Abstract
AIMS
The objectives of this study were to determine the distribution of CYP2C9 variants in Koreans and investigate their association with warfarin dose requirements in patients who received MHVRs.
METHODS
All nine exons, intron–exon junction, and promoter region of CYP2C9 were amplified and directly sequenced in 50 healthy normal Koreans. Additional direct DNA sequencing of the CYP2C9 gene was conducted in 36 of the 267 MHVR patients who required low maintenance warfarin doses without carrying CYP2C9*3 and VKORC1 1173T mutations. The effects of CYP2C9 genetics on warfarin maintenance dose were assessed in 267 MHVR patients.
RESULTS
Thirty-nine single nucleotide polymorphisms (SNPs) including seven previously unidentified SNPs were identified in 50 Koreans by direct DNA sequencing. One of the CYP2C9 haplotypes exhibited an association with warfarin low dose requirement. The adjusted odds ratio for the haplotype between the low dose group and the normal subjects was 2.5 (95% confidence interval 1.05, 6.16). This haplotype consisting of -1565C>T, -1188T>C, IVS3+197G>A, IVS3-334C>T, IVS3-65G>C, IVS4-115A>G, and IVS5-73A>G was found in 15% of 36 MHVR patients who required low warfarin doses, while 4% of 50 normal healthy subjects exhibited this haplotype. One of the SNPs comprising this haplotype, -1565C>T, apparently changed a protein binding pattern as observed in electrophoretic mobility shift assay.
CONCLUSION
The haplotype including -1565C>T, -1188T>C, IVS3+197G>A, IVS3-334C>T, IVS3-65G>C, IVS4-115A>G, and IVS5-73A>G seems to be associated with low warfarin dose requirement and this haplotype could be considered in the development of a warfarin dose prediction model for Asian populations.
Keywords: CYP2C9, single nucleotide polymorphisms, warfarin
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
CYP2C9 single nucleotide polymorphisms (SNPs) are important in safe and effective oral anticoagulation with warfarin use.
Although CYP2C9*2 and *3 are important genetic factors for the warfarin dose, one of the CYP2C9 SNPs, IVS-65G>C, has been suggested to be associated with warfarin sensitivity. However, as of yet, there has been no explanation about the possible mechanism and linkage analysis.
WHAT THIS STUDY ADDS
New information on CYP2C9 SNPs and their occurrences in common haplotype structures in healthy unrelated Koreans and in individuals who require low warfarin dose after mechanical heart valve replacements (MHVRs) were studied.
Additional evidence showed that an Asian dominant haplotype consisting of -1565C>T, -1188T>C, IVS3+197G>A, IVS3-334C>T, IVS3-65G>C, IVS4-115A>G and IVS5-73A>G could be associated with a low warfarin maintenance dose in mechanical heart valve replacement (MHVR) patients.
Introduction
Among the four CYP2C genes, CYP2C9 is the most abundantly expressed in the human liver [1]. The enzyme CYP2C9 is reported to metabolize approximately 16% of human drugs, including the antidiabetic drugs tolbutamide [2] and glipizide [3], the anticonvulsant phenytoin [4], the anticoagulant warfarin [5], the antihypertensive drug losartan [6], the diuretic torasemide [7], and numerous nonsteroidal anti-inflammatory drugs (NSAIDs) [8]. CYP2C9 also metabolizes endogenous substrates, such as arachidonic acid and linoleic acid [9]. CYP2C9 is a genetically polymorphic enzyme responsible for interindividual variation in the metabolism and disposition of many widely used drugs. In particular, genetic polymorphisms in CYP2C9 have caused variable warfarin dose requirements and risk for adverse events, such as thromboembolic or hemorrhagic complications.
At present, more than 30 single nucleotide polymorphisms (SNPs) of CYP2C9 have been reported to the Human P450 Allele Nomenclature Committee. The two most important allele variants, CYP2C9*2 and CYP2C9*3, occur at frequencies of 10.5 and 8.4%, respectively, in Caucasians [10], while these are low in Africans at 2–4 and 1–2%, respectively [11, 12]. In Asians, CYP2C9*2 has not been detected, and CYP2C9*3 has been reported at frequencies of 3–6% [13–15]. CYP2C9*8, *9, and *11 were found in Africans with relatively high frequencies at 3.6, 13, and 5.6%, respectively. However, these SNPs are low or undetected in other racial groups, indicating apparent ethnic differences in the frequencies of CYP2C9 variants [16, 17].
Warfarin is an anticoagulant drug, which is administered as an equal mixture of (S)- and (R)-enantiomers for the treatment of thrombosis and embolism in many disorders. Most of the anticoagulation effect is attributable to the (S)-enantiomer, which is deactivated predominantly by CYP2C9[5]. Frequent blood tests for a stable international normalized ratio (INR) are performed to reduce the unwanted side effects of warfarin. Although CYP2C9*2 and *3 are important genetic factors for the determination of warfarin dose in Caucasians, these alleles are not significant contributing factors in Asians due to the very low allele frequencies. More extensive searches would be required in other ethnic groups in order to obtain a complement of CYP2C9-defective alleles. Ethnic variation in CYP2C9 polymorphisms is apparent, but no discovery studies on CYP2C9 genetic polymorphisms have been carried out in Koreans via direct DNA sequencing. Therefore, we sequenced the CYP2C9 gene in 50 normal healthy subjects and evaluated the allele distributions and frequencies for the first time in a Korean population. An additional purpose of this study was to investigate the influence of CYP2C9 variants on warfarin maintenance dose in a well-characterized Korean cohort of MHVR patients [18] through direct DNA sequencing of 36 out of 267 MHVR patients who required low maintenance warfarin doses.
Methods
DNA samples and subjects
Genomic DNA was prepared from peripheral whole blood using the QIAamp DNA Blood Kit (Qiagen, Valencia, CA, USA) and stored at –20°C until use. Fifty healthy subjects were selected randomly from the south eastern part of the Korean peninsula to investigate genetic polymorphisms of CYP2C9 in Koreans. Genomic DNA samples of 50 Korean subjects were obtained from the DNA Repository of the Pharmacogenomics Research Center, Inje University [19]. All the participants were healthy according to their medical histories, physical examinations and routine laboratory tests. MHVR patients were recruited from the Outpatient Clinic of Busan Paik Hospital between September 2005 and March 2007. Data during visits such as warfarin dose, INRs, bleeding events, co-medications, foods, gender, weight and age were collected. Warfarin dose was adjusted every day based on daily INR for the first 2 weeks in the hospital. Once a stable dose of warfarin was reached, the INR was monitored during intervals of 4–8 weeks. A stable dose was defined as the warfarin dose when INR values were within a therapeutic range (1.7–2.8) three times consecutively. Stable warfarin doses ranged from 1.5 mg day−1 to 9.5 mg day−1. Of the 267 MHVR patients, 36 individuals who required low maintenance doses of warfarin (3 mg day−1) at Paik Hospital, Busan, Korea, were selected as the low dose group and used for full DNA sequencing of the CYP2C9 gene. The low and high dose (5.5 mg day−1) groups were selected from ∼15% of individuals at each end of a normal distribution curve describing data that cluster around the mean.
All participants provided written informed consent prior to the study. The institutional review board (IRB) of Busan Paik Hospital, Inje University College of Medicine, Busan, Korea, approved the protocol. Details on MHVR patients in the present study were described in the previous report [18].
DNA sequencing and variant identification
For direct DNA sequencing of the CYP2C9 gene, primers for the amplification of all exons, exon/intron junctions, 3377 bp from the translation start site, and up to 300 bp after the stop codon at the 3′-untranslated region (3′-UTR) were designed according to a previously reported method with slight modifications [20]. The amplified PCR products were directly sequenced after gel electrophoresis to ensure proper amplification, as described previously [19].
Genotyping
A new pyrosequencing method was developed to detect the presence of IVS4-115A>G. The 646-bp fragment containing an IVS4-115A>G variant was amplified using the following primer sequences: forward, 5′-CTGGTTAGAATTGATCCTCTG-3′, and reverse, 5′-Biotin_GGTAGTTTATATTTCTGTGGGCTC-3′. After amplification of the DNA fragment, a sequencing primer, 5′-AAATTTCCCCATCAAG-3′ was used to detect the IVS4-115A>G variant. PCR was performed using a GeneAmp PCR 9700 instrument (Applied Biosystems, Foster City, CA, USA). Briefly, the PCR reaction included 0.25 µm of each primer and 100 ng of genomic DNA in a 20 µl reaction. PCR was performed using one cycle of initial denaturation (5 min, 94°C); 35 cycles comprising of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s; followed by a final extension at 72°C for 7 min. The reaction mixture was analyzed on a PSQ 96MA Pyrosequencer (Pyrosequencing AB, Uppsala, Sweden). The accuracy of pyrosequencing was verified by direct DNA sequencing using the same genomic DNA. Details of the genotyping method for VKORC1 1173 C>T employing the use of pyrosequencing were described in a previous report [18]. Except for the primer set and sequencing primer, the procedure for pyrosequencing was the same as described previously [21].
Electrophoretic mobility shift assays (EMSA)
Nuclear proteins from human liver tissues were isolated by a NE-PER Nuclear and Cytoplasmic Extraction Reagent (PIERCE, Rockford, USA) system according to the manufacturer's protocol. Liver tissues from Korean donors were obtained from the Busan Paik Hospital (Busan, Korea) affiliated with the tissue repository bank at INJE Pharmacogenomics Research Center (Inje University College of Medicine, Busan, Korea). The approval for the usage of human liver tissues was obtained according to the institutional guideline. [32P]-dATP was incorporated into oligonucleotide primers using T4-polynucleotide kinase (USB, Cleveland, Ohio, USA). The labelled probe (approximately 150 000 counts min−1) was incubated in the presence or absence of nuclear proteins in a 25 mm Hepes buffer (pH 7.8) containing 100 mm KCl, 5 mm MgCl2, 0.3 mm EDTA, 0.5 mm dithiothreitol, 2.5% glycerol and 2% Ficoll 400. Various cold competitors were added to the reaction before the addition of isolated nuclear proteins. After 30 min of incubation at 4°C, the reaction mixture (20 µl) was loaded onto a 5% nondenaturing polyacrylamide gel for electrophoresis in a TAE buffer for 3 h at 100 V. After electrophoresis, the gel was dried and exposed to XAR film (KODAK BioMax, MO, USA). The oligonucleotide probe sequences used were as follows (mutated sequence is underlined): wild-type probe, 5′-CCTCATTCCGGAAATGGGT-3′; variant probe M1 (1565C>T), 5′-CCTCATTCTGGAAATGGGT-3′; and multiple base mutation probe M2, 5′- CCTCAAGAATTCCCTGGGT-3′.
Statistics
The allele frequency, the Hardy-Weinberg equilibrium, 95% confidence interval (CI) and linkage disequilibrium (LD) were analyzed using a SNPAlyse software (Dynacom Co., Yokohama, Japan). Pairwise LD analysis was performed by |D′| and r2 statistics using common SNPs that exhibited more than 0.05 in their allelic frequencies. Haplotype analysis was performed using IIPGA tools (https://innateimmunity.net/IIPGA2/Bioinformatics). Twenty variants identified by direct DNA sequencing in 50 healthy Koreans were applied to select tagging SNPs. The program Tagger was used to select tagging SNPs, which combines the simplicity of pairwise methods with the potential efficiency of multimarker approaches (http://www.broad.mit.edu/mpg/tagger/) [22]. Warfarin dose requirements were determined with stable INR values for three consecutive measurements within 3 month intervals and expressed as the mean mg day−1± standard deviation (SD) [18]. The values between two different genotypes were analyzed using a Wilcoxon rank-sum test. Odds ratios (OR) and 95% CI were estimated using a logistic regression model adjusting for weight, age, aspirin, congestive heart failure/cardiomyopathy, INR-increasing drugs (amiodarone, fluconazole, doxifluridine) and INR-decreasing dietary supplements. All statistical analysis was performed using the SAS program (version 9.1.3; SAS Institute, Cary, NC, USA). A P < 0.05 was considered statistically significant.
Results
Thirty-nine variants of CYP2C9 were identified in 50 Korean individuals and their allele frequencies are presented in Table 1. Of these, 13 SNPs were identified in the 5′-UTR, two in coding regions, 21 in introns, and two in the 3′-UTR. Seven of the 39 SNPs were newly identified in the present study as rare variants (four in the 5′-UTR and three in introns). Two SNPs found in the coding regions included CYP2C9*3 at 6% frequency, and a silent base pair change, g.1425 A>T, in exon 9 at 5% frequency. No significant deviations from Hardy-Weinberg equilibrium were detected for the identified variants. A program at http://www.cbrc.jp/research/db/TFSEARCH.html was used to examine the creation or elimination of transcriptional factor binding elements introduced by the mutation. No significant novel mutations for altered transcriptional binding sites were found in our analysis, except a variant -1565C>T which disrupted a possible binding motif sequence of signal transducers and activator of transcription (STAT) as well as an Elk-1 with 89 and 86% probability, respectively. To predict a possible acceptor or donor site for splicing events, a sequence analysis program at http://www.fruitfly.org/seq_tools/splice.html was used. One variant, IVS4-115A>G was predicted as a possible splicing donor site with 89% probability and this variant was predicted to be a haplotype with -1565C>T, -1188T>C, IVS3+197G>A, IVS3-334C>T, IVS3-65G>C and IVS5-73A>G.
Table 1.
Single nucleotide polymorphisms (SNPs) found in the CYP2C9 gene in a Korean population*
| Subject number (n) | |||||||
|---|---|---|---|---|---|---|---|
| Site | Nucleotide changeand position† | Amino acid change | Nucleotide change and flanking sequence | w/w | w/m | m/m | Minor allele frequency (95% CI) |
| 5-UTR | -3089G>A | TGATTCCAACC(G/A)TATTACATTTTG | 24 | 21 | 5 | 0.31 (0.182, 0.438) | |
| 5-UTR | -2787G>T‡ | GTGTGGGTGCA(G/T)GGAAAGAGGC | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| 5-UTR | -2665∼2664delTG | TCAGTGAC(DEL/TG)TGGAGGGCTTAA | 24 | 21 | 5 | 0.31 (0.182, 0.438) | |
| 5-UTR | -1911T>C | GGACCAAGTTA(T/C)TGCTTTTCTTTGC | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| 5-UTR | -1885C>G | CCTGTATAAAGG(C/G)TTCTCCAAGGCC | 45 | 4 | 1 | 0.06 (0, 0.129) | |
| 5-UTR | -1875G>C‡ | GGCTTCTCCAAG(G/C)CCTTTGACTTAC | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| 5-UTR | -1746C>T‡ | TAGACTGAATTA(C/T)GAAATCCTGAAT | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| 5-UTR | -1565C>T | GCTTCCTCATTC(C/T)GGAAATGGGTC | 46 | 4 | 0 | 0.04 (0, 0.094) | |
| 5-UTR | -1537G>A | TTTATTGTAAGCA(G/A)AGGTAATTGAGA | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| 5-UTR | -1520C>T | ATTGAGAGATT(C/T)AAAAGGGACATG | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| 5-UTR | -1188T>C | ACCTCCCATCTT(T/C)TATTGCATCCAC | 17 | 23 | 10 | 0.43 (0.292, 0.567) | |
| 5-UTR | -981G>A | TGCAGTGATGGA(G/A)AAGGGAGATCC | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| 5-UTR | -641A>T‡ | CTGCCTTCAGGA(A/T)TTTTTTTTAGGG | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 1 | IVS1+83T>C | CCTAGAGGTACA(T/C)GTTACAAGAGG | 48 | 2 | 0 | 0.02 (0, 0.059) | |
| Intron 1 | IVS1-22T>C | CTTCGTTTGCTG(T/C)TATCTCTGTCTA | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 2 | IVS2+73T>C | GACTTACAGAGC(T/C)CCTCGGGCAGA | 48 | 2 | 0 | 0.02 (0, 0.059) | |
| Intron 3 | IVS3+197G>A | GCATGATTGTGC(G/A)TACAGTGTGGG | 41 | 8 | 1 | 0.10 (0.017, 0.183) | |
| Intron 3 | IVS3+239C>T | ATCCCATGTTCTC(C/T)TGAACTTTGCT | 24 | 21 | 5 | 0.31 (0.182, 0.438) | |
| Intron 3 | IVS3+265T>C | TTTTGCTTTCAAA(T/C)AAGAAATGATG | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| Intron 3 | IVS3-334C>T | TCTCAGTGCCTTG(C/T)TGTCTACTGACT | 41 | 8 | 1 | 0.10 (0.017, 0.183) | |
| Intron 3 | IVS3-122T>A‡ | ATGCATGCCGAAC(T/C)CTTTTTTGCTGT | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 3 | IVS3-65G>C | ACTACTATTATCT(G/C)TTAACAAATACA | 46 | 4 | 0 | 0.04 (0, 0.094) | |
| Intron 4 | IVS4-115A>G | TTCCCCATCAAG(A/G)TATACAATATAT | 46 | 4 | 0 | 0.04 (0, 0.094) | |
| Intron 4 | IVS4-50T>C‡ | GGTATATGGTATG(T/C)ATGCTTTTATTAA | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 5 | IVS5-73A>G | ATAACTATGTGA(A/G)TAATTTTGAATTC | 41 | 8 | 1 | 0.10 (0.017, 0.183) | |
| Intron 6 | IVS6+95A>G | TAGAGAAGCTTC(A/G)TTATTTAAACTTT | 48 | 2 | 0 | 0.02 (0, 0.059) | |
| Intron 6 | IVS6+152A>G | ATGGTGATTACA(A/G)TGGGATATCTTGG | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 6 | IVS6+214G>A‡ | TTGGGAGGCTGA(G/A)GTGGGTGGATCA | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Intron 6 | IVS6-137T>A | ATCCTCTCTTTAAG(T/A)TTGCATATACTT | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Exon 7 | 1075A>C | I359L | GGTCCAGAGATAC(A/C)TTGACCTTCTC | 45 | 4 | 1 | 0.06 (0, 0.126) |
| Intron 7 | IVS7+38C>T | CAACTCCATGTTTT(C/T)GAAGTCCCCA | 45 | 5 | 0 | 0.05 (0, 0.11) | |
| Intron 8 | IVS8+53A>T | AGATAACTTTTTTG(A/T)TCCATTGGAAC | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| Intron 8 | IVS8+147C>T | GTGTACACCCTG(C/T)TCATGATACATCC | 24 | 21 | 5 | 0.31 (0.182, 0.438) | |
| Intron 8 | IVS8-112G>A | TTCATCTCTTCTAC(G/A)ATACACTGAAC | 45 | 4 | 1 | 0.06 (0, 0.126) | |
| Intron 8 | IVS8-109A>T | ATCTCTTCTACGAT(A/T)CACTGAACAGT | 17 | 24 | 9 | 0.42 (0.283, 0.557) | |
| Intron 8 | IVS8-98T>A | ATACACTGAACAG(T/A)TATTGCATATTC | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| Exon 9 | 1425A>T | N474N | CAGTTGTCAATGG(A/T)TTTGCCTCTGTG | 46 | 3 | 1 | 0.05 (0, 0.11) |
| 3′UTR | 7G>C | GTCTGAAGAAGA(G/C)CAGATGGCCTG | 49 | 1 | 0 | 0.01 (0, 0.038) | |
| 3′UTR | 396T>A | CTTCTTTTATGCA(T/A)AATGTAGGTCAG | 45 | 4 | 1 | 0.06 (0, 0.126) | |
Parentheses with bold letters indicate the nucleotide change.
The reference sequence used was GenBank accession no. NC_000010.9.
Position is indicated in respect to the start codon ATG of the CYP2C9 gene; the A in ATG is +1 and the next base toward to 5′ is -1.
Variant alleles newly identified in the present study.
Thirty-six subjects who required a low warfarin dose without having CYP2C9*3 and VKORC1 1173C>T mutations [18] were selected for full DNA sequencing of the CYP2C9 gene. The influences of CYP2C9 variants on low warfarin dose requirement were investigated. Of the 24 variants found in the 36 patients, five variants were not found in the normal 50 subjects (Table 2). Among five variants, two were CYP2C9*13 and *14 variants each identified in one individual as a hetrerozygous mutation. The other three variants included two from the promoter region and one from the intron region. These three alleles exhibited no significant consequences in our sequence analysis using molecular software. Twenty SNPs were found only in 50 normal subjects and five SNPs were only observed in the 36 patients. All of these 25 SNPs except CYP2C9*13 and *14 displayed no significant consequences in our software analysis. The degree of linkage disequilibrium (LD) (Figure 1) was analyzed by the Haploview program based on Gabriel's block definition [23] and seven Tagging SNPs (Figure 1B) were determined using SNPs found in 50 normal subjects. Variants with a frequency greater than 5% were included in the Haploview analysis to get a clear boundary of the LD block. The CYP2C9 locus could be divided into two LD blocks between -2665__2664delTG and -1911T>C. Therefore recombination may have occurred between these two boundaries. However, the actual recombination would be a rare occurrence as it was too minor to divide the LD block. There were different frequencies of the haplotype between the group of 36 patients and the 50 healthy normal subjects. The most significant difference between the two groups was from three variants, -1565C>T, IVS3-65G>C and IVS4-115A>G, which were found in perfect LD, exhibiting 4% frequency in the normal group and 15% in the 36 patients (data not shown due to less than 5% frequency). These three variants were predicted to construct a haplotype together with -1188T>C, IVS3+197G>A, IVS3-334C>T, and IVS5-73A>G as reported previously in the Japanese population as CYP2C9*1e (hereafter referred to as CYP2C9*1e) [20]. To study further the association between the CYP2C9*1e and warfarin dose, a genotyping method for the detection of IVS4-115A>G was developed using pyrosequencing (data not shown) as a marker SNP of CYP2C9*1e. The relationship between the haplotype and warfarin sensitivity was investigated (Figure 2). Regardless of the CYP2C9*3,*13, and *14 genotypes, the effect of heterozygous mutation of IVS4-115A>G on warfarin dose sensitivity appeared to be insignificant. However, IVS4-115A>G tended to affect the warfarin dose requirement in patients homozygous for this mutation (P= 0.18) (Figure 2A). The odds ratio for CYP2C9*1e between the low dose vs. the normal dose groups was insignificant (OR 1.75, 95% CI 0.79, 3.87, P= 0.17). However, when the factors affecting warfarin dose were adjusted, the individuals carrying the CYP2C9*1e had a 2.5-fold increased odds of low dose requirement (OR 2.54, 95% CI 1.05, 6.16, P= 0.04). Patients homozygous for the IVS4-115A>G mutation showed warfarin dose requirement levels similar to the individuals carrying CYP2C9*3, *13 and *14 genotypes (Figure 2B). Since one of the SNPs in CYP2C9*1e, -1565C>T, was predicted to alter possibly a transcriptional binding site, gel shift assay was performed (Figure 3). A representative of three different experiments was presented. All experiments consistently indicated a specific binding pattern to wild-type probe only and shifted bands disappeared after the addition of only wild-type cold competitor. Mutated cold competitor (1565T) addition did not change any shifted bands. The probe containing a single mutation at 1565C>T did not exhibit any binding shift as shown in the wild-type. A computer search revealed that this region contained a consensus binding motif (TTCCNNNAA) for STAT proteins [24]. Addition of a cold competitor known as the core sequence of STAT binding sequence (5′-TGGGGCCAGATTCCGAGAAGACAGCAT-3′) [24] did not affect the band shift or density, suggesting that the binding is likely to originate from the other nuclear proteins rather than the STAT proteins (data not shown).
Table 2.
Genetic polymorphisms of the CYP2C9 gene in 36 patients who required for low warfarin dose*
| Subject number (n) | |||||||
|---|---|---|---|---|---|---|---|
| Site | Nucleotide change and position† | Amino acid change | Nucleotide changeand flanking sequence | w/w | w/m | m/m | Allelic frequency (95%CI) |
| 5-UTR | -3089G>A | TGATTCCAACC(G/A)TATTACATTTTG | 23 | 9 | 4 | 0.24 (0.097, 0.375) | |
| 5-UTR | -2665∼-2664delTG | TCAGTGAC(DEL/TG)TGGAGGGCTTAA | 23 | 9 | 4 | 0.24 (0.097, 0.375) | |
| 5-UTR | -1924A>G‡ | GTTTCATGAGTC(A/G)GGGACCAAGTT | 35 | 0 | 1 | 0.03 (0, 0.082) | |
| 5-UTR | -1565C>T§ | GCTTCCTCATTC(C/T)GGAAATGGGTC | 26 | 9 | 1 | 0.15 (0.035, 0.27) | |
| 5-UTR | -1322T>C‡ | AAGTATTGACAT(T/C)AATGATCTAGTA | 35 | 1 | 0 | 0.01 (0, 0.052) | |
| 5-UTR | -1188T>C§ | ACCTCCCATCTT(T/C)TATTGCATCCAC | 14 | 16 | 6 | 0.39 (0.23, 0.548) | |
| Intron | IVS1+83T>C | CCTAGAGGTACA(T/C)GTTACAAGAGG | 34 | 2 | 0 | 0.03 (0, 0.082) | |
| Intron | IVS1+125T>C‡ | CTTTGAAAGGCT(T/C)TTGTTGCCTTTT | 35 | 1 | 0 | 0.01 (0, 0.052) | |
| exon2 | 269T>C‡ | L90P | CCCTGATTGATC(T/C)TGGAGAGGAGT | 35 | 1 | 0 | 0.01 (0, 0.052) |
| Intron | IVS1-22T>C | CTTCGTTTGCTG(T/C)TATCTCTGTCTA | 35 | 1 | 0 | 0.01 (0, 0.052) | |
| Intron | IVS2+73T>C | GACTTACAGAGC(T/C)CCTCGGGCAGA | 34 | 2 | 0 | 0.03 (0, 0.082) | |
| exon3 | 374G>A‡ | R125H | AGGAGATCCGGC(G/A)TTTCTCCCTCAT | 35 | 1 | 0 | 0.01 (0, 0.052) |
| Intron | IVS3+197G>A§ | GCATGATTGTGC(G/A)TACAGTGTGGG | 26 | 9 | 1 | 0.15 (0.026, 0.252) | |
| Intron | IVS3+239C>T | ATCCCATGTTCTC(C/T)TGAACTTTGCT | 25 | 8 | 4 | 0.24 (0.1, 0.38) | |
| Intron | IVS3-334C>T§ | TCTCAGTGCCTTG(C/T)TGTCTACTGACT | 26 | 9 | 1 | 0.15 (0.035, 0.27) | |
| Intron | IVS3-65G>C§ | AACTACTATTATCT(G/C)TTAACAAATAC | 26 | 9 | 1 | 0.15 (0.035, 0.27) | |
| Intron | IVS4-115A>G§ | TTTCCCCATCAAG(A/G)TATACAATATA | 26 | 9 | 1 | 0.15 (0.035, 0.27) | |
| Intron | IVS4-50T>C | TGGTATATGGTATG(T/C)ATGCTTTTATTA | 35 | 1 | 0 | 0.01 (0, 0.052) | |
| Intron | IVS5-73A>G§ | ATAACTATGTGA(A/G)TAATTTTGAATTC | 26 | 9 | 1 | 0.15 (0.035, 0.27) | |
| Intron | IVS6+95A>G | TAGAGAAGCTTC(A/G)TTATTTAAACTTT | 34 | 2 | 0 | 0.03 (0, 0.082) | |
| Intron | IVS6+152A>G | ATGGTGATTACA(A/G)TGGGATATCTTGG | 34 | 2 | 0 | 0.03 (0, 0.082) | |
| Intron | IVS7+38C>T | CAACTCCATGTTTT(C/T)GAAGTCCCCA | 34 | 2 | 0 | 0.03 (0, 0.082) | |
| Intron | IVS8+147C>T | GTGTACACCCTG(C/T)TCATGATACATCC | 24 | 8 | 4 | 0.24 (0.1, 0.38) | |
| Intron | IVS8-109A>T | ATCTCTTCTACGAT(A/T)CACTGAACAGT | 14 | 16 | 6 | 0.39 (0.23, 0.548) | |
Parentheses with bold letters indicate the nucleotide change in the flanking sequence.
The reference sequence used was GenBank accession no. NC_000010.9.
Position is indicated in respect to the start codon ATG of the CYP2C9 gene; the A in ATG is +1 and the next base toward to 5′ is −1.
Unidentified CYP2C9 variants in the 50 normal subjects are indicated in bold letters.
CYP2C9 variants exhibited significantly different allele frequencies compared with the 50 normal healthy subjects.
Figure 1.
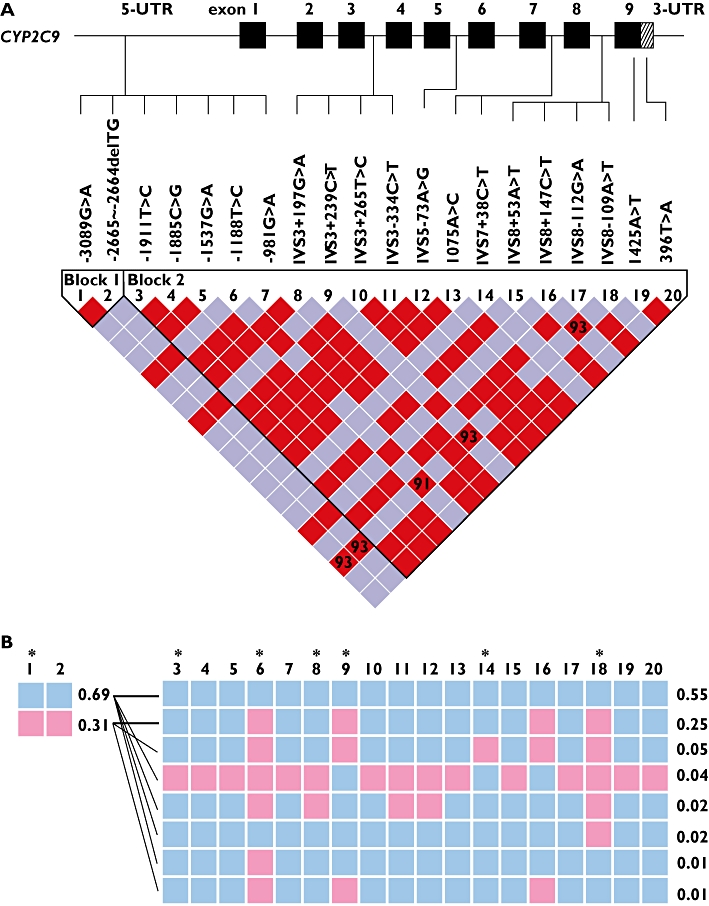
Linkage disequilibrium (LD) map of CYP2C9 single nucleotide polymorphisms obtained from normal healthy subjects (n= 50). CYP2C9 variants at >5% were included in the LD analysis using the statistics |D′| and r2 values [23]. A) Sequential arrangement of CYP2C9 SNPs along with their locations in the CYP2C9 gene. The red colour depicts a significant linkage between the pair of SNPs. Numbers inside the square are the D′ value multiplied by 100. B) CYP2C9 SNPs and their occurrence in common haplotype structures in two blocks. Frequency of each haplotype is shown at the edge. The blue square indicates the most common allele and the pink colour represents the variant allele. The selected tag SNPs are indicated by the star symbol. The thick black lines between haplotypes indicate the most common crossings from block 1 to block 2 and thinner lines mark the less common crossings.
Figure 2.
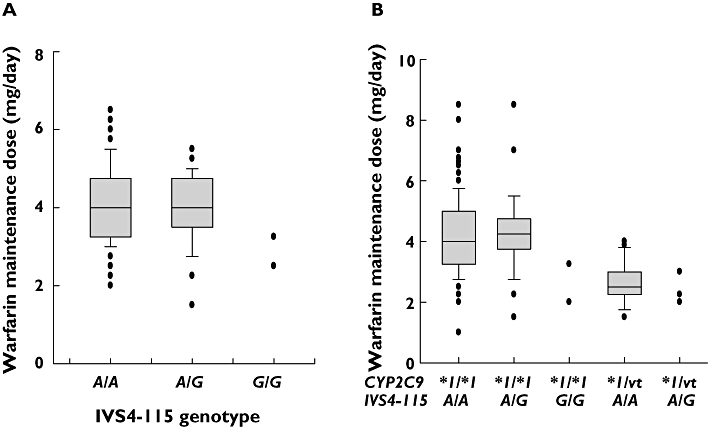
Influence of the CYP2C9*1e haplotype on warfarin maintenance dose. A) All patients with the IVS4-115A>G genotype only, which was used for the detection of CYP2C9*1e. B) Patients stratified by CYP2C9 variants and IVS4-115A>G. All CYP2C9 variants (vt) were heterozygous for mutations of *3,*13, and *14. Boxes extend from the 25th to the 75th percentiles and vertical lines extending from the 10th to 90th percentiles.
Figure 3.
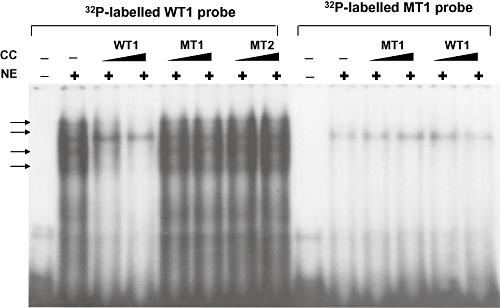
Electrophoretic mobility shift assays (EMSA) of the oligonucleotide containing a -1565C>T change with human liver nuclear extract (NE). EMSAs demonstrate the binding of human liver nuclear proteins (indicated by arrows) to wild-type oligonucleotide probe containing -1565C only. EMSAs were performed as described in Methods. Oligonucleotides for wild-type (WT) and MT-1 (-1565T) were labelled with [32P]-dATP. Oligonucleotides were incubated with nuclear extract at 4°C for 30 min, followed by 5% polyacrylamide gel electrophoresis. Nonradiolabeled cold competitors (CC) were added at 20 and 50-fold excess to determine the specificity of binding complex.
Discussion
The fixed mean daily dose of warfarin in Asians has been reported as low compared with Caucasians [25, 26]. This is consistent with Asians having lower INR values for anticoagulation than Caucasians [27, 28]. This phenotype is not fully explained with the current genotypes of CYP2C9 and VKORC1 even after adjustment for low body weight and other factors present in Asian people, suggesting that unidentified genetic factors or other environmental factors are involved in Asians. Although CYP2C9*2 and *3 are important genetic factors for predicting warfarin dose in Caucasians, these alleles are not critical factors for Asians due to their having very low frequencies. Furthermore, there are still large inter-individual variations in warfarin doses in people with CYP2C9*1/*1 genotype.
Full DNA sequencing of the CYP2C9 gene in 50 individuals from a Korean population exhibited similar variant frequencies compared with other Asian reports [17, 20]. A set of seven tag SNPs of CYP2C9 were determined for the first time in Koreans. It is suggested that these tag SNPs would be required to track all important haplotypes in the CYP2C9 gene in Koreans. Although no novel mutations were detected in our sequencing analysis, two variants were quite intriguing: one in the 5′-UTR (-1565C>T) disrupted a palindromic sequence motif in the promoter region and the other in intron 4 (IVS4-115A>G) had a possible implication in splicing events (predicted at 89% by a splicing prediction program (http://www.fruitfly.org/seq_tools/splice.html). These two variants were found to construct a haplotype with five other intron variants having 4% frequency in 50 randomly selected subjects, and 15% frequency in patients who required low warfarin maintenance doses. These variants were previously reported as a haplotype, known as CYP2C9*1e, which includes -1565C>T, -1188T>C, IVS3+197G>A, IVS3-334C>T, IVS3-65G>C, IVS4-115A>G, and IVS5-73A>G [20]. Interestingly, a SNP in the CYP2C9*1e haplotype, IV3-65G>C, has been reported to be associated with warfarin low dose requirement in Taiwanese Chinese [29] and Han Chinese [30]. Frequency of IVS3-65G>C in the normal population is somewhat inconsistent, since Wang et al. reported a 4.5% frequency in healthy normal Chinese subjects (n= 995) [30] and Blaisdell et al. reported a 4.3% frequency in 23 Asian samples obtained from Human Genetic Cell Repositories sponsored by the National Institute of health housed at the Coriell Institute [17]. However healthy Taiwanese subjects (n= 69) showed 10.1% frequency [29] and normal Japanese subjects (n= 263) exhibited 11.8% frequency [20]. Further study would be necessary to determine the allele frequency in Asian populations. Although the frequency of this allele has been shown to be inconsistent in the literature, association of this haplotype with wafarin low dose requirement appears to be consistent as shown in two independent studies [29, 30] and in the present study. Since this haplotype has not been identified in Caucasians [31], it appears to exist preferentially in Asian populations. As reported in the previous study [29], this haplotype was not found with the CYP2C9*3 variant, suggesting that this haplotype segregated differently from the CYP2C9*3 variant. A sequence analysis program for splicing prediction indicated a possible splicing donor site for variant IVS4-115A>G with 89% probability. However, since CYP2C9 is a large gene spanning more than 50 kb, predicting any obvious changes in splice sites is currently impossible. In our gel shift assay the oligonucleotide probe containing a -1565C>T mutation disrupted its specific binding to nuclear proteins, suggesting that there might be a regulation mechanism related to this mutation (Figure 3). Shintani et al. reported that the promoter construct having a -1565C>T mutation exhibited approximately 15% decreased luciferase activity compared with the wild-type with a slight decreased intrinsic clearance of phenytoin [32]. These data would be supportive of the interpretation of warfarin sensitivity, but our assay of transcriptional activity did not show significant changes between the wild-type and variant promoters in luciferase assays (data not shown) in HepG2 cells. It could be possible that HepG-2 cells may not contain enough transcriptional machinery associated with the bound protein in the present gel-shift assay.
Among the CYP2C9 polymorphisms, CYP2C9*2 and *3 appear to cause functional consequences in most clinical drug responses as well as side effects. CYP2C9*3 (I359L) exhibited a significant decrease in activity for most CYP2C9 substrates in vivo, resulting in approximately 50% of the wild-type activity in heterozygous individuals, with a statistically insignificant impact in some cases, and an approximately 5- to 10-fold reduction in homozygous individuals with statistically significant potency. In the present study, one of our goals was to find CYP2C9 variants or haplotypes responsible for the low warfarin dose requirement. Therefore, individuals carrying CYP2C9*3 were excluded in the resequencing of CYP2C9, since CYP2C9*3 is a commonly known allele for low dose requirement of warfarin in many population studies. Although CYP2C9*3 is a common cause for the low dose requirement in Asia, the possibility of new variants in VKORC1 in addition to CYP2C9*1e in the low dose group cannot be ruled out. In order to compare the distribution of CYP2C9*1e haplotype in the low and high dose groups, one of the variants comprising CYP2C9*1e haplotype, IVS4-115A>G, was genotyped. The low dose group (<3 mg day−1, n= 36) exhibited 10 alleles of IVS4-115G which comprised of three alleles with CYP2C9*1/*3 and seven alleles with CYP2C9*1/*1, resulting in 14% frequency of the IVS4-115A>G. All these 10 alleles were of the VKORC1 1173 T/T genotype. The high dose group (>5.5 mg day−1, n= 34) showed five individuals having a heterozygous mutation of IVS4-115A>G, resulting in 7% frequency of the IVS4-115A>G. Interestingly, all these five individuals were found to have the VKORC1 1173 C/T genotype which has been shown to require a higher warfarin dose than the VKORC1 1173 T/T genotype, suggesting that the high doses of warfarin in these patients may be due to the influence of the VKORC1 genotype and not the CYP2C9 genotype.
Our observations revealed that two individuals having homozygous mutation of CYP2C9*1e showed a low warfarin dose requirement compared with the wild type, similar to the extent of the CYP2C9*3 heterozygous mutation. This phenotype/genotype relationship is in accordance with previous reports [29, 30]. Although we suggested possible changes of splicing and promoter function caused by the haplotype CYP2C9*1e, the present study does not provide a clear cellular mechanism on how CYP2C9*1e causes warfarin sensitivity. However, in addition to our results, the association of CYP2C9*1e with warfarin dose was repeated in two other independent studies in geographically different regions. Further evaluation with different substrates and more subjects with this genotype are necessary to derive a decisive answer on the consequence of CYP2C9*1e in Asians. The clinical relevance of these variants with respect to warfarin dose could be valuable markers in Asian populations.
Acknowledgments
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Ministry of Education, Science and Engineering (MOEST; No. R13-2007-023-00000-0) and by a grant from the Korea Health 21 R&D Project, Ministry for Health, Welfare and Family Affairs (A030001), Republic of Korea. This work was supported by a grant from Inje University, 2007.
Competing interests
There are no competing interests to declare.
REFERENCES
- 1.Goldstein JA, de Morais SM. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics. 1994;4:285–99. doi: 10.1097/00008571-199412000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Miners JO, Birkett DJ. Use of tolbutamide as a substrate probe for human hepatic cytochrome P450 2C9. Methods Enzymol. 1996;272:139–45. doi: 10.1016/s0076-6879(96)72017-7. [DOI] [PubMed] [Google Scholar]
- 3.Kidd RS, Straughn AB, Meyer MC, Blaisdell J, Goldstein JA, Dalton JT. Pharmacokinetics of chlorpheniramine, phenytoin, glipizide and nifedipine in an individual homozygous for the CYP2C9*3 allele. Pharmacogenetics. 1999;9:71–80. doi: 10.1097/00008571-199902000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Bajpai M, Roskos LK, Shen DD, Levy RH. Roles of cytochrome P4502C9 and cytochrome P4502C19 in the stereoselective metabolism of phenytoin to its major metabolite. Drug Metab Dispos. 1996;24:1401–3. [PubMed] [Google Scholar]
- 5.Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, Gelboin HV, Gonzalez FJ, Trager WF. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5:54–9. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- 6.Stearns RA, Chakravarty PK, Chen R, Chiu SH. Biotransformation of losartan to its active carboxylic acid metabolite in human liver microsomes. Role of cytochrome P4502C and 3A subfamily members. Drug Metab Dispos. 1995;23:207–15. [PubMed] [Google Scholar]
- 7.Miners JO, Rees DL, Valente L, Veronese ME, Birkett DJ. Human hepatic cytochrome P450 2C9 catalyzes the rate-limiting pathway of torsemide metabolism. J Pharmacol Exp Ther. 1995;272:1076–81. [PubMed] [Google Scholar]
- 8.Rendic S. Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002;34:83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- 9.Bylund J, Ericsson J, Oliw EH. Analysis of cytochrome P450 metabolites of arachidonic and linoleic acids by liquid chromatography-mass spectrometry with ion trap MS. Anal Biochem. 1998;265:55–68. doi: 10.1006/abio.1998.2897. [DOI] [PubMed] [Google Scholar]
- 10.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 11.Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 2C9 in a Caucasian and a black African population. Br J Clin Pharmacol. 2001;52:447–50. doi: 10.1046/j.0306-5251.2001.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, Cha IJ, Shin JG. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol. 2001;51:277–80. doi: 10.1046/j.1365-2125.2001.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SL, Huang J, Lai MD, Tsai JJ. Detection of CYP2C9 polymorphism based on the polymerase chain reaction in Chinese. Pharmacogenetics. 1995;5:37–42. doi: 10.1097/00008571-199502000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Nasu K, Kubota T, Ishizaki T. Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics. 1997;7:405–9. doi: 10.1097/00008571-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 16.Allabi AC, Gala JL, Horsmans Y, Babaoglu MO, Bozkurt A, Heusterspreute M, Yasar U. Functional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among black Africans. Clin Pharmacol Ther. 2004;76:113–8. doi: 10.1016/j.clpt.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, Xi T, Mohrenweiser H, Ghanayem B, Goldstein JA. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–37. doi: 10.1097/01.fpc.0000114759.08559.51. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS, Lee SS, Oh M, Jang YJ, Kim EY, Han IY, Cho KH, Shin JG. Effect of CYP2C9 and VKORC1 genotypes on early-phase and steady-state warfarin dosing in Korean patients with mechanical heart valve replacement. Pharmacogenet Genomics. 2009;19:103–12. doi: 10.1097/FPC.0b013e32831a9ae3. [DOI] [PubMed] [Google Scholar]
- 19.Lee SJ, Lee SS, Jeong HE, Shon JH, Ryu JY, Sunwoo YE, Liu KH, Kang W, Park YJ, Shin CM, Shin JG. The CYP3A4*18 allele, the most frequent coding variant in asian populations, does not significantly affect midazolam disposition in heterozygous individuals. Drug Metab Dispos. 2007;35:2095–101. doi: 10.1124/dmd.107.016733. [DOI] [PubMed] [Google Scholar]
- 20.Maekawa K, Fukushima-Uesaka H, Tohkin M, Hasegawa R, Kajio H, Kuzuya N, Yasuda K, Kawamoto M, Kamatani N, Suzuki K, Yanagawa T, Saito Y, Sawada J. Four novel defective alleles and comprehensive haplotype analysis of CYP2C9 in Japanese. Pharmacogenet Genomics. 2006;16:497–514. doi: 10.1097/01.fpc.0000215069.14095.c6. [DOI] [PubMed] [Google Scholar]
- 21.Lee SS, Lee SJ, Gwak J, Jung HJ, Thi-Le H, Song IS, Kim EY, Shin JG. Comparisons of CYP2C19 genetic polymorphisms between Korean and Vietnamese populations. Ther Drug Monit. 2007;29:455–9. doi: 10.1097/FTD.0b013e31811f383c. [DOI] [PubMed] [Google Scholar]
- 22.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 23.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 24.Yu CR, Ortaldo JR, Curiel RE, Young HA, Anderson SK, Gosselin P. Role of a STAT binding site in the regulation of the human perforin promoter. J Immunol. 1999;162:2785–90. [PubMed] [Google Scholar]
- 25.Takahashi H, Kashima T, Nomizo Y, Muramoto N, Shimizu T, Nasu K, Kubota T, Kimura S, Echizen H. Metabolism of warfarin enantiomers in Japanese patients with heart disease having different CYP2C9 and CYP2C19 genotypes. Clin Pharmacol Ther. 1998;63:519–28. doi: 10.1016/S0009-9236(98)90103-5. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi H, Kashima T, Nomoto S, Iwade K, Tainaka H, Shimizu T, Nomizo Y, Muramoto N, Kimura S, Echizen H. Comparisons between in-vitro and in-vivo metabolism of (S)-warfarin: catalytic activities of cDNA-expressed CYP2C9, its Leu359 variant and their mixture versus unbound clearance in patients with the corresponding CYP2C9 genotypes. Pharmacogenetics. 1998;8:365–73. doi: 10.1097/00008571-199810000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Leung AY, Chow HC, Kwong YL, Lie AK, Fung AT, Chow WH, Yip AS, Liang R. Genetic polymorphism in exon 4 of cytochrome P450 CYP2C9 may be associated with warfarin sensitivity in Chinese patients. Blood. 2001;98:2584–7. doi: 10.1182/blood.v98.8.2584. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi T. Optimal intensity of warfarin therapy for secondary prevention of stroke in patients with nonvalvular atrial fibrillation: a multicenter, prospective, randomized trial. Japanese Nonvalvular Atrial Fibrillation-Embolism Secondary Prevention Cooperative Study Group. Stroke. 2000;31:817–21. doi: 10.1161/01.str.31.4.817. [DOI] [PubMed] [Google Scholar]
- 29.Chern HD, Ueng TH, Fu YP, Cheng CW. CYP2C9 polymorphism and warfarin sensitivity in Taiwan Chinese. Clin Chim Acta. 2006;367:108–13. doi: 10.1016/j.cca.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Wang TL, Li HL, Tjong WY, Chen QS, Wu GS, Zhu HT, Hou ZS, Xu S, Ma SJ, Wu M, Tai S. Genetic factors contribute to patient-specific warfarin dose for Han Chinese. Clin Chim Acta. 2008;396:76–9. doi: 10.1016/j.cca.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 31.King BP, Khan TI, Aithal GP, Kamali F, Daly AK. Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14:813–22. doi: 10.1097/00008571-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Shintani M, Ieiri I, Inoue K, Mamiya K, Ninomiya H, Tashiro N, Higuchi S, Otsubo K. Genetic polymorphisms and functional characterization of the 5′-flanking region of the human CYP2C9 gene: in vitro and in vivo studies. Clin Pharmacol Ther. 2001;70:175–82. doi: 10.1067/mcp.2001.117367. [DOI] [PubMed] [Google Scholar]


