SUMMARY
We have developed the first red fluorescent protein, named rsTagRFP, which possesses reversibly photoswitchable absorbance spectra. Illumination with blue and yellow light switches rsTagRFP into a red fluorescent state (ON state) or non-fluorescent state (OFF state), respectively. The ON and OFF states exhibit absorbance maxima at 567 and 440 nm, respectively. Due to the photoswitchable absorbance, rsTagRFP can be used as an acceptor for a photochromic Förster resonance energy transfer (pcFRET). The photochromic acceptor facilitates determination of a protein-protein interaction by providing an internal control for FRET. Using pcFRET with EYFP as a donor we observed an interaction between epidermal growth factor receptor and growth factor receptor-binding protein 2 in live cells by detecting the modulation of both the fluorescence intensity and lifetime of the EYFP donor upon the ON-OFF photoswitching of the rsTagRFP acceptor.
INTRODUCTION
Green fluorescent protein (GFP) from Aequorea victoria jellyfish, its mutants and homologues attract a attention due to their usefulness as genetically encoded probes for in vivo labeling (Nienhaus and Wiedenmann, 2009). A unique property of GFP-like proteins is their ability to form a chromophore within the β-barrel secondary structure without any assistance of external enzymes and cofactors other than molecular oxygen. Beside common fluorescent proteins (FPs) of various colors, there are so called photoactivatable FPs, both found in nature and engineered, that are capable of dramatic fluorescence increase in response to specific light illumination (Lukyanov et al., 2005). In particular, several reversibly photoswitchable FPs (rsFPs) have been recently developed. They include cyan mTFP0.7 (Henderson et al., 2007), green Dronpa (Ando et al., 2004) and its derivatives (Ando et al., 2007; Stiel et al., 2007; Andresen et al., 2008), red asFP595 (Lukyanov et al., 2000) and its KFP derivative (Chudakov et al., 2003), rsCherry and rsCherryRev (Stiel et al., 2008), and green-to-red Iris-FP (Adam et al., 2008). These proteins display reversible photoswitching between fluorescent ON state and non-fluorescent OFF state. All rsFPs share a similar photoswitching mechanism that involves cis-trans isomerizaition of the chromophores, accompanied by a change in the protonation state and chromophore flexibility, a loss of planarity, an interaction with protein β-barrel, or a combination of any of the above (Lukyanov et al., 2005; Adam et al., 2008; Stiel et al., 2008; Nienhaus and Wiedenmann, 2009). The increased flexibility of the chromophore and the deviations in its planarity in the OFF state lead to a nonradiative decay process and to a reduced fluorescence quantum yield.
Förster resonance energy transfer (FRET) microscopy that employs genetically encoded cyan and yellow FPs has become a widely used technique to study protein–protein interactions in cells (Ciruela, 2008). FRET pairs that include red FPs as acceptors have appeared in recent years (Goedhart et al., 2007; van der Krogt et al., 2008). The accurate determination of FRET, however, is difficult and can be accompanied by artifacts (Piston and Kremers, 2007). There are several approaches proved to be useful in detecting intracellular FRET, including two- or three-channel ratiometric imaging corrected for excitation and emission crosstalk, acceptor photobleaching, fluorescence lifetime imaging microscopy (FLIM), spectral imaging, and fluorescence polarization imaging (Jares-Erijman and Jovin, 2006). Each of these approaches has some limitations. For example, ratiometric imaging is the simplest method, but it requires reference images from separate control cells. While for sensors with a 1:1 donor:acceptor ratio the ratiometric FRET changes can be detected with a dual-ratio imaging, a three-channel FRET imaging with several corrections is required for protein-protein interactions with variable donor to acceptor stochiometries. The acceptor photobleaching approach is phototoxic for cells, does not allow continuous monitoring of FRET, and reversible photoswitching effects should be taken into account (Sinnecker et al., 2005). FLIM, as well as spectral and fluorescence polarization FRET imaging, all require additional instrumentation. Moreover, the latter two provide a reduced signal-to-noise ratio.
The new approach for accurate intracellular measurements of FRET efficiency is based on photochromic FRET (pcFRET) (Giordano et al., 2002; Mao et al., 2008). In this technique the fluorescence emission of a donor is modulated by reversible changes of the absorbance spectrum of the photochromic acceptor. In other words, the donor fluorescence can be measured in the “presence” (a large overlap between donor emission and acceptor absorbance spectra) and in the “absence” (a small or no overlap) of the acceptor. The advantage of pcFRET over the other FRET assays is that it provides an accurate and repeated FRET quantification for the same FRET pair within the same live cell without the need for corrections based on reference images acquired from separate control cells. Up to date, however, pcFRET has been demonstrated for photoswitchable chemical dyes only (Mao et al., 2008).
Monomeric rsFPs characterized to have reversible changes in absorbance spectrum are currently limited to cyan mTFP0.7 and green Dronpa variants, so there is restricted selection of potential FRET donors. The best characterized monomeric rsFPs in the red region, rsCherryRev and rsCherry, have limited brightness, low contrast, and complex switching behavior, and change the quantum yield rather than the absorbance spectrum upon photoswitching (Stiel et al., 2008; Subach et al., 2009). Therefore, a monomeric rsFP with photoswitching from dark to red states, with high brightness and contrast, and different absorbance spectra in ON and OFF states is very desirable.
Here we applied rational and random mutagenesis to monomeric red FP, TagRFP (Merzlyak et al., 2007), in order to develop its reversibly photoswitchable variant, named rsTagRFP, suitable for pcFRET applications. We demonstrate reversible pcFRET for the EYFP-rsTagRFP pair in a model system and further apply this approach to visualize an interaction between epidermal growth factor receptor (EGFR) and growth factor receptor-binding protein 2 (Grb2) in live mammalian cells.
RESULTS
Development of the reversibly photoswitchable red fluorescent protein
Recently reported TagRFP (Merzlyak et al., 2007) and its more photostable derivative TagRFP-T (Shaner et al., 2008) are among the brightest currently available monomeric red fluorescent proteins. This feature makes TagRFP an attractive template for designing reversibly photoswitchable red FPs. It has been shown that amino acids at positions 148, 165 and 203 (numbering follows the alignment with EGFP, see Figure S1) around the chromophore-forming Tyr66 are important for cis-trans chromophore isomerization in asFP595, its derivatives (Chudakov et al., 2003; Quillin et al., 2005), and rsCherryRev (Stiel et al., 2008). We performed saturating mutagenesis of the TagRFP gene at these positions and screened bacterial colonies of the resulting library of mutants using bright-field and fluorescence stereomicroscope. First, we marked colonies that demonstrated high red fluorescence and photoswitching contrast during repeated cycles of illumination with 445/25 nm and 592/21 nm arrays of high-power light-emitting diodes. From those we selected the colonies that maximally changed their coloration in daylight, from pink to yellow, due to absorbance spectrum change after photoswitching. We first found two mutants, TagRFP/N148A/S165G and TagRFP/N148T/S165G/H203I, which exhibited reversible photoswitching and absorbance changes but had dim red fluorescence in the ON state and rather low switching contrast. To improve these properties we further subjected these mutants to several rounds of random mutagenesis.
Bacterial libraries consisting of up to 7×107 clones were subjected to screening using a fluorescence activated cell sorter (FACS) both in the ON and OFF states, followed by selection of appropriate clones on Petri dishes as described above. After each round, 25–30 clones were analyzed by sequencing. A mixture of enhanced variants was then used as a template for the next round of molecular evolution. After several rounds of random mutagenesis, we found a TagRFP/F84W/I125L/N148A/S165G/F181L/K189N/Y200F variant, which was named rsTagRFP (Figure S1).
Characterization of the purified rsTagRFP
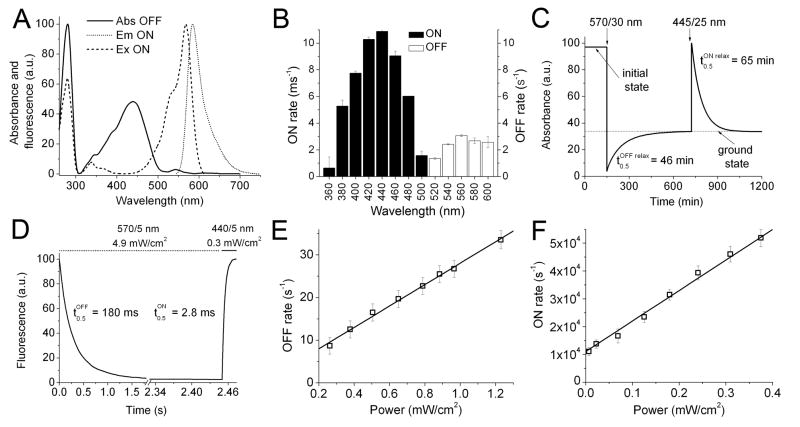
We further characterized the rsTagRFP purified protein and compared its properties with those of rsCherryRev, mCherry and parental TagRFP. Illumination with 445/25 nm light switched rsTagRFP into the ON state with absorbance/emission peaks at 567/585 nm (Figure 1A and Table 1). Illumination with 570/30 nm light switched rsTagRFP into the OFF state with a new absorbance maximum at 440 nm. Interestingly, the 440 nm absorbing form in the OFF state had fluorescence emission at 585 nm; however, its brightness was more than 200-fold lower than that of the 567 nm absorbing form in the ON state (Table 1 and Figure S2A). The fluorescence intensity of the 567 nm absorbing form in the OFF state dropped 20-fold comparing to the ON state. Changes of the ON and OFF absorbance bands upon rsTagRFP photoswitching, calculated as a change of integrated absorbance in 567/100 (for the ON state) and 440/100 nm (for the OFF state) spectral ranges, were 22-fold and 3.4-fold, respectively. Fluorescence contrast between the ON and OFF states was 20-fold, that is similar to the absorbance change for the 567 nm band. It is consistent with the similar quantum yields of the 567 absorbing form in the ON and OFF states (Table 1). In contrast, rsCherryRev had the same absorbance spectra in both the ON and OFF states and the maximally achievable red fluorescence contrast was 3-fold (Table 1). Molar extinction coefficient of rsTagRFP in the ON state measured at pH 7.5 was 36,800 M−1cm−1, and quantum yield was 0.11 (Figure S2B,C). The molecular brightness of rsTagRFP was 18 times greater than that of rsCherryRev and consisted of 24% of that of mCherry. Using photoswitching light intensities sufficient to achieve 20-fold contrast of rsTagRFP, the photostability of rsTagRFP was similar to that of rsCherryRev (Figures S3A,B), which only exhibited 1.5-fold contrast. Increasing the rsCherryRev photoswitching contrast to 3-fold with the larger durations of switching OFF light, however, resulted in the substantially higher photobleaching rate of rsCherryRev (Figure S3C).
Figure 1.
Spectral and photochromic properties of the purified rsTagRFP protein. (A) Absorbance spectrum after irradiation with 570/30 nm light (OFF state; solid line), excitation (dashed line) and emission (dotted line) spectra after irradiation with 445/25 nm light (ON state). (B) Initial rates of switching into ON and OFF states achieved at different wavelengths. The ON and OFF rates were corrected to the power of light. (C) Relaxation kinetics from OFF and ON states in the darkness. After purification, the protein (assumed as an initial state) was switched OFF with 570/30 nm light (82 mW/cm2). After the protein reached a ground state in the darkness, it was switched ON with 445/25 nm light (180 mW/cm2). The absorbance in ON state was normalized to 100%. (D) Time courses of the rsTagRFP red fluorescence during switching OFF and ON with 570/5 (4.9 mW/cm2) and 440/5 nm (0.3 mW/cm2) light, respectively. The fluorescence in the ON state was normalized to 100%. (E, F) Dependencies of the initial rates of switching OFF (E) and ON (F) from the power of 570/30 and 436/20 nm light, respectively. Power density was measured at the back focal plane of a 20× objective lens. See also Figure S1.
Table 1.
Spectral and photochemical characteristics of rsTagRFP in comparison with other relevant red fluorescent proteins.
| Protein | rsTagRFP | rsCherryRev | mCherry | ||||
|---|---|---|---|---|---|---|---|
| State | OFF | ON | OFF | ON | |||
| Absorbance maximum, nm | 440 (major) | 567 (minor) | 440 (minor) | 567 (major) | 572 | 572 | 587 |
| Change of absorbance (fold) and its direction a | 3.4 (increase) | 22 (decrease) | 3.4 (decrease) | 22 (increase) | none | none | |
| Extinction coefficient, M−1cm−1 b | 15,300 | 1,600 | 4,500 | 36,800 | 41,800 | 42,300 | 78,000 |
| Emission maximum, nm | 585 | 585 | 585 | 585 | 608 | 608 | 610 |
| Quantum yield c | 0.0013 | 0.11 | 0.0049 | 0.11 | 0.0017 | 0.0051 | 0.22 |
| Brightness relative to TagRFP, % | 0.12 | 1.0 | 0.13 | 24 | 0.41 | 1.3 | 100 |
| Observed fluorescence contrast (fold) d | 20 | 3 | none | ||||
| pKa | 6.6 | 5.5 | 4.6 | ||||
| Maturation half-time at 37°C (min) | 43 | 42 | 15 | ||||
All characteristics were measured in our lab using the same conditions and buffers except for those of mCherry, which were taken from the original paper (Shaner et al., 2004). The characteristics for the ON and OFF states of rsTagRFP and rsCherryRev were determined at the maximally and minimally achievable levels of the red fluorescence for the ON and OFF states, respectively, during the first switching cycle for rsTagRFP or after several cycles for rsCherryRev; rsTagRFP was cyclically irradiated with 445/25 nm light (180 mW/cm2 for 30 s) and 570/30 nm light (82 mW/cm2 for 2 min); rsCherryRev was cyclically irradiated with 436/20 nm (at 16 mW/cm2 for 0.5 s) and 570/30 nm (at 82 mW/cm2 for 12 s) light. Power density was measured at the back focal plane of a 20× objective lens for rsTagRFP or a 100× oil objective lens for rsCherryRev. Brightness is a product of the extinction coefficient and quantum yield.
Change of absorbance was integrated in ±50 nm range from the maximum.
Extinction coefficients of the rsTagRFP and rsCherryRev chromophore absorbance peaks were calculated using the absorbance at 280 nm determined for the respective proteins (31,700 and 35,900 M−1cm−1, respectively) as described (Pace et al., 1995).
Quantum yield was determined with 540 nm excitation using a values of 0.48 and 0.22 for quantum yields of parental TagRFP (Merzlyak et al., 2007) and mCherry (Shaner et al., 2004) as the references.
Fluorescence contrast is the maximal contrast observed in experiment consisting of the cycling irradiation of rsTagRFP with the 436/20 nm (at 16 mW/cm2 for 0.6 s) and 570/30 nm (at 82 mW/cm2 for 0.5 s) light; power density was measured at the back focal plane of a 60× oil objective lens; the red fluorescence was detected using 570/30 and 615/40 nm filters. ND, not determined.
rsTagRFP was switched ON and OFF with light in the spectral regions of 360–500 and 520–600 nm, respectively (Figure 1B). The maximal switching ON or OFF rates were observed at 440 and 560 nm. These wavelengths coincided with the absorbance maxima of rsTagRFP in the ON and OFF states. The majority of the newly synthesized rsTagRFP molecules (~97%) were found in the ON state (Figure 1C). After a single OFF-ON switching cycle the rsTagRFP sample relaxed to a ground state, having about one third of molecules in the ON state at room temperature. The OFF and ON states were thermally stable in the dark with the relaxation half-times to the ground state of 46 and 65 min, respectively (Figure 1C).
The half-times of the switching rsTagRFP OFF and ON with 570/5 nm (4.9 mW/cm2) and 440/5 nm (0.3 mW/cm2) light were 180 ms and 2.8 ms, respectively (Figure 1D). Taking into account the difference in the light power, switching ON was ~1000-fold faster than switching OFF. The initial rates of switching OFF and ON both displayed linear dependencies on the light powers (Figures 1E,F), suggesting the one-photon photoconversion process. Red fluorescence reversibly switched ON and OFF many times.
The light power densities required for the rsTagRFP photoswitching are comparable to those for Dronpa (Ando et al., 2004), rsCherry and rsCherryRev (Table S1). Recently, a photoconversion of several conventional FPs, including mKate, has been demonstrated (Kremers et al., 2009). However, it required substantially greater light powers and illumination durations (for example, see the relative photoswitching characteristics of mKate in Table S1).
rsTagRFP had a maturation half-time of 43 min at 37°C, that was similar to the maturation half-time of rsCherryRev and 1.5-fold smaller than the maturation half-time of TagRFP (Table 1 and Figure 2A). The apparent pKa for rsTagRFP was 6.6 (Table 1 and Figure 2B). The absorbance spectrum of rsTagRFP at physiological pH of 7.0–8.0 displayed a major peak at 567 nm (Figure 2C). This absorbance at 567 nm gradually decreased with acidification below pH 7.0, and the absorbance peak at 440 nm dominated at pH of 4.0–5.0. These two peaks possibly correspond to anionic and neutral forms of phenolic hydroxyl of the red chromophore (Yampolsky et al., 2008). The photoswitching fluorescence contrast of rsTagRFP also decreased with acidification (Figure 2D) that can hinder pcFRET application of the protein in acidic compartments such as lysosomes. The pH modulation of the rsTagRFP contrast, however, could be applied to measure intracellular pH.
Figure 2.
Biochemical properties of the purified rsTagRFP protein. (A) Maturation kinetics for rsTagRFP compared to that of rsCherryRev and parental TagRFP at 37°C. (B) Equilibrium pH dependence of the rsTagRFP fluorescence in the ON state. (C) Equilibrium pH dependence of the rsTagRFP absorbance spectra. (D) The pH dependence of the photoswitching contrast of rsTagRFP. The fluorescence contrast was determined using a cycling irradiation of rsTagRFP with 436/20 nm (at 16 mW/cm2 for 0.6 s) and 570/30 nm (at 82 mW/cm2 for 0.5 s) light. The power densities were measured at the back focal plane of a 60× oil objective lens.
Application of rsTagRFP to the photochromic FRET
To test rsTagRFP performance as a pcFRET acceptor, we constructed a rsTagRFP-EYFP fusion protein (Figure 3A). EYFP was chosen as donor because of an efficient overlap between the EYFP emission and rsTagRFP absorbance in the ON state (Figure 3B) and expected minimal effect of EYFP excitation light on the rsTagRFP states. Hence, when rsTagRFP is switched ON with 436/20 nm light, there is a an efficient FRET from the EYFP donor. In contrast, when rsTagRFP is switched OFF with 570/30 nm light, the overlap between the EYFP emission and rsTagRFP absorbance disappears and, consequently, FRET does not occur. We observed multiple cycles of reversible quenching of EYFP fluorescence as the result of the rsTagRFP photoswitching both in the purified rsTagRFP-EYFP fusion protein (Figure 3C) and in bacterial colonies expressing the fusion (Movie S1). The reversible quenching of EYFP fluorescence constituted 19–20%. Leakage of rsTagRFP fluorescence into the EYFP emission channel (535/40 nm) was negligible and did not affect the degree of the quenching. Protease cleavage of the EYFP-rsTagRFP fusion protein led to disappearance of the modulation in EYFP emission channel confirming that this effect is dependent on pcFRET (Figure 3D). EYFP alone did not demonstrate any photochromism using the same photoswitching conditions (Figure 3E). Because rsCherryRev does not change its absorbance spectrum during ON-OFF switching, we did not observe modulation in EYFP quenching upon switching the rsCherryRev-EYFP fusion protein (Figure 3F).
Figure 3.
Characterization of the photochromic FRET using fusion proteins. (A) Schematic representation of an energy transfer from EYFP donor to rsTagRFP acceptor in the ON (rsTagRFP is red) and OFF (rsTagRFP is blue) states in the EYFP-rsTagRFP fusion. (B) Overlap between the absorbance spectra of rsTagRFP in the ON and OFF states and the fluorescence emission spectrum of EYFP. (C–F) Cycling irradiation of the EYFP-rsTagRFP fusion (C), the EYFP-rsTagRFP fusion after cleavage resulting in a physical separation of the proteins (D), EYFP alone (E), and the EYFP-rsCherryRev fusion (F) using the 436/20 nm (at 16 mW/cm2 for 0.6 s) and 570/30 nm (at 82 mW/cm2 for 0.5 s) light. The power densities were measured at the back focal plane of a 60× oil objective lens. The EYFP fluorescence was corrected for the leakage of rsTagRFP signal into the EYFP emission channel (480/40 nm). The fluorescence intensities were normalized to those observed in the first switching cycle for rsTagRFP in the ON state. See also Figure S3 and Movie S1.
Detection of the protein-protein interaction in live cells
To demonstrate the utility of rsTagRFP as a switchable pcFRET acceptor, we next studied the interaction between EGFR and Grb2 that is induced upon stimulation with epidermal growth factor (EGF) in mammalian cells. It has been shown that EGF-activated EGFR interacts with Grb2 at plasma membrane and in endosomes (Sorkin et al., 2000; Yamazaki et al., 2002).
We transiently co-expressed EGFR-EYFP and Grb2-rsTagRFP chimeric proteins in HeLa cells. Intracellular distributions of the chimeras in resting cells were different. As expected, the EGFR-EYFP signal mainly localized at the plasma membrane whereas the Grb2-rsTagRFP fluorescence was evenly distributed throughout the cell with somewhat higher accumulation in nuclei (Figure 4A). Addition of 100 ng/ml EGF to the medium led to a rapid accumulation of Grb2-rsTagRFP at the plasma membrane. Then both EGFR-EYFP and Grb2-rsTagRFP colocalized on endosomes, similar to previous observations with EGFR and Grb2 tagged with other FPs (Sorkin et al., 2000; Yamazaki et al., 2002). To induce intensity-based pcFRET, illumination conditions were chosen to minimize irreversible photobleaching of rsTagRFP during OFF-ON cycles (Figure 4B). This is particularly important for prolonged time series since acceptor lost can lead to an underestimate of the FRET efficiency. As expected, Grb2-rsTagRFP photoswitching did not affect the EGFR-EYFP emission in the absence of EGF (Figures 4B,C,D), confirming a lack of interaction between the two fusion proteins in unstimulated cells. In contrast, after EGF addition, Grb2-rsTagRFP photoswitching resulted in a striking and reversible ~20% modulation of the EGFR-EYFP fluorescence both at the plasma membrane and on endosomes, strongly suggesting the occurrence of pcFRET due to the EGFR-Grb2 interactions at these locations. Importantly, fluorescence intensity of the EGFR-EYFP donor in the EGF-stimulated cells returned to the pre-stimulated level upon turning the Grb2-rsTagRFP acceptor OFF (Figure 4B) demonstrating a nearly complete elimination of FRET between EYFP and rsTagRFP in the OFF state.
Figure 4.
Interaction between EGFR-EYFP and Grb2-rsTagRFP in live HeLa cells visualized using intensity-based pcFRET. (A) Epifluorescence images of cells in YFP (left) and TX2 (right) filter sets in the same field of view before and, at different time points, after addition of EGF. (B) Modulation of the total red (red line) and yellow (orange line) fluorescence intensities in the cells during the ON-OFF switching cycles of Grb2-rsTagRFP acceptor. The fluorescence intensities were normalized to those observed in the first switching cycle for rsTagRFP in ON state. (C, D) Changes of the EGFR-EYFP fluorescence in response to the Grb2-rsTagRFP switching. (C) Graph shows a relative increase of the EGFR-EYFP fluorescence intensity upon the Grb2-rsTagRFP switching OFF at various time points before and after EGF stimulation at time zero. To calculate these values, background-subtracted EGFR-EYFP fluorescence intensities in selected regions of interest at plasma membrane (time points −2, 2, and 4 min) or endosomes (time points >4 min) were compared for pairs of images taken when rsTagRFP was in ON or OFF states. (D) Pseudocolor images of EGFR-EYFP fluorescence (upper row: Grb2-rsTagRFP is ON; bottom row: Grb2-rsTagRFP is OFF) at various time points before and after EGF stimulation at time zero. See also Figure S4.
We also tested another pair of interacting proteins, IκBα and p65, from the NF-κB signaling module (Schmid et al., 2000; Hoffmann et al., 2006). IκBα-EYFP and rsTagRFP-p65 chimeric proteins were transiently co-expressed in HeLa cells and analyzed by fluorescence microscopy. As it was expected, photoconversion of rsTagRFP-p65 into the OFF state led to a substantial (6–7%) intensity increase of IκBα-EYFP yellow fluorescence (Figure S4). This reciprocal modulation of the donor and acceptor due to pcFRET can be performed repeatedly.
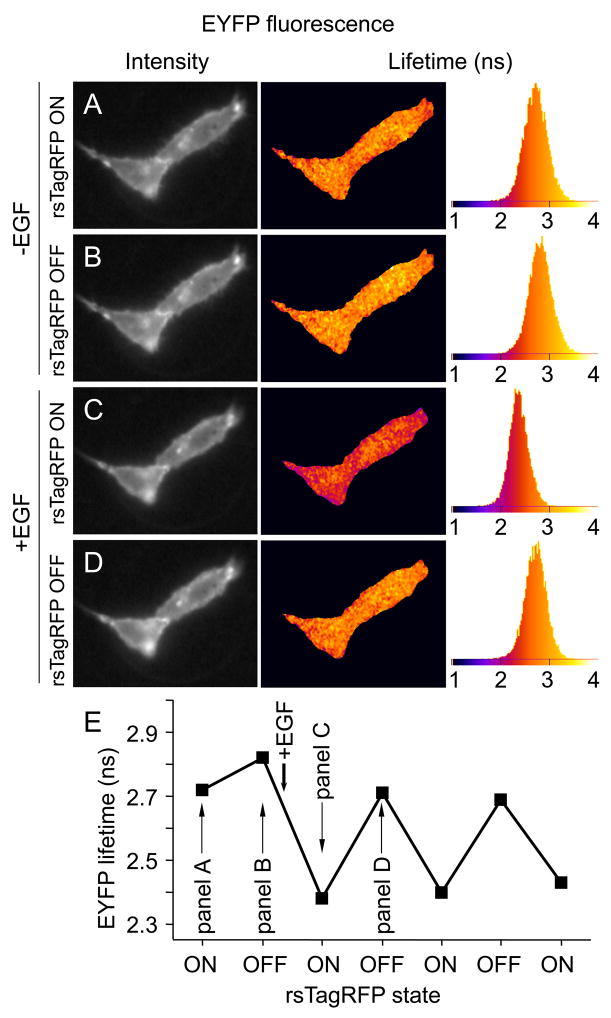
To provide additional evidence for the switchable pcFRET, we next measured the EGFR-EYFP and Grb2-rsTagRFP interaction in live cells, using the FLIM technique. Because fluorescence lifetime is proportional to fluorescence quantum yield, FLIM provides a direct measure for FRET independent of expression levels or excitation intensities (Wallrabe and Periasamy, 2005). However, for measuring EYFP lifetimes with rsTagRFP switched OFF, it proved necessary to perform dual 514 and 577 excitation (see Experimental procedures). To study the effect of dual excitation on the EYFP lifetime measurement, single transfected cells expressing EGFR-EYFP were examined. Upon single 514 nm excitation (97 μW), we measured an EYFP phase lifetime of 2.78±0.06 ns (n=4) when, and upon dual 514 nm (97 μW) and 577 nm (1.55 mW) excitation, the EYFP phase lifetime was 2.79±0.03 ns (n=3), demonstrating that dual excitation had no significant effect on the EYFP fluorescence lifetime. Upon co-expression of Grb2-rsTagRFP, without EGF, the lifetime of the EGFR-EYFP fluorescence was 2.71±0.14 ns with rsTagRFP switched OFF and 2.81 ±0.14 with rsTagRFP switched ON, likely reflecting slight FRET (<3%) in the absence of EGF and rsTagRFP switched on (Figure 5 and Movie S2). Stimulation with EGF, however, resulted in a substantial decrease of the EGFR-EYFP fluorescence lifetime to 2.40±0.03 ns (14% FRET) when Grb2-rsTagRFP was in the ON state (Figure 5). Cyclical photoswitching of rsTagRFP into the OFF state resulted in a repeatable increase in the donor lifetime to a value of 2.70±0.03 ns (3% FRET), approaching the control EYFP lifetime. Considering the (inverse) dependence of the latter residual FRET value on the intensity of co-excitation with 577 nm during the FLIM experiment, the residual FRET detected with rsTagRFP in the OFF state is due to incomplete OFF switching of rsTagRFP under the employed conditions.
Figure 5.
FLIM-based pcFRET of EGFR-EYFP and Grb2-rsTagRFP chimeras co-expressed in live HeLa cells. For FLIM measurements EGFR-EYFP was excited at 514 nm. First, the EYFP lifetime was measured in the absence of EGF, with rsTagRFP switched ON after a 2 s exposure using 470 nm light (panel A). Subsequently, the EYFP lifetime was measured still in the absence of EGF, but with rsTagRFP switched OFF after a 2 s exposure with 470 nm light (panel B). After addition of EGF, a substantial decrease in the EYFP fluorescence lifetime due to FRET could be detected with rsTagRFP switched ON (C), and the lifetime decrease was reversed after switching rsTagRFP OFF with 577 nm (D). ON-OFF switching was repeated in the presence of EGF. In AD from left to right the DC intensity image, the pseudocolored EYFP phase lifetime-image and lifetime-histogram with ns time scale are depicted. In E panel, the average over a whole cell EYFP phase lifetime is plotted for the seven FLIM experiments with approximately 2 min steps between the subsequent FLIM measurements. See also Movie S2.
DISCUSSION
The primary goal of this study was to develop a red FP with a reversibly switchable absorbance. Until now such rsFPs have been available in the cyan-green emission region only. Therefore, a red rsFP with red-shifted absorbance peak could be potentially very useful as an acceptor for the pcFRET. Our directed molecular evolution approach has resulted in the development of such an rsTagRFP, whose absorbance switches ON and OFF at 567 nm with cyan and yellow light, respectively.
A mutant with reversible switching phenotype and minimal number of mutations compared to TagRFP contained N148A and S165G substitutions. According to the previously solved crystal structure of mKate, a far-red derivative of TagRFP, the residues at the positions 148 and 165 form hydrogen bonds with the hydroxyl group of Y66 of the chromophore in cis- and trans-configuration, respectively (Pletnev et al., 2008). These hydrogen bonds appear to be important for stabilizing the cis- and trans-configuration of the chromophore. Both A148 and G165 are not capable of forming the hydrogen bonds with Y66 and, thus, allow the rsTagRFP chromophore to undergo the cis-trans isomerization. The different absorbance maxima during photoswitching are possibly caused by additional reversible chromophore protonation (OFF state) and deprotonation (ON state). The latter hypothesis is supported by the pH dependence of the absorbance spectrum (Figure 2C). Moreover, similar photoswitching mechanism have been described for other rsFPs including Dronpa (Ando et al., 2004; Mizuno et al., 2008), mTFP0.7 (Henderson et al., 2007) and asFP595 (Quillin et al., 2005). Five other substitutions in rsTagRFP such as F84W, I125L, F181L, K189N, and Y200F possibly improve the protein folding, fluorescence brightness, and photoswitching contrast.
As we have shown for the EYFP-rsTagRFP pair, two images of the same cell with rsTagRFP acceptor in the ON and OFF states are enough to detect and confirm the FRET signal. Another advantage of pcFRET approach is that it only requires a digital imaging widefield microscope or standard confocal microscope. The FRET measurements can be repeated many times to increase the accuracy of the FRET determination. The low intensity switching ON blue light and moderate intensity switching OFF yellow light appears to be non-phototoxic for live cells. A Förster radius for the EYFP-rsTagRFP pair with rsTagRFP in the ON state was estimated as described before (Galperin et al., 2004; Merzlyak et al., 2007) and equaled to 5.3 nm. This value is comparable to the Förster radii of 4.9 and 5.4 nm calculated for the popular ECFP-EYFP and mCerulean-mVenus FRET pairs (Rizzo et al., 2006), and enables detection of protein-protein interactions.
Multiple ligands, such as EGF, bind to and activate EGFR localized on the cell surface (Herbst, 2004). EGFR activation initiates intracellular signaling that involves activation of Ras proteins and also results in EGFR internalization by means of clathrin-dependent endocytosis (Sorkin and von Zastrow, 2009). Grb2 binds to the activated EGFR and acts as an adaptor to recruit the E3 ubiquitin ligase Casitas B lineage lymphoma protein to start internalization. EGFR remains ligand-bound, phosphorylated, and active until a late stage of its endosomal trafficking. pcFRET made it possible to monitor EGFR-Grb2 interaction in the same cells at different time points before and after EGF stimulation. No significant difference between the pcFRET efficiencies at the plasma membrane and in the endosomal compartment was observed (Figure 4C). We also found that EGF addition led to a maximal FRET efficiency in less than 2 min without substantial FRET changes during at least the next 30 min (Figures 4C,D). Lastly, our data confirm that the EGFR-Grb2 complexes last through the late stages of endocytosis. The EGFR-Grb2 interaction has been previously observed in the same compartments with a three-channel FRET imaging using the EGFR-EYFP and Grb2-ECFP fusion proteins (Sorkin et al., 2000).
Recently, several fluorescence microscopy techniques have been invented to overcome the diffraction barrier (reviewed in (Hell, 2009)). In particular, reversible saturable optically linear fluorescence transitions (RESOLFT) microscopy (Hofmann et al., 2005), PALM with independently running acquisition (PALMIRA) microscopy (Egner et al., 2007), and photoactivation-localization microscopy with two-color stroboscopic illumination (S-PALM) microscopy (Flors et al., 2007) employ fluorescent probes that can be reversibly photoswitched between a dark OFF state and a fluorescent ON state. Unlike a stimulated emission depletion (STED) microscopy (Hell and Wichmann, 1994), which requires very high power depletion light (>109 W/cm2), the RESOLFT, PALMIRA and S-PALM techniques achieve the subdifraction resolution using significantly lower light intensities. RESOLFT, PALMIRA and S-PALM have been demonstrated with rsFPs such as asFP595 (Hofmann et al., 2005), Dronpa and its derivatives (Dedecker et al., 2007; Flors et al., 2007; Geisler et al., 2007), and rsCherryRev (Stiel et al., 2008). Although beyond the scope of the current study, we believe that rsTagRFP possesses ideal properties to be used as additional red color in these super-resolution microscopy approaches.
SIGNIFICANCE
Compared to the available monomeric red rsFP, rsCherryRev, the rsTagRFP protein has a substantially larger brightness. Unlike rsCherryRev, rsTagRFP has the higher contrast that is independent of the switching cycle (Stiel et al., 2008). Moreover, the rsTagRFP photoconversion is accompanied by dramatic absorbance changes that have allowed for its application in pcFRET with EYFP as a donor. The rsTagRFP-EYFP pair demonstrated a high potential of the pcFRET imaging to study subcellular protein interactions. In future applications, rsTagRFP would enable monitoring of the transport of tagged proteins between different compartments in a single cell, as it has been shown for Dronpa (Ando et al., 2004) using a low power excitation light. Furthermore, a combination of rsTagRFP with Dronpa makes it possible to study multidirectional trafficking of two proteins in the same cell at various times and in different conditions.
EXPERIMENTAL PROCEDURES
Mutagenesis and screening of libraries
The TagRFP (Merzlyak et al., 2007) gene was amplified using polymerase chain reaction (PCR) as BglII-EcoRI fragments and inserted into pBAD/His-B vector (Invitrogen). Site specific mutagenesis of TagRFP was performed using QuickChange Mutagensis Kit (Stratagene). For simultaneous mutagenesis at several sites, including saturated mutagenesis at several positions, an overlap-extension approach was applied (Ho et al., 1989). The rsCherryRev gene (Stiel et al., 2008) was designed on the basis of mCherry (Shaner et al., 2004) using overlap extension. Random mutagenesis performed with GeneMorph II Random Mutagenesis Kit (Stratagene) resulted in the mutation frequency up to 16 mutations per 1,000 base pairs. After mutagenesis a mixture of the mutants was electroporated into LMG194 host cells (Invitrogen).
Typical mutant libraries consisted of about 106–107 independent clones. LMG194 cells were grown at 37°C overnight in a rich medium (RM) supplemented with Ampicillin. Protein expression in the libraries was induced overnight with 0.002% Arabinose. Next morning bacterial cells were washed with phosphate buffered saline (PBS) containing 1 mM EDTA and then diluted with PBS to optical density of 0.02 at 600 nm. Libraries were then photoswitched into the OFF state with an LED array of 36 light-emitting diodes illuminating at 592 nm with 21 nm peak half-width. LED array provided uniform 18 mW/cm2 light flux in area of about 10 cm in diameter at the distance of 5 cm from the array. Typically, photoswitching consisted of up to 30 min of exposure for bacterial libraries in suspension, while mixed on ice for fluorescence activated cell sorting (FACS) screening, or for colonies on dishes. Under these illumination conditions, the viability of bacteria was greater than 90%. After photoswitching into the OFF state the libraries were screened using MoFlo (Dako) cell sorter equipped with Argon, Krypton and Argon-Krypton mixed-gas lasers. Typically about 10 sizes of each library were sorted using 568 nm excitation line and 580 nm LP emission filter (Chroma). The collected non-red bacterial cells were pelletted down, resuspended in ice chilled PBS and photoswitched into the ON state with an LED array of 36 light-emitting diodes illuminating at 445 nm with 25 nm peak half-width. LED array provided uniform 180 mW/cm2 light flux in area of about 10 cm in diameter at the distance of 5 cm from the array. Typically, photoswitching consisted of up to 1 min of exposure for bacterial libraries in suspension, while mixed on ice for FACS screening, or for colonies on dishes. Under these illumination conditions, the viability of bacteria was greater than 90%. After photoswitching into the ON state, the brightest red fluorescent bacterial cells were collected using MoFlo cell sorter with the setting described above. The collected bacterial cells were rescued in SOC medium at 37°C for several hours, and then grown on LB/Ampicillin Petri dishes supplemented with 0.02% arabinose overnight at 37°C. The next day, the dishes were analyzed with Leica MZ16F fluorescence stereomicroscope equipped with custom 570/30 nm exciter and 615/40 nm emitter filters (Chroma) after repeated photoswitching with 445/25 and 592/21 nm LED arrays. Petri dishes were also photographed in daylight to select colonies that change their reflection color from pink to yellow during repeated photoswitching with 445/25 and 592/21 nm LED arrays. Approximately 1–2×102 of the brightest red clones with ability to be repeatedly photoconverted between differently absorbing ON and OFF states, having the highest contrast and minimal blue and green fluorescence, were further analyzed via bacterial streaks on new Petri dishes. Typically 20–40 best photoswitchable candidate clones were sequenced, purified and characterized in more detail.
Characterization of the purified proteins
The rsCherryRev, mCherry, TagRFP and rsTagRFP mutant proteins with polyhistidine tags were expressed in LMG194 bacterial cells grown in RM medium supplemented with 0.002% arabinose overnight at 37°C and then purified using Ni-NTA agarose (Qiagen). For spectroscopy, the photoswitching of purified proteins was performed with 445/25 nm LED array (180 mW/cm2) and using Olympus IX81 inverted microscope equipped with 200W metal-halide arc source, 20× objective lens, 570/30 nm excitation filter in 50 μl quartz microcuvette (Starna Cells) at room temperature The excitation and emission spectra of recombinant proteins were measured with FluoroMax-3 spectrofluorometer (Jobin Yvon). Hitachi U-3010 spectrophotometer was used for absorbance measurements.
To determine extinction coefficients, we relied on measuring absorbances at 280 nm (Pace et al., 1995). For determination of quantum yields, fluorescence intensities of photoswitchable variants were compared to those of two reference standards: parental TagRFP (quantum yield is 0.48 (Merzlyak et al., 2007)) and mCherry (quantum yield is 0.22 (Shaner et al., 2004)) (see Figure S2B,C). The measurements were performed at different protein concentrations using the same excitation wavelength. The quantum yield values were calculated according to the following equation: , where the subscripts ST and X denote the standard and tested samples, respectively; Φ is the fluorescence quantum yield; Grad is the gradient of the plot of integrated fluorescence intensity vs. absorbance; and η is the refractive index of the solvent. In order to minimize possible re-absorption effects, the absorbance of the samples did not exceed 0.1 at and above the excitation wavelength. The pH titrations were performed using a series of buffers (100 mM NaOAc, 300 mM NaCl for pH 2.5–5.0, and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–10.5).
Dependence of initial photoswitching ON and OFF rates on different light wavelengths was measured using purified protein at concentration of 2 mg/ml in aqueous (PBS) drops in oil, using Olympus IX81 inverted microscope equipped with monochromator (Jobin Yvon, Horiba), having 5 nm bandwidth, 150 W Xenon lamp, a 60× 1.35 NA oil objective lens, 570/30 nm excitation, 615/40 nm emission filters. All filters were from Chroma. The light from monochromator was directed to the light-guide using a 20× 0.5 NA objective (UPlanFL, Olympus). The initial photoswitching rate corresponded to the slope of the initial linear part on the progress curve. The ON and OFF rate values were corrected for the power of light measured with power meter 3A-P-SH-V1 (OPHIR) and normalized to the rate values at 440 and 560 nm, respectively.
Relaxation kinetics from the OFF and ON states in the darkness was measured in a cuvette with the protein according to absorbance change at 565 nm. Low intensity probing light of the Hitachi U-3010 spectrophotometer with slits 0.5 nm was used. After purification, the protein was assumed to be in the initial state and was switched into the OFF state by illumination with 570/30 nm light at the power density of 82 mW/cm2 at the back focal plane of a 20× objective lens. After the protein reached the ground state in the darkness, it was switched into the ON state by illumination with 445/25 nm LED array at the power density of 180 mW/cm2. Absorbance was normalized to the maximal one achieved after illumination with 445/25 nm LED array.
Time course of the red fluorescence change during switching into the OFF and ON states with 570/5 and 440/5 nm light respectively was measured using conditions similar to those for the relaxation kinetics measurements. The red fluorescence intensity of rsTagRFP switched into the ON state was normalized to the initial point being 100%.
Dependence of the initial rates of switching into the OFF and ON states on the power of 570/30 and 436/20 nm light, respectively, were measured using the purified protein at concentration of 2 mg/ml in aqueous (PBS) drops in oil using an Olympus IX-81 inverted microscope equipped with 200 W metal-halide lamp, a 60× 1.35 NA oil objective lens, 570/30 nm excitation, 615/40 nm emission filters and neutral density filters (2.9, 6, 11.8, 25, 21.4, 30.7, 41.1, 52.9, 71.8, and 78.5% transmission).
Cycling illumination of EYFP, rsCherryRev, rsTagRFP, rsCherryRev-EYFP, and rsTagRFP-EYFP proteins was performed on Olympus IX-81 inverted microscope equipped with 200 W metal-halide lamp and 60× oil objective lens. The switching cycle included repeated illumination of drops with 436/20 and 570/30 nm light flashes for 0.6 and 0.5 s, respectively. The power densities at the back focal plane of an objective lens were 16 and 82 mW/cm2 for the 436/20 and 570/30 filters, respectively. The rsTagRFP and EYFP fluorescence was detected with red (ex. 570/30, em. 615/30) and green (ex. 480/40, em. 535/40) filter sets, respectively. EYFP fluorescence was either corrected or non-corrected for the leakage of rsTagRFP or rsCherryRev signal into the EYFP channel. To physically separate fluorescent proteins, the purified rsTagRFP-EYFP construct was cleaved with 0.2 mg/ml of trypsin for 5 h at 37°C.
To study protein maturation, LMG194 bacterial cells were grown at 37°C overnight in RM medium supplemented with Ampicillin. Next morning cells were diluted to optical density 1.0 at 600 nm, and 0.2% Arabinose was added. Upon induction of protein expression the bacterial cultures were grown at 37°C in 50 ml tubes filled to the brim and tightly sealed to restrict oxygen availability. After 2 hours the cultures were centrifuged in the same tightly closed tubes. After opening the tubes, the proteins were purified using Ni-NTA resin within 30 min of all procedures and buffers at or below 4°C. Protein maturation occurred in PBS at 37°C; and red fluorescence of the protein was measured using the spectrofluorometer.
Mammalian plasmids and cell culture
prsTagRFP-N1 and prsTagRFP-C1 plasmids, which are analogous to pEGFP-N1 and pEGFP-C1 (Clontech), were constructed. To generate pEGFR-EYFP plasmid, the SacII-BsrGI fragment containing EYFP gene was cut out from pEYFP-N1 vector (Clontech) and swapped with mRFP1 in the pEGFR-mRFP1 plasmid (Galperin et al., 2004). To construct pGrb2-rsTagRFP plasmid, the BamHI-HindIII fragment containing Grb2 gene was cut out of pGrb2-EYFP vector (kindly provided by A. Sorkin) and inserted into the respective sites of the prsTagRFP-N1 plasmid. Plasmid prsTagRFP-p65 was constructed by cloning p65 coding sequence into prsTagRFP-C1 using HindIII and BamHI sites analogously to the pEGFP-p65 plasmid described in (Schmid et al., 2000). To generate pIκBα-EYFP plasmid, the AgeI-NotI fragment containing EYFP gene was cut out from pEYFP-N1 vector (Clontech) and swapped with Dendra2 in the pIκBα-Dendra2 plasmid (Zhang et al., 2007).
HeLa cells were transfected with vectors, encoding target fusions with fluorescent proteins, using FuGene reagent (Roche) and grown in DMEM (Invitrogen) supplemented with 600 mg/l L-glutamine, 50,000 units/l of penicillin, 50 mg/l streptomycin and 10% fetal bovine serum (Sigma) at 37°C in a 5% CO2 atmosphere in glass-bottom dishes. Live cells were imaged at 37°C 24 h after transfection.
Microscopy of mammalian cells
For wide field fluorescence microscopy, a Leica AF6000 LX imaging system based on a DMI 6000B inverted microscope equipped with an HCX PL APO BL 63× 1.4 NA oil objective lens, a Photometrics CoolSNAP HQ CCD camera, and a 120W HXP short arc lamp (Osram) light source was used. A YFP filter set (ex. BP504/12, em. BP542/27) was used to detect EYFP fluorescence; a GFP filter set (ex. BP470/40, em. BP525/50) was used to turn ON rsTagRFP; a TX2 filter set (ex. BP560/40, em. BP645/75) was used to visualize and turn OFF rsTagRFP. Binning 4×4 was used to minimize exposure (and thus undesirable rsTagRFP switching ON) in the YFP channel.
Fluorescence lifetime imaging was performed using the wide-field frequency domain approach on the instrument described (Van Munster and Gadella, 2004), using an RF-modulated image intensifier II18MD (Lambert Instruments) coupled to a CCD camera (Photometrics) as detector. A 63× oil objective lens (Plan Apochromat 1.4 NA) was used. Modulation frequency was set to 75.1 MHz. Twelve phase images with an exposure time of 100–200 ms were acquired in a random recording order to minimize artifacts related to possible photobleaching (van Munster and Gadella, 2004). For EYFP lifetime imaging, the 514 nm argon-ion laser line was used for excitation, which was coupled to the microscope through multimode fiber optics and passed onto the sample with an 80/20 mirror (Chroma). For EYFP FLIM measurements with rsTagRFP in the OFF state, the sample was additionally excited with yellow light from a mercury arc lamp filtered through a 577 DF10 filter (Chroma). The modulated 514 nm laser light from the fiber output and the yellow mercury line were combined with a 50/50 beamsplitter at the laser entry port of the microscope. The EYFP fluorescence emission was filtered by an FF01 534/20 nm (Semrock) emission filter. For FLIM acquisition the excitation power was 97 μW/cm2 514 nm with optional 1.55 mW/cm2 577 nm light to secure rsTagRFP in the OFF state. Every FLIM measurement was followed by a reference measurement to determine the phase and modulation of the excitation light. For reference measurements and for EYFP lifetime measurements with rsTagRFP in the ON state, the additional 577 nm excitation was blocked. The reference was calibrated as described (Merzlyak et al., 2007). From the phase sequence, an intensity (DC) image and the phase and modulation lifetime image was calculated using Matlab macros. From this data, the lifetime of individual cells was determined using ImageJ (http://rsb.info.nih.gov/ij/). Subsequently, average phase and modulation lifetimes (± standard deviation) were calculated. For the presentation of lifetime maps, a 3×3 smooth filter was applied to the lifetime data. The false-color lifetime maps and 1D and 2D histograms were generated by an ImageJ macro. Switching ON/OFF rsTagRFP was performed using another 100W mercury arc lamp connected to an automatic arc lamp/laser switch. For switching rsTagRFP ON the light from this lamp filtered through a 470 DF30 filter (Chroma), was passed onto the sample, using the same 80/20 beam splitter (13.3 mW/cm2 at the back focal plane). Switching rsTagRFP OFF was performed by passing on the light for 2 s from the same arc lamp but filtered through the 577 DF10 filter onto the sample (15.6 mW/cm2 at the back focal plane). FLIM measurements were done immediately after the rsTagRFP switching.
Supplementary Material
Acknowledgments
We thank A. Sorkin for providing plasmids, J. Zhang for assistance with flow cytometry, and L. Van Weeren for transfecting cells for FLIM. This work was supported by the NIH grant GM073913 (to V.V.V.), and in part by the Molecular and Cell Biology program of Russian Academy of Sciences, Rosobrazovanie grant P256, and President of the Russian Federation’s grant MD-2780.2009.4 (to K.A.L.).
Footnotes
Supplemental Data include four Figures, one Table and two Movies, and can be found with this article online.
References
- Adam V, Lelimousin M, Boehme S, Desfonds G, Nienhaus K, Field MJ, Wiedenmann J, McSweeney S, Nienhaus GU, Bourgeois D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc Natl Acad Sci USA. 2008;105:18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Flors C, Mizuno H, Hofkens J, Miyawaki A. Highlighted generation of fluorescence signals using simultaneous two-color irradiation on Dronpa mutants. Biophys J. 2007;92:L97–99. doi: 10.1529/biophysj.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- Andresen M, Stiel AC, Folling J, Wenzel D, Schonle A, Egner A, Eggeling C, Hell SW, Jakobs S. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat Biotechnol. 2008;26:1035–1040. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- Chudakov DM, V, Belousov V, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov KA. Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. J Biol Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- Ciruela F. Fluorescence-based methods in the study of protein-protein interactions in living cells. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Dedecker P, Hotta J, Flors C, Sliwa M, Uji-i H, Roeffaers MB, Ando R, Mizuno H, Miyawaki A, Hofkens J. Subdiffraction imaging through the selective donut-mode depletion of thermally stable photoswitchable fluorophores: numerical analysis and application to the fluorescent protein Dronpa. J Am Chem Soc. 2007;129:16132–16141. doi: 10.1021/ja076128z. [DOI] [PubMed] [Google Scholar]
- Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, Medda R, Andresen M, Stiel AC, Jakobs S, Eggeling C, Schonle A, Hell SW. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C, Hotta J, Uji-i H, Dedecker P, Ando R, Mizuno H, Miyawaki A, Hofkens J. A stroboscopic approach for fast photoactivation-localization microscopy with Dronpa mutants. J Am Chem Soc. 2007;129:13970–13977. doi: 10.1021/ja074704l. [DOI] [PubMed] [Google Scholar]
- Galperin E, V, Verkhusha V, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- Geisler C, Schonle A, Middendorff CV, Bock H, Eggeling C, Egner A, Hell SW. Resolution of λ/10 in fluorescence microscopy using fast single molecule photo-switching. Appl Phys A Mater Sci Process. 2007;88:223–226. [Google Scholar]
- Giordano L, Jovin TM, Irie M, Jares-Erijman EA. Diheteroarylethenes as thermally stable photoswitchable acceptors in photochromic fluorescence resonance energy transfer (pcFRET) J Am Chem Soc. 2002;124:7481–7489. doi: 10.1021/ja016969k. [DOI] [PubMed] [Google Scholar]
- Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS One. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Henderson JN, Ai HW, Campbell RE, Remington SJ. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc Natl Acad Sci USA. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Photoconversion in orange and red fluorescent proteins. Nat Methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, Savitsky AP, Markelov ML, Zaraisky AG, Zhao X, Fang Y, Tan W, Lukyanov SA. Natural animal coloration can Be determined by a nonfluorescent green fluorescent protein homolog. J Biol Chem. 2000;275:25879–25882. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
- Mao S, Benninger RK, Yan Y, Petchprayoon C, Jackson D, Easley CJ, Piston DW, Marriott G. Optical lock-in detection of FRET using synthetic and genetically encoded optical switches. Biophys J. 2008;94:4515–4524. doi: 10.1529/biophysj.107.124859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Mal TK, Walchli M, Kikuchi A, Fukano T, Ando R, Jeyakanthan J, Taka J, Shiro Y, Ikura M, Miyawaki A. Light-dependent regulation of structural flexibility in a photochromic fluorescent protein. Proc Natl Acad Sci USA. 2008;105:9227–9232. doi: 10.1073/pnas.0709599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhaus GU, Wiedenmann J. Structure, dynamics and optical properties of fluorescent proteins: perspectives for marker development. Chemphyschem. 2009;10:1369–1379. doi: 10.1002/cphc.200800839. [DOI] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Anstrom DM, Shu X, O’Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 A resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal. 2006;12:238–254. doi: 10.1017/S1431927606060235. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiel AC, Andresen M, Bock H, Hilbert M, Schilde J, Schonle A, Eggeling C, Egner A, Hell SW, Jakobs S. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys J. 2008;95:2989–2997. doi: 10.1529/biophysj.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW, Jakobs S, Wahl MC. 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem J. 2007;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krogt GN, Ogink J, Ponsioen B, Jalink K. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS One. 2008;3:e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Munster EB, Gadella TW., Jr phiFLIM: a new method to avoid aliasing in frequency-domain fluorescence lifetime imaging microscopy. J Microsc. 2004;213:29–38. doi: 10.1111/j.1365-2818.2004.01249.x. [DOI] [PubMed] [Google Scholar]
- van Munster EB, Gadella TW., Jr Suppression of photobleaching-induced artifacts in frequency-domain FLIM by permutation of the recording order. Cytometry A. 2004;58:185–194. doi: 10.1002/cyto.a.20013. [DOI] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. J Cell Sci. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- Yampolsky IV, Kislukhin AA, Amatov TT, Shcherbo D, Potapov VK, Lukyanov S, Lukyanov KA. Synthesis and properties of the red chromophore of the green-to-red photoconvertible fluorescent protein Kaede and its analogs. Bioorg Chem. 2008;36:96–104. doi: 10.1016/j.bioorg.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.