Abstract
Interstrand cross-links (ICLs) are among the most cytotoxic DNA lesions to cells because they prevent the two DNA strands from separating, thereby precluding replication and transcription. Even though chemotherapeutic cross-linking agents are well established in clinical use, and numerous repair proteins have been implicated in the initial events of mammalian ICL repair, the precise mechanistic details of these events remain to be elucidated. This review will summarize our current understanding of how ICL repair is initiated with an emphasis on the context (replicating, transcribed or quiescent DNA) in which the ICL is recognized, and how the chemical and physical properties of ICLs influence repair. Although most studies have focused on replication-dependent repair because of the relation to highly replicative tumor cells, replication-independent ICL repair is likely to be important in the circumvention of cross-link cytotoxicity in non-dividing, terminally differentiated cells that may be challenged with exogenous or endogenous sources of ICLs. Consequently, the ICL repair pathway that should be considered ‘dominant’ appears to depend on the cell type and the DNA context in which the ICL is encountered. The ability to define and inhibit distinct pathways of ICL repair in different cell cycle phases may help in developing methods that increase cytotoxicity to cancer cells while reducing side-effects in non-dividing normal cells. This may also lead to a better understanding of pathways that protect against malignancy and aging.
INTRODUCTION
Interstrand cross-links (ICLs) present a unique problem to the repair apparatus of the cell because this type of lesion involves both strands of DNA. Failure to remove these lesions from DNA ultimately leads to cell death because the covalently linked DNA strands prevent strand separation and ultimately block replication and transcription. Much of what we know about DNA ICL repair derives from studies carried out in bacteria and yeast, whereas the mechanisms of ICL repair in mammalian cells, is less well understood.
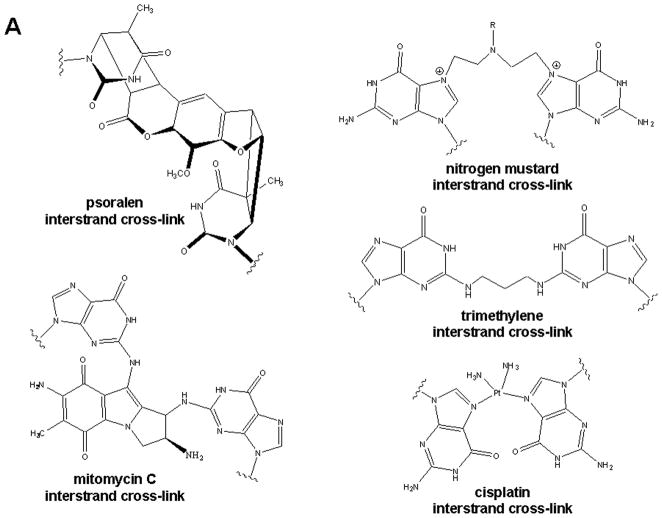
A number of different types of bifunctional alkylating agents and other bifunctional compounds are capable of reacting with DNA to create ICLs. These cross-linking agents, such as nitrogen mustards, nitrosoureas, mitomycin C, psoralen, and platinum compounds, have been commonly used in chemotherapy for many decades. Many of these are used in the clinic as anti-cancer treatment agents, however because delivery is non-specific, ICLs are created in normal cells as well (McHugh et al. 2001). Furthermore, elegant work by a number of groups has shown that ICLs can arise through endogenous sources (Summerfield and Tappel 1984; Caulfield et al. 2003; Kozekov et al. 2003; Sczepanski et al. 2008; Stone et al. 2008). Each of these cross-linking sources creates a different cross-link structure with unique structural characteristics and it is becoming increasingly evident that the chemical and physical structure of an ICL can affect both DNA helix structure at the site of the cross-link and the manner in which the ICL is repaired. The chemical structures of the ICLs described in this review are shown in Figure 1. Several reviews are available that discuss how ICLs affect DNA helix structure (Rajski and Williams 1998; Noll et al. 2004; Noll et al. 2006).
Figure 1.
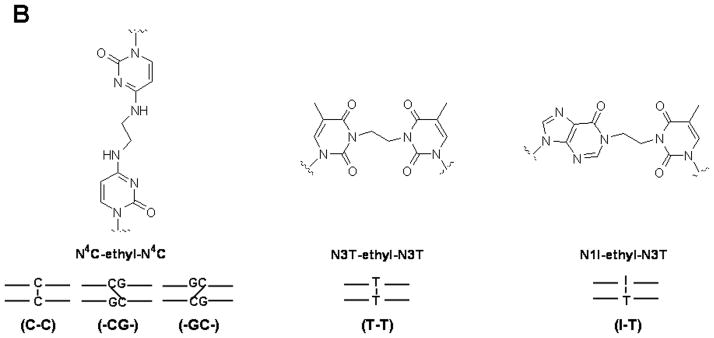
A) Chemical structures of common chemotherapeutic interstrand cross-links. B) Alkyl interstrand cross-link mimics. The N4C-ethyl-N4C cross-link preserves the normal Watson-Crick base-pair with a guanine, in contrast to the N3T-ethyl-N3T and N1I-ethyl-N3T cross-links, which block the hydrogen bond face of the adducted bases. Schematics of the duplexes and the abbreviations used for each cross-link are shown.
Studies in wild-type and repair-deficient mammalian cell lines in which the cells are treated with cross-linking agents have identified some of the pathways that are involved in ICL repair. Most of these studies have focused on S phase repair to uncover mechanisms of chemoresistance that arise in rapidly dividing cancer cells (McHugh et al. 2001). Certain studies have implied that ICL repair occurs only during S phase when a replication fork encounters an ICL (Akkari et al. 2000). There is now significant evidence that repair of ICLs occurs in a G1 context in vivo in yeast (McHugh and Sarkar 2006) and in mammalian cells [(Wang et al. 2001; Zheng et al. 2003; Muniandy et al. 2009) and our unpublished results]. As discussed below, the experiments that suggest ICL repair only occurs in S phase may have overlooked more subtle G1 type repair pathways that are not as dominant relative to S phase repair pathways in rapidly dividing cells. G1 type ICL repair would be, however, the only means of defense against chemotherapeutic insult or endogenously generated ICLs in non-dividing cells. Additionally, it is important to consider repair in a G1 context, as there is evidence that exposure to exogenous and endogenous sources of ICLs promotes aging (Grillari et al. 2007).
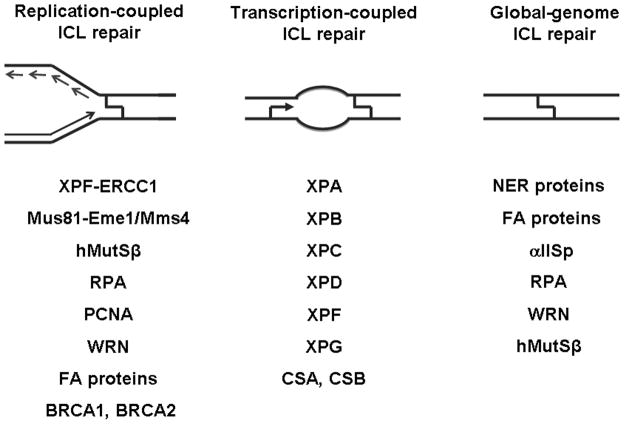
This review will focus on the initial steps of ICL repair and consider proteins that are implicated in this process. In doing so, we will consider ICL repair in three different contexts: repair coupled to DNA replication; transcription-coupled repair; and repair in DNA that is undergoing neither replication nor transcription. We will first examine proteins implicated in the initial steps of replication-coupled ICL repair, as repair in this context has been the focus of many groups. These experiments have been done for the most part through bulk treatment of cells with bifunctional alkylating agents. Experiments in mammalian cells and cell extracts using DNA substrates that contain defined cross-link lesions have provided a more detailed picture of the initial events of the repair process. We will then explore the more limited field of G1 ICL repair starting with transcription-coupled repair of ICLs in cultured mammalian cells, which is comprised mainly of experiments that utilize a site-specific plasmid reporter system. We will then discuss more mechanistic studies that have used purified proteins and mammalian-based extract systems to examine how ICLs are processed and the proteins implicated in the initial steps of repair. Most of the substrates used in these mechanistic studies do not have mammalian origins of replication or promoters and therefore mimic a global genomic DNA repair type context.
Initial processing of ICL repair appears complex, as different repair proteins are utilized depending on the context in which the ICL is recognized. Although numerous repair proteins have been implicated, the precise mechanistic details of these events remain to be elucidated. Nevertheless, a picture is beginning to emerge that the context in which an ICL is recognized and the chemical and physical properties of ICLs influence the initial events of repair.
REPLICATION-COUPLED ICL REPAIR
Very early investigations by Cole and coworkers used plasmids that contained ICLs to examine ICL repair in E. coli (Cole 1973; Cole and Sinden 1975). The plasmids were capable of replication, and these studies showed that repair of the ICLs depends upon nucleotide excision repair (NER) and homologous recombination (HR). Biochemical studies (reviewed in (Noll et al. 2006)) revealed that UvrABC is able to make incisions on either side of the ICL in vitro. Such incisions “unhook” the cross-link from one of the DNA strands, and it is this step that is critical for further processing and repair of the cross-link.
Studies in which budding yeast, Saccharomyces cerevisiae, were treated with psoralen plus UV light or nitrogen mustards, agents that produce ICLs in genomic DNA, showed that mutants defective in NER or HR were extremely sensitive to these agents ((Noll et al. 2006) and references therein). The identities of the proteins that initially recognize the cross-link are unclear. Cycling cells treated with cross-linking agents accumulate double-strand breaks (DSBs). Structurally, these DSBs are not the same as those caused by ionizing radiation. Rather they are replication-induced fork breaks with a DSB on one end and a remaining three stranded, cross-linked structure. DSBs are the result of a stalled replication fork at an ICL. It is this encounter that is most likely the mechanism by which ICLs are initially detected in replicating DNA.
Early studies in dividing mammalian cells suggested that DNA polymerase encounters with ICLs during replication were an important mechanism leading to their removal (Vos and Hanawalt 1987). Experiments in synchronized human skin fibroblasts treated with 4′-hydroxymethyl-4,5′,8-trimethylpsoralen followed by UV irradiation suggested that ICLs elicited a cell cycle check point and were repaired in S phase, but not in G1 or G2 (Akkari et al. 2000). However, recent evidence using an elegant in vitro system has shown that a site-specific ICL can elicit a check point response in the absence of replication (Ben-Yehoyada et al. 2009). The method of Akkari et al. used to measure cross-link removal during replication required separation of BrdU labeled DNA as a measure of replicated DNA versus parental DNA. However, because a repair synthesis step is required to remove the ICL, repaired DNA that may not have undergone replication could still have incorporated BrdU, thus complicating interpretation.
Unlike the situation in E. coli and S. cerevisiae, ICL processing during replication in mammalian cells is dependent upon only some of the proteins involved in NER, namely XPF-ERCC1. In addition, various studies have implicated a role for a number of other proteins in the initial steps of ICL repair. The following sections will consider the evidence implicating these proteins. A more comprehensive discussion by Legerski (Legerski 2010) regarding S phase ICL repair can be found in this issue.
XPF-ERCC1
XPF-ERCC1 is a structure-specific endonuclease that is capable of making 5′ incisions at the junction between double-stranded DNA and a 3′ single-stranded region, and is responsible for the 5′ incision in the NER pathway (Sijbers et al. 1996; Bessho et al. 1997). Studies in which human cells were treated with various cross-linking agents showed that cells deficient in proteins that are part of the NER pathway were sensitive to these agents (Friedberg et al. 1979; Cleaver 1980; Fujiwara 1982). Later studies employing mutants of Chinese hamster ovary (CHO) cells showed that cell lines deficient in ERCC1 were approximately 90 times more sensitive than wild-type cells to the cross-linking agents mitomycin C and diepoxybutane (Hoy et al. 1985). In contrast, cells deficient in XPD, one of the NER helicases and a subunit of the TFIIH transcription factor, were only moderately (3–4 times) sensitive.
More extensive studies by De Silva and coworkers (De Silva et al. 2000) examined the roles of NER, HR and non-homologous end-joining (NHEJ) in the repair of ICLs created by the nitrogen mustard mechlorethamine (HN2). Cell survival experiments showed that cells deficient in the NER proteins XPF or ERCC1, and cells deficient in HR proteins, XRCC2 and XRCC3, were 12–26 times more sensitive to HN2 than their wild-type parents. In contrast, mutants deficient in the XPB or XPD helicases, or XPG, an endonuclease that makes incisions on the 3′ side of DNA lesions were only slightly sensitive. The NHEJ-deficient mutant, XRCC5, showed sensitivities similar to those of wild-type cells. These results, which were similar to previous experiments employing other cross-linking agents, including mitomycin C, cyclophosphamide and diepoxybutane (Hoy et al. 1985; Caldecott and Jeggo 1991; Andersson et al. 1996; Damia et al. 1996), further supported the notion that XPF-ERCC1 and the HR proteins, XRCC2 and XRCC3, play important roles in ICL repair.
To gain a better understanding of the roles of these proteins in repair, a modified Comet assay was used to monitor ICL unhooking in HN2-treated cells (De Silva et al. 2000). Wild-type and XPB- and XPD-deficient hamster cells were able to unhook approximately 85% of the ICLs 48 hrs after treatment with HN2, whereas XPF- and ERCC1- deficient cells were only able to unhook 15% of the ICLs over this same period of time. XRCC2 and XRCC3 HR mutants were able to unhook the ICLs to the same extent as their isogenic parents. These results suggested that XPF-ERCC1 is involved in the critical initial step of replicative ICL processing, cross-link unhooking.
Pulsed field gel electrophoresis (PFGE) was used to monitor formation of DSBs in HN2-treated hamster cells (De Silva et al. 2000). As was the case in S. cerevisiae (McHugh et al. 2000), significant levels of DSBs were observed when exponentially growing wild-type cells were treated with HN2 for 1 hr. These DSBs resulted from formation of ICLs rather than monoadducts because treatment of the cells with the monoalkylating agent 2-chlorethlamine, HN1, elicited no formation of DSBs. Considerably lower levels (6- to 8-fold less) of DSBs were seen in non-dividing cells treated with the same concentrations of HN2. This result suggests that significant amounts of ICL repair takes place during S phase, possibly due to the presence of stalled replication forks. However, mechanisms of repair in G1 phase would not be expected to arise through stalled replication fork generated DSBs. Furthermore, G1 type repair pathways that have been characterized thus far involve NER followed by translesion synthesis (TLS), which does not involve the formation of DSBs. Therefore, the DSBs monitored by PFGE may have been a specific measure of S phase ICL repair resulting from replication fork encounter with ICLs.
Wild-type cells treated with HN2 for 1 hr and then allowed to recover for 24 hrs showed complete repair of the DSBs (De Silva et al. 2000). Similar repair was observed in the XRCC5 cells, demonstrating that NHEJ was not responsible for repair. In contrast, both the XRCC2 and XRCC3 cells showed greater than 60% DSBs after 24 hrs, demonstrating that HR was involved in the repair of these lesions.
DSB formation and repair was also examined in HN2-treated NER mutant cell lines. All of the mutants, XPG, XPB, XPF and ERCC1 showed the same levels of DSB formation and repair as in the wild-type AA8 cells. Thus, despite the implication that XPF-ERCC1 is involved in the initial cross-link unhooking step, this endonuclease appeared not to be responsible for the formation of DSBs.
The effect of ERCC1 on DSB formation was monitored by observing γ-H2AX foci formation in primary mouse embryo fibroblasts (MEFs) treated with mitomycin C (Niedernhofer et al. 2004). γ-H2AX foci, the phosphorylated form of histone H2AX, accumulates at the site of DSBs. Elevated levels of γ-H2AX foci were observed in MEFs treated with mitomycin C suggesting that like nitrogen mustard ICLs, the ICLs created by this agent induce formation of DSBs. Similar levels of γ-H2AX foci were observed in ERCC1 −/− cells, a result that suggests that ERCC1 does not contribute to the formation of these lesions and is consistent with the ERCC1-independent formation of DSBs in nitrogen mustard treated cells (De Silva et al. 2000). Although ERCC1 was not responsible for DSB formation, the breaks persisted for a much longer time than in wild-type cells. XPF-ERCC1 is therefore required for subsequent resolution of the DSBs. The observation that DSBs disappeared at the same rate in XPA −/− cells as in wild-type cells showed that the requirement for ERCC1 in DSB resolution lies outside the role it plays in NER (Busch et al. 1997; Niedernhofer et al. 2001). It is still not clear whether the resolution of the DSB by XPF-ERCC1 is due to actual unhooking of the ICL or recombinational repair, for which it has a known role (Niedernhofer et al. 2004; Ahmad et al. 2008; Al-Minawi et al. 2008; Al-Minawi et al. 2009). It remains possible that XPF-ERCC1 has a role in both of these steps of ICL repair.
DSB formation was found to be cell-cycle dependent. Wild-type MEFs arrested in G1 showed significantly reduced γ-H2AX foci formation after treatment with mitomycin C compared to proliferating cells. When the cells were released from G1, an increased number of foci were observed. However, it should be noted that during NER repair in G1 phase of the cell cycle, a faint ‘pan-nuclear’ γ-H2AX signal was observed and only during S phase are bright γ-H2AX foci formed (Marti et al. 2006). Therefore, the method of measuring γ-H2AX foci may occlude observation of G1 repair pathways, one of which is known to involve the global genome (GG-) NER pathway (Muniandy et al. 2009).
Although the modified Comet assays have implicated XPF-ERCC1, but not other NER proteins, in the unhooking step involving HN2 generated ICLs, the situation appears to be different in the case of ICLs formed by the cancer chemotherapeutic agent cis-diamminedichloroplatinum(II) (cisplatin) (De Silva et al. 2002). Cisplatin forms intrastrand cross-links with -GpG-, -ApG- and -GpNpG- sequences in DNA, and to a much lesser extent G-G ICLs. Cell survival assays showed that XPF cells were 37–40-fold more sensitive to treatment with cisplatin than were wild-type AA8 cells. When treated with cisplatin, XPD-, XPB- or XPG-deficient cells were only 1.3- to 3.1-fold more sensitive than the parental cell line. This result is similar to that observed when these cells are treated with nitrogen mustards; however cisplatin only forms 1% ICLs. When ICL unhooking was examined using the Comet assay, XPF-, ERCC1-, XPB- and XPG-deficient cells treated for 1 hr with cisplatin all showed little or no unhooking ability, even after 72 hrs. This result suggested that the differences in sensitivities of XPF and ERCC1 cells to cisplatin versus that seen in other NER mutants are not due to differences in their abilities to unhook the platinum cross-link, and implicates all of the NER proteins in the unhooking step. In a separate series of experiments it was shown that there were no differences in the abilities of the NER mutant cells to remove platinum intrastrand cross-links (De Silva et al. 2002). Thus, the extreme sensitivity of XPF- and ERCC1-deficient cells to cisplatin implied an additional role for these proteins independent of the NER pathway in the processing of the cisplatin lesion. This extreme sensitivity may possibly be explained through its role in recombinational repair (Ahmad et al. 2008; Al-Minawi et al. 2008). The observation that like XPF and ERCC1 mutants, XRCC2 and XRCC3 mutants were also extremely sensitive (38–50-fold) to cisplatin treatment (De Silva et al. 2002) demonstrates that recombinational repair pathways play an important role in cisplatin adduct repair and supports the notion that XPF-ERCC1 is involved in recombinational repair of cisplatin adducts as well.
Efforts have been made to gain a better understanding at the molecular level of the role of XPF-ERCC1 in ICL repair by carrying out biochemical studies in mammalian cell extracts with a defined, site-specific ICL substrate. As discussed in detail below, Legerski’s group has carried out extensive studies that have also implicated XPF-ERCC1 and HR pathways. While the plasmids that carried the ICLs did not contain mammalian replication origins, the involvement of XPF-ERCC1, HR, and mismatch repair proteins, which cooperate with replication machinery, indicates a replicative ICL repair pathway may be involved.
Mu et al. did not detect recombinant XPF-ERCC1-mediated endonucleolytic incisions in their cross-linked linear DNA substrates (Mu et al. 2000). Similar observations were made by two other groups (Kuraoka et al. 2000; Fisher et al. 2008). Kuraoka et al. created a substrate designed to mimic a stalled replication fork (Kuraoka et al. 2000). This Y-shaped substrate consisted of a duplex joined to two non-complementary single-stranded tails. A psoralen cross-link was positioned at the junction between the duplex and the tails. An incision was observed on the 5′ side of the cross-link when this substrate was incubated with recombinant XPF-ERCC1 in the absence of RPA. When the cross-link was moved away from the junction into the interior region of the duplex, incisions were observed on both the 5′ and 3′ sides of the cross-link. These incisions unhook the cross-link. Bessho and coworkers investigated processing by XPF-ERCC1 of a similar psoralen cross-linked Y-shaped substrate containing a psoralen cross-link positioned in the stem of the Y at the three-way junction formed by the stem and two duplex tails (Fisher et al. 2008). When this substrate was incubated with recombinant XPF-ERCC1, an incision was observed 3–4 nucleotides (nt) away from the 5′ side of the cross-link. An additional incision was observed 5–10 nt away on the 3′ side of the cross-link. These two incisions unhook the cross-link and the 3′ incision also creates a DSB. The results of these experiments suggest a model in which a stalled replication fork creates a Y-shaped junction that allows XPF-ERCC1 to cleave sequentially on the 3′ and 5′ side of the cross-link, although this has yet to be shown in vivo.
Repair of ICLs in replicating DNA involves the formation of DSBs. Bessho has described a system that may provide further insight into the role of XPF-ERCC1 in ICL-induced DSB formation (Bessho 2003). Plasmid DNA was prepared that contained a single psoralen ICL positioned 205 nt away from an SV40 origin of replication. This cross-linked substrate was incubated in a HeLa nuclear extract supplemented with SV40 T antigen and the incorporation of radioactive nucleotides was monitored. Not surprisingly, DNA synthesis was inhibited 80–90% compared to a non-cross-linked control, and multiple termination sites were observed, with the longest representing termination one nt before the cross-link. Significant, however, was the observation of DSBs formed near the site of the cross-link. It remains to be seen if DSBs are created in extracts deficient in XPF-ERCC1 or Mus81-Eme1, another endonuclease implicated in ICL unhooking (see discussion below).
A system in Xenopus extracts originally described by Lu et al. (Lu et al. 2005) has recently been employed with plasmids with mammalian origins of replication that contained either a non-distorting nitrogen mustard ICL mimic or a cisplatin ICL (Raschle et al. 2008). The results showed that repair of the ICL required convergence of replication forks on either side of the cross-link. The forks initially terminated 20–24 nt ahead of the ICL. Interestingly, the forks approached 4 nt closer to the cisplatin ICL than was the case for the mustard mimic ICL. This difference most likely reflects the greater distortion imparted to the helix by the cisplatin ICL and suggests that cross-link structure may affect repair even during replication. Subsequently, one of the stalled replication forks approached to within one nt from the cross-link. This approach up to the cross-link preceded any incisions made near the site of the cross-link. Incisions made at the site of the cross-link resulted in cross-link unhooking and generation of a DSB. The identity of the proteins responsible for unhooking and DSB incisions remains to be determined.
While it is clear that XPF-ERCC1 is important for ICL repair, its role in the unhooking step has been questioned (Bergstralh and Sekelsky 2008). XPF-ERCC1 is now known to have a role in recombinational pathways implicated in ICL repair (Ahmad et al. 2008; Al-Minawi et al. 2008; Al-Minawi et al. 2009). It has been suggested that an alternative interpretation of the modified Comet assay is that failed recombination intermediates result in DNA that migrates with a similar retarded mobility as cross-linked DNA, thus making it appear as if XPF-ERCC1 were responsible for unhooking (Bergstralh and Sekelsky 2008). These recombination intermediates would not be covalently linked, but topological constraints may prevent their denaturation, thus mimicking the behavior of cross-linked DNA in this assay. If the modified Comet assay does indeed measure recombination intermediates, it appears that it is only certain types. For instance, while the XRCC2 and XRCC3 mutants did not generate longer tail moments, the XRCC3 mutant did and therefore this mutant appeared to be unable to unhook platinum cross-links (De Silva et al. 2002). Considering XRCC1-3 are all involved in recombination, this suggests that only certain specific failed recombination intermediates may migrate with a slower mobility in this assay.
It appears that XPF-ERCC1 is important in recombinational ICL repair and possibly the initial unhooking steps during replication. The diverse roles of this endonuclease and the many pathways used to repair ICLs have made teasing out the exact roles of XPF-ERCC1 in ICL repair complicated. The reader should consult the article in this issue by Nairn (Nairn 2010) for further discussion of this topic.
Mus81-Eme1/Mms4
Mus81-Eme1 also known as Mus81-Mms4 is a heterodimeric, structure-specific endonuclease related to XPF-ERCC1 that has been implicated in the repair of DNA ICLs (Osman and Whitby 2007; Ciccia et al. 2008). Like XPF-ERCC1-deficient cells, cells deficient in Mus81-Eme1 are extremely sensitive to agents that create ICLs (Dendouga et al. 2005). Mus81−/− mice are viable, but were found to be hypersensitive to the ICL agent mitomycin C (McPherson et al. 2004). Biochemical characterization of Mus81-Eme1 suggested that it may play a role in the processing of replication forks (Ciccia et al. 2003). The endonuclease was shown to be capable of cleaving Y-shaped substrates that mimic replication forks. This activity combined with the hypersensitivity of Mus81 −/− cells to interstrand cross-linking agents suggested that Mus81-Eme1 might be responsible for cross-link-induced DSB formation.
When mouse embryonic stem cells were exposed to mitomycin C for 24 hrs, DSBs were not observed by PFGE (Hanada et al. 2006). However, DSBs were observed in ERCC1-deficient cells under the same conditions. Similar effects were seen when the cells were treated with cisplatin. Wild-type cells cultured continuously with mitomycin C accumulated in S phase, and showed increased DSB formation, whereas the DNA from similarly treated Mus81-deficient cells, which also accumulated in S phase, did not form DSBs. Furthermore, when DNA replication was blocked by addition of thymidine, mitomycin C treatment failed to elicit DSBs as monitored by γ-H2AX foci formation. These results, in combination with the structure-specific endonuclease activity of Mus81-Eme1, led Hanada et al. to propose that during replication, Mus81-Eme1 acts on ICL-induced stalled replication fork structures to first create a DSB which then initiates additional incisions by an unknown nuclease, possibly XPF-ERCC1, that results in unhooking of the cross-link (Hanada et al. 2006; Osman and Whitby 2007).
It appears that during S phase, replication fork encounter with an ICL leads to a DSB, involving Mus81-Eme1, which must be resolved through recombinational pathways that involve XPF-ERCC1. However, the question remains if and how XPF-ERCC1 is involved in the initial unhooking step and whether other nucleases are involved in unhooking.
BRCA1, BRCA2, and WRN
BRCA1, an E3 ubiquitin ligase, is involved in the maintenance of the genome, particularly repair of DSBs (Boulton 2006). BRCA1-deficient cells are hypersensitive to agents that create DNA ICLs. A recent study has explored the interaction between BRCA1 and WRN and their role in ICL repair (Cheng et al. 2006). A modified Comet assay was used to monitor the repair of psoralen ICLs in wild-type HeLa cells and in cells treated with siRNAs to knockdown BRCA1 and/or WRN. The results suggested that BRCA1 and WRN were both involved in repair of the psoralen cross-links. The observation that knockdown of both proteins resulted in the same level of repair as knockdown of either mutant alone showed that both proteins function in the same repair pathway.
Immunoprecipitation experiments demonstrated that BRCA1 interacts with WRN. An additional protein partner of BRCA1, BARD1, was also found to interact with WRN. The helicase activity of WRN on a forked substrate was stimulated 4.5-fold in the presence of BRCA1/BARD1.
To determine which activity of WRN was responsible for ICL repair in cells, Werner syndrome cells complemented with either wild-type WRN, helicase-deficient WRN, exonuclease-deficient WRN, or helicase- and exonuclease-deficient WRN. The cells were treated with 8-methoxypsoralen/365 nm light and cross-link unhooking was assessed using the modified Comet assay (Cheng et al. 2006). The Comet tail moment decreased with time after exposure in cells complemented with wild-type WRN or exonuclease-deficient WRN suggesting that cross-link unhooking had taken place. In contrast, the comet tail moment remained unchanged in cells complemented with helicase- plus exonuclease-deficient WRN or in helicase-deficient WRN cells. These results, which demonstrate that the WRN helicase, but not its nuclease activity is required for ICL processing, are in agreement with the CRS assays of Zhang et al. (Zhang et al. 2005) described below.
Taken together, the studies described above suggest that WRN helicase activity, which can be stimulated by BRCA1, may be involved in initial ICL processing. This processing most likely occurs at stalled replication forks that are formed during DNA replication.
BRCA2 interacts directly with RAD51 and is involved in HR and DSB repair (Boulton 2006). Cipak et al. investigated the role of BRCA2 in ICL replication-coupled repair in cell extracts derived from a BRCA2-deficient human cell line, CAPAN-1 (Cipak et al. 2006). The DNA substrate for these studies contained an SV40 origin of replication and a single psoralen ICL. Replication was observed to take place past the site of the cross-link, indicating that unhooking of the cross-link had occurred. In contrast, incubation in the CAPAN-1 extract resulted in an approximately 60% decrease in replication past the ICL. Replication was restored when CAPAN-1 extracts were complemented with BRCA2, however DSB intermediates still accumulated. These results, which appear to be in agreement with genetic evidence, suggest that unlike BRCA1, BRCA2 is not involved in the initial steps of ICL processing, but rather in subsequent HR-mediated DSB repair.
Fanconi anemia proteins
Fanconi anemia (FA) is an autosomal recessive genetic disorder that is marked by chromosome instability. Patients who suffer from FA have congenital abnormalities and are prone to developing cancer, particularly leukemia. Cells from FA patients are extremely sensitive to agents that create DNA ICLs (Carreau et al. 1999) and this observation suggests that FA proteins, of which there are 15, are necessary for repair of ICLs.
The exact role of the FA complex of proteins in ICL repair is still quite unclear [for a discussion see the following recent reviews (Kennedy and D’Andrea 2005; Niedernhofer et al. 2005; Andreassen and Ren 2009; Thompson and Hinz 2009). It appears that the FA complex may sense stalled replication forks created by ICLs; help recruit repair proteins to this site; and remodel the stalled replication fork to allow repair to proceed (Niedernhofer et al. 2005; Thompson and Hinz 2009).
A substrate containing a single psoralen ICL and a weak mammalian origin of replication was used to further examine the mechanism of recombinational ICL repair (Zhang et al. 2007). The assay was designed such that repair by a nucleotide excision repair/translesion synthesis mechanism would not be monitored. Instead, a mutation was placed to prevent reporter expression unless recombination with a homologous, also mutated, plasmid took place. Therefore, the only repair signals observed were through repair of the psoralen cross-link by recombinational mechanisms. It was found that the cross-linked plasmid alone did not induce recombinational repair when incubated as an intact plasmid. However, a DSB introduced next to the ICL-induced recombinational repair of the ICL. The observation that this repair pathway relied on XPF-ERCC1, MutSβ, REV3 and components of the FA pathway, demonstrated that these factors were involved in a recombination-dependent ICL repair pathway. As discussed in a later section, in cell-free assays a psoralen ICL-induced recombination into a homologous undamaged plasmid which relied upon MutSβ, FA components, XPF-ERCC1, XRCC2 and XRCC3 (Li et al. 1999; Zhang et al. 2002). However, the substrates used in this assay did not contain DSBs, which were shown to be required for repair in the recombination repair assay (Zhang et al. 2007).
Experiments in various FA cell lines and extracts suggest that FA proteins are not required for the initial steps that process the cross-link. Thus, for example, the Comet assay was used to assess the kinetics of ICL unhooking in cells that were treated with psoralen and UV light (Rothfuss and Grompe 2004). In these experiments FANCA and FANCD2 cell lines showed essentially the same kinetics of unhooking as wild-type cells. Furthermore, DSB formation in FANCC and FANCD2 cells was similar to that in wild-type cells as determined by γH2AX foci formation. However, certain FA proteins such as FANCJ and FANCM have helicase and nuclease activities, and therefore it remains possible that some of these proteins are involved in the initial steps of repair. It has recently been shown that FANCD2 is monoubiquitinated independent of processing by XPF-ERCC1 (Bhagwat et al. 2009). However, nucleolytic incisions by XPF-ERCC1 were required to form stable chromatin bound monoubiquitinated FANCD2 foci. This is the first data to connect nucleolytic processing of ICLs to the FA pathway. It remains unclear whether the formation of chromatin-bound FANCD2 requires XPF-ERCC1 for unhooking or recombination.
SIGNIFICANCE OF G1 ICL REPAIR PATHWAYS
Most studies have focused on S phase ICL repair pathways because of the significance of bifunctional alkylating agents in cancer chemotherapy. However, most of the cells in mammals are non-dividing, terminally differentiated cells. There is evidence that the use of cross-linking chemotherapeutics induce premature aging in long-term survivors. Acquired premature progeroid syndrome or APPS, the term given to the long-term side-effects of such chemotherapies, is characterized by impaired cognitive, visual and musculoskeletal functions (Grillari et al. 2007). Furthermore, premature aging syndromes have been observed in humans who have mutated proteins implicated in ICL repair, such as XPF-ERCC1, WRN, Fanconi anemia proteins, CSA, and CSB (Mitchell et al. 2003; Grillari et al. 2007). Considering the large amount of evidence that shows ICLs arise through natural exogenous and endogenous sources (Summerfield and Tappel 1984; Caulfield et al. 2003; Kozekov et al. 2003; Sczepanski et al. 2008; Stone et al. 2008), G1 type ICL repair would be the only means of defense against these lesions in non-dividing cells. The formation of ICLs in terminally differentiated cells is believed to contribute to malignancy associated with aging (Grillari et al. 2007). Even in dividing cells, repair in a G1 context would theoretically help reduce the burden of replication fork collapse as a dividing cell enters S phase. Therefore, G1 repair pathways may also contribute to ICL chemoresistance in tumor cells. Determining the pathways of S phase as well as G1 phase ICL repair may be important in developing therapeutic strategies that can be used to increase the effectiveness of killing tumor cells while reducing the side effects in benign, terminally differentiated cells. Also, an understanding of ICL repair in G1 may contribute to our understanding of the basic mechanisms of aging. In the following section, two different forms of G1 ICL repair will be discussed in detail: transcription-coupled ICL repair and repair of ICLs in quiescent DNA. The NER pathway appears to be important in both these contexts. A further discussion by Wood (Wood 2010) of the involvement of NER in ICL repair can also be found in this issue.
A recombination-independent ICL repair pathway involving NER followed by TLS was first observed in E. coli by Loechler and colleagues (Berardini et al. 1997; Berardini et al. 1999). Furthermore, the presence of a replication-independent ICL repair pathway essential for cell viability was identified in the G1 phase of the cell cycle in S. cerevisiae (McHugh and Sarkar 2006; Sarkar et al. 2006). This study suggested that recombination-independent repair in yeast in G1 is initiated by the GG-NER pathway. Very recent studies have also shown that G1 repair of ICLs in cultured mammalian cells involves the GG-NER pathway (Muniandy et al. 2009). These studies demonstrate that a NER/TLS pathway of ICL repair operates in E. coli, yeast and mammalian cells.
TRANSCRIPTION-COUPLED ICL REPAIR
Transcribed regions of a non-dividing cell have been found to have ‘domain-associated’ repair meaning that in euchromatic regions undergoing transcription, both the transcription-coupled and global genome nucleotide excision repair pathways (TC-NER and GG-NER) are more active than in other regions of the genome (Nouspikel et al. 2006). An ICL imposes an absolute block to transcription and therefore it is imperative for a non-diving cell to repair this lesion when it resides in an actively transcribed gene. Thus, it is not surprising that transcription-coupled ICL repair mechanisms have evolved.
Transcription-coupled nucleotide excision repair
Fundamentally, there are two mechanisms by which transcription can initiate repair. The first mechanism results from the formation of open chromatin structures surrounding a transcribed region, which permits access to repair factors. The second mechanism is initiated by the presence of a stalled elongating RNA polymerase (Laine and Egly 2006; Episkopou et al. 2009). The first mechanism would not discriminate between the two DNA strands and would also involve XPC as a recognition factor, whereas the second mechanism would be XPC-independent, CSA- and CSB-dependent, and would repair lesions preferentially on the transcribed strand. As mentioned above, domain associated repair is more efficient in open regions of the genome and involves both the GG- and TC-NER pathways. These two distinct mechanisms of transcriptionally-derived recognition may explain why the global-genome recognition complex, XPC-hHR23B, was found to be involved in repair of certain ICLs such as a mitomycin C ICL (Zheng et al. 2003) embedded in a reporter plasmid with a strong transcriptional promoter, and was required for removal of psoralen ICLs from the genome of cultured cells (Muniandy et al. 2009). However, repair of psoralen ICLs (Wang et al. 2001) and alkyl ICLs (our unpublished results), when biased towards transcription-coupled repair using the plasmid reporter system, only partially requires XPC because the level of repair in XPC cells is higher than in CSB cells. Existing evidence suggests that TC-NER is central to initiating repair of ICLs during transcription. Although transcription-coupled repair in mammalian cells was initially identified by Hanawalt and colleagues in the mid 1980s with the observation of strand-preferential removal of pyrimidine dimers in the transcriptionally active DHFR gene (Bohr et al. 1985; Mellon et al. 1986), the mechanistic details of TC-NER are still being uncovered.
There are two genes that are specific to TC-NER in mammalian cells: CSA and CSB. These two genes are named after Cockayne Syndrome, a condition resulting from defects in either of these two genes. The CSA protein resides in a complex with DDB1, Cullin4A, and Roc1, and displays E3 ubiquitin ligase activity (Groisman et al. 2003; Groisman et al. 2006). CSA also physically associates with RNA polymerase II in a UV-dependent manner (Groisman et al. 2003; Groisman et al. 2006). Additionally, knockdown of CSA results in deficient NER, suggesting ubiquitination is important to carry out TC-NER (Groisman et al. 2003).
The CSB protein is a member of the SWI2/SNF2 family of ATP-dependent chromatin remodeling factors and, like other SWI/SNF family members, exerts DNA-dependent ATPase and nucleosome remodeling activities (Citterio et al. 2000). However, CSB does not exhibit the classical helicase activity common to this protein family (Selby and Sancar 1997; Citterio et al. 2000). CSB is the first repair protein identified as having a direct role in modulating nucleosome structure (Citterio et al. 2000). Furthermore, CSB has been shown to stimulate RNA synthesis in vitro and in vivo (Balajee et al. 1997; Selby and Sancar 1997; Groisman et al. 2006; Laine and Egly 2006) and to interact with transcriptional complexes as well as the NER repair factors XPG and XPA (Laine and Egly 2006). Still, the role of CSB during general transcription remains a controversial issue (Citterio et al. 2000; Groisman et al. 2006). CSB knockout mice, and some patients deficient in CSB, display a mild phenotype suggesting that CSB is not required for general transcription (Groisman et al. 2006). It was recently shown that CSB is a substrate of the CSA ubiquitin ligase complex. CSB was shown to be degraded at a late stage of the repair process in a proteasomal- and CSA-dependent manner, which ultimately leads to the recovery of RNA synthesis after transcription-coupled repair (Groisman et al. 2006). In addition, CSB was determined not to participate in the removal of the stalled elongating RNA polymerase II (Selby and Sancar 1997). Other in vitro pull down experiments suggested that the stalled polymerase is not removed during assembly of the TCR complex (Fousteri et al. 2006). However, TFIIH is likely to be partially responsible for the ATP-dependent removal of the stalled RNA polymerase, as the release of the stalled polymerase was observed to be less efficient in the absence of TFIIH (Laine and Egly 2006). Although the initial events are becoming clearer in the context of TC-NER, the molecular details still remain largely unknown specifically in relation to ICL repair.
Transcription and the repair of interstrand cross-links
The connection between ICLs and transcription was first described in the early 1990’s. In addition to showing that the efficiency of ICL repair increased in actively transcribed regions, Hanawalt and colleagues showed that the formation of cross-links was more efficient in transcribed regions of an active gene (Islas et al. 1991). Thus, chromatin structure of specific genomic regions was determined to be a critical factor in mammalian ICL formation and repair (Islas et al. 1991; Dronkert and Kanaar 2001; Laine and Egly 2006; Fousteri and Mullenders 2008).
Hanawalt’s initial cross-linking studies were done using a renaturing agarose gel electrophoresis technique to examine the processing of psoralen ICLs in the expressed dihydrofolate reductase (DHFR) gene and the unexpressed FMS proto-oncogene in human fibroblasts (Vos and Hanawalt 1987; Islas et al. 1991). These studies determined that approximately 92% of the fragments containing the expressed dihydrofolate reductase gene were cross-linked, whereas only 37% of the unexpressed FMS proto-oncogene fragments were cross-linked. After 24 hrs, 90% of the cross-links in the DHFR gene were removed compared to nondetectable levels of cross-link removal in the FMS proto-oncogene (Islas et al. 1991). Similarly, Bohr and colleagues, showed that cisplatin ICLs are repaired more efficiently in transcribed regions of the DHFR gene at low cross-link concentrations in an ERCC1-dependent manner in Chinese hamster ovary cells (Jones et al. 1991; Larminat et al. 1993; Larminat and Bohr 1994). Another study detected nitrogen mustard ICL formation at higher levels in the overexpressed c-MYC proto-oncogene compared to that of the weakly expressed N-RAS gene and non-transcribed regions in a human tumor cell line (Futscher et al. 1992). The nitrogen mustard cross-links in the c-MYC gene disappeared more rapidly suggesting that nitrogen mustard ICLs are produced and processed faster in transcribed regions (Futscher et al. 1992). Open chromatin structure at sites of transcription may explain these observations, as other lesions have been shown to form and be repaired specifically in openly transcribed domains (Nouspikel et al. 2006). Conversely, equal repair was identified in active and in inactive regions at very high concentrations of ICLs in multiple studies in hamster cells (Jones et al. 1991; Larminat et al. 1993). These experiments suggest that when overwhelmed with ICLs, both transcription-coupled and global genome ICL repair pathways become active.
A separate study using psoralen ICLs in the constitutively expressed adenine phosphoribosyltransferase (APRT) gene in hamster cells demonstrated that 90% of the mutations arose from monoadducts remaining on the non-transcribed strand, suggesting the transcribed strand was preferentially incised (Sage et al. 1993). Evidence of strand specificity for the transcribed strand has been shown for psoralen ICLs in the DHFR gene (Islas et al. 1994). The strand preference in transcription-coupled repair is not surprising since the first description of TC-NER demonstrated a repair preference for removal of pyrimidine dimers from the transcribed strand of the DHFR gene (Mellon et al. 1987).
The majority of studies of transcription-coupled ICL repair have been carried out in mammalian cells using plasmids that contained a single site-specific cross-link located between a CMV promoter and a luciferase reporter gene. This system is biased towards transcription-coupled repair because the lesion is placed downstream of a constitutive mammalian transcriptional promoter. Host-cell reactivation (HCR) assays are used to study repair of the ICL as signaled by expression of the luciferase reporter gene. There has been little work to date on the mechanistic details of transcription-coupled ICL repair due to the inherent difficulty in monitoring transcription-coupled repair in cell extracts (Laine and Egly 2006).
Using a psoralen cross-linked substrate, Li and coworkers demonstrated the presence of a transcription-driven, recombination-independent ICL repair pathway in mammalian cells. The repair efficiency was determined to be approximately 50% as shown by the reactivation of a cross-linked plasmid compared to an undamaged plasmid (Wang et al. 2001). Repair efficiencies of the psoralen cross-linked plasmids were significantly reduced to less than 5% in XPA-, XPB-, XPD-, XPG- and were surprisingly, equally sensitive in XPF- and ERCC1-deficient cells. This indicates that during transcription-coupled ICL repair, which involves NER/TLS, there does not appear to be a special role for XPF-ERCC1 outside of NER. The repair efficiency was only partially reduced in an XPC-deficient cell line indicating that, in this system, the GG- NER pathway is only slightly involved and supports the notion that the plasmid reporter method of measuring repair is transcriptionally biased. Consistent with this result, as discussed below, a significant decrease in repair of various cross-links was observed in cells mutated in CSA or CSB. These results suggest that although both transcription-coupled and global-genome NER pathways play important roles in initial processing of ICLs independent of replication, in a highly transcribed environment, TC-NER appears to be the dominant pathway involved. The precise mechanistic role of NER proteins in recombination-independent ICL repair has not yet been determined, although it is believed to be involved in the initiation step of ICL repair (see discussion below). Recovered plasmids from these experiments had high rates of mutation, demonstrating the repair pathway(s) involved were error-prone. Also, evidence showed that polymerase eta was partially involved in the pathway, although it was not essential, suggesting the involvement of other translesion polymerases (Wang et al. 2001; Zheng et al. 2003).
Experiments were also carried out on a plasmid containing a single site-specific mitomycin C cross-link. Similar to the psoralen plasmid experiments, the repair efficiency of the mitomycin C cross-linked plasmid was approximately 50% of a non-damaged plasmid in wild-type cells (Zheng et al. 2003). This ICL was also repaired in a recombination-independent manner using both the transcription-coupled and GG-NER pathways as evidenced by dramatic reductions, approximately 10-fold less than wild-type cells, in repair efficiencies in both XPC-and CSA- and CSB-deficient cells. Why both the GG- and TC-NER pathways were equally involved in repair of the mitomycin C, but not the psoralen ICL, was not clear, although the chemical and physical properties of the cross-linked DNA may be responsible.
Recovered plasmids from Li and colleagues’ experiments showed there was strand bias for mutations on the transcribed strand, suggesting that the transcribed strand was preferentially incised during transcription-coupled ICL repair, consistent with the early results by Hanawalt and colleagues.
Another research group using a HCR assay demonstrated that repair of mitomycin C cross-linked plasmids is NER-dependent, which also confirms Li’s results that NER proteins are essential for transcription-coupled ICL repair (Ahn et al. 2004).
HCR assays using reporter plasmids containing a site-specific psoralen or mitomycin C cross-link were also performed in a human mismatch repair mutant cell line, Hec59, defective for MSH2. The results for both cross-linked plasmids showed significant increases in the level of recombination-independent repair in the absence of MSH2, suggesting MSH2 was involved in a competing pathway possibly involving recombination (Zheng et al. 2006).
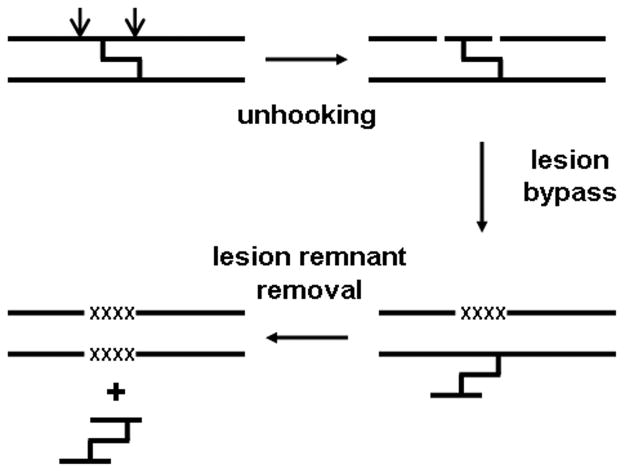
Li and coworkers’ previous studies showed that recombination-independent repair in mammalian cells is error-prone. They further tested REV1 and REV3 to determine whether these proteins were involved in this repair pathway and whether they are responsible for the high mutation rate (Shen et al. 2006). Assays performed in REV1 and REV3 knockout cells demonstrated these proteins are the principle lesion-bypass components of this pathway, while polymerase-eta (XPV) deficient cells were able to repair the ICL much more efficiently (Wang et al. 2001; Zheng et al. 2003; Shen et al. 2006). This study also showed that the initiation of lesion bypass is controlled by the monoubiquitination of PCNA at lysine 164. Additionally, characterization of the plasmids recovered from the REV1 and REV3 knockout cells showed that mutations were greatly decreased in the knockout cells, indicating that the high level of mutagenesis was predominantly due to REV1 and REV3 (Shen et al. 2006). A model for recombination-independent ICL repair in mammalian cells is shown in Figure 2.
Figure 2.
Recombination-independent interstrand cross-link repair.
Structural factors can determine the effects of DNA damage on transcription, specifically important structural factors that can influence transcription-coupled repair are size and shape of damage as well as local DNA sequence and structure (Scicchitano et al. 2004). It is not known if cross-link-induced distortions affect where the RNA polymerase stalls and thus initiate repair, but recent evidence suggests that cross-link structure affects both transcription and the subsequent repair synthesis step [(Smeaton et al. 2009) and our unpublished results]. ICLs that block the hydrogen bond face of the cross-linked nucleotide, such as the synthetic T-T and I-T mismatched cross-links shown in Figure 1B, which mimic BCNU type cross-links, can impede bacterial and viral purified replicative polymerases as well as polymerases present in mammalian cell extracts (Smeaton et al. 2009). However, when cross-links that do not interfere with the hydrogen bond face, such as the synthetic C-C mismatched cross-link shown in Figure 1B, which mimics nitrogen mustard type ICLs, efficient bypass was observed (Smeaton et al. 2009). We have also observed significant effects of blocking the hydrogen bond face on repair efficiency using the HCR assay in mammalian cells (Hlavin and Miller, unpublished results). Furthermore, preliminary evidence suggests that a distorted -GC- cross-link is repaired with much higher efficiency than a non-distorted -CG- cross-link in mammalian cells (Smeaton and Miller, unpublished results), a result similar to that previously observed in E. coli (Noll et al. 2005). Taken together, these data indicate that cross-link-induced distortions and the chemical structure of an ICL play an important role in determining how ICLs are initially recognized and processed independent of replication.
It is clear that many questions remain surrounding the details of TC-ICL repair. Further research in this area might examine whether the classical mechanisms of NER act during ICL repair and, if so, how the bubble structure is formed in the presence of an ICL. It will be interesting to determine how the structure and distortion induced by an ICL affects TC-ICL repair, with specific attention paid to the position of the stalled RNA polymerase relative to the site of the cross-link. The location of the stalled polymerase may have significant effects on the recruitment of repair proteins to the site of damage and thus influence repair efficiency.
ICL REPAIR IN QUIESCENT DNA
In non-dividing cells, monoadducts that lie within transcribed areas of the genome have domain associated repair and both GG-NER and TC-NER are active (Nouspikel et al. 2006). The necessity of removing an adduct from the transcribed strand is apparent. However, most monoadducts placed in the non-transcribed strand do not appreciably block transcription. If, however, a second lesion in the transcribed strand is located near a lesion in the non-transcribed strand, the repair machinery would lack a faithful template due to the presence of the first lesion, a situation that could result in mutation. However, ICLs present an absolute block to transcription, and therefore repair by a GG-ICL type repair pathway would be a highly advantageous means of reducing the number of ICLs encountered by elongating RNA polymerases. As discussed below, extract-based data has implicated a number of different GG-ICL type repair pathways
In a non-dividing cell, it is not apparent that monoadduct lesions that lie outside of transcribed domains would cause harm to the cell. In contrast, an ICL, which tethers both strands of the DNA duplex, may cause unique forms of damage to the cell when in quiescent areas of the genome relative to monoadducts. For instance, tethering of the strands may have an important impact on DNA topology and supercoiling that must be dealt with regardless of where in the genome the ICL resides.
There is significant evidence, based on studies with substrates that lack mammalian origins of replication or promoters, that shows defined ICL substrates are processed by seemingly many different pathways in mammalian cell extracts. While the HCR assay is biased toward transcription-coupled ICL repair, there is evidence that the GG-NER pathway plays a role in reactivation of the reporter plasmids. This conclusion is based, in some cases, on a dependence on XPC for repair in the HCR assay, especially with certain cross-links like mitomycin C (Zheng et al. 2003). Furthermore, it has been shown recently in cells that a pathway of G1 repair of ICLs involves the GG-NER pathway independent of transcription (Muniandy et al. 2009).
Mechanistic studies of ICL repair
Methods used to assess ICL repair in cells, whether done by bulk treatment and Comet/survival assays or through the use of site-specific reporter plasmids provide important information on the pathways used to repair ICLs. However, these assays are not able to provide mechanistic insight into the details of processing or identify the proteins required at each step in the repair process. Because the precise mechanistic details of ICL repair are unknown, cell-free extracts have been used to examine how ICLs are processed. Some studies have been carried out using purified enzymes that have been implicated in ICL recognition and incision (Kuraoka et al. 2000; Mustra et al. 2001; Mustra et al. 2007; Fisher et al. 2008; Zhao et al. 2009). However, reconstitution of the initial steps of recognition and unhooking of a cross-link from quiescent DNA has yet to be realized with recombinant proteins.
It is known that multiple pathways of ICL repair exist and, consistent with this, work in extracts has uncovered seemingly numerous pathways of GG-type ICL repair processing. However, it appears that the processing observed and the proteins required in extract-based studies may depend largely on the ICL substrate used, the assay used, and the assay conditions.
Site-specific ICLs used in mechanistic studies are embedded in a short DNA duplex. The short cross-linked duplex is inserted into either a longer linear duplex or a circular DNA. This is necessary in order to provide a substrate of sufficient length to examine repair processing by pathways such as NER (Huang and Sancar 1994). The properties of the substrate can make a large difference in the observed repair signals. For instance, as discussed above, there have been studies in extracts using substrates in which a mammalian origin of replication was located upstream of an ICL (Cipak et al. 2006; Raschle et al. 2008). However, there has been no work to date using transcriptional-based ICL substrates in cell extracts. This is largely due to the fact that both transcription and repair in cell extracts occur with very low efficiency (Laine and Egly 2006). Most mechanistic studies have used substrates that contain neither an origin of replication nor a transcriptional promoter and thus mimic repair from a global genomic context.
While this section will focus on studies using ICL substrates that do not contain mammalian origins of replication or transcriptional promoters, the conditions used in an assay may still dramatically affect the precise repair process monitored. For instance, when an ICL substrate is incubated in a cell extract, even without an origin or promoter, the presence of an undamaged homologous donor substrate may influence the ICL pathway being monitored (Bessho et al. 1997; Li et al. 1999; Zheng et al. 2006; Smeaton et al. 2008).
Recognition of ICLs – Studies with purified porteins
Studies using purified proteins have examined the ability of proteins implicated in ICL repair to specifically recognize ICL substrates (Kuraoka et al. 2000; Mustra et al. 2001; Mustra et al. 2007; Fisher et al. 2008; Zhao et al. 2009). Other studies have focused on the ability of the XPF-ERCC1 nuclease to cleave cross-linked substrates because of its apparent special role in ICL repair (Kuraoka et al. 2000; Fisher et al. 2008). However, as was discussed above, the activity of purified XPF-ERCC1 appears to occur exclusively on substrates that mimic stalled replication forks. Furthermore, there is no evidence that XPF-ERCC1 plays a special role outside of NER during replication-independent ICL repair [Hlavin and Miller, unpublished results and (Wang et al. 2001; Zheng et al. 2003; Muniandy et al. 2009)]. Therefore, this section will focus on recognition of ICLs by other proteins implicated in ICL repair, as the activity of purified XPF-ERCC1 on defined substrates was described above.
Many different proteins have been shown to bind with some specificity to cross-linked DNA. The damage recognition components of the NER pathway are XPC-hHR23B and XPA-RPA. XPA has been shown to have some level of specificity towards a mitomycin C cross-linked substrate and undergo a conformational change upon binding to the lesion (Mustra et al. 2001; Mustra et al. 2007). Both XPC-hHR23B and XPA-RPA were shown to bind selectively to an ICL that was placed in a triplex-forming oligonucleotide (TFO) (Thoma et al. 2005). The protein HMGB1 was shown to bind cooperatively to psoralen TFOs with RPA and may coordinate the action of NER factors at ICL sites (Reddy et al. 2005). It was also shown that MutSβ, which as discussed below is implicated in ICL repair (Zhang et al. 2002; Wu et al. 2005), also specifically recognizes psoralen TFOs (Zhao et al. 2009). Some cooperation was observed between XPA and MutSβ, however none was detected between MutSβ and XPC-hHR23B. It is not clear whether these proteins cooperate or compete to process the ICL through different pathways. However, the HCR data, which has shown that MMR mutants promote the use of NER/TLS error-prone pathways, seems to suggest that competition for entering different pathways is likely (Zheng et al. 2006).
It has also been demonstrated that the nuclear matrix protein nonerythroid α spectrin (α IISp) specifically binds to psoralen cross-linked DNA and associates with Fanconi anemia proteins (McMahon et al. 2001). It has been demonstrated that α IISp interacts with the nuclear matrix (Bachs et al. 1990) and this observation is consistent with data demonstrating that the nuclear matrix may serve as an important locus for ICL repair (Atanassov et al. 2005).
Mechanistic ICL repair experiments in mammalian cell extracts
While no incisions were observed when recombinant XPF-ERCC1 was incubated with ICL substrates embedded in completely duplexed DNA (Kuraoka et al. 2000), extract-based assays have detected XPF-ERCC1 specific incisions that were unique to its role in NER (Kumaresan et al. 1995; Kumaresan and Lambert 2000; Mu et al. 2000; Zhang et al. 2000; Zhang et al. 2002), NER-dependent incisions (Bessho et al. 1997; Smeaton et al. 2008), and XPF-ERCC1-independent incisions (Smeaton et al. 2008). It is therefore still unclear how XPF-ERCC1 is involved in the initial incision stages of ICL processing.
There have been many different types of extract-based assays that have provided important mechanistic insights into ICL repair. Two commonly used assays are the incision/unhooking assay and the repair synthesis assay. In the incision/unhooking assay, a radiolabel placed at a specific position within the substrate is used to assess the exact locations of nucleolytic incision events. The labeled substrate is incubated with a whole cell extract and the incision products are analyzed by gel electrophoresis. Alternatively, in the repair synthesisassay, α-32P dNTPs are used to monitor nucleotide incorporation into a non-labeled substrate during ICL processing. The non-labeled substrate is incubated with a cell extract and 32P incorporation is used as a measure of repair synthesis. The type of assay used to study ICL repair must be considered when interpreting data. For instance, the incision/unhooking assay directly measures nucleolytic action on ICL substrates, and requires, at a minimum, recognition and incision proteins, and possibly proteins that couple the two events. The repair synthesis assay requires proteins for recognition, multiple incisions resulting in unhooking, repair synthesis and in some assays also ligation. Furthermore, it is likely that the unhooking and repair synthesis processes are coupled through the use of yet other proteins, as is the case with repair processes such as NER (Staresincic et al. 2009). Therefore, a productive repair signal from a repair synthesis assay requires many more proteins and steps than does an incision/unhooking assay. These two types of assays provide complementary information about the details of the different mechanistic steps in the ICL repair process.
Extract-based assays have been employed because the suite of proteins required for ICL repair and the steps in which they act remains for the most part unknown. There are, however, many complications that arise when working with extracts. For instance, different types of cell extract preparations may have widely different activities due to isolation of different proteins. Furthermore, even the same type of extract preparation may have different levels of activity when made at different times. Such variation can arise from many sources including quality and concentration of the extract and the amount of sheared genomic DNA that co-purifies with the extract. In addition, many proteins can potentially interfere with ICL processing. For instance, nucleic acid binding proteins present in a crude cell extract could interfere with repair protein binding. To circumvent this problem, non-damaged competitor DNA is commonly added to reduce non-specific binding to the damaged substrate. Extracts also have phosphatases that can remove a terminal 32P label, and therefore the label is usually placed internally in the substrate. There are also non-specific nucleases (exo and endo) that can nick and degrade substrates. This is controlled for by using a non-damaged substrate that is otherwise identical to the damaged one and assaying for random nuclease activity. DNA polymerase-mediated non-specific nick translation can also occur in cell extracts, but this is usually controlled for by using a non-damaged substrate as in the case of the incision assay. There are proteins, such as Ku70/80, that bind to the ends of linear DNA and can interfere with DNA repair processing of linear substrates. This unwanted binding can be alleviated by using circular substrates or modifying the ends of the linear substrate with a biotin/streptavidin conjugate (Mason et al. 2008). Exonuclease activity on linear substrates can be minimized through the use of nuclease resistant non-ionic methylphosphonate linkages at the terminal ends of a linear substrate. When combined with biotin/streptavidin, these modifications increase NER and ICL repair signals in mammalian cell extracts (Mason et al. 2008; Smeaton et al. 2008).
Incision of ICLs in extracts
Using chromatin-associated protein extracts from human cells, Kumaresan, Lambert and colleagues studied processing of a site-specific 4,5′,8-trimethylpsoralen (HMT) psoralen ICL embedded in a ~140 linear substrate (Kumaresan et al. 1995; Kumaresan and Lambert 2000; Kumaresan et al. 2002; Kumaresan et al. 2007). They observed incisions on the 5′ and 3′ sides of the psoralen ICL. On the furan-adducted strand, incisions occurred at the 5th and 6th phosphodiester 5′ to the ICL, but on the pyrone-adducted strand, incisions occurred at the 13th and 14th phosphodiester 5′ to the ICL (Kumaresan et al. 1995). This difference in incision location based on which side of the psoralen ring is processed is consistent with the notion that the structure of an ICL can influence repair processing. In contrast, incisions on the 3′ side of the ICL were observed at the 4th or 5th phosphodiester bonds on both of the pyrone- and furan-adducted strands. These incisions were divalent metal ion (Mg2+)-dependent and were later shown to require ATP (Kumaresan and Lambert 2000).
In these experiments the 32P label was placed 5′ of the ICL. After incubation with cell extract, the ICL was reversed by irradiation with short wavelength UV light. This procedure allowed visualization of both 5′ and 3′ incisions within a single labeled substrate. Under these conditions, if both incisions were coupled on the same substrate, only the 5′-most incision would be observed. Since both 5′ and 3′ incisions were observed, this result indicates that the incisions are, at least part of the time, not coupled. It is not clear whether coupled incisions occur using chromatin-associated extracts that lead to the production of an unhooked product.
The XPF-ERCC1 nuclease, but not other NER proteins, was shown to be responsible for the incisions observed (Kumaresan and Lambert 2000; Kumaresan et al. 2002; Kumaresan et al. 2007). XPF-ERCC1 was shown to cooperate with proteins in the Fanconi anemia pathway and the scaffolding protein α IISp. Chromatin associated protein extracts derived from Fanconi anemia complementation groups A, B, C, D2, F and G were all defective in the production of XPF-ERCC1-mediated 5′ and 3′ incisions (Kumaresan and Lambert 2000; Kumaresan et al. 2007). In FA-A cells, the level of XPF-ERCC1 was normal, however in many of the cell lines including FA-A, there were reduced levels of α IISp. The incisions observed require the use of α IISp, as antibodies against α IISp inhibited incision. Purified α IISp enhanced the XPF-ERCC1-mediated incision and was shown to bind specifically to a psoralen cross-link with Fanconi anemia proteins (McMahon et al. 2001). Furthermore, α IISp co-localized with XPF and FANCA at sites of damage (Sridharan et al. 2003). As previously mentioned, because α IISp interacts with the nuclear matrix (Bachs et al. 1990) and there is evidence that ICL repair takes place at the nuclear matrix (Atanassov et al. 2005), it is believed that α IISp may serve as a scaffold for the assembly of ICL repair factors.
Cross-link repair synthesis assays in mammalian cell extracts
Extensive efforts have been made to gain a better understanding at the molecular level, of the proteins involved in ICL repair by carrying out a cross-link-induced repair synthesis (CRS) assay developed by Legerski’s group (Li et al. 1999). The assay employed a damaged plasmid containing a single psoralen ICL (CLT), a non-damaged control plasmid (CT) whose sequence was identical to that of CLT, and a slightly larger non-damaged donor plasmid (DT) (Li et al. 1999). All three plasmids were derived from the same parental plasmid and therefore contained homologous sequences. The plasmids used did not contain origins of replication or any mammalian promoters. These assays, which were carried out in human or rodent whole cell extracts, examined DNA synthesis by monitoring incorporation of radioactive nucleotides into plasmid DNA. Surprisingly, incubation of the CLT and DT plasmids in HeLa whole cell extract resulted in incorporation of radioactivity into both plasmids, however incubation of monoadducted plasmid with DT did not. This incorporation was approximately 20- to 30-fold greater than the incorporation observed when the DT was incubated with the CT plasmid or when the plasmids were incubated by themselves. These results are consistent with a mechanism whereby the psoralen ICL entered a recombination type repair pathway, a possibility that was confirmed by failure to observe incorporation in XRCC2 and XRCC3 mutant extracts (Li et al. 1999). Furthermore, it was shown that when an ICL was placed in direct repeat sequences, a single strand annealing pathway was responsible for ICL repair (Zheng et al. 2006). Certain XPF mutant cell lines tested were not defective in the CRS assay, indicating that certain mutations may affect the function of XPF in NER and ICL repair separately (Zhang et al. 2000). It was also shown that RPA and PCNA play crucial roles in productive CRS (Li et al. 2000).
Consistent with the results of Hartley and colleagues, the CRS assay was only slightly diminished in extracts derived from XPA-, XPC- or XPG-deficient cells. However, very little incorporation was observed when the CLT and DT plasmids were incubated in hamster whole cell extracts deficient in XPF or ERCC1 (Li et al. 1999; Zhang et al. 2000). This result is consistent with the genetic evidence that implicates a role for XPF-ERCC1 in ICL repair outside of NER.
The observed incorporation into both the damaged plasmid and the DT plasmid suggested some type of recombination had occurred. However, when donor plasmids with reduced degrees of homology were used, significant levels of incorporation into the donor plasmid were still observed. Furthermore, HeLa whole cell extracts immunodepleted of hRad51, a protein required for HR showed no decrease in the level of incorporation into the DT plasmid. Nevertheless, incorporation into the DT plasmid was reduced significantly in extracts derived from XRCC2- and XRCC3- deficient cells. Taken together, these results suggested that HR was likely not involved in the repair of the ICLs in the extracts. Rather the authors suggested that some form of break-induced replication (BIR), which only requires limited extents of homology might be responsible for the repair synthesis seen in the CRS assay. The previously discussed role of XPF-ERCC1 in ICL-induced recombination pathways may also explain the pronounced effect observed in XPF-ERCC1 cells over other NER mutants.
The authors went on to use the CRS assay to analyze incisions. The complication with such an analysis is that labeled repair synthesis products are actually being monitored, and therefore many of the initial incision sites may go unobserved due to nick translation, repair synthesis, and ligation. However, the authors did observe a repair synthesis product that terminated directly 3′ to the adducted psoralen ICL (Zhang et al. 2002). This observation along with the detection of DNA repair synthesis 5′ of the ICL indicated unhooking of the ICL had taken place. The authors also observed DSBs surrounding the site of the psoralen ICL. XPF-ERCC1, but not other NER factors, was required for these incisions. Furthermore, there was a dependency on MutSβ, but not MutSα or MutL, to specifically recognize psoralen ICLs and create the incisions surrounding the ICL. It was later found through extensive fractionation efforts that the proteins CDC5L, PRP19/PSO4, WRN, PLRG1 and SPF27 were also required in the CRS assay (Zhang et al. 2005). It was found that the helicase and not the exonuclease function of WRN was required for repair, consistent with observations by others (Cheng et al. 2006). Interestingly, many of these factors are also essential for pre-mRNA splicing and have appeared to evolve a dual role in DNA repair processing. It was also shown that knockdown of CDC5L or PRP19 decreased luciferase expression in the transcription-based luciferase reporter assay using a site specific psoralen cross-linked plasmid. Both the CRS assay and reporter assay rely on a number of ICL repair processing steps for a repair signal to be observed. This data clearly indicates a role for these factors in ICL repair processing, but at which mechanistic step they function remains a question. Interestingly, ICL unhooking was not observed when MutSβ, RPA, PCNA, Pso4 and WRN were reconstituted in vitro, a result that suggests that additional factors are required for the unhooking step (Zhang et al. 2005).
NER-dependent dual 5′ incisions
Another approach, similar to that used by Lambert and colleagues, has used linear substrates to monitor not only incision, but repair synthesis events. Work by Bessho, Sancar and colleagues demonstrated the surprising finding that the mammalian NER pathway makes dual incisions 22–28 nt 5′ to the site of the ICL (Bessho et al. 1997). These dual 5′ incisions release an undamaged oligonucleotide but do not result in removal of the ICL. Both mammalian cellular extracts as well as a purified, reconstituted NER system carried out the same dual 5′ incision reaction. We have also observed the same dual 5′ incision phenomena with psoralen ICLs and with alkyl cross-links of various chemical structures (Smeaton et al. 2008). Consistent with the known response of NER to distortions, we have found that the level of the dual 5′ incisions correlates with the level of distortion induced by the ICL, but not the chemical structure of the ICL itself. For instance, when a N4C-ethyl-N4C cross-link was placed into a -CG-sequence context, the resulting structure was found to be very similar to B-form DNA and demonstrated very little distortion as measured by NMR and X-ray structure analysis (Noll et al. 2005; Swenson et al. 2007). However, when placed in a -GC- sequence context, this cross-link induced significant distortion to the DNA duplex including increased bending and dynamics in the DNA as well as a small unpaired region surrounding the ICL site (Noll et al. 2005; Smeaton et al. 2008). The level of dual 5′ incisions was much greater in substrates containing the distorted -GC- ICL versus those containing the non-distorting -CG- ICL. It is therefore of considerable interest to determine, in the absence of fork encounter, which properties of the ICL are responsible for recognition.
The NER apparatus makes a bubble of approximately 25 nt that surrounds the site of a monoadduct lesion in DNA (Evans et al. 1997). The inability of the NER machinery to make incisions on either side of an ICL may simply be that the same open complex cannot form around an ICL, which inherently blocks DNA duplex opening. A remaining question is whether the dual 5′ incisions occur in vivo. It has been shown that removal of a psoralen cross-link from the genome of intact cells requires XPC and the GG-NER pathway and not transcription during G1 phase (Muniandy et al. 2009). It remains possible that the NER apparatus in cell extracts can only create dual 5′ incisions, but an additional factor allows the NER machinery to make dual incisions bracketing the ICL leading to unhooking in vivo. It also remains a formal possibility that the dual 5′ incisions do occur in vivo and are necessary for the recruitment of other proteins that unhook the ICL.
It was found that after the dual 5′ incisions occur, the gap that remains undergoes a futile repair synthesis process (Mu et al. 2000). Repair synthesis occurred up to but not past the site of the psoralen cross-link, a result that is consistent with our own observations (Smeaton et al. 2009). This leaves a nick just 5′ of the ICL which may be subject to polymerase exonucleolytic degradation of the newly synthesized DNA. This results in a futile process of fill-in and resection. However, a portion of the molecules religated to regenerate the original cross-linked duplex. The encounter and blockage of the polymerase at the site of the ICL due to repair synthesis after dual 5′ incisions may initiate a repair response that results in removal of the ICL.
Unhooking of ICLs in mammalian cell extracts
In addition to the dual 5′ incisions, we have recently observed an ICL unhooking activity in mammalian cell extracts that is carried out by unidentified proteins (Smeaton et al. 2008). Cross-link unhooking was competitively inhibited by cross-linked linear DNA duplexes, but not by the corresponding non-damaged duplex, a result that showed the unhooking reaction was specific for ICL-damaged DNA (Smeaton et al. 2008). To further support this notion, the level of unhooking was shown to be significantly dependent upon the level of distortion induced by the ICL. Similar to the dual 5′ incisions, the level of unhooking was 10-fold greater for the distorted -GC- ICL compared to the non-distorting -CG- ICL. Cross-links with intermediate levels of distortion were found to undergo intermediate levels of unhooking. These results indicated that the level of helix distortion induced by a cross-link may significantly influence ICL recognition and repair. We also found that although the dual 5′ incisions and cross-link unhooking reactions were responsive to distortions, they did not depend on the chemical structure of the ICL.
Incisions were observed on the 5′ and 3′ sides of an ICL embedded in the middle of a 150 bp linear DNA duplex. The incisions, which required ATP and Mg2+, were located between 1 and 2 nt away from the ICL on the 5′ side of alkyl ICLs and 4 nt 5′ to a psoralen ICL. In agreement with the results of Sancar (Bessho et al. 1997), we did not observe dual 3′ incisions mediated by the NER pathway. However we did observe incisions ranging from the 3rd to 7th nt 3′ of an alkyl ICL and 4th to 8th nt 3′ of a psoralen ICL. In the studies of Bessho and colleagues (Bessho et al. 1997), there does appear to be faint signals from a single 5′ incision and multiple 3′ incisions similar to those that we have observed. There are two explanations for why the incision signals we observed were of higher intensity than those seen by Bessho et al. First, in the work by Bessho, dNTPs were employed in the ICL incision reactions. We found that addition of dNTPs to the extracts significantly decreased the 5′ and 3′ incision signals (Smeaton et al. 2008). We attributed this reduction to the likelihood that fill-in and ligation will obscure the incision signals. Second, the cross-linked duplexes we employed contained methylphosphonate and biotin/strepavidin end modifications as described above, and these modifications increased the intensities of the incision signals (Mason et al. 2008).
Sequencing gel analysis of incisions also complicates the ability to observe an unhooked product. This is due to the fact that cross-linked molecules migrate with anomalous mobilities on denaturing gels because they can reanneal with high efficiency. Under denaturing conditions, cross-linked molecules adopt a variety of structures ranging from completely duplexed to completely denatured. It is known that a fully duplexed ICL substrate will run faster than a denatured ICL substrate on a denaturing gel (Bessho et al. 1997; Smeaton et al. 2008). Therefore, on denaturing sequencing gels with substrates on the order of ~150 bp, the ICL substrate itself runs as a large smear rather than a clean band. On a sequencing gel, a single-stranded 150mer migrates with a mobility that is in the center of the large smear generated by the ICL substrate. This phenomena likely accounts for why both we and others did not initially observe an unhooked product, as it was masked by the smear of unprocessed ICL on sequencing gels. The use of a 6% non-sequencing polyacrylamide gel run at elevated temperature (60°C) allowed greater denaturation of the ICL substrate and thus efficient separation of the cross-linked and single stranded 150mer (Smeaton et al. 2008). Using this assay system we demonstrated that the single, NER-independent 5′ and 3′ incisions were coupled and resulted in an unhooked product (Smeaton et al. 2008). Unhooking occurred efficiently in all NER cells lines tested, including an ERCC1 knockout line and in the UV41, XPF mutant line, results that indicated that XPF-ERCC1 is not involved in the ICL unhooking we observed. Like the 5′ and 3′ incisions, the ICL unhooking reaction required ATP and metal ions. The unhooked product was found to be a single-stranded 150mer that contained the remnant of the cross-link. Fragments resulting from DSBs were not observed in these reactions. We later showed that a subsequent NER-independent repair synthesis reaction occurs that fills in the gap created by unhooking of the cross-link with the repair synthesis patch occurring between the observed sites of NER-independent incisions. This result supports the notion that the 5′ and 3′ incisions give rise to the unhooked product whose removal is coupled to a repair synthesis step (Smeaton et al. 2008; Smeaton et al. 2009).
The positions of the 5′ and 3′ incisions suggested that an oligonucleotide 4–7 bases in length would remain attached to the cross-link remnant of the unhooked product. However, careful analysis of the unhooked product showed that it contained only a single nt attached to the tethered ICL (Smeaton et al. 2009). This result could be due to a number of reasons. First, it is possible that the 5′ and 3′ incisions were not both generated by endonucleases. A single endonucleolytic incision followed by exonuclease activity that degraded past the cross-link site would result in unhooking with a single base tethered to the ICL. Kinetically, we observed that the 5′ and 3′ incisions were closely coupled, a result that is consistent with two coupled endonucleolytic cuts or one endonucleolytic cut closely coupled with exonuclease degradation (Smeaton et al. 2008). Such coupling argues against a mechanism in which nicking is followed by random exonuclease action. Furthermore, we observe a single base product in a time frame in which non-specific nucleases do not degrade the unprotected excised damaged oligonucleotide that is generated by NER in our mammalian based extracts (Smeaton et al. 2008). It has been demonstrated that when a polymerase encounters an unhooked, tethered ICL, if an oligonucleotide remains attached, this serves as a potent block to bypass by E. coli pol IV (Kumari et al. 2008). However, if only a single nt resides on the tethered ICL, bypass is observed with higher efficiency. Therefore, if unhooking occurs by two endonucleolytic cuts, there is likely a coupled exonucleolytic step to remove the oligonucleotide in order to promote polymerase bypass.
Surprisingly the unhooking we observed in whole cell extracts occurred completely independent of NER proteins, including XPF-ERCC1 and XPC (Smeaton et al. 2008). The lack of dependence on XPC, the NER damage recognition protein, indicates that, at least in extracts, the observed dual 5′ incisions and the formation of a 5′ open complex are not required for unhooking. It has been shown, however, that the GG-NER pathway is absolutely required for the removal of psoralen ICLs from the genome of G1 phase cultured mammalian cells (Muniandy et al. 2009). Muniandy, Seidman and colleagues used a unique method of laser pulsing to form psoralen ICLs with high efficiency in specific regions of the genome. These authors found that removal of the psoralen ICL was dependent on the GG-NER pathway and was not affected by transcription. This observation clearly demonstrates that ICL repair from latent DNA does occur in mammalian G1 phase cells.
The factors required for the ICL unhooking reaction that we have observed in mammalian whole cell extracts remain unknown. A small scale, rapid method was developed to make Manley based whole cell extracts that are competent for NER and ICL unhooking (Smeaton et al. 2007). This method allowed us to test the unhooking ability of a large panel of DNA repair mutant cell lines implicated in ICL repair (Smeaton and Miller, unpublished results). As mentioned, all NER mutant extracts tested were able to unhook the cross-link. We found that efficient unhooking occurred in cell extracts derived from lines defective in mismatch repair factors including two separate lines defective in MutSβ. Extracts derived from MutL or MutSα mutant cells were also able to unhook the cross-link. We also observed efficient unhooking in a WRN helicase mutant extract. These results indicate that the unhooking activity we observe is distinct from that observed by Legerski and colleagues in the CRS assay. Furthermore, we have observed efficient unhooking in extracts deficient in FANCG, which along with the lack of dependence on XPF-ERCC1, argues that the processing we observe is different from that of Lambert and colleagues. We have also found that ICL unhooking occurs efficiently in MUS81 mutant extracts. Extracts deficient in XRCC2 and XRCC3 were able to unhook the cross-link, which is not surprising considering HR is downstream of unhooking. A recently identified nuclease, SLX1-SLX4 (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009), was shown to be involved in ICL repair and to interact with XPF-ERCC1 and MUS81-EME1, demonstrating it helps coordinate repair proteins at sites of ICL repair. This protein was shown to have Holiday junction resolvase activity and to be involved in recombination. Furthermore, it was shown that depletion of SLX1 or SLX4 does not affect the production of DSBs during S phase repair of ICLs, but rather is required for resolving them. Therefore, it seems unlikely that this factor is responsible for the initial unhooking step, but is rather acting downstream in the ICL repair process. However, its coordination with two important ICL repair nucleases may mean that even in G1 phase, this complex may coordinate with yet other nucleases to sites of ICL damage.
NER and unhooking during G1 phase ICL repair
Repair of ICLs from genomic DNA in G1 phase of mammalian cells was found to be dependent on the GG-NER pathway (Muniandy et al. 2009) and repair of reporter plasmids without origins of replication that contain a site specific ICL also requires the entire complement of the NER machinery, with the primary route being through the TC-NER pathway [(Wang et al. 2001; Zheng et al. 2003) and Hlavin and Miller, unpublished results]. Therefore, it is apparent that there is a major requirement for the entire NER pathway in vivo for replication-independent ICL repair. An important question that remains is whether in vivo the NER pathway is able to unhook the cross-link, or if the NER-dependent dual 5′ incisions observed in extracts also occur in vivo and serve as an absolute signal for ICL removal in the absence of fork encounter. Whether there is a relation in vivo between the dual 5′ incisions and the unhooking we observe is unknown. It is possible that some form of bubble structure is generated by helicase activity next to an ICL to stimulate repair even in the absence of a stalled polymerase. The NER-dependent dual 5′ incisions creates a bubble of ~25 nt 5′ of the ICL, which may stimulate removal of the ICL by creating a structure required for unhooking in vivo. NER dual 5′ incisions were not required for the unhooking we observe in whole cell extracts. However, in an extract, proteins are mixed together, while in a living cell, proteins are compartmentalized. Thus, it remains possible that the unhooking activity we observe in extracts is related to NER and that in intact cells, but not in extracts, the NER dual 5′ incisions are an absolute requirement for recruitment of the unhooking factors.
Overall, mechanistic studies have shown that the level of distortion induced by an ICL can influence the initial recognition and processing step, whereas the chemical structure of an ICL can influence the repair synthesis step (Smeaton et al. 2008; Smeaton et al. 2009). Furthermore, it appears that there are not only different pathways of ICL repair depending on the phase of the cell cycle, but that even during the G1 phase many different pathways may act to repair ICLs.
CONCLUSION
It should be apparent that repair of ICLs is a complicated process and that the results of experiments designed to probe the repair mechanisms appear in some cases to be contradictory. The use of defined ICL substrates has significantly advanced our understanding of the molecular details of ICL processing, although the pathways involved and their mechanisms of action have yet to be completely elucidated. The results thus far demonstrate the importance of multiple pathways in order to alleviate the extreme cytotoxicity of exogenously and endogenously derived ICLs throughout the cell cycle.
There is mounting evidence that cross-link structure and cross-link-induced distortions affect the position of DNA polymerase stalling (Raschle et al. 2008); can affect repair in a transcription-biased HCR assay (our unpublished results); and can significantly affect on unhooking and subsequent repair synthesis steps (Smeaton et al. 2008; Smeaton et al. 2009). An understanding of how the chemical nature of ICLs affect repair may influence the design of more effective bifunctional chemotherapeutic agents.
Finally, as evidenced by the above discussions and summarized in Figure 3, the context in which an ICL is encountered, such as during replication, transcription or in quiescent DNA, may dictate which ICL repair proteins or pathways are involved. Thus, the ICL repair pathways that should be considered ‘dominant’ appear to depend on the context in which the ICL is encountered.
Figure 3.
Diagram of three contexts of interstrand cross-link recognition: replication-coupled ICL repair, transcription-coupled ICL repair, and global-genome ICL repair. The proteins implicated in ICL repair for each specific DNA context are listed below each diagram.
From a practical point of view, the ability to define distinct pathways of ICL repair in S phase and G1 may also help develop methods to increase cancer cytotoxicity while reducing side-effects in non-dividing cells by selectively inhibiting S phase specific ICL repair pathways. Furthermore, G1 type ICL repair pathways may play an essential role in protection against endogenous ICLs and therefore gaining further insight of G1 ICL repair may lead to a better understanding of some of the pathways that protect against malignancy and aging.
Acknowledgments
The work from our laboratory that was described in this review was supported by a grant from the National Cancer Institute (CA082785, P.S.M.). E.M.H. was supported by a training grant from the National Cancer Institute (T32 CA09110) and M.B.S. was supported by a predoctoral fellowship from the American Heart Association (103527).
References
- Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28(16):5082–92. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B, Kang D, Kim H, Wei Q. Repair of mitomycin C cross-linked DNA in mammalian cells measured by a host cell reactivation assay. Mol Cells. 2004;18(2):249–55. [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20(21):8283–9. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, et al. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36(1):1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35(1):128–35. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson BS, Sadeghi T, Siciliano MJ, Legerski R, Murray D. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer Chemother Pharmacol. 1996;38(5):406–16. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Ren K. Fanconi Anemia Proteins, DNA Interstrand Crosslink Repair Pathways and Cancer Therapy. Curr Cancer Drug Targets. 2009;9:101–117. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov B, Gospodinov A, Stoimenov I, Mladenov E, Russev G, Tsaneva I, Anachkova B. Repair of DNA interstrand crosslinks may take place at the nuclear matrix. J Cell Biochem. 2005;96(1):126–36. doi: 10.1002/jcb.20518. [DOI] [PubMed] [Google Scholar]
- Bachs O, Lanini L, Serratosa J, Coll MJ, Bastos R, Aligue R, Rius E, Carafoli E. Calmodulin-binding proteins in the nuclei of quiescent and proliferatively activated rat liver cells. J Biol Chem. 1990;265(30):18595–600. [PubMed] [Google Scholar]
- Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci U S A. 1997;94(9):4306–11. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35(5):704–15. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181(9):2878–82. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36(12):3506–13. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24(2):70–6. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278(7):5250–4. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol Cell Biol. 1997;17(12):6822–30. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem. 1997;272(6):3833–7. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D’Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 participates in the Fanconi anemia pathway of crosslink repair. Mol Cell Biol. 2009 doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–69. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem Soc Trans. 2006;34(Pt 5):633–45. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- Busch DB, van Vuuren H, de Wit J, Collins A, Zdzienicka MZ, Mitchell DL, Brookman KW, Stefanini M, Riboni R, Thompson LH, et al. Phenotypic heterogeneity in nucleotide excision repair mutants of rodent complementation groups 1 and 4. Mutat Res. 1997;383(2):91–106. doi: 10.1016/s0921-8777(96)00048-1. [DOI] [PubMed] [Google Scholar]
- Caldecott K, Jeggo P. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat Res. 1991;255(2):111–21. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
- Carreau M, Alon N, Bosnoyan-Collins L, Joenje H, Buchwald M. Drug sensitivity spectra in Fanconi anemia lymphoblastoid cell lines of defined complementation groups. Mutat Res. 1999;435(1):103–9. doi: 10.1016/s0921-8777(99)00041-5. [DOI] [PubMed] [Google Scholar]
- Caulfield JL, Wishnok JS, Tannenbaum SR. Nitric oxide-induced interstrand cross-links in DNA. Chem Res Toxicol. 2003;16(5):571–4. doi: 10.1021/tx020117w. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34(9):2751–60. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278(27):25172–8. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–87. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13(8):729–33. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20(20):7643–53. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. DNA damage, repair systems and human hypersensitive diseases. J Environ Pathol Toxicol. 1980;3(4 Spec No):53–68. [PubMed] [Google Scholar]
- Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc Natl Acad Sci U S A. 1973;70(4):1064–8. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RS, Sinden RR. Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 1975;5B:487–95. doi: 10.1007/978-1-4684-2898-8_10. [DOI] [PubMed] [Google Scholar]
- Damia G, Imperatori L, Stefanini M, D’Incalci M. Sensitivity of CHO mutant cell lines with specific defects in nucleotide excision repair to different anti-cancer agents. Int J Cancer. 1996;66(6):779–83. doi: 10.1002/(SICI)1097-0215(19960611)66:6<779::AID-IJC12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20(21):7980–90. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30(17):3848–56. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol Cell Biol. 2005;25(17):7569–79. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486(4):217–47. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Episkopou H, Kyrtopoulos SA, Sfikakis PP, Fousteri M, Dimopoulos MA, Mullenders LH, Souliotis VL. Association between transcriptional activity, local chromatin structure, and the efficiencies of both subpathways of nucleotide excision repair of melphalan adducts. Cancer Res. 2009;69(10):4424–33. doi: 10.1158/0008-5472.CAN-08-3489. [DOI] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16(21):6559–73. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138(1):78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J Biol Chem. 2008;283(3):1275–81. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18(1):73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23(4):471–82. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Ehmann UK, Williams JI. Human diseases associated with defective DNA repair. Adv Radiat Biol. 1979;8:86–174. [Google Scholar]
- Fujiwara Y. Defective repair of mitomycin C crosslinks in Fanconi’s anemia and loss in confluent normal human and xeroderma pigmentosum cells. Biochim Biophys Acta. 1982;699(3):217–25. doi: 10.1016/0167-4781(82)90110-5. [DOI] [PubMed] [Google Scholar]
- Futscher BW, Pieper RO, Dalton WS, Erickson LC. Gene-specific DNA interstrand cross-links produced by nitrogen mustard in the human tumor cell line Colo320HSR. Cell Growth Differ. 1992;3(4):217–23. [PubMed] [Google Scholar]
- Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 2007;35(22):7566–76. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20(11):1429–34. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113(3):357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25(20):4921–32. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 1985;45(4):1737–43. [PubMed] [Google Scholar]
- Huang JC, Sancar A. Determination of minimum substrate size for human excinuclease. J Biol Chem. 1994;269(29):19034–40. [PubMed] [Google Scholar]
- Islas AL, Baker FJ, Hanawalt PC. Transcription-coupled repair of psoralen cross-links but not monoadducts in Chinese hamster ovary cells. Biochemistry. 1994;33(35):10794–9. doi: 10.1021/bi00201a029. [DOI] [PubMed] [Google Scholar]
- Islas AL, Vos JM, Hanawalt PC. Differential introduction and repair of psoralen photoadducts to DNA in specific human genes. Cancer Res. 1991;51(11):2867–73. [PubMed] [Google Scholar]
- Jones JC, Zhen WP, Reed E, Parker RJ, Sancar A, Bohr VA. Gene-specific formation and repair of cisplatin intrastrand adducts and interstrand cross-links in Chinese hamster ovary cells. J Biol Chem. 1991;266(11):7101–7. [PubMed] [Google Scholar]
- Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125(1):50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Hang B, Lambert MW. Human endonucleolytic incision of DNA 3′ and 5′ to a site-directed psoralen monoadduct and interstrand cross-link. J Biol Chem. 1995;270(51):30709–16. doi: 10.1074/jbc.270.51.30709. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Hwang M, Thelen MP, Lambert MW. Contribution of XPF functional domains to the 5′ and 3′ incisions produced at the site of a psoralen interstrand cross-link. Biochemistry. 2002;41(3):890–6. doi: 10.1021/bi011614z. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Lambert MW. Fanconi anemia, complementation group A, cells are defective in ability to produce incisions at sites of psoralen interstrand cross-links. Carcinogenesis. 2000;21(4):741–51. doi: 10.1093/carcin/21.4.741. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Sridharan DM, McMahon LW, Lambert MW. Deficiency in incisions produced by XPF at the site of a DNA interstrand cross-link in Fanconi anemia cells. Biochemistry. 2007;46(50):14359–68. doi: 10.1021/bi7015958. [DOI] [PubMed] [Google Scholar]
- Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J Biol Chem. 2008;283(41):27433–7. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275(34):26632–6. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. Embo J. 2006;25(2):387–97. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22(8):430–6. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Larminat F, Bohr VA. Role of the human ERCC-1 gene in gene-specific repair of cisplatin-induced DNA damage. Nucleic Acids Res. 1994;22(15):3005–10. doi: 10.1093/nar/22.15.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larminat F, Zhen W, Bohr VA. Gene-specific DNA repair of interstrand cross-links induced by chemotherapeutic agents can be preferential. J Biol Chem. 1993;268(4):2649–54. [PubMed] [Google Scholar]
- Legerski R. Mechanisms of S phase repair of interstrand crosslinks. Environ Mol Mutag 2010 [Google Scholar]
- Li L, Peterson CA, Lu X, Wei P, Legerski RJ. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol Cell Biol. 1999;19(8):5619–30. doi: 10.1128/mcb.19.8.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peterson CA, Zhang X, Legerski RJ. Requirement for PCNA and RPA in interstrand crosslink-induced DNA synthesis. Nucleic Acids Res. 2000;28(6):1424–7. doi: 10.1093/nar/28.6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang N, Vasquez K, Barton M, Legerski R. Repair of psoralen interstrand cross-links in Xenopus laevis egg extracts is highly mutagenic. Biochem Biophys Res Commun. 2005;336(1):69–75. doi: 10.1016/j.bbrc.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci U S A. 2006;103(26):9891–6. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason TM, Smeaton MB, Cheung JC, Hanakahi LA, Miller PS. End modification of a linear DNA duplex enhances NER-mediated excision of an internal Pt(II)-lesion. Bioconjug Chem. 2008;19(5):1064–70. doi: 10.1021/bc7004363. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5(10):1044–7. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20(10):3425–33. doi: 10.1128/mcb.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2(8):483–90. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- McMahon LW, Sangerman J, Goodman SR, Kumaresan K, Lambert MW. Human alpha spectrin II and the FANCA, FANCC, and FANCG proteins bind to DNA containing psoralen interstrand cross-links. Biochemistry. 2001;40(24):7025–34. doi: 10.1021/bi002917g. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304(5678):1822–6. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986;83(23):8878–82. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–9. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15(2):232–40. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20(7):2446–54. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser localized DNA interstrand crosslinks in G1 phase mammalian cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35(1):116–27. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Mustra DJ, Warren AJ, Hamilton JW. Preferential binding of human full-length XPA and the minimal DNA binding domain (XPA-MF122) with the mitomycin C-DNA interstrand cross-link. Biochemistry. 2001;40(24):7158–64. doi: 10.1021/bi002820u. [DOI] [PubMed] [Google Scholar]
- Mustra DJ, Warren AJ, Wilcox DE, Hamilton JW. Preferential binding of human XPA to the mitomycin C-DNA interstrand crosslink and modulation by arsenic and cadmium. Chem Biol Interact. 2007;168(2):159–68. doi: 10.1016/j.cbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Nairn RS. Multiple roles of ERCC1-XPF in mammalian interstrand crosslink repair. Environ Mol Mutag. 2010 doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. Embo J. 2001;20(22):6540–9. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123(7):1191–8. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24(13):5776–87. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106(2):277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll DM, Noronha AM, Wilds CJ, Miller PS. Preparation of interstrand cross-linked DNA oligonucleotide duplexes. Front Biosci. 2004;9:421–37. doi: 10.2741/1246. [DOI] [PubMed] [Google Scholar]
- Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, flexibility, and repair of two different orientations of the same alkyl interstrand DNA cross-link. Biochemistry. 2005;44(18):6764–75. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26(23):8722–30. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 2007;6(7):1004–17. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Rajski SR, Williams RM. DNA Cross-Linking Agents as Antitumor Drugs. Chem Rev. 1998;98(8):2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44(11):4188–95. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24(1):123–34. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage E, Drobetsky EA, Moustacchi E. 8-Methoxypsoralen induced mutations are highly targeted at crosslinkable sites of photoaddition on the non-transcribed strand of a mammalian chromosomal gene. Embo J. 1993;12(2):397–402. doi: 10.1002/j.1460-2075.1993.tb05671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo J. 2006;25(6):1285–94. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scicchitano DA, Olesnicky EC, Dimitri A. Transcription and DNA adducts: what happens when the message gets cut off? DNA Repair (Amst) 2004;3(12):1537–48. doi: 10.1016/j.dnarep.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Sczepanski JT, Jacobs AC, Greenberg MM. Self-promoted DNA interstrand cross-link formation by an abasic site. J Am Chem Soc. 2008;130(30):9646–7. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94(21):11205–9. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272(3):1885–90. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281(20):13869–72. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86(5):811–22. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, McGregor Mason T, Noronha AM, Wilds CJ, Miller PS. Distortion-dependent unhooking of interstrand cross-links in mammalian cell extracts. Biochemistry. 2008;47(37):9920–30. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, Noronha AM, Murphy SP, Wilds CJ, Miller PS. Effect of cross-link structure on DNA interstrand cross-link repair synthesis. Chem Res Toxicol. 2009;22(7):1285–97. doi: 10.1021/tx9000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;35(22):e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan D, Brown M, Lambert WC, McMahon LW, Lambert MW. Nonerythroid alphaII spectrin is required for recruitment of FANCA and XPF to nuclear foci induced by DNA interstrand cross-links. J Cell Sci. 2003;116(Pt 5):823–35. doi: 10.1242/jcs.00294. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. Embo J. 2009;28(8):1111–20. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, et al. Interstrand DNA cross-links induced by alpha, beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41(7):793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield FW, Tappel AL. Detection and measurement by high-performance liquid chromatography of malondialdehyde crosslinks in DNA. Anal Biochem. 1984;143(2):265–71. doi: 10.1016/0003-2697(84)90662-6. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138(1):63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson MC, Paranawithana SR, Miller PS, Kielkopf CL. Structure of a DNA repair substrate containing an alkyl interstrand cross-link at 1.65 A resolution. Biochemistry. 2007;46(15):4545–53. doi: 10.1021/bi700109r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33(9):2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat Res. 2009;668(1–2):54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos JM, Hanawalt PC. Processing of psoralen adducts in an active human gene: repair and replication of DNA containing monoadducts and interstrand cross-links. Cell. 1987;50(5):789–99. doi: 10.1016/0092-8674(87)90337-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21(3):713–20. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ Mol Mutag. 2010 doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6(6):551–7. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem. 2005;280(49):40559–67. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6(11):1670–8. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol Cell Biol. 2002;22(7):2388–97. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang X, Peterson C, Li L, Legerski R. Differential processing of UV mimetic and interstrand crosslink damage by XPF cell extracts. Nucleic Acids Res. 2000;28(23):4800–4. doi: 10.1093/nar/28.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37(13):4420–9. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wang X, Legerski RJ, Glazer PM, Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair (Amst) 2006;5(5):566–74. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23(2):754–61. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]