Abstract
The two major cellular sites for membrane protein degradation are the proteasome and the lysosome. Ubiquitin attachment is a sorting signal for both degradation routes. For lysosomal degradation, ubiquitination triggers the sorting of cargo proteins into the lumen of late endosomal multivesicular bodies (MVBs)/endosomes. MVB formation occurs when a portion of the limiting membrane of an endosome invaginates and buds into its own lumen. Intralumenal vesicles are degraded when MVBs fuse to lysosomes. The proper delivery of proteins to the MVB interior relies on specific ubiquitination of cargo, recognition and sorting of ubiquitinated cargo to endosomal subdomains, and the formation and scission of cargo-filled intralumenal vesicles. Over the past five years, a number of proteins that may directly participate in these aspects of MVB function and biogenesis have been identified. However, major questions remain as to exactly what these proteins do at the molecular level and how they may accomplish these tasks.
Keywords: endosome, lysosome, ubiquitin, ESCRT, downregulation, peptidase
INTRODUCTION
The primary responsibility of endosomes is to separate proteins that will recycle to other cellular locales from those that will be degraded in lysosomes. In this way endosomes regulate the composition of the cell surface and thus play a pivotal role in a vast array of biological functions. Maturation from early to late endosomes entails the removal of both proteins and lipids via tubulo-vesicular elements that sort components back to the trans-Golgi network and plasma membrane. Integral membrane proteins that have not been recycled, as well as soluble lumenal materials, are instead delivered to late endosomes and lysosomes (Gruenberg & Maxfield 1995). Stable lysosomal membrane proteins such as LAMP-1 remain on the limiting membrane of endosomes. However, proteins destined for degradation are incorporated into intralumenal vesicles (ILVs) that bud from the limiting membrane, giving multivesicular endosomes or multivesicular bodies (MVBs) their characteristic appearance. Fusion of MVBs with lysosomes initiates the degradation of ILVs and their contents, owing to the unique composition of ILVs, which renders them susceptible to lysosomal hydrolases. Targeting of protein cargoes into this degradative pathway is highly regulated and largely dependent on posttranslational modification by ubiquitin (Ub). Here we survey recent advances and remaining questions about this complicated sorting reaction.
MULTIVESICULAR BODIES FOR DEGRADATION AND STORAGE
MVBs were first noted in early electron microscopy studies as unique membrane-enclosed structures with ILVs (Palade 1955, Sotelo & Porter 1959). Without functional data, however, these structures were simply referred to as MVBs. Subsequently, the invagination of the limiting membrane of MVBs was observed, suggesting that these structures might mediate some form of autophagy (Hirsch et al. 1968). Later studies following internalized proteins defined MVBs as part of the endocytic pathway and also defined ILVs as a degradation route for proteins such as the epidermal growth factor receptor (EGFR) (Felder et al. 1990, Gorden et al. 1978, Haigler et al. 1979, van Deurs et al. 1993). MVBs are now understood to perform a variety of functions within the endocytic pathway and, as such, can have different compositions and morphologies. However, MVBs contain endocytic markers such as Rabs, LAMPs, and endocytosed tracers, which distinguishes them from other organelles with internal membranes, such as autophagic bodies or multilamellar lysosomes. Protein sorting into MVBs and delivery to the lysosomal lumen are the major mechanisms for degrading post-Golgi integral membrane proteins in all eukaryotic cells. This serves as a mechanism for destroying damaged proteins as well as proteins that undergo downregulation or clearing from the cell surface as a part of a regulatory process. Although the proteasome, a large chambered protease composed of several catalytic and regulatory subunits, can degrade integral membrane proteins in the early secretory pathway through the process of endoplasmic reticulum–associated degradation (ERAD), the proteasome does not appear to play a direct role in degrading proteins that travel past the Golgi, despite indications from early studies using proteasome inhibitors. Rather, one of the effects of proteasome inhibitors is to inhibit MVB protein sorting by an unknown mechanism (Hammond et al. 2003, Longva et al. 2002, Rocca et al. 2001, van Kerkhof et al. 2001).
MVBs are much more dynamic and versatile than they were once recognized to be. For instance, the ability of cells to upregulate MVB formation and degradative capacity is exemplified by studies showing that growth factor stimulation can increase inward budding and MVB formation (White et al. 2006). Reticulocytes also serve as a dramatic example of upregulation of the MVB-exosome pathway. The differentiation of reticulocytes into erythrocytes is accompanied by massive destruction of cytosolic proteins, organelles, and integral membrane proteins and the generation of MVBs. Proteins that normally recycle, like the transferrin receptor (TfR), are instead incorporated into MVBs for eventual secretion at the cell surface (Pan & Johnstone 1983). Protein sorting into ILVs does not always destine the protein for degradation, however. Proteins such as class II MHC and tetraspanins accumulate in MVBs/late endosomes and compose a specialized class II MHC compartment (MIIC). One fate that the ILVs of MIICs face is to be secreted from the cell upon fusion of the MVB limiting membrane with the plasma membrane. These ILVs are precursors of exosomes, which are produced by a wide variety of cells, including neurons, epithelial cells, mast cells, tumor cells, melanocytes, and cells within the hematopoietic lineage (van Niel et al. 2006a).
Class II MHC–containing ILVs back-fuse to the limiting endosomal membrane, thereby taking the class II MHC out of storage and facilitating antigen presentation at the cell surface (Chow & Mellman 2005). This phenomenon has been observed when immature dendritic cells are stimulated to mature, which results in the appearance of deep tubular endosomal structures concomitant with the loss of internal membranes. This ability to back-fuse has been hypothesized for other processes, including the recycling route of the tetraspanin CD63, which is typically enriched in ILVs of late endosomes (Kobayashi et al. 2000), as well as the efflux of certain viruses and toxins (see sidebar: Viruses and Multivesicular Bodies).
MVBs also give rise to more specialized, nondegradative endosomal compartments with stable contents. For instance, von Willebrands Factor (vWF) is packaged into the ILVs of both alpha granules of platelets and the Weibel-Palade bodies of endothelial cells. The association of vWF with ILVs appears to modify the activity of vWF when it is released via an exosomal pathway (Heijnen et al. 1999). Similarly, azurophilic granules of neutrophils are derived from MVBs and contain ILVs enriched with the tetraspanin CD63. Secretory granules of mast cells are also derived from the endosomal system and contain class II MHC–enriched ILVs that are secreted upon calcium stimulation (Raposo et al. 1997). Furthermore, melanosomes are derived from endosomes, and some of the intralumenal scaffolding proteins (for example, Pmel17/Silver) that immobilize pigment are proteolytically processed upon their sorting to ILVs, yet these compartments are different from degradative MVBs (Theos et al. 2006).
Two interesting questions arise from these observations: Are these MVB-like structures derived from a common precursor, and how do they avoid fusion with lysosomes? One possibility is that distinct early endosomal populations, yet to be accurately defined, contribute to these differences. Additionally, distinct ILVs may form as a consequence of different lipid and/or protein cargo composition. For instance, the lipid bis(monoacylglycero)phosphate/lysobisphosphatidic acid (BMP/LBPA) accumulates in internal membranes that are distinct from those membranes mediating the degradation of cell surface receptors. Different ILVs may be cordoned off into endosomal regions capable of forming distinct endosomal carrier vesicles (Gu & Gruenberg 1999). Recent evidence suggests that many of these different types of MVB and ILV populations may arise from common pathways. For instance, ubiquitination serves as a potent signal for sorting a variety of cell surface receptors and other proteins into the ILVs of MVBs, yet it was not known until recently whether this degradative pathway was used for generating other compartments like the ILVs of the MIIC. The MIIC in immature dendritic cells is characterized by a multivesicular appearance in which class II MHC and the tetraspanin CD63 are found within ILVs. Although the MIIC acts as a storage compartment, it does so only transiently because class II MHC proteins are eventually degraded by lysosomal hydrolases (Villadangos et al. 2005). Interestingly, class II MHC is ubiquitinated in immature dendritic cells, and this ubiquitination is responsible for both its sequestration from the plasma membrane (Shin et al. 2006) and its residence within ILVs of MVBs/late endosomes (van Niel et al. 2006b), suggesting a common sorting mechanism with the Ub-dependent degradative sorting pathway of MVBs. Also, the fact that class II MHC is coenriched with markers of exosomes within the MIIC of dendritic cells implies that exosomes are derived from the same common ILV origin. Indeed, at least some of the proteins found trapped in exosomes are thought to constitute part of the endosomal protein machinery responsible for sorting proteins along the endocytic/degradative pathway, e.g., clathrin, Tsg101, and Alix (Thery et al. 2001, Wubbolts et al. 2003). Furthermore, exosomes contain ubiquitinated proteins, implying their common origin with ILVs that carry ubiquitinated protein cargo for degradation (Buschow et al. 2005). Even ILV pathways that use an Ub-independent pathway, which generate highly specialized MVB-like organelles like melanosomes, apparently use the same endosomal subdomains enriched in the same machinery that controls the movement of Ub-dependent cargo into the degradative MVB/ILV pathway. Pmel17, however, appears to enter ILVs in a manner that does not depend on either ubiquitination or the degradative MVB sorting machinery (Theos et al. 2006). Clearly, it would be interesting to investigate the differences between these pathways and whether common mechanisms exist to unite them.
SORTING SIGNALS FOR ENTRY INTO THE MULTIVESICULAR BODY LUMEN
Ubiquitin as a Sorting Signal
The default pathway for internalized membrane proteins, lipids, and fluid is recycling to the cell surface (Mayor et al. 1993). In contrast, protein transport into the MVB interior requires positive sorting signals. Inducing lysosomal delivery of recycling proteins by blocking their retrieval from endosomal compartments without a positive signal for ILV entry results in delivery to the limiting membrane, where only lumenal epitopes are degraded (Urbanowski & Piper 2001). Although internalization from the cell surface may serve as the rate-limiting step for the degradation of some proteins, for other proteins the MVB sorting step may be the rate-limiting and most physiologically relevant step. Ubiquitination causes sorting toward lysosomes from a number of compartments, including the Golgi (where Ub serves as a signal to divert proteins to endosomes), the cell surface (where Ub promotes internalization), and the MVB (where Ub mediates incorporation into ILVs). Ub works as a mobile sorting signal via its recognition surface, which interacts with a variety of protein sorting machines (Figures 1–3).
Figure 1.
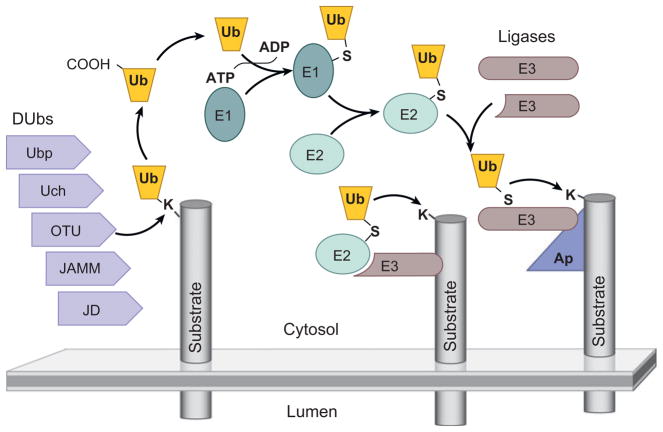
Ubiquitination and endosomal ubiquitin-sorting receptors. The transfer of ubiquitin (Ub) to substrate proteins involves the sequential action of three classes of enzymes: an activating enzyme (E1), a conjugation enzyme (E2), and a ligase (E3) (Hershko & Ciechanover 1998). Most eukaryotic cells have a single E1 enzyme, which uses ATP to form a high-energy thiol-ester bond with the C-terminal glycine (G76) of Ub E1 and then transfers Ub to one of a handful of E2 enzymes, which also forms a thiol-ester bond with Ub G76. E2s can bind RING-finger-type E3 ligases that bridge the substrate protein with E2, thus facilitating the transfer of Ub from E2 to the substrate. E2 can also transfer Ub to HECT-type E3 ligases, which also form a thiol-ester bond and directly transfer Ub onto substrate proteins. Ub is typically attached to lysine side chains (K) of substrate proteins and forms an isopeptide bond. E3s can sometimes interact directly with their substrate or use other adaptor proteins (Ap) to target their substrates. Ub is removed from proteins by Ub-specific proteases or deubiquitinating enzymes (DUbs). The major DUb families are grouped on the basis of the similarities of their active site (Ubp, Ub peptidase; Uch, Ub C-terminal hydrolase; OTU, otubain; JAMM, Jab1/MPN domain metalloenzyme; JD, Josephin domain).
Figure 3.
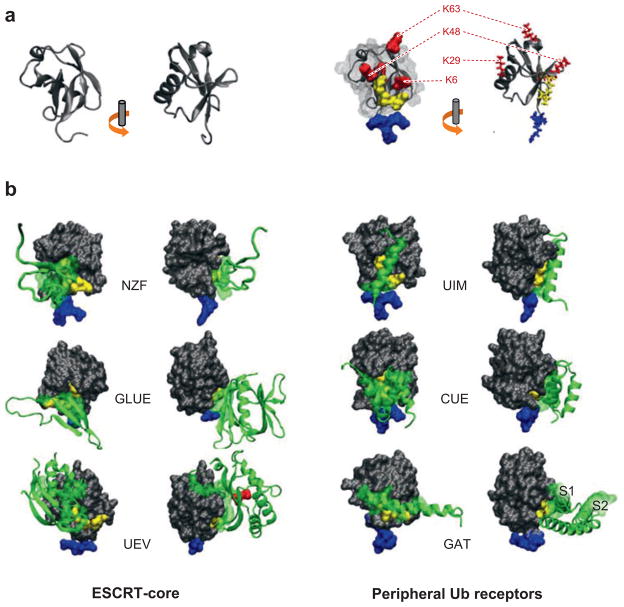
Recognition of ubiquitin. (a) Ub is depicted as a cartoon in two orientations. The flexible C terminus containing G75 and G76 is oriented at the bottom and colored blue. Some, but not all, of the lysines of Ub are labeled in red, and the hydrophobic patch on the surface of Ub composed of L8, I44, and V70 is shown in yellow. (b) The corresponding orientations of Ub interfaced with the indicated Ub-binding domains. On the left are the domains found in the endosomal sorting complexes required for transport (ESCRT)-I/II supercomplex; on the right are Ub-binding motifs found in peripheral Ub receptors. The GAT domain contains two separable Ub-binding sites (S1 and S2), which are shown in the side orientation. For the face-on view of the GAT domain and the GRAM-like Ub binding on Eap45 (GLUE) domain, part of the domain is removed to highlight the interface with Ub. All Ub-binding motifs interact to some degree with the I44 hydrophobic patch, and none interact with the alpha-helix region on the back of Ub. The red, highlighted residue in the Ub-E2-like variant domain (UEV) structure corresponds to M85.
Ub as an MVB sorting signal was first described in yeast and was inferred from many studies demonstrating a tight correlation between the lysosomal degradation of a variety of membrane proteins and their ubiquitination (Hicke & Dunn 2003). Direct evidence that Ub directed MVB sorting was provided by mutating the lysine residues of MVB cargo such as Cps1 and Phm5, which are normally ubiquitinated. These mutations did not alter the lysosomal targeting of these proteins but caused them to accumulate at the limiting vacuolar/lysosomal membrane rather than within the vacuole lumen (Katzmann et al. 2001, Reggiori & Pelham 2001). Demonstrating that Ub was a sufficient signal was accomplished by fusing Ub in frame to proteins not normally sent to the vacuole lumen for degradation (Reggiori & Pelham 2001, Urbanowski & Piper 2001). Many studies with proteins in both yeast and animal cells have now confirmed the universal nature of Ub as an MVB targeting signal.
Although certain Ub-dependent functions involve the formation of long polyubiquitin chains, MVB targeting does not; the fusion of a single Ub is sufficient to direct ILV targeting, and there are few long polyubiquitin chains on MVB cargo proteins (Hicke & Dunn 2003, Dupre et al. 2001). Many efficiently downregulated proteins are modified by several Ubs, yet these either are linked directly to substrate proteins via several lysine residues (multiple monoubiqitination) (Haglund et al. 2003) or may be present in short oligo-Ub chains consisting of lysine 63 (K63)-linked Ubs (Hicke & Dunn 2003, Dupre et al. 2001). These extra Ubs may be important for increased MVB sorting efficiency because the attachment of a single Ub may not be efficient in some contexts (Raiborg et al. 2002). Alternatively, the K63-Ub linkage may provide specific sorting information; however, all the Ub-binding domains present in proteins that participate in the MVB sorting process have been structurally defined as mono-Ub-binding domains. Although these domains have multiple sites for Ub binding (Figure 3) and have higher affinity for Ub chains, to date there are no direct data showing that a specific K63 linkage is required for their association. However, distinct linkages may signify as-yet-unappreciated subtleties in sorting outcomes.
Ub is recognized by an expanding cohort of endosomal proteins, which may act as Ub-sorting receptors responsible for binding and directing cargo toward ILVs (Figure 2). The identification of these Ub-binding proteins was greatly facilitated by both directed two-hybrid screens and bioinformatics studies, which defined likely Ub interaction domains (Hicke & Dunn 2003, Hurley et al. 2006). Ub has five β-sheets, mostly contained on the front side, and an α-helix down the back side. All the endosomal Ub-binding proteins recognize the front face of Ub and, in particular, a hydrophobic patch consisting of the residues L8, I44, and V70, suggesting that these residues may compete in a sequential hand-off reaction during the sorting process. Figure 3 divides the known endosomal Ub-binding proteins into two groups: the peripheral Ub-binding receptors and the ESCRT-core Ub-binding proteins. The former include the vacuolar sorting protein (Vps) 27/Hrs-Hse1/STAM complex, which contains Ub-interacting motif (UIM) domains; Golgi-associated, Gamma ear–containing Arf binding (GGA) proteins [which contain a GGA and Target of myb1, or TOM1 (GAT) domain]; and the TOM1/Tollip complex (which contains a GAT and a CUE domain). All the peripheral components localize to endosomes and interact with clathrin. Interestingly, all these components harbor a Vps27, Hrs, STAM (VHS) domain, whose function in this context remains unknown. Immuno-EM analysis has shown that Hrs is concentrated in flat clathrin subdomains on endosomes, which are implicated in the formation of MVBs (Sachse et al. 2002). It is likely that the other peripheral components are localized similarly. The ESCRT-core Ub-binding proteins include Vps23/Tsg101 and Vps36/EAP45, subunits of ESCRT-I and ESCRT-II. Both Vps23 and Tsg101 bind Ub similarly, using a Ub E2 variant (UEV) domain (Sundquist et al. 2004, Teo et al. 2004b). Mammalian Vps36/EAP45 uses a GLUE domain to bind both phosphorylated phosphatidylinositols and Ub (Alam et al. 2006, Hirano et al. 2006, Teo et al. 2006). Several residue differences in the yeast Vps36 GLUE domain are predicted to prevent its Ub binding. However, inserted into the GLUE domain are two new zinc finger (NZF) domains, one of which can bind Ub (Alam et al. 2004). All the Ub-binding modules discussed have low affinity for Ub, with the highest at ~10 μM for the GAT domain, which has two Ub-binding motifs (Bilodeau et al. 2004). In vivo, Ub-binding proteins may cluster or multimerize to increase their avidity for Ub. Alternatively, this low affinity may permit reversible binding and sequential interactions between the Ub-binding receptors and ubiquitinated cargoes to facilitate Ub-dependent MVB sorting.
Figure 2.
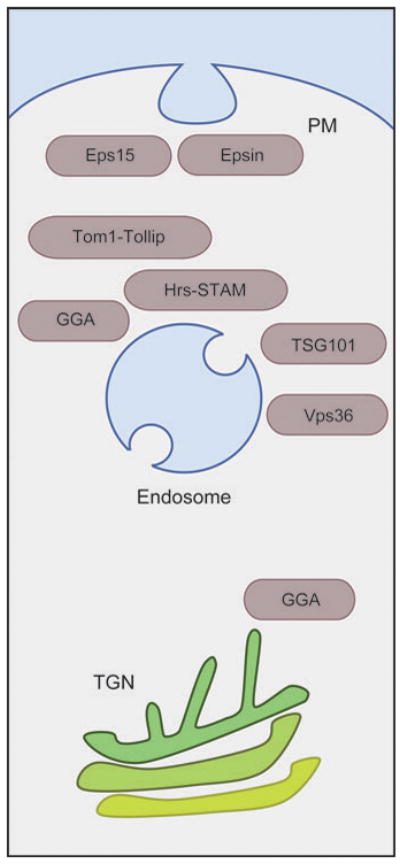
Ub-binding proteins involved in protein trafficking throughout the post-Golgi/endocytic system. Proteins such as Eps15, Epsin, Tom1, Tollip, GGA, Hrs, STAM, TSG101, and Vps36/Eap45 (see text) may work as receptors for Ub-cargo and catalyze a number of distinct transport steps that convey cargo toward the lysosome for degradation. These transport steps include internalization from the plasma membrane (PM), vesicle formation at the trans-Golgi network (TGN), and lumenal vesicle formation at the endosome.
Virtually all the endosomal Ub-binding proteins discussed above undergo some level of ubiquitination in mammalian cells. This phenomenon is strictly correlated to the ability of these proteins to bind monoubiquitin and has been referred to as coupled monoubiqitination (Woelk et al. 2006). This modification is thought to lead to inactivation of these proteins because of an intramolecular interaction between the attached Ub and the Ub-binding domain. Indeed, ubiquitinated forms of epsin and GGA3 are no longer able to interact with some of their binding partners (Chen et al. 2003, Yogosawa et al. 2006), and when Ub is fused in frame to Hrs, Hrs-Ub is incapable of exerting dominant-negative effects upon overexpression (Hoeller et al. 2006). The mechanism responsible for monoubiquitination of these proteins relies on the ability of the Ub-binding proteins to associate with other ubiquitinated proteins undergoing ubiquitination (Woelk et al. 2006). It is not yet clear whether coupled ubiquitination signifies a precise form of regulation that switches off protein function, stimulates the formation of large complexes of other Ub-binding proteins, or simply represents the concomitant hazard these proteins face by engaging Ub-cargo.
The majority of Ub is removed from cargo just prior to the delivery of cargo into ILVs. In yeast the Doa4/Ubp4 deubiquitinating enzyme (DUb) performs this function. Loss of Doa4 drastically reduces Ub levels in the cell and thus inhibits a variety of Ub-dependent processes (Amerik et al. 2000). However, cargo proteins with in-frame fusions of Ub are normally sorted to the vacuole interior in the presence and absence of Doa4 (Reggiori & Pelham 2001). These data highlight two key points: (a) There is tremendous flux of Ub through MVBs, and (b) the removal of Ub from cargo is not required for the delivery of Ub-cargo into ILVs because Ub is degraded with cargoes when Doa4 function is lost. Interestingly, histological analysis has shown that Ub accumulates within lysosomes of aging neurons in the brain, suggesting that the removal of Ub from lysosomal cargo may falter later in life (Mayer et al. 1992). This may lead indirectly to a variety of pathogenic consequences owing to depletion of the pool of available Ub, similar to what has been proposed for other neurodegenerative pathologies (Almeida et al. 2006).
A Doa4-like DUb dedicated to recycling Ub from MVB cargo proteins has yet to be identified in animal cells. Two candidate enzymes are UBPY/USP8, a Ubp family member similar in domain organization to Doa4, and AMSH, a Jab/MPN1 domain metalloenzyme (JAMM) domain peptidase. Loss of UBPY also leads to severe changes in endosomal morphology, a block in degradation of the MVB substrate proteins EGFR and EGF, and the accumulation of ubiquitinated proteins on endosomes without a concomitant decrease in free Ub levels (Bowers et al. 2006, Mizuno et al. 2006, Row et al. 2006). These effects mimic the effects of the loss of function of many class E Vps proteins responsible for MVB sorting and suggest that UBPY may provide critical functions other than deubiquitinating cargo (for example, deubiquitinating the Ub-recognition machinery) (Mizuno et al. 2006, Row et al. 2006). AMSH can interact with late-acting MVB sorting machinery, similar to Doa4 (Amerik et al. 2000, Luhtala & Odorizzi 2004, Tsang et al. 2006), and under some conditions, loss of AMSH can slow degradation of EGFR (Ma et al. 2007). However, AMSH may also function at an early step in the MVB sorting pathway via additional interactions with STAM and clathrin; AMSH may provide an editing function to precisely regulate the disposition of cargo when AMSH is recognized by the MVB sorting machinery (Bowers et al. 2006, McCullough et al. 2004). Similarly, in yeast the STAM homolog Hse1 associates with both the HECT Ub ligase Rsp5 and DUbs to regulate the efficiency of cargo sorting (Ren et al. 2007). This earlier role for DUb enzymes suggests that ubiquitination is a dynamic process wherein MVB cargoes likely undergo multiple rounds of ubiquitination/deubiquitination while they travel toward the MVB interior. Typically, only a small fraction of receptors that undergo efficient Ub-dependent lysosomal degradation are ubiquitinated at any one time, likely owing to the balance of both DUbs and ligases they encounter along the way. This underscores the dynamic nature of protein ubiquitination and regulatory opportunities the cell has to specify protein ubiquitination and thus MVB sorting. In animal cells, efficient downregulation of the EGFR requires its continued association with the really interesting new gene (RING) Ub-ligase Cbl during its journey to endosomes (Longva et al. 2002). Other ligases associate with Ub-sorting machinery; these include atrophin-1-interacting protein 4 (AIP4), a HECT-type ligase that associates with Hrs (Marchese et al. 2003) and the murine double minute 2 (MDM2); Tsg101-associated ligase (Tal); Nedd4.1; and Mahogunin ligases that associate with ESCRT-I (Amit et al. 2004, Blot et al. 2004, Li et al. 2001, Yoon Kim et al. 2007). It will be important to establish whether the purpose of these ligases is to modify cargo or cargo sorting machinery.
Non-Ubiquitin Sorting Signals
Although many Ub-dependent MVB cargoes have been described, much less is known about non-Ub signals that sort proteins to ILVs. One protein in yeast that uses both pathways is Sna3, a small protein with two transmembrane domains and cytosolically disposed N and C termini. Although Sna3 is ubiquitinated, nonubiquitinatable forms of Sna3 still sort to the vacuole interior, although they utilize the same endosomal sorting machinery as Ub-cargo. The sorting signals for Sna3 include an N-terminal tyrosine (Y)-containing sequence and a PPAY motif, which interacts with the HECT-type Ub-ligase Rsp5 (McNatt et al. 2007, Oestreich et al. 2007a). The Rsp5-Sna3 interaction is required for MVB sorting even in the absence of Sna3 ubiquitination. Rsp5 ubiquitinates a number of MVB cargoes but also partially localizes to endosomes and interacts with some of the Ub-cargo sorting machinery (Figures 4–6). Thus, binding Rsp5 may work as a sorting signal by bridging Sna3 with MVB protein sorting machinery. However, the enzymatic activity of Rsp5 is required for the sorting of forms of Sna3 that are resistant to ubiquitination, suggesting an additional level of regulation by this ligase.
Figure 4.
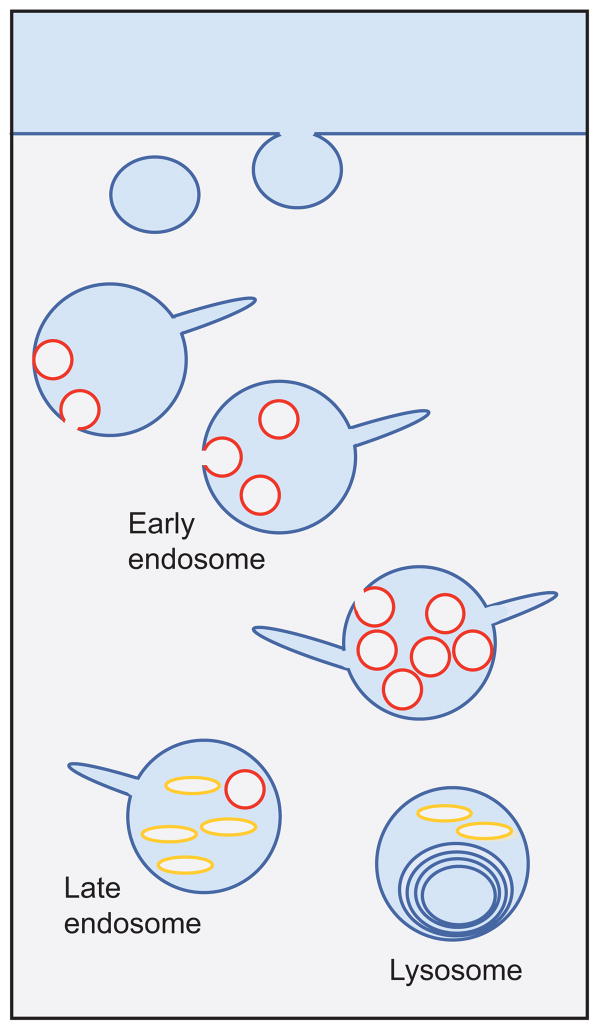
MVB anatomy. Schematic of the endocytic pathway indicating the progression from early endosomes to multivesicular late endosomes and finally to multilamellar lysosomes. Internal vesicle formation occurs during endosome maturation. Pictured are two types of internal membranes: phosphatidylinositol 3-phosphate [PI(3)P] positive (red ) and LBPA positive ( yellow).
Figure 6.
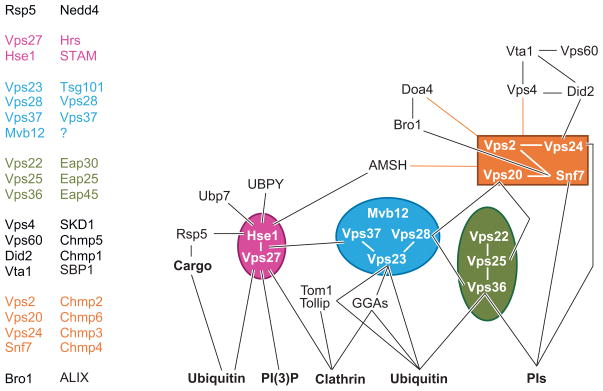
Map of key MVB biogenesis machinery. Schematic representation of protein-protein interactions among components involved in MVB sorting. ESCRT-I, -II, and -III are highlighted in blue, green, and orange, respectively. The Vps27-Hse1 complex is shown in pink. No distinction between yeast and mammalian proteins is shown in an attempt to highlight some of the conserved interactions.
Viral gag proteins represent another cargo that displays a variety of sorting signals directing incorporation into ILV-like particles. Although the ubiquitination of gag correlates with its sorting into virus-like particles, mutant gag proteins that are not ubiquitinated are still capable of budding. This suggests Ub is not required in targeting gag to ILVs. Different viral gag proteins can interact with a variety of machinery that is normally used for sorting Ub-cargo into MVBs, including Nedd4, a homolog of the Rsp5 E3 ligase (Morita & Sundquist 2004). This raises an interesting parallel between gag-Nedd4 ILV targeting in animal cells and Sna3-Rsp5 MVB sorting in yeast and suggests that other ubiquitination-independent ILV targeting processes employed by viruses may also be used by endogenous MVB cargoes.
TfRs enter MVBs for later release as exosomes from reticulocytes as they mature to erythrocytes; however, the TfR does not appear to be ubiquitinated. Instead, its MVB sorting is mediated by a Y-based motif that is usually recognized by the AP-2 adaptor complex to mediate internalization. However, during differentiation, the AP-2 adaptor is degraded. This allows the binding of TfR to the protein Alix, which in turn can interact with other MVB sorting machinery and may mediate the incorporation of TfR into ILVs (Geminard et al. 2004).
The melanosomal protein Pmel17/Silver enters ILVs in a Ub-independent manner and ultimately forms the protein scaffold that holds melanin pigments (Berson et al. 2003). At least one sorting signal for Pmel17 incorporation into ILVs is located on its lumenal domain and may work by helping the protein aggregate into an arrangement conducive to ILV incorporation. Interestingly, MVB sorting of Pmel17 is not altered when the sorting of Ub-cargo is compromised by perturbation of the Ub-dependent MVB sorting pathway (e.g., by altering Hrs, Tsg101, or Vps4 function). Pmel17 does, however, concentrate at the same clathrin-enriched subdomains that participate in sorting of Ub-cargo and is also a constituent of exosomes, implying some level of commonality with the pathway for sorting Ub-cargo (Theos et al. 2006).
MACHINERY
Lipid Sorting
Differential lipid sorting and the formation of lipid subdomains play important roles in the biogenesis of MVBs. One important requirement is to render ILVs susceptible to degradation while leaving the limiting membrane resistant to hydrolases. Additionally, the organization of the protein sorting machinery and perhaps the budding and scission of the ILV may be controlled by lipid composition and localized lipid remodeling. Three interesting lipids that appear to play a role in MVB biogenesis are phosphatidylinositol 3-phosphate [PI(3)P], cholesterol, and BMP/LBPA. Each may play a role in subdomain organization, vesicle formation, and ILV degradation.
PI(3)P is enriched on the cytosolic face of early endosomes, where it recruits a variety of proteins to specialized endosomal subdomains. Several key proteins that compose the MVB sorting machinery bind PI(3)P; the presence of both this lipid and the PI(3)P-binding domains of the sorting machinery is critical for MVB formation (Futter et al. 2001, Odorizzi et al. 1998, Piper et al. 1995, Teo et al. 2006) (Figure 5). Most of the endosomal PI(3)P in yeast and animal cells is actually found within the endosomal lumen associated with ILVs, consistent with the observation that PI(3)P is elevated in yeast mutants in which vacuolar hydrolases are inactivated (Gillooly et al. 2000, Wurmser & Emr 1998). The density of PI(3)P in ILVs is highest within early endosomes and is significantly lower in late endosomes, suggesting that the PI(3)P-containing ILVs are quickly degraded or susceptible to phosphatase activity. PI(3)P is also the precursor for phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], the product of the Fab1/PIKfyve kinase. Loss of PI(3,5)P2 results in enlarged vacuolar structures in both yeast and animal cells and causes defects in the delivery of Cps1 to the vacuole lumen (Odorizzi et al. 1998). Loss of PI(3,5)P2, however, does not affect the proper MVB sorting of many other Ub-dependent and Ub-independent cargos, indicating that PI(3,5)P2 does not play a critical role in the processes of cargo recognition and ILV formation (Odorizzi et al. 1998, Urbanowski & Piper 2001).
Figure 5.
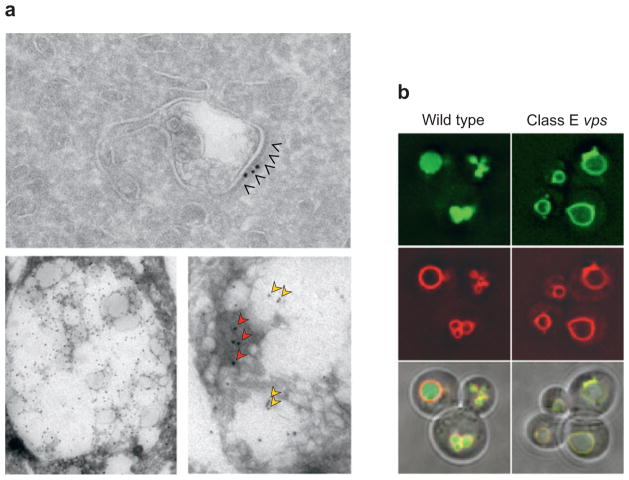
Electron and fluorescence micrographs of MVBs and late endosomes. (a) An early endosome with a tubular extension, which serves as a carrier for recycling proteins such as a transferrin receptor. An electron-dense protein coat can be seen on the surface of endosomes. This subdomain is highlighted by arrowheads and is labeled with colloidal gold for clathrin [from M. Sacshe and J. Klumpermann (Sachse et al. 2002), with permission]. The electron micrographs in the lower panels show late endosomes from BHK cells labeled for phosphatidylinositol 3-phosphate [PI(3)P] singly (left) or double labeled (right) for PI(3)P (red arrows) and for LBPA ( yellow arrows). Provided by Rob Parton, University of Brisbane. (b) Fluorescence micrographs of wild-type and class E vps mutant cells expressing GFP-tagged MVB cargo and labeled with the lipophilic dye FM4-64 (red ), which marks the limiting membrane (provided by A. Oestreich, Mayo Clinic). MVB cargo is localized within the limiting membrane of the vacuole in wild-type cells. The mutant class E vps cells mislocalize the MVB cargo to the limiting membrane of the vacuole and the aberrant class E compartment.
Cholesterol is also implicated in MVB biogenesis owing to its significant enrichment in MVBs (Hornick et al. 1985). Analysis of exosomes reveals that much of the endosomal cholesterol resides in ILVs (Mobius et al. 2003, Wubbolts et al. 2003), although the levels of cholesterol within intralumenal membranes vary among cell types (Subra et al. 2006). Blocking MVB formation with dominant-negative Vps4 results in the accumulation of large, cholesterol-rich endosomes (Bishop & Woodman 2000, Yang et al. 2004). Enrichment of cholesterol and sphingolipids in ILVs may render them susceptible to breakdown because cholesterol is largely depleted by the late endosomal stage. Targeting ILVs for degradation involves proteins such as saposins, which bind to and accelerate sphingolipid degradation (Kolter & Sandhoff 2005), and possibly the endosomal cholesterol efflux pathway, which relies on proteins such as the Niemann-Pick type C (NPC) 1 transporter and NPC2 lipid transfer protein. Cholesterol concentration may also facilitate the budding process either by promoting proper membrane curvature (Wang et al. 2007) or by organizing discrete sudomains enriched in proteins such as tetraspanins (Charrin et al. 2003, Silvie et al. 2006), which are incorporated into ILVs (Escola et al. 1998). Interestingly, inducing accumulation of cholesterol within endosomes with the drug U18666A dramatically stimulates inward budding of endosomes and the formation of MVBs in Dictyostelium (Marchetti et al. 2004). Also, two members of the oxysterol-binding-protein family, Osh6 and Osh7, associate with Vps4, an AAA-ATPase that controls the assembly of the MVB protein sorting machinery (Wang et al. 2005).
BMP/LBPA is enriched in late endosomes and lysosomes and is not detected in earlier endosomal compartments where ILV formation is highly active. Moreover, intralumenal membranes containing BMP/LBPA are distinct from those containing PI(3)P (Gillooly et al. 2000, Kobayashi et al. 2002) and appear to be devoid of activated EGFR destined for lysosomes and devoid of class II MHC within MIIC (Mobius et al. 2003, White et al 2006). LBPA has not been reported to be present in yeast MVBs or vacuoles, which otherwise can accumulate PI(3)P-enriched ILVs. Furthermore, BMP/LBPA ILVs can form in the absence of PI(3)P generation, suggesting that this process is independent of at least some of the protein machinery that controls PI(3)P/cholesterol ILVs used for protein degradation (Matsuo et al. 2004, White et al. 2006). LBPA may help form multivesicular-like structures because BMP/LBPA-containing liposomes spontaneously form inward-budding profiles in a pH-inducible manner (Matsuo et al. 2004). One protein that may control the formation of BMP/LBPA ILVs is Alix. Alix binds to BMP/LBPA, and Alix depletion reduces the accumulation of BMP/LBPA inside late endosomes (Matsuo et al. 2004). In vitro, BMP/LBPA is a very dynamic lipid that promotes fusion and may have interesting functions in vivo by facilitating back-fusion of ILVs with the limiting membrane of late endosomes (Kobayashi et al. 2002). Back-fusion appears important for the entry of both endocytosed anthrax toxin and endocytosed VSV into the cytosol (Abrami et al. 2004, Le Blanc et al. 2005). Both anthrax toxin and the VSV capsid are delivered to the interior of ILVs within early endosomes. Their release from ILVs requires delivery to BMP/LBPA-enriched late endosomes; antibodies to LBPA or down-regulation of Alix retard the efflux of anthrax toxin and VSV capsid from endosomes (Le Blanc et al. 2005). Such a BMP/LBPA-dependent exit strategy may also be required to keep recycling proteins like the cation-independent mannose-6-phosphate receptor out of lumenal membranes (Kobayashi et al. 1998). BMP/LBPA may promote back-fusion directly by modifying the ILV in which anthrax toxin and VSV are first delivered or later intercalating into ILVs and altering their fusion potential. Perturbing BMP/LBPA may also have direct effects by altering the dynamics of the endosomal lumen because antibodies to BMP/LBPA mimic the action of U18666A and loss of NPC1 in that they cause the accumulation of cholesterol in late endosomes.
Lipids impact the MVB sorting reaction at a variety of levels: the recruitment of effector proteins, the formation of ILVs, and even the relative sensitivity of the ILVs to lysosomal hydrolases as compared with the limiting membrane. Furthermore, MVB sorting impacts the metabolism of PI(3)P, cholesterol, and other lipids, highlighting the importance of this pathway beyond the context of protein turnover.
Protein Machinery
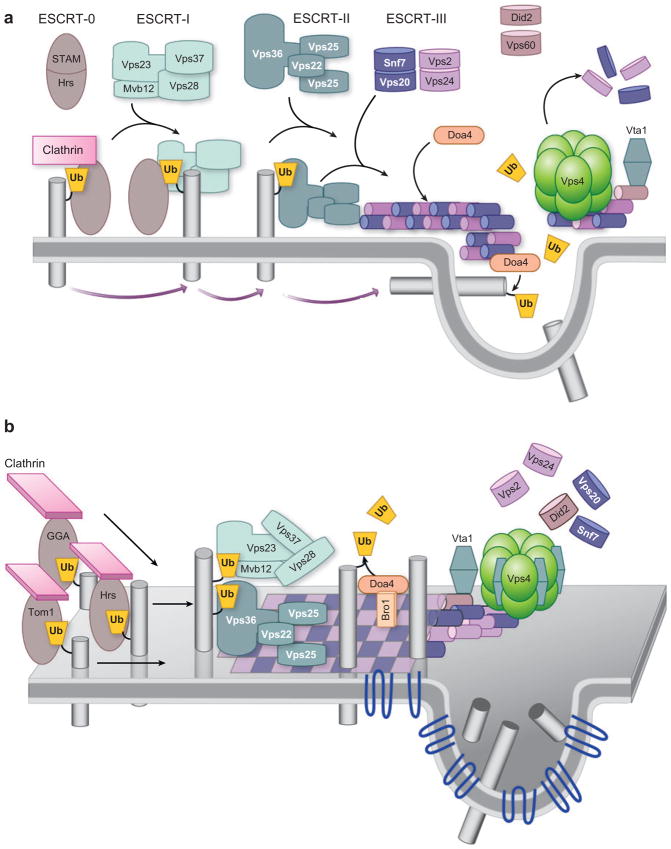
The search for protein machinery that controls MVB biogenesis has focused primarily on two approaches. One is proteomic analysis of exosomes and viruses, which can be constituents of MVBs. These studies reveal that polytopic tetraspanin proteins are major components of ILVs (Escola et al. 1998, Hegmans et al. 2004, Thery et al. 2001, Wubbolts et al. 2003). Other components include Rab GTPases, membrane-binding annexins, and constituents of the actin cytoskeleton. Genetic screens in yeast have identified a number of nonessential proteins defined as class E Vps proteins, originally characterized by a common phenotype (Raymond et al. 1992). Class E vps mutants have a number of defects in the endocytic pathway (Figure 5b): defects in structure in that endosomes are greatly enlarged and form cisternal stacks, defects in the trafficking of late Golgi proteins and vacuolar proteins from endosomes, defects in sorting proteins to the endosomal lumen, and defects in generating ILVs (Odorizzi et al. 1998, Raymond et al. 1992, Rieder et al. 1996). Many of these components have clear homologs in animal cells, which are important for regulating receptor signaling and downregulation. A wealth of structure/function analyses have defined these components as members of so-called ESCRT complexes (Figures 6 and 7). Earlier studies, mostly in yeast, supported the model pictured in Figure 7. Four ESCRT complexes (0, I, II, III) act sequentially to sort cargo and to form a coated subdomain on endosomes that forms the ILV. The Vps27/Hrs-Hse1/STAM complex (ESCRT-0) is first recruited to endosomes by binding PI(3)P and ubiquitinated cargo; in animal cells this leads to colocalization within clathrin-rich subdomains. ESCRT-0 then recruits ESCRT-I to the membrane, followed by recruitment of ESCRT-II. Finally, ESCRT-II recruits ESCRT-III subunits, which can form a large polymer on the surface of endosomes (Katzmann et al. 2002, Raiborg et al. 2003). The AAA ATPase Vps4 works to break apart ESCRT-III and the other ESCRT complexes, resulting in their dissociation from the membrane to complete the cycle. Ubiquitinated cargo is passed sequentially through the ESCRT complexes, first bound by ESCRT-0 via UIM domains, then to the UEV domain of Vps23/Tsg101, and then to Vps36 of the ESCRT-II complex, which somehow conveys Ub-cargo to the forming ILV with the aid of ESCRT-III and associated factors.
Figure 7.
MVB core machinery. (a) Schematic for how class E Vps proteins may control MVB sorting. Pictured is the sequential recruitment of ESCRT complexes that are nucleated by the association of ESCRT-0 (STAM/Hrs) with endosomal membranes by binding phosphatidylinositol 3-phosphate [PI(3)P]. Vps27/Hrs recruits clathrin to form an endosomal subdomain and also binds and activates ESCRT-I, which in turn binds and recruits the ESCRT-II complex to endosomes. ESCRT-II then recruits ESCRT-III, which forms a polymer. This network is then disassembled at the last step of ILV formation. Ub-cargo is sorted in a sequential manner as well: It is first recognized by ESCRT-0 and then passed first to ESCRT-I and then to ESCRT-II before being incorporated into ILVs. At some point, Ub-cargo is deubiquitinated so that Ub is recycled. (b) An alternative model, wherein the Hrs complex is one of many peripheral Ub-sorting receptors that gather Ub-cargoes on endosomes and concentrate them in clathrin-rich subdomains. These receptors then transfer cargo to the ESCRT-I/II supercomplex, which has a variety of Ub-binding sites that can recognize a host Ub-cargo. ESCRT-I/II sits enmeshed in a specialized subdomain enriched in tetraspanins, sphingolipids, and other components that will be incorporated into ILVs. This meshwork is organized by ESCRT-III, which forms a polymer (represented as a flat checkerboard lattice) that can incarcerate cargo and hold ESCRT-I/II. This coated subdomain houses DUbs like Doa4, which can remove Ub without allowing cargo to escape. The ESCRT-III polymer is contained within endosomal subdomains by the Vps4 ATPase. ILV formation is catalyzed by the aggregation of cargo and tetraspanins that form large oligomeric structures. ILV formation is also facilitated by sorting of lipids conducive to membrane curvature.
Both new and old data, however, challenge this model. That ESCRT complexes are sequentially recruited is supported by the findings that loss of ESCRT-0 diminishes the association of ESCRT-I with endosomes and that both Vps27 and Hrs directly bind to the UEV domain of Vps23/Tsg101 via Pro-Ser-Ala-Pro and Pro-Ser-Asp-Pro motifs, respectively (Bache et al. 2003, Bilodeau et al. 2003, Katzmann et al. 2003). Also, ESCRT-I can bind ESCRT-II directly, and this interface is important for the function of ESCRT-I (Gill et al. 2007). ESCRT-II can also bind ESCRT-III (Teo et al. 2006). Overexpression of ESCRT-II suppresses loss of ESCRT-I, consistent with a function of ESCRT-II downstream of ESCRT-I. Lastly, ESCRT-I and -II are required for appropriate assembly of ESCRT-III (Babst et al. 2002b), and ESCRT-III is required for the recruitment of Vps4 to endosomes (Babst et al. 2002a). However, loss of ESCRT-I does not prevent ESCRT-II association with endosomes, although it does hamper ESCRT-III recruitment (Babst et al. 2002b). Loss of ESCRT-0 does not block the association of ESCRT-III with endosomes (Curtiss et al. 2007), and loss of ESCRT-I inhibits the recruitment of clathrin to Hrs-positive endosomes (Doyotte et al. 2005). Although the presence of Vps27 (ESCRT-0) is required for the association of ESCRT-I with membranes, mutating the motifs in Vps27 that bind Vps23 (ESCRT-I) does not alter MVB formation and only partially affects the sorting efficiency of particular cargo (Bilodeau et al. 2003). These data emphasize that, although these recruitment assays are helpful in identifying some of the overall requirements for endosome association and protein interactions, they are performed in mutant cells lacking proper MVB function and thus represent a number of direct and indirect effects. Such observed effects on component localization do not mean they must occur in a transient and sequential mode in normal cells.
The idea that Ub-cargo is transferred from one complex to the next is attractive because ESCRT-0, I, and II each have Ub-binding domains that must compete for binding to the surface of Ub. Evidence that the Ub-binding domains of the Vps27-Hse1 complex bind and sort Ub-cargo is supported by mutations in the three UIM domains of this complex that block intralumenal sorting of Ub-cargo but do not block delivery of Ub-independent cargo or cause defects in other Vps27-dependent functions (Bilodeau et al. 2002). These data suggest that the Ub-binding function of Vps27-Hse1 is dedicated to sorting Ub-cargo and not to some other general function. Such specificity has not been demonstrated for Vps36; loss of its Ub-binding NZF domain results in a general loss of function (Alam et al. 2004). The function of Vps23 and Tsg101 binding to Ub is also unclear. Mutations of the Vps23 UEV domain that fail to bind Ub also largely phenocopy a null mutant (Katzmann et al. 2001); however, the mutation analyzed (M85, Figure 3) is not part of the Ub-binding interface, which suggests a more general defect in the resulting mutant.
Another key idea in the model shown in Figure 7 is that many of the class E Vps proteins are essential in the processes of MVB protein sorting and ILV formation because loss of any component causes a class E vps phenotype. Studies in animal cells have yielded surprises, however, because depletion of individual class E Vps proteins causes varying phenotypes. One possible explanation is that the functional integrity of many class E proteins in yeast is exquisitely sensitive to their interactions with other components such that the loss of one component disrupts the function of the entire class E machine. In animal cells, however, these same proteins have more intermolecular connections that may compensate for the loss of a single component (Bowers et al. 2004, von Schwedler et al. 2003). Many studies in animal cells support some role of the class E proteins in the endocytic pathway. Interpreting many of the initial studies can be complicated because some utilize overexpression experiments that may indirectly perturb the entire network of class E proteins or RNAi-mediated depletion that may leave residual levels of protein behind. Also, many experiments follow the lysosomal degradation of proteins such as the EGFR as a measure of MVB sorting. This is typically done by following the loss of lumenal EGFR epitopes that report on the arrival of EGFR at late endosomal/lysosomal compartments. These methods, however, do not differentiate delivery to the late endosomal limiting membrane from delivery to ILVs, nor do they differentiate the process of sorting ubiquitinated cargo from upstream defects in proper ubiquitination of cargo. Despite these caveats, many of these data conflict with the canonical view gleaned from the yeast data.
Perhaps the closest phenotype to a yeast class E mutant in animal cells comes from loss of ESCRT-I. Depletion of Tsg101 in cell lines blocks degradation of EGFR, promotes recycling of EGF, and prolongs activation of the EGF signaling pathway (Babst et al. 2000, Bowers et al. 2006, Doyotte et al. 2005, Langelier et al. 2006, Razi & Futter 2006). Internalized EGFR is mislocalized to intricate tubular arrays and multicisternal endosomes (Doyotte et al. 2005, Razi & Futter 2006). However, Drosophila photoreceptor cells depleted of Vps28 form MVBs normally, and resting A431 cells depleted of Tsg101 have near-wild-type levels of normal MVBs (Razi & Futter 2006, Sevrioukov et al. 2005).
Loss of ESCRT-II slows degradation of EGFR, although not to the same extent as does loss of Tsg101 (Langelier et al. 2006). However, degradation of EGF and ubiquitinated class I MHC is not affected, nor is the gross morphology of endosomes altered upon ESCRT-II loss (Bowers et al. 2006). Loss of Hrs leads to a number of developmental defects (Lloyd et al. 2002). However, degradation of EGFR and its ligand is only modestly affected (Bache et al. 2003, Kanazawa et al. 2003, Lu et al. 2003, Razi & Futter 2006). Loss of Hrs decreases the number of normal MVBs, and in their place are swollen early endosomes (Bache et al. 2003, Komada & Soriano 1999, Lloyd et al. 2002, Razi & Futter 2006). Although the level of ILVs within swollen endosomes is lower, the latter can receive EGFR (Razi & Futter 2006).
Inhibiting Vps4 results in swollen endosomes, the accumulation of tubular extensions, extensive accumulation of ESCRT components and recycling proteins on endosomal membranes, and a block in EGFR degradation (Bishop & Woodman 2001, Bowers et al. 2006, Langelier et al. 2006, Yoshimori et al. 2000). Yet expression of dominant-negative, ATPase-defective Vps4 inhibits ILV formation by only ~30% (Sachse et al. 2004, Yoshimori et al. 2000). The relevant target of Vps4 is likely to be the ESCRT-III polymer; loss of the mammalian ESCRT-III subunits Vps24 and Vps20/CHMP6 also leads to the accumulation of EGFR in endosomes (Bache et al. 2006, Langelier et al. 2006). However, EM analysis shows that these endosomes contain significant levels of ILVs and that much of the EGFR is sorted into them (Bache et al. 2006).
Taken together, these studies show that the class E proteins play an important role in protein sorting to MVBs but that not all class E proteins work to execute the same step. Furthermore, whereas class E proteins may regulate aspects of ILV formation, endosome maturation, and fusion, they may not work directly to form ILVs. This is consistent with proteomic analysis of ILVs that shows that most of the class E proteins are not incorporated into ILVs (Reggiori & Pelham 2001, Thery et al. 2001, Wubbolts et al. 2003). Finally, ESCRT subunits play a variety of roles in many other processes such as transcription, cytoskeletal organization, cell cycle control, pH sensing, and mRNA localization (Irion & St Johnston 2007, Kullas et al. 2004, Slagsvold et al. 2006, Xu et al. 2004), leaving us to wonder whether the proteins perform diverse tasks or whether the same molecular task is used by different processes.
What functions can we ascribe to the various protein machinery? Figure 7 presents a model that reconciles many of these data. However, we emphasize that this represents only one of many models that can be extrapolated.
The first step is the initial gathering of Ub-cargo by peripheral Ub-sorting receptors such as the Hrs/STAM complex. The Hrs/STAM complex is not part of the core ESCRT constituency because its localization is distinct from ESCRT-I (Bache et al. 2003) and its depletion causes different defects than ESCRT-I depletion (Razi & Futter 2006). Furthermore, Hrs interacts with a variety of proteins that participate in a host of other endosomal processes (Chin et al. 2001, Rayala et al. 2006, Sun et al. 2003, Yan et al. 2005). Finally, the Hrs/STAM complex is not encoded in plant and Dictyostelium genomes, although those organisms do have ESCRT-I, II, and III homologs (Marchetti et al. 2004, Winter & Hauser 2006). The Hrs-STAM complex works in parallel with GGA proteins as well as the Tom1-Tollip complex to bind, retain, and concentrate Ub-cargo on endosomal subdomains (Raiborg et al. 2002). Both GGA3 and the Tom1-Tollip complex localize to endosomes, and loss of GGA3 or Tollip (Brissoni et al. 2006, Puertollano & Bonifacino 2004) results in sorting defects of particular Ub-cargoes. Interestingly, plants and Dictyostelium do have Tom1 homologs. The presence of several Ub-sorting receptors implies that different Ub-cargoes may be carried by different receptors.
These peripheral Ub-sorting receptors concentrate in clathrin-rich subdomains on the endosomal surface. The formation or stability of these domains may be promoted by the ability of these proteins to bind clathrin (Katoh et al. 2006, Raiborg et al. 2002, Raiborg et al. 2006). Clathrin likely plays a role in organizing the protein sorting machinery or in retaining Ub-sorting receptors at the limiting membrane so they do not follow MVB cargo into ILVs. Interestingly, in contrast to animal cells, clathrin does not accumulate on endosomes in class E vps yeast mutants, and deletion of the clathrin-binding site on Vps27 is without effect (Katzmann et al. 2003). However, loss of clathrin binding by Vps27 results in several synthetic effects with mutations in other class E machinery components and in some instances results in the transport of Vps27 into the vacuole lumen (R. Piper, unpublished data).
All these peripheral Ub-receptors interact with Vps23/Tsg101 of ESCRT-I (Bilodeau et al. 2003, Katzmann et al. 2003, Lu et al. 2003, Puertollano 2005, Puertollano & Bonifacino 2004). The revised model suggests both ESCRT-I and ESCRT-II work in the same step as part of a large supercomplex, a speculation supported by the tight connection between yeast Vps28 and ESCRT-II Vps36 (Gill et al. 2007). If such a supercomplex exists in vivo, it likely forms on membranes because biochemical analysis of soluble fractions finds ESCRT-I and ESCRT-II as discrete complexes (Babst et al. 2002b, Katzmann et al. 2001). One of the key observations supporting the view that ESCRT-II acts sequentially downstream of ESCRT-I is that defects in cargo sorting from loss of ESCRT-I subunits are suppressed upon overexpression of ESCRT-II (Babst et al. 2002b, Curtiss et al. 2007). However, these data are also consistent with the view that ESCRT-I and ESCRT-II proteins work together at the same step, and may explain the mild phenotype of ESCRT-II depletion in animal cells (Bowers et al. 2006). The model shown in Figure 7b depicts the ESCRT-I/II complex as a Ub-cargo-binding protein because the complex possesses multiple Ub-binding sites (Alam et al. 2004, Katzmann et al. 2001). The residues on the surface of Ub critical for Vps23/TSG101 binding, but not other Ub-binding modules, are required to confer MVB sorting on cargo (Bilodeau et al. 2003). Multiple Ub-binding domains may be required to bind a variety of cargos that present Ub in different contexts. Interestingly, the GLUE domain of mammalian Vps36 extends below the flexible C terminus of Ub, where it may be influenced by the context of the isopeptide bond, which may cause a bias for binding particular Ub-cargo (Figure 3). Also, deletion of the ESCRT-I subunit Mvb12 causes a selective sorting defect of some Ub-cargoes but not others (Curtiss et al. 2007, Oestreich et al. 2007b). The function of the ESCRT-I/II supercomplex may be both to promote the release of Ub-cargo from peripheral Ub receptors and to help position Ub-cargo for eventual incorporation into MVBs. The exact roles of the Ub-binding domains of ESCRT-I and II are still unclear; these Ub-binding domains may not interact with cargo but instead arbitrate some other regulatory event mediated by coupled monoubiquitination.
At some point, Ub-cargo must be moved to and immobilized in an environment that will allow its eventual incorporation into ILVs, yet still allow the removal of the Ub. Tomographic EM studies reveal that the endosomal subdomains from which ILVs are formed are distinct from the clathrin-enriched subdomains containing the Ub-sorting receptors such as Hrs (Murk et al. 2003). The incarceration of cargo may be mediated in part by ESCRT-III subunits, which polymerize and depolymerize on and off the surface of endosomes. Interestingly, when the ESCRT-III polymer is locked onto endosomes, the Ub-cargo Cps1 appears to enter a microenvironment that makes it less susceptible to lumenal proteolysis (Babst et al. 2002a). EM data indicate that a clathrin-negative protein coat is present at the surface of endosomes where ILVs appear to form and that MVB cargo such as the EGFR moves into these regions after concentrating first in adjacent clathrin-positive bilayered coats (Myromslien et al. 2006, Sachse et al. 2004). Thus, it will be informative to determine whether these regions are selectively enriched in ESCRT-III subunits. Alternatively, the ESCRT-III polymer may work to position the protein sorting machinery because it binds directly to the ESCRT-I component Vps28 (Bowers et al. 2004) and the ESCRT-II components Vps25/Eap20 (Teo et al. 2004a). The yeast ESCRT-III lattice also positions the DUb Doa4 (Odorizzi 2006). If the ESCRT-III polymer could indeed trap cargo prior to incorporation into ILVs, this arrangement of DUb activity would be optimal for recycling Ub without compromising the forward progression of cargo into the ILV. Binding and structural studies on the human ESCRT-III subunit, CHMP3, show that it is composed of a four-helix bundle, with a fifth short α-helix that may regulate dimerization through one of its interfaces (Muziol et al. 2006). Movement of the C-terminal tail may expose these interfaces and trigger a series of homo- and hetero-oligomerizing events with other ESCRT-III subunits owing to conservation of the residues that line the interface. Interestingly, the crystal structure of the CHMP3 dimer reveals a relatively flat complex with an extensive basically charged surface that may promote association with planar membranes (Muziol et al. 2006).
The Vps4 AAA ATPase may work to contain the ESCRT-III polymer, trimming its edges so as not to interfere with the organization of other endosomal subdomains. Indeed, inhibiting Vps4 causes a massive accumulation of protein coats on endosomes (Sachse et al. 2004).
Vps4 is thought to form a double-ring structure of 10–12 subunits, similar to other AAA ATPases such as p97/VCP/Cdc48 (Babst et al. 1998, Scott et al. 2005a). The same C-terminal fifth α-helix region of ESCRT-III subunits that is thought to regulate dimerization also serves as the interaction site for the microtubule interacting and trafficking (MIT) domain of Vps4 (Scott et al. 2005b), implying that Vps4 may transition ESCRT-III subunits from an open conformation ensconced in a polymer to closed monomeric forms (Lin et al. 2005). Exactly how Vps4 activity is regulated is unclear; however, a newly discovered factor, Vta1, binds Vps4 at a 1:1 ratio and stimulates its ATPase activity (Azmi et al. 2006, Lottridge et al. 2006). Vta1 in turn interacts with Did2/Vps46/Fti1 (Lottridge et al. 2006) and Vps60/Mos10 (Azmi et al. 2006, Shiflett et al. 2004). Did2 and Vps60 are likely part of an extended ESCRT-III complex polymer, yet their loss in yeast yields a milder class E phenotype than does loss of ESCRT-III core subunits (Babst et al. 2002a). The ability of Did2 and Vps60 to bridge Vta1 with ESCRT-III subunits may provide a way to regulate Vps4 activity on the basis of the composition or conformational state of the extended ESCRT-III polymer (Lottridge et al. 2006).
After cargo is incarcerated, it must be incorporated into ILVs. How ILVs are actually formed is entirely unclear. At some level lipid sorting must occur and may form the basis of membrane curvature through the uneven distribution of cone-shaped and inverted-cone-shaped lipids in the bilayer. ESCRT-III may play a role in ILV formation, perhaps by holding asymmetric lipid bilayers and membrane-deforming proteins in a planar bilayer owing to the flat shape of ESCRT-III dimers. Rapid membrane deformation and internal budding may then be triggered by the removal of ESCRT-III polymer from this subregion by Vps4. That ILVs are still observed when Vps4-ESCRT-III function is compromised suggests that other factors are also involved. An excellent set of candidates that may affect either the incarceration of cargo or perhaps ILV formation are the tetraspanins, which are greatly enriched in ILVs (Escola et al. 1998). Tetraspanins can interact with a wide range of membrane proteins, many of which are sorted to the MVB interior. Tetraspanins can homo-and hetero-oligomerize into complex webs or tetraspanin-enriched microdomains (TEMs), which also appear to connect with the actin cytoskeletal machinery and gather cholesterol (Charrin et al. 2003, Delaguillaumie et al. 2004, Hemler 2003, Silvie et al. 2006). At the cell surface TEMs can also cluster HIV-1 gag proteins, which in turn recruit ESCRT-I, Vps4, and Alix to these subdomains and may serve as virus budding sites (Booth et al. 2006, Nydegger et al. 2006). If such TEMs formed on endosomes, they would make excellent candidates for subdomains that support ILV formation. Proteomic analysis of ILVs has found equivalents to tetraspanins in yeast (Reggiori & Pelham 2001). One of these proteins is Sna3, which sorts to ILVs in a Ub-independent fashion. Sna3 has two transmembrane domains, is a major component of yeast ILVs, and appears to oligomerize in vivo (D. Katzmann, unpublished results). Although loss of Sna3 does not cause a sorting defect, other Sna3-like candidates exist among other polytopic membrane proteins that localize to the vacuolar lumen and exhibit no known function (Huh et al. 2003). Tetraspanins may also form the machinery that makes the ILVs, because of the ability of tetraspanins to aggregate into higher-order structures; aggregation may be a mechanism for inward budding (Vidal et al. 1997). Not only do tetraspanins form microdomains, but at least some tetraspanin-related proteins deform membranes dramatically, consistent with a role in ILV formation (Wrigley et al. 2000).
VIRUSES AND MULTIVESICULAR BODIES.
The production of various enveloped viruses such as HIV and Ebola depends on some of the same protein machinery that controls MVB formation. Topologically, the budding of viruses out of the cytosol (either from the cell surface or from endosomes) is similar to the budding of endosomes from the limiting membrane to form ILVs. Viral budding is promoted by the gag proteins, which contain a late domain required for the later stages of virus production. The gag protein, in the absence of other viral components, can bud and produce virus-like particles. Late domains can interact with various components of the MVB sorting machinery, including Tsg101, Alix, and the Nedd4 Ub ligase. Each of these components has extensive interactions with other components required for MVB formation and thus provides a way to recruit gags to the budding machinery. Loss of the MVB biogenesis machinery leads to a block in the scission or detachment of viruses from the limiting membrane. Thus, viral budding has been a powerful mechanistic link in the understanding of MVB biogenesis.
STAGES OF PROTEIN SORTING AND INTRALUMENAL VESICLE FORMATION.
The process of sorting ubiquitinated cargo into ILVs can be broken down into various steps:
Ub-cargo binding and concentration by Ub-sorting receptors in subdomains that segregate it from recycling proteins.
Detachment of Ub-sorting receptors from cargo.
Incarceration of cargo so that it is immobilized at or near sites of ILV formation. Deubiquitination may occur here without allowing cargo to escape its degradative fate.
Lipid sorting or segregation into the nascent ILV budding site to effect membrane curvature and/or to ensure that the ILV membrane is vulnerable to degradation by lysosomal hydrolases.
Membrane deformation and inward protrusion, followed by ILV fission.
Degradation of ILV membranes and encased proteins upon the delivery of contents to lysosomes.
The present challenge is to assign the known candidate machinery accurately to these various tasks and identify new machinery that executes the remaining tasks.
SUMMARY POINTS.
Multivesicular bodies (MVBs) are formed from early endosomes by the inward budding of the limiting membrane into the lumen. The resulting intralumenal vesicles (ILVs) can house proteins in temporary storage compartments or serve as a device to deliver the entire protein to the lysosome for degradation.
The major signal for sorting proteins into MVBs for degradation is ubiquitin (Ub), which is covalently attached to membrane protein cargo. Ub is then recognized by a host of protein sorting complexes that ultimately deliver cargo to the MVB lumen.
Candidate protein machinery that controls MVB formation has been identified by analysis of ILVs and through genetic screens in yeast. Some of this machinery is organized in discrete endosomal subdomains that serve as sites for the concentration of ubiquitinated cargo. Other subregions serve as sites for ILV budding. Exactly what machinery is used for protein sorting versus membrane deformation and vesicle formation is unclear. The functions of many of these candidate proteins have yet to be explored. Also, a comprehensive genetic approach to identifying all potential proteins involved in MVB biogenesis has yet to be undertaken.
Acknowledgments
We thank the following investigators for insightful discussions: Markus Babst, Rob Parton, Willem Stoorvogel, Judith Klumpermann, and Paul Luzio. This work was supported by RO1 GM73024 to DJK and by RO1 GM58202 to RCP.
- ILVs
intralumenal vesicles of multivesicular bodies
- MVB
multivesicular body
- Epidermal growth factor receptor (EGFR)
a receptor tyrosine kinase that undergoes ligand-stimulated activation, ubiquitination, internalization, and lysosomal degradation
- MIIC
endosomal compartments enriched in class II MHC protein complexes
- Tsg101
a protein encoded by tumor susceptibility gene found by screening for loss-of-function mutations that caused cells to transform
- ESCRTs (endosomal sorting complexes required for transport)
first described in yeast, they contain several of the class E Vps proteins involved in MVB formation
- GLUE (GRAM-like Ub binding on Eap45)
a divergent pleckstrin homology domain that binds Ub and phosphorylated phosphatidylinositols
- Hrs (hepatocyte growth factor receptor tyrosine kinase substrate)
homolog of yeast Vps27; both binds ubiquitin and localizes to endosomes
- STAM (signal-transducing adaptor molecule)
a binding partner for Hrs that may serve to recognize ubiquitinated cargo
- EAP (RNA polymerase II elongation factor (ELL)-associated proteins)
part of the ESCRT-II complex of mammalian cells
- UEV (ubiquitin E2–like variant domain)
has some homology to known ubiquitin E2 conjugating enzymes
- UIM
ubiquitin-interacting motif
- Doa4
found in a yeast genetic screen for defects in degradation of MAT alpha1; a deubiquitination enzyme required for the removal of ubiquitin from endocytic cargo proteins
- DUb
deubiquitinating enzyme or ubiquitin peptidase that cleaves ubiquitin from substrates attached to the C terminus of ubiquitin
- AMSH
an endosomal deubiquitination enzyme found as an associated molecule with the SH3 domain of STAM
- CHMPs (charged multivesicular body proteins)
originally identified as chromatin-modifying protein; form the extended ESCRT-III group of proteins
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
Contributor Information
Robert C. Piper, Email: robert-piper@uiowa.edu.
David J. Katzmann, Email: katzmann.david@mayo.edu.
LITERATURE CITED
- Abrami L, Lindsay M, Parton RG, Leppla SH, Van Der Goot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–51. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, et al. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–30. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–21. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CG, Takahashi RH, Gouras GK. Beta-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci. 2006;26:4277–88. doi: 10.1523/JNEUROSCI.5078-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–80. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–52. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I, Davies B, Dimaano C, Payne J, Eckert D, et al. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–17. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002a;3:271–82. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002b;3:283–89. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–58. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–93. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–42. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache KG, Stuffers S, Malerod L, Slagsvold T, Raiborg C, et al. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell. 2006;17:2513–23. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol. 2003;161:521–33. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–39. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Allaman MM, Surendhran K, Kearney WR, et al. The GAT domains of clathrin-associated GGA proteins have two ubiquitin binding motifs. J Biol Chem. 2004;279:54808–16. doi: 10.1074/jbc.M406654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–43. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–39. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276:11735–42. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- Blot V, Perugi F, Gay B, Prevost MC, Briant L, et al. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117:2357–67. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–35. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, et al. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281:5094–105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- Brissoni B, Agostini L, Kropf M, Martinon F, Swoboda V, et al. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr Biol. 2006;16:2265–70. doi: 10.1016/j.cub.2006.09.062. [DOI] [PubMed] [Google Scholar]
- Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood Cells Mol Dis. 2005;35:398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Charrin S, Manie S, Thiele C, Billard M, Gerlier D, et al. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol. 2003;33:2479–89. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- Chen H, Polo S, Di Fiore PP, De Camilli PV. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci USA. 2003;100:14908–13. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LS, Raynor MC, Wei X, Chen HQ, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276:7069–78. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- Chow AY, Mellman I. Old lysosomes, new tricks: MHC II dynamics in DCs. Trends Immunol. 2005;26:72–78. doi: 10.1016/j.it.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol Biol Cell. 2007;18:636–45. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaguillaumie A, Harriague J, Kohanna S, Bismuth G, Rubinstein E, et al. Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J Cell Sci. 2004;117:5269–82. doi: 10.1242/jcs.01380. [DOI] [PubMed] [Google Scholar]
- Doyotte A, Russell MR, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “Class E” compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–17. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- Dupre S, Volland C, Haguenauer-Tsapis R. Membrane transport: ubiquitylation in endosomal sorting. Curr Biol. 2001;11:R932–34. doi: 10.1016/s0960-9822(01)00558-9. [DOI] [PubMed] [Google Scholar]
- Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–27. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- Felder S, Miller K, Moehren G, Ullrich A, Schlessinger J, Hopkins CR. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 1990;61:623–34. doi: 10.1016/0092-8674(90)90474-s. [DOI] [PubMed] [Google Scholar]
- Futter CE, Collinson LM, Backer JM, Hopkins CR. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J Cell Biol. 2001;155:1251–64. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geminard C, De Gassart A, Blanc L, Vidal M. Degradation of AP2 during reticulocyte maturation enhances binding of hsc70 and Alix to a common site on TFR for sorting into exosomes. Traffic. 2004;5:181–93. doi: 10.1111/j.1600-0854.2004.0167.x. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, et al. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007;26:600–12. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–88. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden P, Carpentier JL, Cohen S, Orci L. Epidermal growth factor: morphological demonstration of binding, internalization, and lysosomal association in human fibroblasts. Proc Natl Acad Sci USA. 1978;75:5025–29. doi: 10.1073/pnas.75.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Maxfield F. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–63. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Gu F, Gruenberg J. Biogenesis of transport intermediates in the endocytic pathway. FEBS Lett. 1999;452:61–66. doi: 10.1016/s0014-5793(99)00561-x. [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–66. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Haigler HT, McKanna JA, Cohen S. Direct visualization of the binding and internalization of a ferritin conjugate of epidermal growth factor in human carcinoma cells A-431. J Cell Biol. 1979;81:382–95. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond DE, Carter S, McCullough J, Urbe S, Vande Woude G, Clague MJ. Endosomal dynamics of Met determine signaling output. Mol Biol Cell. 2003;14:1346–54. doi: 10.1091/mbc.E02-09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, et al. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164:1807–15. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–99. [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–32. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- Hirsch JG, Fedorko ME, Cohn ZA. Vesicle fusion and formation at the surface of pinocytic vacuoles in macrophages. J Cell Biol. 1968;38:629–32. doi: 10.1083/jcb.38.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–69. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Hornick CA, Hamilton RL, Spaziani E, Enders GH, Havel RJ. Isolation and characterization of multivesicular bodies from rat hepatocytes: an organelle distinct from secretory vesicles of the Golgi apparatus. J Cell Biol. 1985;100:1558–69. doi: 10.1083/jcb.100.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445:554–58. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa C, Morita E, Yamada M, Ishii N, Miura S, et al. Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem Biophys Res Commun. 2003;309:848–56. doi: 10.1016/j.bbrc.2003.08.078. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Imakagura H, Futatsumori M, Nakayama K. Recruitment of clathrin onto endosomes by the Tom1-Tollip complex. Biochem Biophys Res Commun. 2006;341:143–49. doi: 10.1016/j.bbrc.2005.12.156. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–55. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–23. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Beuchat MH, Chevallier J, Makino A, Mayran N, et al. Separation and characterization of late endosomal membrane domains. J Biol Chem. 2002;277:32157–64. doi: 10.1074/jbc.M202838200. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–97. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, et al. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell. 2000;11:1829–43. doi: 10.1091/mbc.11.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphin-golipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 1999;13:1475–85. doi: 10.1101/gad.13.11.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullas AL, Li M, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell. 2004;3:1609–18. doi: 10.1128/EC.3.6.1609-1618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I, Luyet PP, Pons V, Ferguson C, Emans N, et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7:653–64. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liao J, Ruland J, Mak TW, Cohen SN. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc Natl Acad Sci USA. 2001;98:1619–24. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–69. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–54. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci USA. 2006;103:6202–7. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci USA. 2003;100:7626–31. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166:717–29. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YM, Boucrot E, Villen J, Affar EB, Gygi SP, et al. Targeting of AMSH to endosomes is required for EGF receptor degradation. J Biol Chem. 2007;282:9805–12. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003;5:709–22. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Mercanti V, Cornillon S, Alibaud L, Charette SJ, Cosson P. Formation of multivesicular endosomes in Dictyostelium. J Cell Sci. 2004;117:6053–59. doi: 10.1242/jcs.01524. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–34. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Mayer RJ, Laszlo L, Middleton A, Landon M, Hope J, Lowe J. Ubiquitin, lysosomes and neurodegenerative diseases. Biochem Soc Trans. 1992;20:645–48. doi: 10.1042/bst0200645. [DOI] [PubMed] [Google Scholar]
- Mayor S, Presley JF, Maxfield FR. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J Cell Biol. 1993;121:1257–69. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, McKittrick I, West M, Odorizzi G. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol Biol Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E, Kobayashi K, Yamamoto A, Kitamura N, Komada M. A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic. 2006;7:1017–31. doi: 10.1111/j.1600-0854.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- Mobius W, van Donselaar E, Ohno-Iwashita Y, Shimada Y, Heijnen HF, et al. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–31. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Murk JL, Humbel BM, Ziese U, Griffith JM, Posthuma G, et al. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc Natl Acad Sci USA. 2003;100:13332–37. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–30. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Myromslien FD, Grovdal LM, Raiborg C, Stenmark H, Madshus IH, Stang E. Both clathrin-positive and -negative coats are involved in endosomal sorting of the EGF receptor. Exp Cell Res. 2006;312:3036–48. doi: 10.1016/j.yexcr.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–32. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns3P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–58. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, et al. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007a;18:707–20. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich AJ, Davies BA, Payne JA, Katzmann DJ. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol Biol Cell. 2007b;18:646–57. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955;1:59–68. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–78. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–17. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R. Interactions of TOM1L1 with the multivesicular body sorting machinery. J Biol Chem. 2005;280:9258–64. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–51. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–98. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–55. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci. 2006;119:2414–24. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- Raposo G, Tenza D, Mecheri S, Peronet R, Bonnerot C, Desaymard C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol Biol Cell. 1997;8:2631–45. doi: 10.1091/mbc.8.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala SK, Hollander P, Balasenthil S, Molli PR, Bean AJ, et al. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) interacts with PELP1 and activates MAPK. J Biol Chem. 2006;281:4395–403. doi: 10.1074/jbc.M510368200. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17:3469–83. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–86. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Kee Y, Huibregtse JM, Piper RC. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol Biol Cell. 2007;18:324–35. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–99. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca A, Lamaze C, Subtil A, Dautry-Varsat A. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor beta chain to late endocytic compartments. Mol Biol Cell. 2001;12:1293–301. doi: 10.1091/mbc.12.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–24. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Sachse M, Strous GJ, Klumperman J. ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J Cell Sci. 2004;117:1699–708. doi: 10.1242/jcs.00998. [DOI] [PubMed] [Google Scholar]
- Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–28. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–69. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci USA. 2005b;102:13813–18. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov EA, Moghrabi N, Kuhn M, Kramer H. A mutation in dVps28 reveals a link between a subunit of the endosomal sorting complex required for transport-I complex and the actin cytoskeleton in Drosophila. Mol Biol Cell. 2005;16:2301–12. doi: 10.1091/mbc.E04-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10982–90. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shin JS, Ebersold M, Pypaert M, Delamarre L, Hartley A, Mellman I. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature. 2006;444:115–18. doi: 10.1038/nature05261. [DOI] [PubMed] [Google Scholar]
- Silvie O, Charrin S, Billard M, Franetich JF, Clark KL, et al. Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J Cell Sci. 2006;119:1992–2002. doi: 10.1242/jcs.02911. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and nonendosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–26. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Sotelo JR, Porter KR. An electron microscope study of the rat ovum. J Biophys Biochem Cytol. 1959;5:327–42. doi: 10.1083/jcb.5.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2006;89:205–12. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Sun W, Yan Q, Vida TA, Bean AJ. Hrs regulates early endosome fusion by inhibiting formation of an endosomal SNARE complex. J Cell Biol. 2003;162:125–37. doi: 10.1083/jcb.200302083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–89. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004a;7:559–69. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J Biol Chem. 2004b;279:28689–96. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, et al. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev Cell. 2006;10:343–54. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–46. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic. 2001;2:622–30. doi: 10.1034/j.1600-0854.2001.20905.x. [DOI] [PubMed] [Google Scholar]
- van Deurs B, Holm PK, Kayser L, Sandvig K, Hansen SH. Multivesicular bodies in HEp-2 cells are maturing endosomes. Eur J Cell Biol. 1993;61:208–24. [PubMed] [Google Scholar]
- van Kerkhof P, Alves dos Santos CM, Sachse M, Klumperman J, Bu G, Strous GJ. Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol Biol Cell. 2001;12:2556–66. doi: 10.1091/mbc.12.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem (Tokyo) 2006a;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- van Niel G, Wubbolts R, Ten Broeke T, Buschow SI, Ossendorp FA, et al. Dendritic cells regulate exposure of MHC class II at their plasma membrane by oligoubiquitination. Immunity. 2006b;25:885–94. doi: 10.1016/j.immuni.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J Cell Sci. 1997;110:1867–77. doi: 10.1242/jcs.110.16.1867. [DOI] [PubMed] [Google Scholar]
- Villadangos JA, Schnorrer P, Wilson NS. Control of MHC class II antigen presentation in dendritic cells: a balance between creative and destructive forces. Immunol Rev. 2005;207:191–205. doi: 10.1111/j.0105-2896.2005.00317.x. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, et al. The protein network of HIV budding. Cell. 2003;114:701–13. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhang Y, Li H, Chieu HK, Munn AL, Yang H. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO J. 2005;24:2989–99. doi: 10.1038/sj.emboj.7600764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang L, Huang HW. Evidence of Cholesterol Accumulated in High Curvature Regions: Implication to the Curvature Elastic Energy for Lipid Mixtures. Biophys J. 2007;92:2819–30. doi: 10.1529/biophysj.106.097923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter V, Hauser MT. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006;11:115–23. doi: 10.1016/j.tplants.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8:1246–54. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- Wrigley JD, Ahmed T, Nevett CL, Findlay JB. Peripherin/rds influences membrane vesicle morphology. Implications for retinopathies. J Biol Chem. 2000;275:13191–94. doi: 10.1074/jbc.c900853199. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–72. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–42. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–37. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16:2470–82. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Coppens I, Wormsley S, Baevova P, Hoppe HC, Joiner KA. The Plasmodium falciparum Vps4 homolog mediates multivesicular body formation. J Cell Sci. 2004;117:3831–38. doi: 10.1242/jcs.01237. [DOI] [PubMed] [Google Scholar]
- Yogosawa S, Kawasaki M, Wakatsuki S, Kominami E, Shiba Y, et al. Monoubiquitylation of GGA3 by hVPS18 regulates its ubiquitin-binding ability. Biochem Biophys Res Commun. 2006;350:82–90. doi: 10.1016/j.bbrc.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Yoon Kim B, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18:1129–42. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, et al. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell. 2000;11:747–63. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]