Abstract
C-myc and Bcl2 are well characterized oncogenes that are capable of forming G-quadruplex structures. Promoter regions of C-myc and Bcl2 forming G-quadruplex structures are chemically synthesized and G-quadruplex structure is formed in presence of 100 mM potassium ion. Three different porphyrin drugs, namely TMPyP2, TMPyP3, and TMPyP4 are allowed to interact with quadruplex DNA complex and the site and nature of interaction are studied. Drug interactions with quadruplex DNA were carried out in different potassium ionic strengths using fluorescence spectroscopy. It is found that fluorescence hypochromicity decreases with an increase in ionic strength in the case of TMPyP4, TMPyP3, and TMPyP2. Fluorescence titration studies and Job plots indicate that four molecules of TMPyP4, two molecules of TMPyP3 and TMPyP2 are interacting with one molecule of quadruplex DNA.
1. Introduction
Bcl2 (B-cell/lymphoma-2) is a proto-oncogene which encodes for 25 KD protein which has a peculiar function of blocking programmed cell death without affecting proliferation [1]. There are about 25 genes in the Bcl2 family. The Bcl2 gene is known to be responsible for a number of cancers like melanoma, breast, prostate, lung carcinoma, schizophrenia, and autoimmunity [2]. An increase in Bcl2 levels in the cell is reported to lead to poor prognosis [3]. Hence control of the production of Bcl2 in a cell has become a significant area of research. Like Bcl2, C-myc is also an oncogene which has a G-quadruplex forming sequence in its promoter region. The protein product of C-myc proto-oncogene controls a variety of genes that together enhance proliferation of cell [4, 5]. C-myc is overexpressed in many human cancers cells like leukemia, lung, breast, pancreatic, cervical, etc, [6–8]. The role of G-quadruplexes in cancer therapeutics is increasing. Work is under way in many laboratories elsewhere to identify a suitable synthetic compound or macromolecul, which can stabilize the quadruplex complex in vivo [9]. Many synthetic chemicals were tested for their interaction with quadruplex DNA. Few among them are anthraquinones [10], acridines [11], perylenes [12], and triazines [13, 14]. Porphyrins are chemical compounds, until that can interact with quadruplex DNA structure and stabilize them [15, 16]. Today, there has been controversy in the nature and site of poprhyrin binding to quadruplex DNA structure. Kimura et al. [17] have studied the interaction between G-quadruplex-TMPyP4 using 2-aminopurine substituted human telomeric DNA sequence. Han et al. [18] have demonstrated that TMPyP4 is a suitable telomerase inhibitor because of its low toxicity.
Different TMPyP4 binding modes to DNA are reported in the literature, like intercalation between the G-C base pairs [19, 20], binding to major groove [21] and minor groove [22, 23], and binding to the DNA surface [24]. Haq et al. [25] have studied the binding of cationic porphyrin to G-quadruplex DNA using ITC, UV-Vis spectroscopy and have shown that the stoichiometry of porphyrin binding to G-quadruplex DNA depends on the number of intervals between successive G-tetrad planes. Freyer et al. [26] have recently reported that the binding of TMPyP4 to c-myc NHE III promoter is in the ratio of 4 : 1, using microcalorimetric and spectroscopic techniques.
2. Materials and Methods
2.1. Oligonucleotide Synthesis
For this study, two different quadruplex forming oncogenic sequences, corresponding to Bcl2 and C-myc promoter regions were synthesized and their interactions with three different porphyrin drugs were studied. The sequences of oligonuclotides used in the present study are shown below. The structure of G-quadruplex DNA and porphyrin drugs is shown in Figure 1. All the structures shown in Figure 1 are drawn using ACD/ChemSketch (Version 12):
Figure 1.
The structures of G-quadruplex DNA and TMPyP2, TMPyP3, and TMPyP4.
-
27Bcl2 - CGG GCG CGG GAG GAA GGG GGC GGG AGC
-
C-myc - TGG GGA GGG TGG GGA GGG TGG GGA AGG
The 27Bcl2 and C-myc oligonucleotide stock solutions were prepared by dissolving each oligonucleotide into 100 mM KBPES buffer (0.01 M KH2PO4, 0.01 M K2HPO4, 0.001 M EDTA, 0.02–0.6 M KCl, KOH added drop wise to pH 7.0). DNA samples were dialyzed (1000 molecular weight cutoff membrane) against two changes of buffer (1L, 24 hours each) at 4°C. Concentration of all the oligonucleotides was measured using UV-Vis spectrophotometry with molar extinction coefficients determined by using the nearest neighbor calculation for single stranded DNA [27] and the absorbance of thermally denatured constructs extrapolated back to 25°C and/or a total phosphate analysis technique [28]. The extinction coefficient determined for two G-quadruplexes by these techniques was € 254 = 207840 M−1Cm−1 (27Bcl2) and € 260 = 215330 M−1Cm−1 (c-myc), respectively.
The cationic porphyrins (TMPyP4, TMPyP3, and TMPyP2) were purchased from Frontier Scientific (Logan, Utah, USA). All the porphyrin drugs were used without further purification. The porphyrin solutions were prepared by dissolution of each drug into a measured volume of final dialysate from the appropriate oligonucleotide solution preparation as the buffer. The concentration of each porphyrin drug was calculated using the molar extinction coefficient € 424 = 2.26 × 105 M−1Cm−1 [29], € 417 = 2.50 × 105 M−1Cm−1 [30], € 414 = 1.82 × 105 M−1Cm−1 [31] for TMPyP4, TMPyP3, and TMPyP2, respectively.
2.2. CD Experimental Studies
Circular dichroism experiments were performed using a JASCO 815 CD spectropolarimeter (Jasco, Tokyo, Japan). The CD spectra were recorded from 200 to 500 nm for characterizing the G-quadruplex DNA and for studying induced CD by porphyrins. For each CD experiment, 5 × 10−6 M G-quadruplex DNA was used. CD spectra were recorded using 10 × 10−6 M (1 : 2) and 20 × 10−6 M (1 : 4) TMPyP4. CD titrations were done in 100 mM KBPES buffer (pH 7.0) at 25°C. TMPyP4 was added using a calibrated micropipette. Each CD spectrum is recorded three times, and the average of 3 scans is considered.
2.3. Fluorescence Experiments
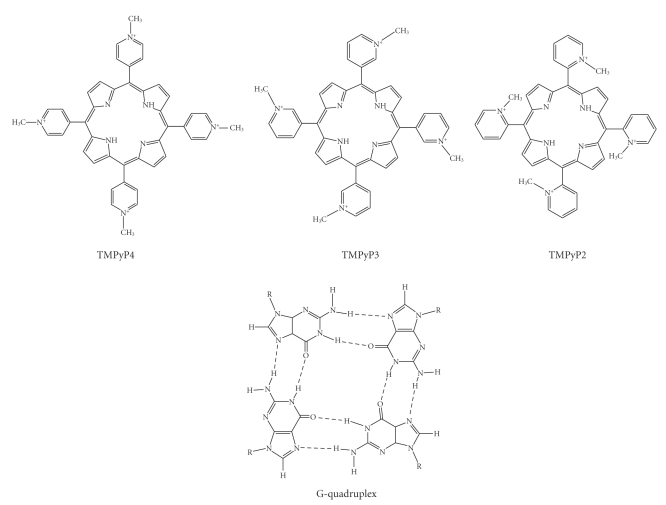
Fluorescence emission spectra were measured at 25°C using a FluoroMax-3 spectrofluorimeter (Horiba Jobin Yvon, Edison, NJ) using a 1 cm path length quartz cuvette. G-quadruplex DNA samples were measured in 2 mL of 100 mM KBPES buffer (pH 7.0). Through out the fluorescence experiment, concentration of G-quadruplex DNA is kept constant (200 nM) and titrated with increasing concentrations of cationic porphyrin (0 to 1200 nM). Fluorescence spectra were recorded after each addition of porphyrin to the fluorescent cuvette. G-quadruplex DNA and porphyrin drug mixture volume were kept constant throughout the fluorescence titration (2 mL). After each titration, the quartz cuvette was thoroughly washed with distilled water and dilute nitric acid (approximately 0.1 N, nitric acid) to remove traces of porphyrin sticking to the walls of quartz cuvette. TMPyP4, TMPyP3, and TMPyP2 were excited at 433 nm, 417 nm, and 424 nm, respectively and emission spectra for each titration were collected from 600 to 800 nm. Each spectrum was recorded three times and the average of three scans was taken. The difference in fluorescence emission maxima before and after the addition of G-quadruplex DNA was plotted against concentration of porphyrin to obtain the porphyrin-G-quadruplex binding curve. In the fluorescence binding curves (Figure 4), the points of intersection of two extrapolated lines (initial and final), at saturation levels, will indicate the stoichiometry of G-quadruplex DNA and porphyrin drugs.
Figure 4.
Fluorescence titration curves. 200 nM quadruplex DNA is titrated with 0–1200 nm concentration of three different porphyrin drugs. Stoichiometry is considered at saturation levels. Hence the slop of initial and end points are considered.1: TMPyP4, 2: TMPyP3, and 3: TMPyP2.
To investigate the effect of ionic strength on fluorescence hypochromicity (% FH), four different KBPES buffer solutions (pH 7.0) with increasing concentration of K(I) ion were prepared. The KBPES (pH 7.0) buffer solutions used in the present study contained 30 mM, 100 mM, 200 mM, and 600 mM of potassium ion.
2.4. UV-Vis Spectroscopic Titrations
UV-Vis spectroscopic titrations were performed using an Agilent 8453 diode array (Agilent, Santa Clara, CA) at 25°C using a 1 cm path length quartz cuvette. Stock solutions of 10 μM porphyrin and 10 μM of G-quadruplex DNA were prepared in 100 mM KBPES buffer (pH 7.0). Two series of solutions were used for the experiments. First two mL of dilute G-quadruplex DNA (10 μM) were placed in a 1 cm path length quartz cuvette, and twenty five × 50 μL injections of porphyrin solution (10 μM) were added manually. In the second part, two mL of dilute cationic porphyrin solution (10 μM) were used and twenty five × 50 μL injections of G-quadruplex DNA solution were added. Absorption spectra were collected from 350 nm to 500 nm. The quartz cells were thoroughly cleaned with distilled water and 0.1 N nitric acid to remove traces of porphyrin that were deposited on the walls of quartz cell. The difference in the absorption maxima of the soret band was plotted versus the prophyrin mole fraction to generate a Job plot [32–34].
UV-Vis absorption titrations were done by adding G-quadruplex DNA stock solution (200–350 nM) in 100 mM KBPES buffer (pH 7.0) to the quartz cuvette containing approximately 1 μM porphyrin solution prepared in the same buffer. G-quadruplex DNA and porphyrin samples were freshly prepared before performing the experiment. UV spectra were collected from 200 nm to 500 nm. The titrations were carried out until the porphyrin soret band remains at a fixed wavelength upon five successive additions of G-quadruplex DNA.
3. Results and Discussion
3.1. CD Studies
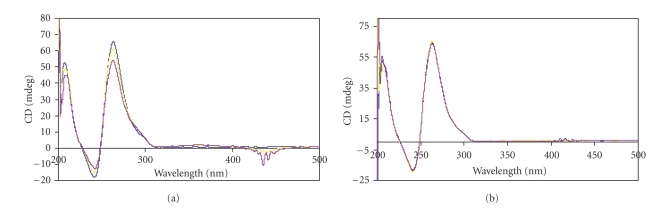
The CD spectrum (Figure 2(a)) shows a prominent positive peak at 265 nm and a small hump at 295 nm, indicating that the 27Bcl2 is forming antiparallel quadruplex structure in solution. On addition of TMPyP4 (at 1 : 2 and 1 : 4 DNA : TMPyP4 ratio) a weak induced CD band (ICD) is seen around 435 nm, indicating that TMPyP4 is efficiently interacting with quadruplex DNA. Whereas Figure 2(b) demonstrates comparatively weaker interaction of TMPyP4 with C-myc quadruplex DNA. Furthermore, the CD spectrum shows a positive peak at 265 nm. This indicates that it is forming parallel quadruplex DNA structure in solution. When (10 × 10−6 M) and (20 × 10−6 M) concentrations of TMPyP4 is added to 27Bcl2 quadruplex DNA (5 × 10−6 M), a negative cotton effect is observed. Small or no cotton effect is seen, when double and quadruple concentrations of drug are added to 5 × 10−6 M, C-myc DNA. A negative cotton effect in the case of 27Bcl2 is observed due to the difference in the conformation of Bcl2 quadruplex DNA. Occurrence of induced CD at 435 nm and negative cotton effect at 265 nm indicates that Bcl2 quadruplex complex has a marginally better interaction efficiency with TMPyP4.
Figure 2.
(a) CD spectra for the 27Bcl2 mid promoter sequence, wavelength in nm is plotted on x-axis, and Ellipticity, θ, is plotted on the y-axis in mdeg. The 5 × 10−6 M quadruplex DNA is considered for each CD experiment. CD spectra are recorded at 1 : 2 and 1 : 4 quadruplex DNA: TMPyP4 stoichiometry. Each spectrum is recorded thrice, and the average of the three runs is considered. (b) CD spectra for the C-myc sequence, wavelength in nm is plotted on x-axis and Ellipticity, θ, is plotted on the y-axis in mdeg. The 5 × 10−6 M quadruplex DNA is considered for each CD experiment. CD spectra are recorded at 1 : 2 and 1 : 4 quadruplex DNA: TMPyP4 stoichiometry. Each spectrum is recorded thrice, and the average of the three runs is considered.
3.2. UV-Titration Studies
UV-Vis spectroscopy is an excellent technique for finding the extent of drug interaction with DNA. In all the drug-DNA interactions, the soret band has shown a red shift. The soret band has shifted from 423 nm to 441 nm (18 nm shift) in case of TMPyP4 and the isobestic point is prominent at 436 nm (UV-Vis spectra are similar for both 27Bcl2 and C-myc G-quadruplex sequences). In case of TMPyP3, the soret band has shifted from 417 nm to 431 nm (13-14 nm shift) and isobestic point is observed at around 426 nm. In the case of TMPyP2 the soret band does not show a shift from 414 nm and an isobestic point is observed at around 422 nm. Percentage of hypochromicity for each G-quadruplex DNA is calculated by following the procedure described by Keating and Szalai [32]. The details of UV-Vis spectral studies are shown in Table 1. Results demonstrate that both quadruplex DNA's can interact with TMPyP3 and TMPyP4 drugs. But among them, TMPyP4 interaction with C-myc and 27Bcl2 quadruplex DNA is more efficient than TMPyP3. TMPyP2 has very little or no interaction. Similar observations have been made by Anantha et al. [35]. They showed that TMPyP4 exhibits both intercalation and external binding. From UV spectral analysis, it is evident that the isobestic point when TMPyP4 interacts with 27Bcl2 quadruplex DNA shows slight deviation at the end (isobestic point is not a tight single point). This indicates that interaction involves multiple steps. Where as with C-myc quadruplex DNA, the isobestic point does not show any deviation (a tight point is observed). Similar observations were made while observing the drug-DNA interaction, fluorimetrically.
Table 1.
UV-Vis absorption titration parameters for TMPyP4, TMPyP3, and TMPyP2 with four different Bcl2 and C-myc G-quadruplex DNA. Soret band hypochromicity is calculated using the formula: % Hypochromicity = [(€ free − € bound)/€ free] × 100. € free and € bound are the extinction coefficient of free and bound porphyrin, respectively.
| S. No. | Name of quadrupelx DNA and cationic porphyrin | Soret band shift (nm) | Isobestic point (nm) | Percentage hypochromicity |
|---|---|---|---|---|
| 1. | 27Bcl2-TMPyP4 | 423–441 | 435.5 | 59.7% |
| 2. | 27Bcl2-TMPyP3 | 417–431 | 426.5 | 41.5% |
| 3. | 27Bcl2-TMPyP2 | No shift | 422.0 | 23.0% |
| 4. | C-myc-TMPyP4 | 423–439 | 432.2 | 52.3% |
| 5. | C-myc-TMPyP3 | 417–429 | 425.3 | 39.5% |
| 6. | C-myc-TMPyP2 | No shift | 421.7 | 14.7% |
3.3. Job Plot
Job plots make it possible to find out the stoichiometry interactions between drugs and quadruplex DNA. From Figure 3, it is evident that four molecules of TMPyP4 are interacting with each molecule of C-myc and 27Bcl2 quadruplex DNA. Similar observations have been made by Freyer et al. [26] only two molecules of TMPyP2 and TMPyP3 are interacting with each quadruplex DNA complex. The variation in the number of drug molecules interacting with quadruplex DNA is probably due to the placement and rotation of methyl groups placed on the porphyrin molecule. In the case of TMPyP4, the N–CH3 group is in the para position and it has about 360° degree of rotation along the axis. Hence the molecules can attain perfect planar structure when it is interacting with quadruplex DNA. Due to this reason, it can intercalate as well as bind externally to quadruplex DNA molecule. In the case of TMPyP3 the N-CH3 group is in meta position and it can have a restricted rotation on its axis. Therefore, this molecule cannot attain perfect coplanarity, like TMPyP4. Hence it cannot intercalate completely in between two G-quartets but prefers to bind externally. With TMPyP2, the N–CH3 groups are in ortho position. The N–CH3 groups have much more restricted rotation on its axis, in case of TMPyP2. Hence, the molecule is incapable of attaining perfect planar structure, exhibits intercalation with quadruplex DNA and shows only external binding. Probably due to this reason, TMPyP4 can intercalate and bind externally, and the drug : quadruplex DNA stoichiometry is 4 : 1. Where as, TMPyP2 and TMPyP3 prefers to bind externally and the stoichiometry is 2 : 1. Details were shown in Table 2.
Figure 3.
Job plots for the titration of the 27Bcl2 mid promoter and C-myc quadruplex DNA on interaction with TMPyP2, TMPyP3, and TMPyP4 drugs. Point of intersection of two lines is indicated by a dashed line.
Table 2.
Data obtained from Job plot of Bcl2 and C-myc G-quadruplex DNA with TMPyP4, TMPyP3, and TMPyP2 porphyrin drugs.
| S. No. | Name of gquadrupelx DNA and porphyrin | Value of mole fraction of porphyrin matching with point of intersection | Stoichiometry between gquadruplex DNA porphyrin |
|---|---|---|---|
| 1. | 27Bcl2-TMPyP4 | 0.78 | 1 : 3.54 |
| 2. | 27Bcl2-TMPyP3 | 0.70 | 1 : 2.33 |
| 3. | 27Bcl2-TMPyP2 | 0.58 | 1 : 1.38 |
| 4. | C-myc-TMPyP4 | 0.76 | 1 : 3.17 |
| 5. | C-myc-TMPyP3 | 0.70 | 1 : 2.33 |
| 6. | C-myc-TMPyP2 | 0.58 | 1 : 1.38 |
3.4. Fluorescence Hypochromicity
In order to study the effect of buffer cationic strength on drug binding to quadruplex DNA, fluorescence titration was carried out in different KBPS buffers with variation in potassium ion concentrations (30 mM, 100 mM, 200 mM, and 600 mM). Results emphasize that fluorescence hypochromicity decreases with increase of buffer salt concentration. It demonstrates that as the positive ion cloud around quadruplex complex increases, it occupies the available binding sites on quadruplex complex surface and reduces the possibility for drug molecules to bind exterior of quadruplex DNA. A decrease in fluorescence intensity of the drug may also be due to quenching by the surrounding water molecules. But if the drug molecule shows an intercalative mode of binding, quenching of fluorescence intensity is comparatively less. Because drug molecules are sandwiched between two G-quartets, the distance between guanine bases in quadruplex and the drug molecules becomes less. As the drug molecules come close to guanines in quadruplex, energy transfer between them takes place, which results in an increase of fluorescence. An increase in fluorescence intensity by energy transfer between DNA bases and the drug molecules, on intercalation, was reported earlier [36, 37]. The extent of fluorescence attenuation is not the same with all the three porphyrin drugs. It is more with TMPyP2, moderate with TMPyP3 and less with TMPyP4. These results indicate that TMPyP4 can show both intercalative as well as end binding to quadruplex complex while TMPyP3, and TMPyP2 prefer to bind externally. From Tables 3(a) and 3(b) it is evident that percentage of fluorescence hypochromicity is comparatively higher (from lower to higher ionic strength), in the case of TMPyP3 and TMPyP2. This indicates that drug molecules are quenched by the surrounding water molecules. But, in thecase of TMPyP4, fluorescence attenuation is gradual. This demonstrates that though there is quenching of TMPyP4 fluorescence by surrounding water molecules, there is some increase in the fluorescence, due to energy transfer between TMPyP4 and guanines in the quadruplex on drug intercalation. Due to this, TMPyP4 could exhibit fluorescence even at 600 mM salt concentration. Where as in case of TMPyP2 and TMPyP3 fluorescence intensity is almost quenched by the surrounding water molecules, as they are binding externally. The results obtained with 27Bcl2 as well as C-myc quadruplex are shown in Tables 3(a) and 3(b), respectively.
Table 3.
(a) Effect of ionic strength on the percentage of fluorescence hypochromicity while 27Bcl2 interacts with porphyrin drugs.
| S. No. | Ionic strength of KBPES buffer (pH7.0) | Stoichiometry DNA : drug | Percentage of fluorescence hypochromicity |
|---|---|---|---|
| 1. | TMPyP4 with 27BCL2 | ||
| 1.a | 30 mM | 1 : 4 | 34.2% |
| 1.b | 100 mM | 1 : 4 | 28.5% |
| 1.c | 200 mM | 1 : 4 | 20.3% |
| 1.d | 600 mM | 1 : 4 | 6.5% |
|
| |||
| 2. | TMPyP3 with 27BCL2 | ||
| 2.a | 30 mM | 1 : 2 | 59.8% |
| 1 : 3 | 51.5% | ||
| 2.b | 100 mM | 1 : 2 | 52.5% |
| 1 : 3 | 49.5% | ||
| 2.c | 200 mM | 1 : 2 | 33.6% |
| 1 : 3 | 16.6% | ||
| 2.d | 600 mM | 1 : 2 | 8.6% |
| 1 : 3 | 4.9% | ||
|
| |||
| 3. | TMPyP2 with 27BCL2 | ||
| 3.a | 30 mM | 1 : 2 | 37.4% |
| 1 : 3 | 40.2% | ||
| 3.b | 100 mM | 1 : 2 | 11.9% |
| 1 : 3 | 12.3% | ||
| 3.c | 200 mM | 1 : 2 | 6.5% |
| 1 : 3 | 4.4% | ||
| 3.d | 600 mM | 1 : 2 | 0.7% |
| 1 : 3 | 0.3% | ||
(b) Effect of ionic strength on the percentage of fluorescence hypochromicity while C-myc interacts with porphyrin drugs.
| S. No. | Ionic strength of KBPES buffer (pH7.0) | Stoichiometry DNA : drug | Percentage of fluorescence hypochromicity |
|---|---|---|---|
| 1. | TMPyP4 with C-myc | ||
| 1.a | 30 mM | 1 : 4 | 39.2% |
| 1.b | 100 mM | 1 : 4 | 32.6% |
| 1.c | 200 mM | 1 : 4 | 29.3% |
| 1.d | 600 mM | 1 : 4 | 5.5% |
|
| |||
| 2. | TMPyP3 with C-myc | ||
| 2.a | 30 mM | 1 : 2 | 52.8% |
| 1 : 3 | 49.5% | ||
| 2.b | 100 mM | 1 : 2 | 47.5% |
| 1 : 3 | 42.2% | ||
| 2.c | 200 mM | 1 : 2 | 29.6% |
| 1 : 3 | 12.6% | ||
| 2.d | 600 mM | 1 : 2 | 6.6% |
| 1 : 3 | 3.9% | ||
|
| |||
| 3. | TMPyP2 with C-myc | ||
| 3.a | 30 mM | 1 : 2 | 42.4% |
| 1 : 3 | 37.2% | ||
| 3.b | 100 mM | 1 : 2 | 12.6% |
| 1 : 3 | 7.2% | ||
| 3.c | 200 mM | 1 : 2 | 7.8% |
| 1 : 3 | 5.4% | ||
| 3.d | 600 mM | 1 : 2 | 0.6% |
| 1 : 3 | 0.2% | ||
3.5. Fluorescence Titration
In this study, 200 nM of G-quadruplex DNA was considered for interaction with three different porphyrin drugs in the range of 0–1200 nM. From Figure 4 it is evident that the binding curve has multiple inflection points. 27Bcl2 shows a comparatively higher number of inflection points, indicating that the interaction occurs in multiple steps. Since it is intended to know the stoichiometry at saturation levels, only slopes of initial and final titrations are considered. The titration demonstrates that with one molecule of G-quadruplex DNA, four molecules of TMPyP4 and two molecules of TMPyP2 and TMPyP3 are interacting. It is also clear that the saturation point is occurring relatively at lower concentration in case of TMPyP2 (as the slope of binding curve is steep), moderate in TMPyP3, and at higher concentration in TMPyP4. The reason for difference in the nature of interaction of three porphyrins with quadruplex DNA is probably due to the orientation of N-CH3 groups on the drug molecule and the nature of binding.
4. Conclusions
From the present study, it can be concluded that cationic porphyrins show different modes of binding to G-quadruplex DNA. The nature of binding depends upon the position of the bulky N+-CH3 groups on porphyrin. Experiments with CD, fluorescence, and UV-Vis spectroscopic techniques indicate that TMPyP4 can bind externally as well as intercalate between the two G-tetrads. Two porphyrins, TMPyP3 and TMPyP2, prefer external binding to G-quadruplex DNA. Job plots and fluorescence binding curves indicate that four molecules of TMPyP4 and two molecules of TMPyP2 and TMPyP3 interact with 27Bcl2/C-myc G-quadruplex DNA. As the ionic strength of the buffer increases, fluorescence hypochromicity decreases. The extent of reduction in fluorescence hypochromicity is high in TMPyP2 and TMPyP3, and moderate in the case of TMPyP4. The variation in fluorescence hypochromicity is probably due to the difference in the mode of drug interaction with quadruplex DNA.
Acknowledgments
N. Nagesh and V. K. Sharma are thankful to Dr. Lalji Singh, Director of CCMB, Hyderabad, India for his encouragement and support given to undertake this work. N. Nagesh and V. K. Sharma contributed equally to this work.
References
- 1.Cohen JJ, Duke RC. Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. Journal of Immunology. 1984;132(1):38–42. [PubMed] [Google Scholar]
- 2.McDonnell TJ, Troncoso P, Brisbay SM, et al. Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Research. 1992;52(24):6940–6944. [PubMed] [Google Scholar]
- 3.Baretton GB, Diebold J, Christoforis G, et al. Apoptosis and immunohistochemical bcl-2 expression in colorectal adenomas and carcinomas: aspects of carcinogenesis and prognostic significance. Cancer. 1996;77(2):255–264. doi: 10.1002/(SICI)1097-0142(19960115)77:2<255::AID-CNCR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and Cellular Biology. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang CV, Resar LMS, Emison E, et al. Function of the c-Myc oncogenic transcription factor. Experimental Cell Research. 1999;253(1):63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 6.Cole MD. The Myc oncogene: its role in transformation and differentiation. Annual Review of Genetics. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- 7.Henriksson M, Selivanova G, Lindström M, Wiman KG. Inactivation of Myc-induced p53-dependent apoptosis in human tumors. Apoptosis. 2001;6(1-2):133–137. doi: 10.1023/a:1009644716727. [DOI] [PubMed] [Google Scholar]
- 8.Boxer LM, Dang CV. Translocations involving c-Myc and c-Myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 9.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. Journal of the American Chemical Society. 2007;129(43):12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D, Thompson B, Cathers BE, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. Journal of Medicinal Chemistry. 1997;40(14):2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 11.Burger AM, Dai F, Schultes CM, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Research. 2005;65(4):1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 12.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry. 1998;37(36):12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 13.Riou JF, Guittat L, Mailliet P, et al. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher TM. Telomerase: a potential therapeutic target for cancer. Expert Opinion on Therapeutic Targets. 2005;9(3):457–469. doi: 10.1517/14728222.9.3.457. [DOI] [PubMed] [Google Scholar]
- 15.Seenisamy J, Bashyam S, Gokhale V, et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-Myc G-quadruplex structure. Journal of the American Chemical Society. 2005;127(9):2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 16.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. Journal of the American Chemical Society. 1999;121(15):3561–3570. [Google Scholar]
- 17.Kimura T, Kawai K, Fujitsuka M, Majima T. Detection of the G-quadruplex-TMPyP4 complex by 2-aminopurine modified human telomeric DNA. Chemical Communications. 2006;(4):401–402. doi: 10.1039/b514526k. [DOI] [PubMed] [Google Scholar]
- 18.Han H, Langley DR, Rangan A, Hurley LH. Selective interactions of cationic porphyrins with G-quadruplex structures. Journal of the American Chemical Society. 2001;123(37):8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 19.Guliaev AB, Leontis NB. Cationic 5,10,15,20-tetrakis (N-methylpyridinium-4-yl) porphyrin fully intercalates at 5′-CG-3′ steps of duplex DNA in solution. Biochemistry. 1999;38(47):15425–15437. doi: 10.1021/bi9913808. [DOI] [PubMed] [Google Scholar]
- 20.Marzilli LG, Banville DL, Zon G, Wilson WD. Pronounced 1H and 31P NMR spectral changes on meso-tetrakis(N-methylpyridinium-4-yl)porphyrin binding to tetradecaoligo-deoxyribonucleotides: evidence for symmetric, selective binding to 5′CG3′ sequences. Journal of the American Chemical Society. 1986;108(14):4188–4192. [Google Scholar]
- 21.Ohyama T, Mita H, Yamamoto Y. Binding of 5,10,15,20-tetrakis(N-methylpyridinium-4-yl)-21H,23H-porphyrin to an at-rich region of a duplex DNA. Biophysical Chemistry. 2005;113(1):53–59. doi: 10.1016/j.bpc.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 22.Kim JO, Lee YA, Yun BH, Han SW, Kwag ST, Kim SK. Binding of meso-tetrakis(N-methylpyridinium-4-yl)porphyrin to AT oligomers: effect of chain length and the location of the porphyrin stacking. Biophysical Journal. 2004;86(2):1012–1017. doi: 10.1016/S0006-3495(04)74176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford KG, Neidle S. Perturbations in DNA structure upon interaction with porphyrins revealed by chemical probes, DNA footprinting and molecular modelling. Bioorganic and Medicinal Chemistry. 1995;3(6):671–677. doi: 10.1016/0968-0896(95)00052-i. [DOI] [PubMed] [Google Scholar]
- 24.Lee S, Jeon SH, Kim BJ, Han SW, Jang HG, Kim SK. Classification of CD and absorption spectra in the Soret band of H2TMPyP bound to various synthetic polynucleotides. Biophysical Chemistry. 2001;92(1-2):35–45. doi: 10.1016/s0301-4622(01)00181-8. [DOI] [PubMed] [Google Scholar]
- 25.Haq I, Trent JO, Chowdhry BZ, Jenkins TC. Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin. Journal of the American Chemical Society. 1999;121(9):1768–1779. [Google Scholar]
- 26.Freyer MW, Buscaglia R, Kaplan K, Cashman D, Hurley LH, Lewis EA. Biophysical studies of the c-Myc NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophysical Journal. 2007;92(6):2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fassman GD. Handbook of Biochemistry and Molecular Biology. Cleveland, Ohio, USA: CRC; 1975. Nucleic acids. [Google Scholar]
- 28.Plum GE. Current Protocols in Nucleic Acid Chemistry. Vol. 7.3. New York, NY, USA: John Wiley & Sons; 2000. Determination of oligonucleotide molar extinction coefficients; pp. 1–17. [Google Scholar]
- 29.Pasternack RF, Huber PR, Boyd P, et al. Aggregation of meso-substituted water-soluble porphyrins. Journal of the American Chemical Society. 1972;94(13):4511–4517. doi: 10.1021/ja00768a016. [DOI] [PubMed] [Google Scholar]
- 30.Kalyanasundaram K. Photochemistry of water-soluble porphyrins: comparative study of isomeric tetrapyridyl- and tetrakis(N-methylpyridiniumyl)porphyrins. Inorganic Chemistry. 1984;23(16):2453–2459. [Google Scholar]
- 31.Hambright P, Gore T, Burton M. Synthesis and characterization of new isomeric water-soluble porphyrins. Tetra(2-N-methylpyridyl)porphine and tetra(3-N-methylpyridyl)porphine. Inorganic Chemistry. 1976;15(9):2314–2315. [Google Scholar]
- 32.Keating LR, Szalai VA. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry. 2004;43(50):15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 33.Likussar W, Boltz DF. Theory of continuous variations plots and a new method for spectrophotometric determination of extraction and formation constants. Analytical Chemistry. 1971;43(10):1265–1272. [Google Scholar]
- 34.Ingham KC. On the application of Job’s method of continuous variation to the stoichiometry of protein-ligand complexes. Analytical Biochemistry. 1975;68(2):660–663. doi: 10.1016/0003-2697(75)90666-1. [DOI] [PubMed] [Google Scholar]
- 35.Anantha NV, Azam M, Sheardy RD. Porphyrin binding to quadruplexed T4G4 . Biochemistry. 1998;37(9):2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- 36.Lubitz I, Borovok N, Kotlyar A. Interaction of monomolecular G4-DNA nanowires with TMPyP: evidence for intercalation. Biochemistry. 2007;46(45):12925–12929. doi: 10.1021/bi701301u. [DOI] [PubMed] [Google Scholar]
- 37.Suh D, Chaires JB. Criteria for the mode of binding of DNA binding agents. Bioorganic and Medicinal Chemistry. 1995;3(6):723–728. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]