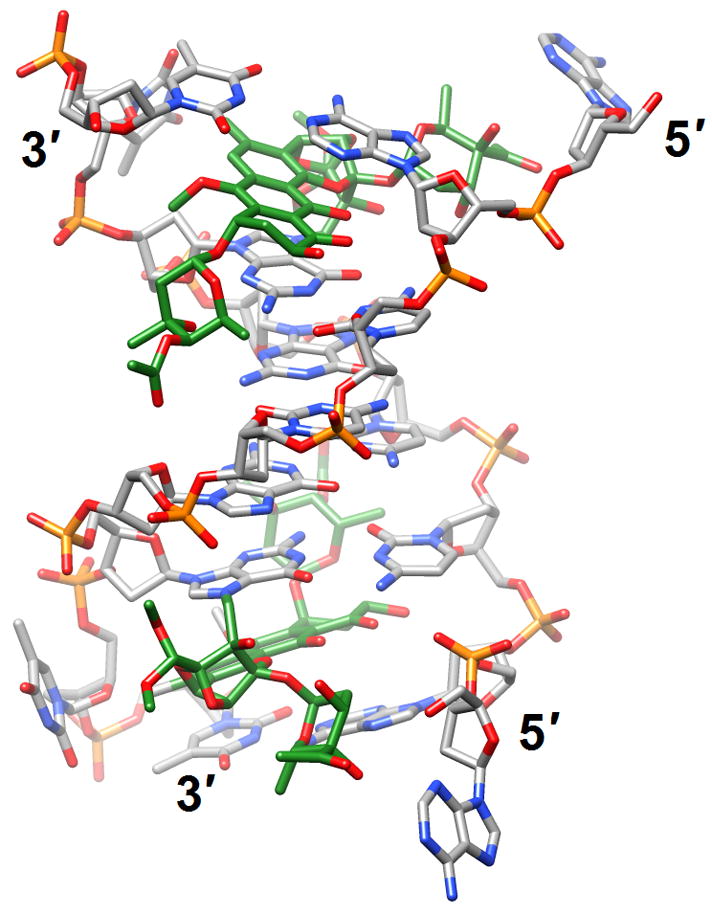
Effects of guanine adducts on DNA conformation. (a) A C2-(2-Naphthyl)-substituted pyrrolo[2,1-c][1,4]benzodiazepines (carbon atoms highlighted in green) covalently attached to G in the duplex [d(AATCTTTAAAGATT)]2 [46] (2k4l; only the underlined portion of the duplex is shown). (b) Trioxacarcin A molecules (carbon atoms highlighted in green) covalently attached to Gs and intercalated into the duplex [d(AACCGGTT)]2, thereby forcing 3′-adjacent Ts into an extra-helical position [52] (3c2j).
Three further investigations focused on guanine adducts, namely 1,N2-etheno-2′-deoxyguanosine (1,N2-εdG), 1R,2S,3R,4S-benzo[c]phenanthrene-N)-dG (B[c]P-N2-dG), and α-OH-PdG, a product of the reaction between acrolein and guanine. At neutral pH the 1,N2-εdG lesion was found to stack with flanking base pairs but the cytosine on the opposite strand was extrahelical and located inside the minor groove [47] (2k1y). Upon lowering of the pH, a second form emerged in which cytosine was presumably protonated and engaged in a Hoogsteen pair with 1,N2-εdG. The B[c]P-N2-dG adduct was studied inside a duplex that contained a (CG)3 iterated repeat, 5′-d(…CGCX CG…)-3′:5′-d(…CG CGCG…)-3′, where X= B[c]P-N2-dG [48] (2rou). Structure refinements using MD simulations and restrained by inter-proton distances and torsion angles obtained from NMR data revealed that the B[c]P moiety intercalated on the 3′-side of the adducted base, with its terminal ring pointing into the major groove. Both flanking base pairs maintained Watson-Crick hydrogen bonds but the intercalation resulted in local unwinding of the helix. In the third NMR structure determination, 3D-models based on restrained MD simulations of the duplex with α-OH-PdG showed the adducted G in a syn conformation, stabilized by intra-residue hydrogen bonds [49] (2kd9 and 2kda). The adducted residue exhibited various hydrogen-bonding patterns to cytosine from the opposite strand that was protonated under the conditions used in the NMR experiments (ca. pH 6).
To shed light on the ability of the trans-4-hydroxynonenal (HNE)-derived exocyclic 1,N2-dG adduct with 6S,8R,11S- but not with 6R,8S,11R-stereochemistry to form inter-strand N2-dGxN2-dG cross-links in the 5′-CpG-3′ DNA sequence context, the structures of both were assessed by NMR inside the duplex 5′-d(GCTAGCXAGTCC)-3′:5′-d(GGACTCGCTAGC)-3′, (X=6S,8R,11S- or 6R,8S,11R-HNE-dG) [50] (2k8t and 2kau). Although the cyclic hemiacetals of either configuration were located in the minor groove, the reactive one was oriented in the 5′-direction whereas the one unable to cross-link was oriented in the 3′-direction. This stereochemical difference most likely provides the 6S,8R,11S-HNE-dG isomer with a kinetic advantage and helps explain its ability to cross-link Gs across strands. When the same reactive isomer was mismatched opposite dA, it maintained the ring-closed 1,N2-HNE-dG form. In acidic solution, the adducted nucleoside adopted predominantly the syn conformation - with the HNE moiety located in the major groove - and paired with the protonated adenine [51] (2kar and 2kas). At basic pH the adducted nucleoside assumed mainly the anti conformation, with the HNE moiety intercalated and its aliphatic chain oriented toward the minor groove. This resulted in the displacement of the complementary A in the opposite strand and formation of a bulge. An unrelated adduct was also observed to extrude a base from a DNA duplex. In the crystal structure of the 2:1 complex between trioxacarcin A and [d(AACCGGTT)]2, the antibiotic was covalently bound to N7 of G (underlined) and intercalated on the 3′-side of the adducted G, thereby flipping out the adjacent T (italicized) [52] (3c2j; Figure 4b).
The ability of the above enal-derived 1,N2-dG adducts to induce inter-strand DNA cross-links in the 5′-CpG-3′ but not in the 5′-GpC-3′ sequence context was further probed by an NMR investigation of synthetic trimethylene cross-links placed at either step [53•] (2knk and 2knl). In the CpG sequence cross-linked and neighboring base pairs exhibited only minor deviations in terms of stacking interactions. Conversely, the stacking of the cross-linked pairs in the GpC sequence was greatly perturbed. This differential stacking may explain the higher stability of the trimethylene cross-link at CpG steps and account for the sequence selectivity of cross-link formation by the native 1,N2-dG adducts.
NMR was also used to study the effects on the DNA duplex geometry by intra-strand Pt-GG cross-links with trans-R,R-diaminocyclohexane ligands formed by oxaliplatin {cis-[Pt(DACH)(oxalate)] (DACH = trans-R,R-1,2-diaminocyclohexane), an FDA-approved drug for the treatment of for colorectal cancer} as well as the influence of flanking bases on the helix distortions [54] (2k0t, 2k0u, 2k0v and 2k0w). Unlike oxaliplatin, the cationic platinum(II) complex, cis-diammine(pyridine)chloroplatinum(II), cis-[Pt(NH3)2(py)Cl]+ (cDPCP), bound DNA mono-functionally, as revealed by the crystal structure of cDPCP bound to N7 of a single G in a dodecamer duplex [55••] (3co3). Despite the absence of an intra-strand cross-link, the large shift and slide observed in the base-pair step to the 5′-side of the Pt adduct is reminiscent of the structural perturbations seen with cisplatin intra-strand cross-links. Remarkably, cDPCP-DNA adducts are removed by the nucleotide excision repair (NER) machinery much less efficiently than the bifunctional platinum-DNA cross-links.
A potential limitation of the structural data discussed in this section is that the perturbations caused by DNA adducts and their recognition were evaluated in the absence of mismatch repair proteins, DNA damage-recognition proteins or trans-lesion DNA polymerases. Although we are aware of the growing database of 3D-structures of such proteins in complex with damaged DNA, we have intentionally limited this review to 3D-structure determinations of DNA alone.
Metal ion-DNA interactions
The crystal structures of tetragonal and trigonal forms of the rhodium complex delta-Rh(bpy)2(chrysi)3+ (chrysi=5,6-chrysenequinone diimine) bound to the DNA duplex [d(CGGAAATTACCG)]2 with A:A mismatches (underlined bases) revealed metalloinsertion from the minor groove side at mismatch sites [56] (3gsj and 3gsk; Figure 5). Despite both adenosines being ejected into the major groove at the coordination sites, the duplex underwent only limited conformational changes. Thus, the helical rise, the geometry of flanking base pairs, and the sugar puckers of adjacent residues were basically unaltered. In the tetragonal form, a third rhodium complex was found to intercalate into the central base-pair step. The crystal structure of another DNA-intercalator complex was determined by single wavelength anomalous dispersion (SAD) based on coordinated Mn2+ ions [57] (3go3). In the duplex [d(ACGTACGT)]2 echinomycin molecules were found to wrap their aromatic moieties around base pairs at CpG steps. As a result the helix was unwound, thus enabling binding of Mn2+ to N7 of all four guanines. Although C:G pairs exhibited significant buckling, they remained in the standard Watson-Crick form, whereas the As and Ts in the center formed Hoogsteen pairs. The N7 atom of G was also confirmed as the preferred coordination site for Mn2+ in the crystal structure of the DNA duplex [d(CGTTAATTAACG)]2 [58] (3eil). In the crystal metal ions link the guanine major groove edges to phosphates, a coordination mode that contrasts with those previously observed with Mg2+, Ca2+ and Ni2+. The authors speculated that sequences with TTAA repeats occur so rarely in all genomes because they may be susceptible to formation of closely spaced thymine dimers (see introduction).
Figure 5.
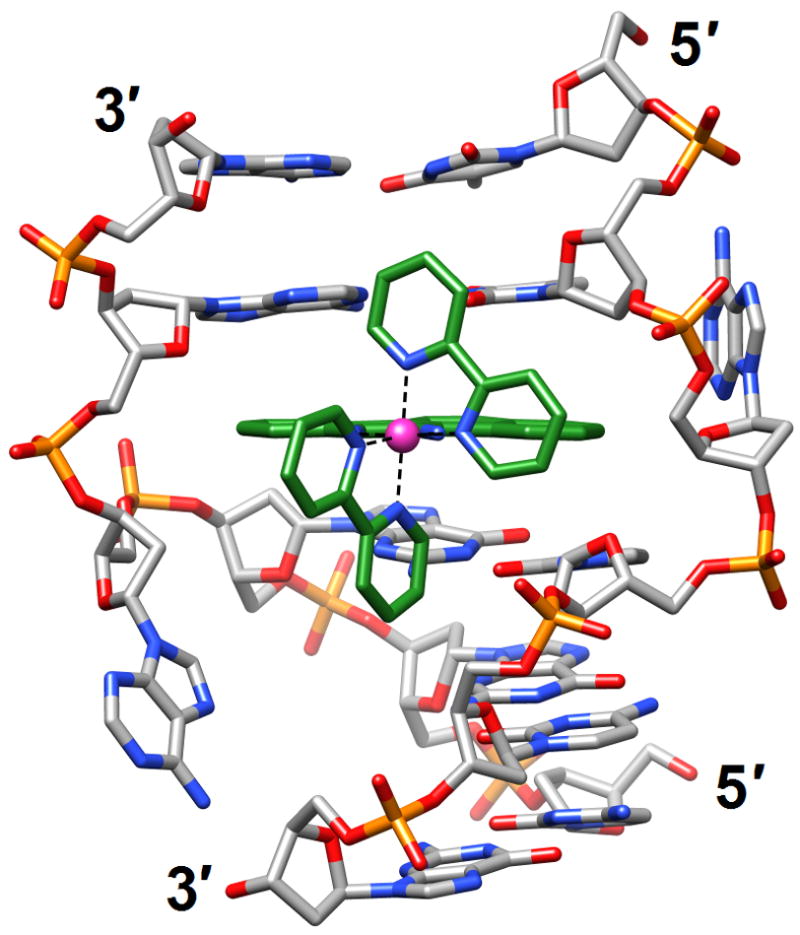
A delta-Rh(bpy)2(chrysi)3+ complex intercalated into the DNA duplex [d(CGGAAATTACCG)]2 [56] (3gsj). The metal ion is shown as a pink sphere and carbon atoms are highlighted in green. Adenosines underlined in the sequence are ejected into the major groove by the metalloinsertion.
Chemically modified nucleic acids
The preparation of chemically modified nucleic acids (CNAs) and investigations of their stability, activity and structure continue to be a very active field of research, in line with the potential application of such systems in diagnostics, drug discovery, nucleic acid etiology, genomics, material science, nanotechnology, RNA interference and target validation as well as for probing protein-nucleic acid interactions and for the generation of derivatives for crystallographic phasing. The use of ODNs as antisense oligonucleotides (AONs) for interfering with biological information transfer was initiated by two studies on the inhibition of viral replication and viral RNA translation by Zamecnik and Stephenson [59,60]. Paul C. Zamecnik, a pioneer of AON applications among many other scientific achievements died at the end of 2009 [61].
There are numerous structure determinations of CNAs and for in-depth reviews and discussion of this topic, please see the updates by Egli and Pallan [62,63•]. Recent publications of chemical modifications and studies of their structural consequences concern all basic components of DNA, sugar, phosphate and base, and we will discuss them in this order. ODNs with cyclohexene nucleosides (CeNA) in place of the natural 2′-deoxyribonucleoside residues display enhanced thermal and enzymatic stabilities. The crystal structure of the DDD with a single CeNA-A per strand displayed only minor deviations from the B-form structure of the native duplex, but the spine of water molecules in the minor groove was partially disrupted in the vicinity of the more hydrophobic CeNA residues [64] (2p8d). A more drastic change caused by a modification of the sugar moiety was observed in the structure solved by NMR of a fully-4′-thio-modified DDD duplex [65•] (2rmq; Figure 6a). Remarkably, replacement of the furanose ring oxygen with sulfur triggered conversion of the standard B-DNA conformation to an RNA-like A-form. This transformation is quite unexpected but sheds light on some of the interesting properties of 4′-thio-DNA, such as enhanced RNA affinity and resistance to nucleases. It also provided the solution to a long-standing puzzle, initially reported by Walker more than a decade ago [66], namely the curious inability by RNase H to cleave RNA paired to 4′-thio-DNA. In light of the structure determination by Matsugami et al. [65•], it is now clear that the geometry of the RNA:4′-thio-DNA hybrid resembles a canonical A-form, a duplex type that the enzyme is competent to bind but not to cleave [62].
Figure 6.
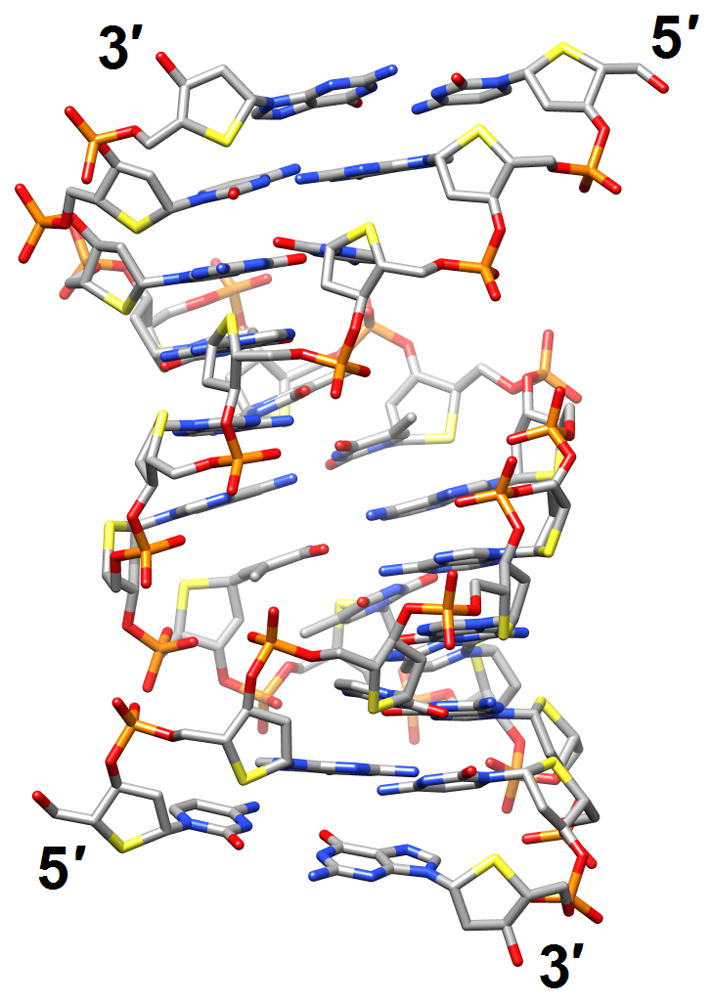
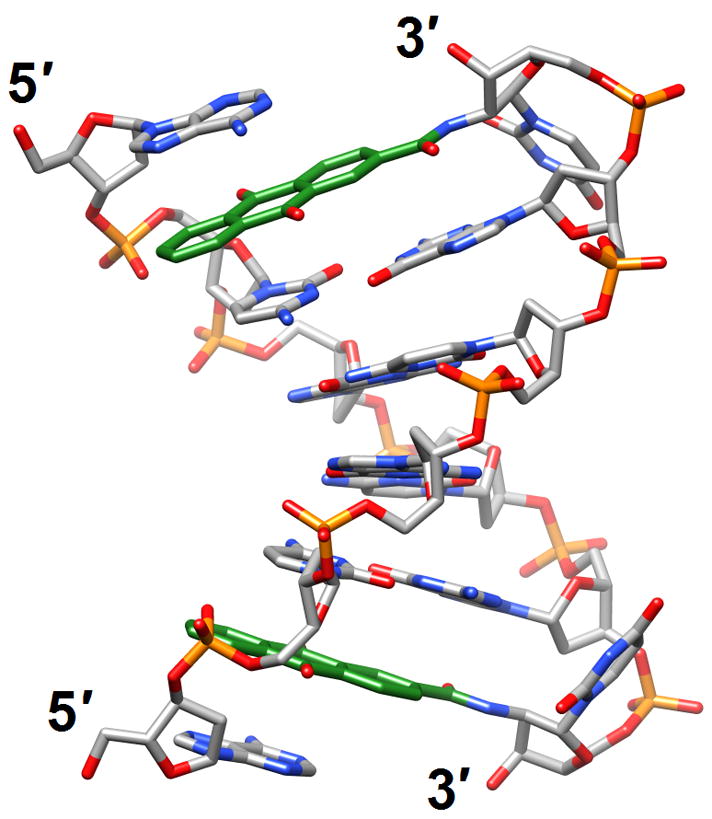
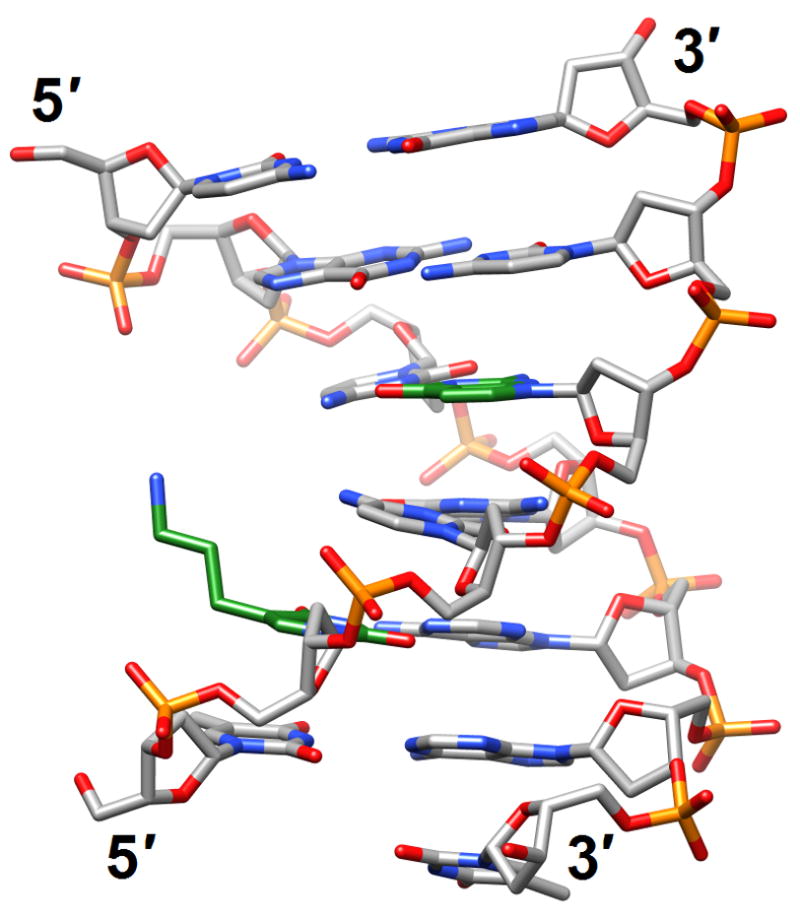
Structural consequences of chemical modification. (a) A-form geometry of the duplex formed by 4′-thio-DNA [65•] (2rmq). Sulfur atoms are highlighted in yellow. (b) Extensive stacking interactions and hydrogen bonding are at the origin of the dramatic stability increases with DNA and RNA duplexes capped by the 3′-terminal 2′-anthraquinoylamido-2′-deoxyuridine residue [71] (2kk5). Carbon atoms of the aromatic moiety are highlighted in green. (c) Orientation of the tethered cation from 5-(3-aminopropyl)-2′-dU8 relative to 7-deaza-G10 (N+2) in the NMR structure of a modified DDD [72] (2qef). Carbon atoms of 5-(3-aminopropyl)-uracil and 7-deaza-guanine are highlighted in green.
By comparison, replacement of the 2′-oxygen atom with sulfur in 2′-thiomethyl-modified DNA didn’t fundamentally alter the preference of the sugar for a so-called C3′-endo (North), RNA-like conformation. In crystal structures of the DDD with either one or both Ts replaced by 2′-SMe-U, the duplex underwent a conformational transition as a result of the North pucker of central residues [67] (3ey2 and 3ey3). The minor groove of the duplex was significantly wider than that in the native DDD and, together with a likely local stiffening of the backbone, this provided an explanation for the inability of the restriction enzyme Eco RI to cleave at the recognition site GAATTC contained in the sequence of the dodecamer. Another modification that leads to an Alike conformation of the backbone is so-called tricyclo-DNA (tc-DNA). Oligo-tc-DNAs exhibit higher stability of self-pairing and RNA affinity than DNA and in the crystal structure of a DDD with single tc-DNA-A residues per strand, modified nucleosides displayed an A-type C2′-exo sugar pucker [68] (2rf3). Relative to B-form DNA, the conformations of the β and γ backbone torsion angles in tc-DNA were swapped, a feature that is most likely a direct consequence of the presence of the cyclopropane moiety in this modification. The steric bulk of the sugar in close vicinity of the phosphate as seen in the tc-DNA structure poses a challenge for nucleases and explained the increased ability of tc-DNA to evade degradation by nucleases. The consequences of so-called locked nucleic acid (LNA) residues with a [C2′]-O2′-CH2-[C4′] modification were assessed in the context of the telomeric sequence 5′-dG-lG-dG-lG-dT4-dG-lG-dG-lG (lG=LNA-G) [69] (2wcn). The resulting bimolecular G-quadruplex displayed a folding topology that is distinct from the native form in that each stretch of guanines exhibited a sharp V-shaped turn that pairs the first and fourth residues in the same tetrad. Once again, this study demonstrated how subtle changes in the sequence and chemistry can affect the folding topology and loop structure of G-quadruplexes.
The formacetal backbone modification removes the negative charge of the phosphate group, but incorporated into RNA leads to a slight thermodynamic stabilization of the duplex. However, inside DNA the formacetal linkage results in a destabilization of ca. 1.5°C per modification. In the crystal structure of an A-form DNA single formacetal modifications per strand fit seamlessly into the duplex, but exhibited poor hydration relative to phosphate groups [70•] (3hr3). Osmotic stressing studies using several small molecule cosolutes demonstrated that more water was released from formacetal-modified RNAs upon melting than from modified DNAs. Together with the stability and structural data these results indicated that RNA may tolerate non-ionic modifications better than DNA. Moving on to another modification, it was reported that addition of a 3′-terminal 2′-anthraquinoylamido-2′-deoxyuridine (Uaq) residue enhanced the melting temperature of a duplex by up to 18°C and also resulted in a dramatic difference in the thermodynamic stability between matched and mismatched bases at the adjacent pair. Thus, a single mismatched base facing the 3′-terminus led to a loss of a much as −24°C (ΔTm) in the case of RNA and of up to −30°C in DNA. The structure of a DNA duplex with 3′-terminal Uaq residues solved by NMR revealed that the cap interacts with the 5′-terminus of the target strand via stacking and hydrogen bonding [71] (2kk5; Figure 6b). The lower stability observed for terminal mismatch pairs adjacent to Uaq is most likely due to the loss of optimal stacking interactions.
A more benign nucleobase modification, 7-deaza-dG, was incorporated into the DDD in place of G either alone or in combination with 5-(3-aminopropyl)-2′-deoxyuridine (replaces T) to probe the mechanism underlying the formation of inter-strand N+2 DNA cross-links by nitrogen mustards [72] (2qef and 2qeg). 7-Deaza-dG placed at the underlined position in the dodecamer sequence d(CGCGAATTCGCG) prevented binding of a Mg2+ normally coordinated at one end of the duplex in crystal structures of the DDD [5,7]. Indeed, the absence of the metal ion reduced the degree of bending into the major groove. Replacement of a T (italicized in the above sequence) with 5-(3-aminopropyl)-2′-deoxyuridine restored the bend even in the presence of 7-deaza-G at the N+2 G, thereby supporting the idea that a cationic moiety, either tethered or in the form of a metal ion, affects the bend [4]. Moreover, these findings raise the possibility that tethered N7-dG aziridinium ions, the active species involved in the formation of inter-strand 5′-GNC-3′ cross-links, modulate the electrostatics of the major groove to position the ionic moiety near the edge of the N+2 G (Figure 6c). Like 7-deaza G, the 2,4-difluorotoluene analog (F) of T is supposedly more hydrophobic than its native counterpart. However, the crystal structure of a B-form DNA duplex with A:F revealed that the F analog is able to form hydrogen bonds, and therefore provides an exception to the rule that organic fluorine is a poor hydrogen bond acceptor [73•] (3i8d). More importantly, this observation cast doubt on the steric hypothesis of DNA replication, i.e. that shape is more important than hydrogen bonding for accurate replication by some polymerases. Another unexpected finding related to a hydrophobic base analog concerns the stable phenyl-cyclohexyl recognition in the center of a DNA duplex [74••]. Incorporation of the phenyl-cyclohexyl-C-nucleoside into DNA actually led to a gain in the thermodynamic stability and increases the repertoire of molecular entities that are tolerated in the interior of a double helix.
Several studies concerned the synthesis and structural analysis of CNAs with selenium replacing oxygen in the base or sugar moieties [75–79] (3bm0, 3hg8, 3hgd, 3ifi, 3ijk, 3ijn and 3hgd). Selenium is a useful modification for crystallographic phasing via SAD or MAD and for dissecting protein-nucleic acid interactions, but Se-containing nucleosides are also being explored for their potential anti-tumor activity [80] and the 4-Se-TTP analog could find applications in diverse areas due to the fact that is colored [81]. Recently, the synthesis and incorporation into ODNs of tellurium-modified nucleosides (2′-TeMe-T, 2′-TePh-T) for structural and functional investigations has also been reported [82] (3fa1).
Artificial pairing systems
Glycol nucleic acid (GNA [83••]; the analog is also referred to as glycerol nucleic acid [84]) contains an acyclic three-carbon propylene glycol moiety in place of DNA’s 2′-deoxyribose and arguably constitutes the simplest version of a phosphodiester-based nucleic acid system. GNA displays stable self-pairing and the S-enantiomer cross-pairs with RNA. However, S- and R-GNA strands do not pair with each other and neither pairs with DNA. The crystal structure of an S-GNA octamer with two synthetic Cu2+ base-pair mimics revealed a ribbon-like right-handed duplex with a strongly inclined backbone and a large shift between adjacent base pairs [83••] (2jja; Figure 7a). The duplex featured 16 base pairs per turn (average twist 23°) and a helical rise of 3.8 Å, with an average distance between adjacent phosphates of 5.4 Å. Although the GNA duplex is significantly unwound compared with A- and B-form duplexes, the strong backbone inclination and the short spacing between intra-strand phosphates are reminiscent of RNA. The similar geometries help explain the pairing between S-GNA and RNA. Because R-GNA is left-handed it is unable to pair with right-handed RNA. The surprisingly high stability of GNA duplexes is likely the result of extensive inter-strand base stacking and favorable, hydrophobic intra-strand contacts between nucleobase and backbone atoms [83••]. It is also consistent with a significantly lower entropic penalty for duplex formation compared with DNA [85]. Given the lack of a cyclic sugar in the GNA backbone, this increased conformational preorganization of single strands is rather unexpected.
Figure 7.
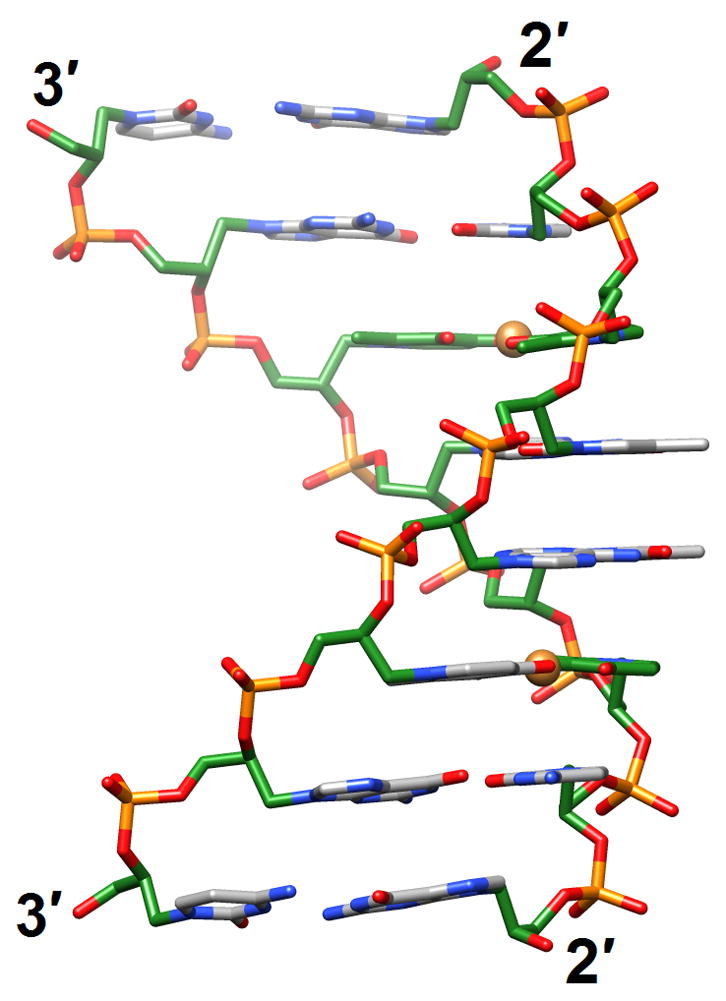
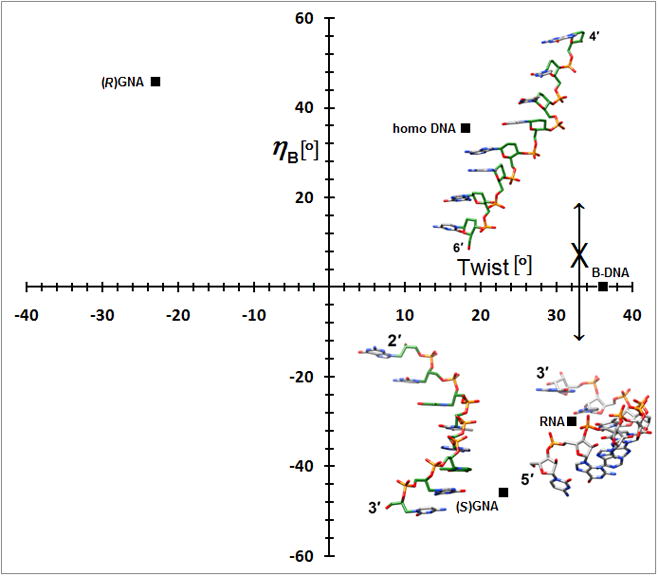
A simpler nucleic acid and the structural origins of cross-pairing between different nucleic acid systems or the absence thereof. (a) Crystal structure of S-GNA [83••] (2jja). Carbon atoms of the glycol and 2,6-dipicolinate (Dipic) as well as pyridine (Py) moieties are highlighted in green and copper ions that induce pairing between Dipic and Py are orange. (b) The backbones in both RNA and S-GNA are negatively inclined relative to the base-pair planes [87•] and both are right-handed: Pairing occurs. Unlike in RNA, the backbone-base inclination angle ηB in homo-DNA is positive: No pairing.
A second crystal structure of a GNA duplex provided evidence that the analog exhibits considerable variations in its geometry [86]. Thus, the duplex [3′-g(GCGCGC)-2′]2 displayed a compressed helical rise (2.6 Å) and twisting more like B-DNA (35.7°; 10 residues per turn), whereas average slide (−3.4 Å) and intra-strand phosphate-phosphate distance were virtually the same as in the structure determined earlier. Like DNA, GNA may be able to adapt to the geometry of the conformationally more rigid RNA upon pairing. GNA also illustrates the usefulness of a parameter recently derived from the crystal structure of homo-DNA, the backbone-base inclination angle that serves as a determinant of nucleic acid self- and cross-pairing [87•] (2h9s). Like GNA and RNA, homo-DNA has a strongly inclined backbone, but unlike in S-GNA or RNA where the angle between the backbone and the base planes is negative, it is positive in homo-DNA. Nucleic acid systems with sizeable inclination angles of opposite sign are unable to pair (Figure 7b). Although R-GNA and homo-DNA have positively inclined backbones, the former is left-handed and the latter is right-handed, and the opposing twists also prevent pairing between the two. Despite a more strongly inclined backbone in S-GNA relative to RNA, the difference is not substantial enough to prevent pairing between the two. However, DNA that adapts to RNA by switching to the A-form is apparently unable to undergo an even more pronounced inclination between its backbone and base planes and, therefore, is unable to cross-pair with S-GNA.
DNA as a nanomaterial
We will end this review by highlighting several breakthroughs in harnessing DNA base-pairing and the cohesion and predictable interactions, geometry and dimension provided by it to construct nanoscale structures and lattices. In the first two examples, gold nanoparticles were fused to thiol-terminated ODNs for generating single crystals of nanoparticles [88,89]. In order to avoid aggregation the lengths and sequences as well as the nature and flexibility of the linkers between ODNs and gold nanoparticles were varied systematically [90]. Moreover, powder diffraction and in situ small angle X-ray scattering (SAXS) were used to monitor the progress in crystallization and direct the experiments in the right direction. The use of SAXS is particularly noteworthy and coincides with a renaissance of this technique in all areas of structural biology where its use, particularly in combination with X-ray, NMR and/or EM has provided insights into the shape and dynamics of molecular assemblies that would have remained hidden if each technique had been applied in isolation. In the case of the DNA-mediated nanoparticle crystals, size was a limiting factor and prevented the use of conventional single-crystal X-ray crystallography. Taking the construction of a DNA-based 3D-lattice to an even higher level, Seeman and coworkers reported the 4Å-resolution structure of a self-assembled triangular DNA in a crystal [91••] (3gbi). This was the crowning event of many years of systematic studies to control the self-assembly of the genetic brick (DNA) and mortar (base-pairing and stacking) into a well-ordered macromolecular 3D-crystalline lattice. Dramatic progress has not only been made in terms of constructing but also in characterizing DNA nanostructures, as exemplified by the recent high-resolution analysis of such a structure by cryoEM that revealed individual strands and grooves despite the relatively small size of the target (<100 kDa) [92]. Overall, DNA remains exciting as a template, tool and target and almost 60 years after the discovery of its structure we can still look forward to a bright future.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of special interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh S-H, Srinivasan AR, Schneider B. The nucleic acid database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.Subirana JA, Messeguer X. The most frequent short sequences in non-coding DNA. Nucleic Acids Res. 2010;37 doi: 10.1093/nar/gkp1094. in press. A wake up call to the nucleic acid structural community in that it provides evidence that the 3D structures of most of the commonly encountered sequence motifs remain uncharacterized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wing R, Drew H, Takano T, Broka C, Takano S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 4.Allemann RK, Egli M. DNA bending and recognition. Chem Biol. 1997;4:643–650. doi: 10.1016/s1074-5521(97)90218-0. [DOI] [PubMed] [Google Scholar]
- 5.Tereshko V, Minasov G, Egli M. The Dickerson-Drew B-DNA dodecamer revisited at atomic resolution. J Am Chem Soc. 1999;121:470–471. [Google Scholar]
- 6.Tereshko V, Minasov G, Egli M. A “ghydrat-ion”h spine in a B-DNA minor groove. J Am Chem Soc. 1999;121:3590–3595. [Google Scholar]
- 7.Egli M. DNA-cation interactions: quo vadis? Chem Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 8•.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. A comprehensive analysis of the 3D structures of protein-DNA complexes reveals that arginine insertion into the minor groove is common, consistent with the enhanced negative electrostatic potential at that site, and represents a general mechanism for indirect readout. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman S. Promoter quadruplexes. Chem Eng News. 2009 Nov. 2:28–30. [Google Scholar]
- 11••.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. Makes the case that supercoiling that is induced by transcription has dynamic effects on DNA in the MYC promoter element, converting it from duplex DNA to non-duplex DNA structures. The latter control both turning on and off of transcription and the rate of transcription firing and are amenable to small-molecule targeting. [DOI] [PubMed] [Google Scholar]
- 12••.Neidle S. The structures of quadruplex nucleic acids and their complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. The most recent update on the diverse topologies of G-rich quadruplex structures and the structures determined for ligand-quadruplex complexes thus far. Emphasizes the role played by loop sequences in determining fold. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G- tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J Am Chem Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balkwill GD, Garner TP, Williams HE, Searle MS. Folding topology of a bimolecular DNA quadruplex containing a stable mini-hairpin motif within the diagonal loop. J Mol Biol. 2009;385:1600–1615. doi: 10.1016/j.jmb.2008.11.050. [DOI] [PubMed] [Google Scholar]
- 22.Martino L, Virno A, Pagano B, Virgilio A, Di Micco S, Galeone A, Giancola C, Bifulco G, Mayol L, Randazzo A. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J Am Chem Soc. 2007;129:16048–16056. doi: 10.1021/ja075710k. [DOI] [PubMed] [Google Scholar]
- 23.Hounsou C, Guittat L, Monchaud D, Jourdan M, Saettel N, Mergny JL, Teulade-Fichou M. G-quadruplex recognition by quinacridines: a SAR, NMR, and biological study. ChemMedChem. 2007;2:655–666. doi: 10.1002/cmdc.200600286. [DOI] [PubMed] [Google Scholar]
- 24.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J Mol Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J Am Chem Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 26•.Campbell NH, Patel M, Tofa AB, Ghosh R, Parkinson GN, Neidle S. Selectivity in ligand recognition of G-quadruplex loops. Biochem. 2009;48:1675–1680. doi: 10.1021/bi802233v. Crystal structures of complexes between a series of disubstituted acridine ligands and a bimolecular DNA G-quadruplex reveal that the nature of the loop has a significant influence on ligand selectivity for particular quadruplex folds. [DOI] [PubMed] [Google Scholar]
- 27.Alzeer J, Vummidi BR, Roth PJ, Luedtke NW. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew Chem Int Ed Engl. 2009;48:9362–9365. doi: 10.1002/anie.200903685. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen B, Boykin DW, Wilson WD. DNA minor groove interactions of antiparasitic diamidines: reevaluation of the crescent-shape concept in groove-binding. In: Lee M, Strekowski L, editors. Synthetic and biophysical studies of DNA binding compounds. 2007. pp. 39–66. [Google Scholar]
- 29.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arafa RK, Ismail MA, Munde M, Wilson WD, Wenzler T, Brun R, Boykin DW. Novel linear triaryl guanidines, N-substituted guanidines and potential prodrugs as antiprotozoal agents. Eur J Med Chem. 2008;43:2901–2908. doi: 10.1016/j.ejmech.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen B, Neidle S, Wilson WD. A role for water molecules in DNA-ligand minor groove recognition. Acc Chem Res. 2009;42:11–21. doi: 10.1021/ar800016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tevis DS, Kumar A, Stephens CE, Boykin DW, Wilson WD. Large, sequence-dependent effects on DNA conformation by minor groove binding compounds. Nucleic Acids Res. 2009;37:5550–5558. doi: 10.1093/nar/gkp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Moreno T, Pous J, Subirana JA, Campos JL. Coiled coil conformation of a pentamidine-DNA complex. Acta Cryst D. 2010;66 doi: 10.1107/S0907444909055693. in press. Reports a novel coiled coil structure adopted by a complex between the DNA duplex d(ATATATATAT)2 and pentamidine, with a mixture of Watson-Crick and Hoogsteen base pairs and formation of cross-links between neighboring DNAs. [DOI] [PubMed] [Google Scholar]
- 34•.Glass LS, Nguyen B, Goodwin KD, Dardonville C, Wilson WD, Long EC, Georgiadis MM. Crystal structure of a trypanocidal 4,4′-bis(imidazolinylamino)diphenylamine bound to DNA. Biochem. 2009;48:5943–5952. doi: 10.1021/bi900204w. The ligand that is predicted to be highly twisted in its energy-minimized state adopts a planar crescent shape when bound in the minor groove of the DNA. The structure supports the notion of an induced fit resulting from conformational changes in both the ligand and the DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc Natl Acad Sci USA. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. The crystal structure of an 8-ring cyclic polyamide bound to DNA reveals widening of the minor groove along with a severe bend toward the major groove, supporting the idea that allosteric perturbation of the DNA helix provides a molecular basis for disruption of transcription factor-DNA interfaces by small molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masliah G, Rene B, Zargarian L, Fermandjian S, Mauffret O. Identification of intrinsic dynamics in a DNA sequence preferentially cleaved by topoisomerase II enzyme. J Mol Biol. 2008;381:692–706. doi: 10.1016/j.jmb.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Narayana N, Weiss MA. Crystallographic analysis of a sex-specific enhancer element: Sequence-dependent DNA structure, hydration, and dynamics. J Mol Biol. 2009;385:469–490. doi: 10.1016/j.jmb.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pous J, Urpi L, Subirana JA, Gouyette C, Navaza J, Campos JL. Stabilization by extra-helical thymines of a DNA duplex with Hoogsteen base pairs. J Am Chem Soc. 2008;130:6755–6760. doi: 10.1021/ja078022+. [DOI] [PubMed] [Google Scholar]
- 39.Girard E, Prangé T, Dhaussy A-C, Migianu-Griffoni E, Lecouvey M, Chervin J-C, Mezouar M, Kahn R, Fourme R. Adaptation of the base-paired double-helix molecular architecture to extreme pressure. Nucleic Acids Res. 2007;35:4800–4808. doi: 10.1093/nar/gkm511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilton DJ, Ghosh M, Chary KVA, Akasaka K, Williamson MP. Structural change in a B-DNA helix with hydrostatic pressure. Nucleic Acids Res. 2008;36:4032–4037. doi: 10.1093/nar/gkn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santini GP, Cognet JA, Xu D, Singarapu KK, Herve du Penhoat C. Nucleic acid folding determined by mesoscale modeling and NMR spectroscopy: solution structure of d(GCGAAAGC) J Phys Chem B. 2009;113:6881–6893. doi: 10.1021/jp8100656. [DOI] [PubMed] [Google Scholar]
- 42•.Viladoms J, Escaja N, Frieden M, Gomez-Pinto I, Pedroso E, Gonzalez C. Self-association of short DNA loops through minor groove C:G:G:C tetrads. Nucleic Acids Res. 2009;37:3264–3275. doi: 10.1093/nar/gkp191. Describes the structures of short DNAs that self-associate under formation of symmetric dimers stabilized by a novel kind of minor groove base tetrad that differs from the familiar patterns in G-rich quadruplexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khuu P, Ho PS. A rare nucleotide base tautomer in the structure of an asymmetric DNA junction. Biochem. 2009;48:7824–7832. doi: 10.1021/bi900829b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda T, Ito Y, Shibata T, Mikawa T. A non-canonical DNA structure enables homologous recombination in various genetic systems. J Biol Chem. 2009;284:30230–30239. doi: 10.1074/jbc.M109.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bang J, Bae SH, Park CJ, Lee JH, Choi BS. Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites. J Am Chem Soc. 2008;130:17688–17696. doi: 10.1021/ja8038272. [DOI] [PubMed] [Google Scholar]
- 46.Antonow D, Barata T, Jenkins TC, Parkinson GN, Howard PW, Thurston DE, Zloh M. Solution structure of a 2:1 C2-(2-naphthyl) pyrrolo[2,1-c][1,4]benzodiazepine DNA adduct: Molecular basis for unexpectedly high DNA helix stabilization. Biochem. 2008;47:11818–11829. doi: 10.1021/bi801225q. [DOI] [PubMed] [Google Scholar]
- 47.Zaliznyak T, Lukin M, Johnson F, de los Santos C. Solution structure of duplex DNA containing the mutagenic lesion 1,N(2)-etheno-2′-deoxyguanine. Biochem. 2008;47:4606–4613. doi: 10.1021/bi7022514. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Schnetz-Boutaud NC, Kroth H, Yagi H, Sayer JM, Kumar S, Jerina DM, Stone MP. 3′-Intercalation of a N2-dG 1R-trans-anti-benzo[c]phenanthrene DNA adduct in an iterated (CG)3 repeat. Chem Res Toxicol. 2008;21:1348–1358. doi: 10.1021/tx7004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaliznyak T, Bonala R, Attaluri S, Johnson F, de los Santos C. Solution structure of DNA containing alpha-OH-PdG: the mutagenic adduct produced by acrolein. Nucleic Acids Res. 2009;37:2153–2163. doi: 10.1093/nar/gkp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Wang H, Qi N, Lloyd RS, Rizzo CJ, Stone MP. The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2′-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5′-CpG-3′ sequence. Biochem. 2008;47:11457–11472. doi: 10.1021/bi8011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H, Wang H, Lloyd RS, Rizzo CJ, Stone MP. Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA. Chem Res Toxicol. 2009;22:187–200. doi: 10.1021/tx800320m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfoh R, Laatsch H, Sheldrick GM. Crystal structure of trioxacarcin A covalently bound to DNA. Nucleic Acids Res. 2008;36:3508–3514. doi: 10.1093/nar/gkn245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Huang H, Dooley PA, Harris CM, Harris TM, Stone MP. Differential base stacking interactions induced by trimethylene interstrand DNA cross-links in the 5′-CpG-3′ and 5′-GpC-3′ sequence contexts. Chem Res Toxicol. 2009;22:1810–1816. doi: 10.1021/tx900225c. The sequence selectivity of enal-mediated cross-linking can be attributed to differential base stacking and in turn stabilities at the CpG and GpC base-pair steps as revealed by NMR experiments in solution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharyya D, Ramachandran S, Sharma S, Pathmasiri W, King CL, Baskerville-Abraham I, Boysen G, Swenberg JA, Campbell SL, Dokholyan NV, Chaney SG. Flanking bases influence the nature of DNA distortion by oxaliplatin 1,2-intrastrand (GG) cross-links. J Mol Biol. 2010 doi: 10.1371/journal.pone.0023582. Vol:submitted. (According to information on the last author’s website; states in e-mail on 12/30/09 that paper will hopefully be published soon) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D’Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects. Proc Natl Acad Sci USA. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. Although cDPCP binds DNA monofunctionally the crystal structure of an adducted DNA displays distortions that are more characteristic of those induced by cisplatin intrastrand cross-links. The drug effectively blocks transcription from adducted DNA templates and, remarkably, hampers removal by the nucleotide excision repair apparatus to a larger degree than bifunctional platinum-DNA cross-links. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode. Biochem. 2009;48:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfoh R, Cuesta-Seijo JA, Sheldrick GM. Interaction of an echinomycin-DNA complex with manganese ions. Acta Cryst F. 2009;65:660–664. doi: 10.1107/S1744309109019654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millonig H, Pous J, Gouyette C, Subirana JA, Campos JL. The interaction of manganese ions with DNA. J Inorg Biochem. 2009;103:876–880. doi: 10.1016/j.jinorgbio.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Zamecnik PC, Stephenson ML. Inhibition of rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephenson ML, Zamecnik PC. Inhibition of rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isselbacher KJ, Paul C. Zamecnik (1912–2009) Science. 2009;326:1359. doi: 10.1126/science.1184725. [DOI] [PubMed] [Google Scholar]
- 62.Egli M, Pallan PS. Insights from crystallographic studies into the structural and pairing properties of nucleic acid analogs and chemically modified DNA and RNA oligonucleotides. Annu Rev Biophys Biomol Struct. 36:281–305. doi: 10.1146/annurev.biophys.36.040306.132556. [DOI] [PubMed] [Google Scholar]
- 63•.Egli M, Pallan PS. Crystallographic studies of chemically modified nucleic acids: a backward glance. Chem Biodiv. 2010;7:60–89. doi: 10.1002/cbdv.200900177. A comprehensive overview of crystal structures of chemically modified nucleic acids and artificial pairing systems and insights into the consequences of modifications for stability and function as well as lessons for the design of nucleic acids with tailor-made properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robeyns K, Herdewijn P, Van Meervelt L. Influence of the incorporation of a cyclohexenyl nucleic acid (CeNA) residue onto the sequence d(CGCGAATTCGCG) Nucleic Acids Res. 2008;36:1407–1414. doi: 10.1093/nar/gkm1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Matsugami A, Ohyama T, Inada M, Inoue N, Minakawa N, Matsuda A, Katahira M. Unexpected A-form formation of 4′-thioDNA in solution, revealed by NMR, and the implications as to the mechanism of nuclease resistance. Nucleic Acids Res. 2008;36:1805–1812. doi: 10.1093/nar/gkn011. Small change – big effect: Replacement of the intra-ring oxygen by sulfur in the oligo-2′-deoxyribose context flips a B-form duplex to the A-form. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker RT. 4′-Thio-2′-deoxyribonucleosides, their chemistry and biological properties - a review. R Soc Chem Spec Publ (Anti-Infectives) 1997;198:203–237. [Google Scholar]
- 67.Pallan PS, Prakash TP, Li F, Eoff RL, Manoharan M, Egli M. A conformational transition in the structure of a 2′-thiomethyl-modified DNA visualized at high resolution. Chem Commun (Camb) 2009:2017–2019. doi: 10.1039/b822781k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pallan PS, Ittig D, Heroux A, Wawrzak Z, Leumann CJ, Egli M. Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles. Chem Commun (Camb) 2008:883–885. doi: 10.1039/b716390h. [DOI] [PubMed] [Google Scholar]
- 69.Nielsen JT, Arar K, Petersen M. Solution structure of a locked nucleic acid modified quadruplex: introducing the V4 folding topology. Angew Chem Int Ed Engl. 2009;48:3099–3103. doi: 10.1002/anie.200806244. [DOI] [PubMed] [Google Scholar]
- 70•.Kolarovic A, Schweizer E, Greene E, Gironda M, Pallan PS, Egli M, Rozners E. Interplay of structure, hydration and thermal stability in formacetal modified oligonucleotides: RNA may tolerate nonionic modifications better than DNA. J Am Chem Soc. 2009;131:14932–14937. doi: 10.1021/ja904926e. The hydrophobic formacetal linker has little effect on RNA hydration but decreases DNA hydration, which suggests that it is an excellent mimic of the phosphate linkage in RNA and possibly suitable for probing RNA-protein interactions and in RNAi applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patra A, Richert C. High fidelity base pairing at the 3′-terminus. J Am Chem Soc. 2009;131:12671–12681. doi: 10.1021/ja9033654. [DOI] [PubMed] [Google Scholar]
- 72.Wang F, Li F, Ganguly M, Marky LA, Gold B, Egli M, Stone MP. A bridging water anchors the tethered 5-(3-aminopropyl)-2′-deoxyuridine amine in the DNA major groove proximate to the N+2 C. G base pair: implications for formation of interstrand 5′-GNC-3′ cross-links by nitrogen mustards. Biochem. 2008;47:7147–7157. doi: 10.1021/bi800375m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Pallan PS, Egli M. Pairing geometry of the hydrophobic thymine analogue 2,4-difluorotoluene in duplex DNA as analyzed by X-ray crystallography. J Am Chem Soc. 2009;131:12548–12549. doi: 10.1021/ja905739j. The geometry of F:A pairs in the crystal structure of a modified DNA duplex is consistent with formation of a hydrogen bond between fluorine and the exocyclic amino group of A, challenging the view that the 2,4-difluorotoluene base analog is entirely hydrophobic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74••.Kaufmann M, Gisler M, Leumann CJ. Stable cyclohexyl-phenyl recognition in the center of a DNA duplex. Angew Chem Int Ed Engl. 2009;48:3810–3813. doi: 10.1002/anie.200900153. Reports the unexpected finding of increased thermodynamic stability as a result of incorporation of a hydrophobic and non-aromatic entity into the core of a DNA duplex. [DOI] [PubMed] [Google Scholar]
- 75.Hassan AE, Sheng J, Jiang J, Zhang W, Huang Z. Synthesis and crystallographic analysis of 5-Se-thymidine DNAs. Org Lett. 2009;11:2503–2506. doi: 10.1021/ol9004867. [DOI] [PubMed] [Google Scholar]
- 76.Sheng J, Salon J, Gan J, Huang Z. Synthesis and crystal structure study of 2′-Se-adenosine-derivatized DNA. Science in China. 2010;53:78–85. [Google Scholar]
- 77.Salon J, Sheng J, Gan J, Huang Z. Synthesis and crystal structure of 2’f-Se-modified guanosine containing DNA. J Org Chem. 2010:75. doi: 10.1021/jo902190c. in press. [DOI] [PubMed] [Google Scholar]
- 78.Sheng J, Huang Z. Selenium derivatization of nucleic acids for X-ray crystal structure and function studies. Chem Biodiv. 2010;7 doi: 10.1002/cbdv.200900200. in press. [DOI] [PubMed] [Google Scholar]
- 79.Hassan AEA, Sheng J, Zhang W, Huang Z. High fidelity of base pairing by 2-selenothymidine in DNA. J Am Chem Soc. 132 doi: 10.1021/ja909330m. in press. [DOI] [PubMed] [Google Scholar]
- 80.Lin L, Sheng J, Momin RK, Du Q, Huang Z. Facile synthesis and anti-tumor cell activity of Se-containing nucleosides. Nucleosides, Nucleotides and Nucleic Acids. 2009;28:56–66. doi: 10.1080/15257770802581765. [DOI] [PubMed] [Google Scholar]
- 81.Caton-Williams J, Huang Z. Synthesis and DNA polymerase incorporation of colored 4-selenothymidine triphosphate with a single atom substitution. Angew Chem Int Ed Engl. 2008;47:1723–1725. doi: 10.1002/anie.200705213. [DOI] [PubMed] [Google Scholar]
- 82.Sheng J, Hassan A, Huang Z. Synthesis of the first tellurium-derivatized oligonucleotides for structural and functional studies. Chem Europ J. 2009;15:10210–10216. doi: 10.1002/chem.200900774. [DOI] [PubMed] [Google Scholar]
- 83••.Schlegel MK, Essen L-O, Meggers E. Duplex Structure of a minimal nucleic acid. J Am Chem Soc. 2008;130:8158–8159. doi: 10.1021/ja802788g. Reports the crystal structure of the arguably simplest version of a nucleic acid with a phosphate backbone and reveals how the glycol or glycerol backbone can support highly stable Watson-Crick-like duplex formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang RS, McCullum EO, Chaput JC. Synthesis of two mirror image 4-helix junctions derived from glycerol nucleic acid. J Am Chem Soc. 2008;130:5846–5847. doi: 10.1021/ja800079j. [DOI] [PubMed] [Google Scholar]
- 85.Schlegel MK, Xie X, Zhang L, Meggers E. Insight into the high duplex stability of the simplified nucleic acid GNA. Angew Chem Int Ed Engl. 2009;48:960–963. doi: 10.1002/anie.200803472. [DOI] [PubMed] [Google Scholar]
- 86.Schlegel MK, Essen L-O, Meggers E. Atomic resolution structure of the simplified nucleic acid GNA. Chem Commun (Camb) 2010 doi: 10.1039/b916851f. in press. [DOI] [PubMed] [Google Scholar]
- 87•.Pallan PS, Lubini P, Bolli M, Egli M. Backbone-base inclination as a fundamental determinant of nucleic acid self- and cross-pairing. Nucleic Acids Res. 2007;35:6611–6624. doi: 10.1093/nar/gkm612. Defines a new geometric parameter that describes the inclination between backbone and base-pair planes and serves as a predictor for the strand polarity in nucleic acid self-pairing and the ability of native and artificial nucleic acids to cross-pair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 89.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 90.Richert C, Meng M, Müller K, Heimann K. The third dimension: DNA-driven formation of nanoparticle crystals. Small. 2008;4:1040–1042. doi: 10.1002/smll.200800326. [DOI] [PubMed] [Google Scholar]
- 91••.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C, Seeman NC. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. Delivers proof in the form of a crystal structure of an object composed of DNA strands that it is possible to design and self-assemble a well-ordered macromolecular 3D crystalline lattice with precise control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]