The Chlamydia-encoded protein Tarp is phosphorylated in the host cell cytoplasm and is a multivalent hub for antiapoptotic signaling molecules.
Abstract
Many bacterial pathogens translocate effector proteins into host cells to manipulate host cell functions. Here, we used a protein microarray comprising virtually all human SRC homology 2 (SH2) and phosphotyrosine binding domains to comprehensively and quantitatively assess interactions between host cell proteins and the early phase Chlamydia trachomatis effector protein translocated actin-recruiting phosphoprotein (Tarp), which is rapidly tyrosine phosphorylated upon host cell entry. We discovered numerous novel interactions between human SH2 domains and phosphopeptides derived from Tarp. The adaptor protein SHC1 was among Tarp’s strongest interaction partners. Transcriptome analysis of SHC1-dependent gene regulation during infection indicated that SHC1 regulates apoptosis- and growth-related genes. SHC1 knockdown sensitized infected host cells to tumor necrosis factor–induced apoptosis. Collectively, our findings reveal a critical role for SHC1 in early C. trachomatis–induced cell survival and suggest that Tarp functions as a multivalent phosphorylation-dependent signaling hub that is important during the early phase of chlamydial infection.
Introduction
The obligate intracellular pathogen Chlamydia trachomatis is the cause of preventable blindness (trachoma; Wright et al., 2008) and is responsible worldwide for up to 90 million cases per year of sexually transmitted disease (Brunham and Rey-Ladino, 2005). C. trachomatis exhibits a unique biphasic developmental cycle, which is initiated by the infectious elementary bodies (EBs). Once engulfed by the host cell, EBs differentiate into actively replicating reticulate bodies (RBs) within a protective vacuole called the inclusion. RBs eventually redifferentiate to form EBs, which are released ready to infect new cells.
Gram-negative bacteria such as Chlamydia are known to express and secrete host interactive proteins via type III or type IV secretion systems into the host cell cytoplasm to modulate host cell processes (Hueck, 1998; Christie et al., 2005). Some of these effector proteins become phosphorylated upon host cell entry, such as the translocated intimin receptor (Tir) of enteropathogenic Escherichia coli (EPEC; Kenny et al., 1997) and the cytotoxicity-associated gene A (CagA) of Helicobacter pylori (Asahi et al., 2000). C. trachomatis uses a type III system to secrete numerous effector proteins (Valdivia, 2008). One of these, the translocated actin-recruiting phosphoprotein (Tarp), becomes tyrosine phosphorylated upon entry of C. trachomatis into the host cell (Clifton et al., 2004). To date, several host cell kinases have been implicated in Tarp phosphorylation including SRC, ABL, and SYK (Elwell et al., 2008; Jewett et al., 2008; Mehlitz et al., 2008), and the number of Tarp phosphorylation sites varies between Chlamydia species and serovars (Jewett et al., 2008).
Although Chlamydia species continue to be refractory to genetic manipulation (Heuer et al., 2003), Tarp’s function has been assessed using the heterologous type III secretion system of Yersinia pseudotuberculosis (Clifton et al., 2004). This study revealed a critical role of Tarp in the actin-driven uptake of bacteria by host epithelial cells. Tyrosine phosphorylation of Tarp, however, appears to be uncoupled from actin polymerization because nonphosphorylated Chlamydophila pneumonia Tarp still induces actin polymerization (Clifton et al., 2005). Instead, actin polymerization is thought to be stimulated through oligomerization of a WAVE2-like actin-binding domain in the C terminus of the protein (Jewett et al., 2006), and Tarp phosphorylation takes place at multiple N-terminal motifs (Jewett et al., 2008). Phosphotyrosine-containing motifs are known to interact with SRC homology 2 (SH2) or phosphotyrosine binding (PTB) domains of signaling proteins (Schlessinger and Lemmon, 2003). Phosphorylation of Tyr179 and Tyr189 of Tarp has previously been implicated in recruiting the RAC guanine nucleotide exchange factor VAV2 and the regulatory subunit of phosphatidylinositol 3-kinase (PI3K; Lane et al., 2008). These interactions are thought to participate in a redundant invasion mechanism. Paradoxically, however, cell entry is independent of Tarp phosphorylation (Clifton et al., 2005; Jewett et al., 2006). A more comprehensive analysis of phosphotyrosine-mediated interactions is therefore necessary to establish Tarp’s functional repertoire.
A prerequisite of Chlamydia replication is the pathogen’s ability to prevent apoptosis of its host cell. C. trachomatis–infected cells become resistant to numerous pro-apoptotic stimuli, including staurosporine, etoposide, TNF, FAS antibody, and granzyme B/perforin (Fan et al., 1998). Anti-apoptotic activity is thought to be conveyed by the proteolytic degradation of the pro-apoptotic BH3-only proteins BIM/BOD and PUMA and by the mitochondrial sequestration of BAD during chlamydial infection (Fischer et al., 2004). Alternatively, recent studies have implicated the anti-apoptotic BCL-2 family member MCL-1 as a key factor in preventing apoptosis (Rajalingam et al., 2008). In addition, Chlamydia subverts the function of the pro-apoptotic PKCδ by increasing diacylglycerol levels in the chlamydial inclusion membrane (Tse et al., 2005). Together, these observations suggest that Chlamydia prevents host cell apoptosis through a variety of mechanisms, likely acting sequentially as infection proceeds (Fan et al., 1998; Perfettini et al., 2002; Rajalingam et al., 2008).
An important pathway modulating host cell apoptosis and survival is the RAS/RAF/MEK/ERK MAPK signaling cascade. C. trachomatis infection leads to extracellular signal-regulated kinase (ERK) activation, followed by the downstream activation of cytosolic phospholipase A2 (cPLA2; Su et al., 2004), the induction of interleukin-8 (IL-8; Buchholz and Stephens, 2008), TNF receptor 1 (TNFR1) shedding (Paland et al., 2008), and stabilization of MCL-1 (Rajalingam et al., 2008). Recent work has revealed that MEK (MAPK/ERK kinase)/ERK activation is independent of RAS/RAF during mid and late C. trachomatis infection (Gurumurthy et al., 2010). Upon activation, MEK1/2 phosphorylates ERK1/2 at specific tyrosine and threonine residues, which then directly phosphorylates a variety of transcription factors including c-JUN, c-MYC, and nuclear factor κB (NF-κB). The SRC homology containing protein SHC1 represents a molecular adapter, linking extracellular signals to mitogenic responses (Pelicci et al., 1992). SHC1 exists as three isoforms, all encoded by a single gene locus (Luzi et al., 2000). The isoforms share a common domain composition: a C-terminal SH2 domain, an N-terminal PTB domain, and a central collagen-homology domain (CH1) harboring several phosphorylation sites (Ravichandran, 2001). Interaction at these sites with growth factor receptor-bound protein 2 (GRB2), in conjunction with the RAS exchange factor SOS, activates the RAS/RAF/MEK/ERK pathway to induce a mitogenic response (van der Geer et al., 1996).
Here, we used a systematic approach to discover and quantify phosphorylation-dependent interactions between the bacterial effector protein Tarp and host cell SH2/PTB domain–containing proteins. We found that Tarp interacts with a variety of SH2 domain–containing proteins, which suggests it functions as a multivalent signaling hub. SHC1 was identified as one of the strongest interaction partners of phosphorylated Tarp derived from both chlamydial serovariants L2 and D. Knockdown of SHC1 sensitized C. trachomatis-infected cells to apoptosis at an early stage of the chlamydial developmental cycle and blocked induction of cell growth–related gene expression. SHC1-mediated cell survival and gene regulation was found to be controlled through both MEK/ERK-dependent and -independent signaling. Thus, C. trachomatis confers resistance to pro-apoptotic stimuli upon host cells during the early stages of infection. This facet of apoptosis resistance appears to be mediated by Tarp’s interactions with SHC1.
Results
Peptide design of Tarp phosphorylation sites
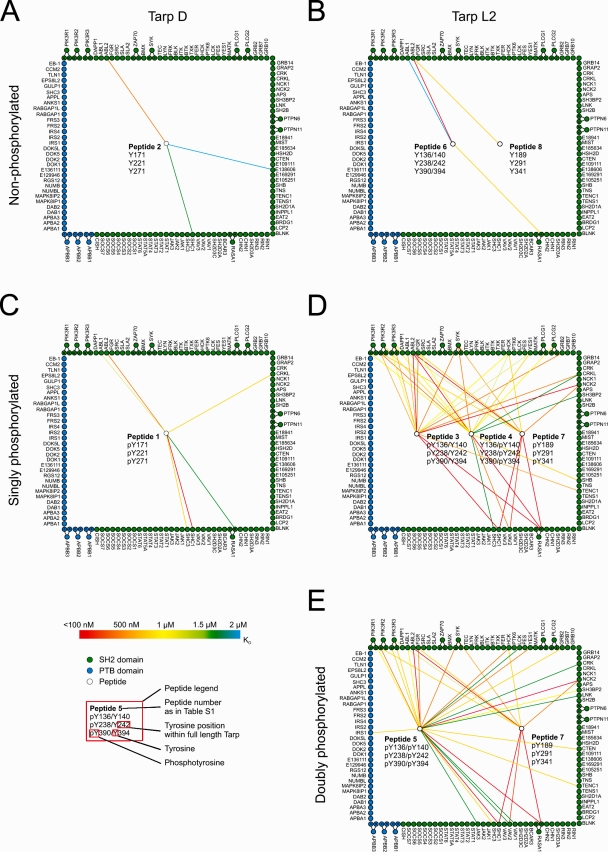
To identify the cellular interaction partners of Tarp, we synthesized fluorescently labeled peptides representing the N-terminal sites of tyrosine phosphorylation on Tarp. C. trachomatis D Tarp features three identical ENIYE motifs at tyrosine residues Y171, Y221, and Y271 (Fig. 1 A), whereas C. trachomatis L2 Tarp features nine sites of tyrosine phosphorylation at residues Y136, Y140, Y189, Y239, Y242, Y291, Y341, Y390, and Y394 (Fig. 1 B). Six of these nine C. trachomatis L2 phosphorylation sites—Y136/140, Y238/242, and Y390/394—are double-phosphorylation sites of the primary sequence ENIYENIYESI (Fig. 1 B). Eight peptides, each comprising 18 amino acids surrounding the respective phosphorylation sites, were synthesized in their non-, singly, or doubly phosphorylated states (Table S1). A fluorescent dye was coupled to the N terminus of each peptide for detection purposes (see Materials and methods). These peptides were then used to investigate interactions with human SH2 and PTB domains.
Figure 1.
Quantitative protein interaction mapping. Quantitative data were obtained by probing SH2/PTB domain microarrays with fluorescently labeled peptides derived from Tarp. The resulting data were used to construct quantitative protein interaction maps. White circles within the square represent peptides derived from C. trachomatis. Peptide allocation can be derived from Table S1 and the peptide legend, which shows phosphorylation status and position within the full-length protein. For example, peptide 2 in A mimics the three identical ENIYE repeats of serovar D Tarp in the nonphosphorylated state. Green and blue circles around the square represent individual SH2 and PTB domains, respectively. Circles outside the square indicate tandem domains (the corresponding spot on the protein array contains both domains), which are connected to their individual domains (i.e., full-length SYK is in possession of two SH2 domains located either N or C terminally). A color-coded line is used to reflect the measured interaction strength between each domain–peptide pair (n = 2). Exact values are provided in Table S1. Interactions with KD ≥ 2 µM are omitted. (A and B) Interaction maps of nonphosphorylated Tarp D and L2. (C and D) Interaction maps of singly phosphorylated Tarp D and L2. (E) Interaction map of doubly phosphorylated Tarp from serovariant L2.
Quantitative interactome analysis
To identify and quantify interactions between Tarp-derived peptides and human signaling proteins, we probed protein microarrays comprising 133 SH2 and PTB domains in duplicate with eight concentrations of each Tarp-derived peptide, and then fit the observed fluorescence data, Fobs, to an equation that describes saturation binding, as described previously (Jones et al., 2006). This enabled us to measure binding affinities between every peptide and every recombinant domain. From the resulting quantitative dataset, we constructed a graphical representation of biophysical interactions with phospho-sites on the D and L2 serovariants of Tarp (Fig. 1). Three Tarp phosphorylation states were defined: nonphosphorylated (Fig. 1, A and B), singly phosphorylated (Fig. 1, C and D), and doubly phosphorylated (Fig. 1 E). Because not all SH2/PTB domain–containing proteins are expressed at appreciable levels in every cell type or at every subcellular location, these diagrams should be viewed as quantitative maps of potential Tarp interactions in the context of particular cell expression patterns.
Nonphosphorylated peptides derived from both Tarp D and Tarp L2 exhibited very few interactions with human SH2 and PTB domains, as anticipated. Most notably, the tyrosine kinase ABL2 (ARG) and the GTPase-activating protein RASA1 interacted with Tarp peptides (Fig. 1, A and B; and Table S1). By probing the arrays with the singly phosphorylated version of the Tarp D peptide, the affinity of its interaction with ABL2 increased and several new interactions appeared, including a high affinity interaction with the SH2 domain of SHC1 (KD < 200 nM) and interactions with the SH2 domains of CRKL, ABL1, SHC3, and RASA1. Even more dramatic changes were observed with phosphorylated Tarp L2 peptides. Numerous high affinity interactions occurred, most notably with the SH2 domains of ABL1, ABL2, FGR, SYK, HCK, YES1, GRB2, CRKL, NCK2, SHB, TENC1, RASA1, VAV2, and SHC1; and with all three isoforms of the regulatory subunit of PI3K (Fig. 1 D). Interestingly, the incremental effect of introducing the doubly phosphorylated peptide was minimal: only minor changes in affinities for some interaction partners (e.g., VAV2, PI3KR2, SYK, and HCK) were observed (Fig. 1 E and Table S1). This observation is consistent with our previous findings that doubly phosphorylated peptides derived from a variety of human receptor tyrosine kinases behave similarly as compared with their singly phosphorylated counterparts (Jones et al., 2006). Notably, no PTB domains recognized any of the Tarp-derived peptides, except the PTB domain of IRS1, which recognized the doubly phosphorylated peptide derived from Tarp L2 (Fig. 1 E). Our data suggest that Tarp proteins may have been evolutionarily optimized to recognize SH2 domains that preferentially bind sequences C-terminally of phosphotyrosines (Sudol, 1998).
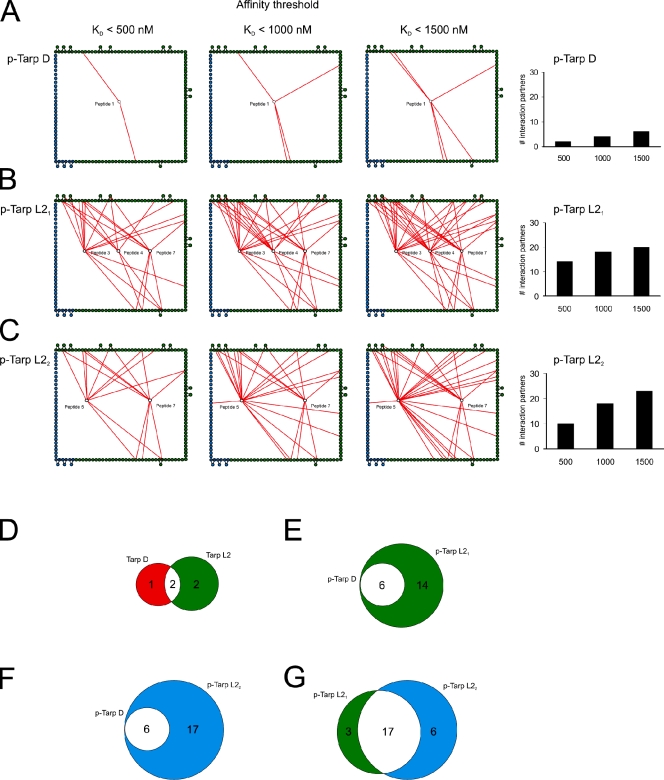
To highlight differences in binding affinities, the interaction maps for Tarp D, Tarp L21 (singly phosphorylated), and Tarp L22 (doubly phosphorylated) were prepared using three different affinity thresholds (Fig. 2, A–C). Tarp D exhibited high-affinity interactions (KD < 500 nM) with two proteins (Fig. 2 A), increasing to six as the threshold was relaxed to 1,500 nM (Fig. 2 A). In contrast, Tarp L21 (singly phosphorylated) exhibited promiscuous binding even at the highest affinity threshold (Fig. 2 B). Surprisingly, hyperphosphorylation of Tarp L2 decreased the number of high-affinity interactions, but the overall number of interaction partners for Tarp L22 sharply increased as the affinity threshold was relaxed (Fig. 2 C). Comparison of the different sets of interaction partners for both Tarp serovariants (Tarp D and L2) revealed two common interaction partners in the nonphosphorylated state (Fig. 2 D), and a core set of six proteins in the phosphorylated state (Fig. 2 E). This set included the adaptor protein SHC1, which bound both serovariants with high affinity. Tarp L21 and L22 were both found to interact with all of the phospho–Tarp D binding partners (Fig. 2, E and F), which suggests that signaling initiated by the Tarp D serovariant constitutes a subset of the signaling initiated by Tarp L2. Tarp L21 and L22 were very similar, with 17 interaction partners in common (Fig. 2 G).
Figure 2.
Quantitative analysis of the phospho-interactome. Interaction networks of the phospho-states from Fig. 1 were binned using three different affinity thresholds (KD < 500 nM, KD < 1000 nM, and KD < 1500 nM). In A–C, only peptide–domain interactions below the stated threshold are shown (red lines). Edge nodes are colored as in Fig. 1. p-Tarp D refers to phosphorylated Tarp from C. trachomatis serovariant D; p-Tarp L21 and L22 refer to singly and doubly phosphorylated Tarp from serovariant L2, respectively. (A) Tarp D is less promiscuous than Tarp L2 and has few interaction partners at all affinity thresholds. (B) Tarp L21 exhibits highly promiscuous binding even at the highest affinity threshold (KD < 500 nM). (C) Tarp L22 has fewer high-affinity interaction partners (KD < 500 nM) than Tarp L21, but the number of interactors increases sharply as the affinity threshold is relaxed. To the right, the number of Tarp interaction partners at each affinity threshold is depicted in bar diagrams. Multiple domains from proteins interacting with more than one phospho-site are only counted once. (D–G) Venn diagrams showing the number of overlapping interactions (white) between nonphosphorylated Tarp D (red) and L2 (green) (D), singly phosphorylated Tarp D (red) and L21 (green) (E), singly phosphorylated Tarp D (green) and doubly phosphorylated L22 (blue) (F), and singly phosphorylated Tarp L21 (green) and doubly phosphorylated L22 (blue) (G).
To identify signaling pathways that are potentially activated by Tarp, we used Ingenuity Pathway Analysis (IPA) software and the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/). Signaling pathways associated with at least two Tarp interaction partners were used to construct a qualitative model of Tarp-mediated signaling. Both serovariants of Tarp were found to interact with proteins commonly associated with EGF receptor (EGFR) signaling and MAPK signaling (Fig. S1, A–C). Interestingly, Tarp L2 also interacted with proteins associated with immune signaling pathways of B and T cells, as well as with innate immune responses of natural killer cells, macrophages, and neutrophils (Fig. S1, B and C). Connection of Tarp L2 with these additional signaling pathways derives mainly from interactions mediated by its doubly phosphorylated state (Fig. S1, B and C).
Overall, we found that phosphorylation of Tarp enables multiple high-affinity interactions with human SH2 domains; that a core set of proteins involved in MAPK signaling is able to interact with both serovariants of Tarp; and that signaling initiated by Tarp D likely constitutes a subset of the signaling initiated by Tarp L2.
Validation of interactions between Tarp and selected host cell proteins
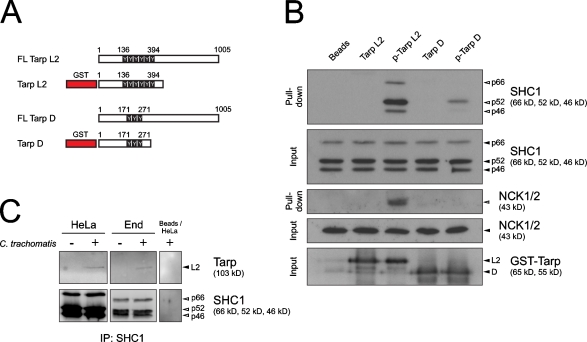
To test whether the identified protein interactions have relevance for chlamydial infection, we selected the strongest interaction partner of both serovariants of Tarp, SHC1, and a serovariant L2-specific interaction partner, NCK2, for further experimental validation. Heterologously expressed GST fused to Tarp from C. trachomatis serovariants D and L2 (Fig. 3 A) was coupled to beads, phosphorylated in vitro by SRC kinase, and incubated with host cell lysate. Truncated versions of N-terminal Tarp were used to avoid the actin-nucleating activity of the C-terminal region (Jewett et al., 2006) and additional interaction of cellular proteins with any noncharacterized Tarp motifs. Both phosphorylated Tarp D and Tarp L2 bound SHC1 and showed a strong preference for the SHC1p52 isoform (Fig. 3 B). In contrast, only phosphorylated Tarp L2 was capable of precipitating NCK2 (Fig. 3 B). This is consistent with the array data because peptides derived from phospho-sites on Tarp L2, but not Tarp D, were recognized by the SH2 domain of NCK2. We focused on SHC1, the common interaction partner of both serovariants, and found, in a coimmunoprecipitation experiment, that endogenous SHC1 and Tarp L2 coprecipitated from two separate C. trachomatis–infected cell types (Fig. 3 C). These experiments show that the interactions with SHC1 and NCK2 highlighted by the protein microarrays are also observed with full-length proteins.
Figure 3.
SHC1 is an interaction partner of Tarp D and L2. (A) Graphical representation of Tarp D and L2 and the respective GST–Tarp fusion proteins including all phospho-sites. Truncated versions of Tarp were used to avoid the actin-nucleating activity of the C terminus (Jewett et al., 2006) and any unknown binding regions. (B) Western blot showing pull-down of SHC1 and NCK2 from HeLa cell lysate using bead-coupled phospho-GST-Tarp. The fusion protein was phosphorylated in vitro with recombinant human SRC. Input shows Tarp loading (bead coupled, black arrowheads) and SHC1 and NCK2 loading in HeLa cell lysate (black arrowheads). Pulled-down SHC1 and NCK2 are indicated by open arrowheads. SHC1 was only pulled down in lanes where Tarp was phosphorylated (indicated as p-Tarp). NCK2 was only pulled down with p-Tarp L2. As a control, beads were phosphorylated in the absence of GST-Tarp and did not pull down SHC1. (C) Western blot showing coimmunoprecipitation of SHC1 (white arrowheads) and Tarp (black arrowhead) after infection of HeLa or End1/E6E7 cells (C. trachomatis L2, MOI 500) for 60 min.
SHC1 activation and influence on MEK/ERK signaling during C. trachomatis cell entry
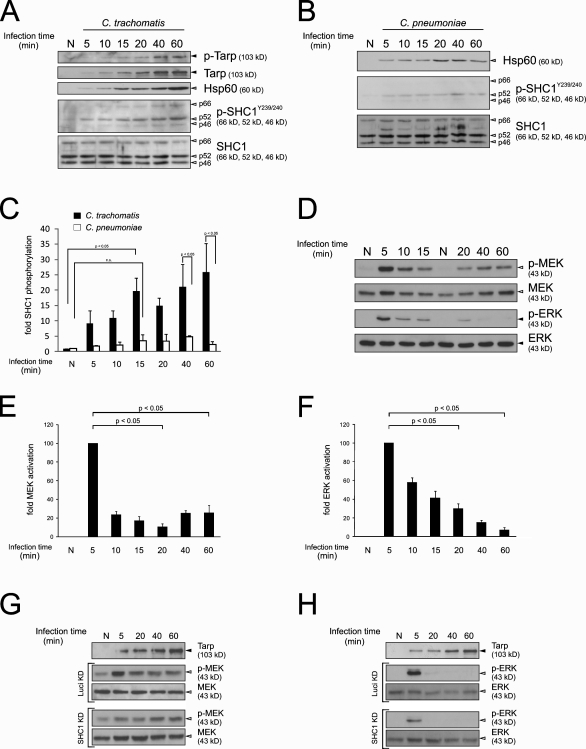
Next, we assessed the influence of Tarp–SHC1 interaction on major downstream signaling pathways by investigating the activation of the MEK/ERK cascade upon phosphorylation of SHC1. As previously shown, recruitment of SHC1 to endogenous human receptors leads to its phosphorylation on Tyr239/240 and Tyr317 (van der Geer et al., 1996). To test SHC1 activation during C. trachomatis infection, time course experiments were performed in HeLa cells. All three SHC1 isoforms were phosphorylated on Tyr239/240 during invasion of C. trachomatis (Fig. 4 A) for up to 5 h postinfection (pi; Fig. S2 A). In contrast, during infection with C. pneumoniae, which translocates a Tarp homologue (Cpn0572) lacking the phosphorylation sites (Clifton et al., 2005), SHC1 phosphorylation was only marginally increased (Fig. 4 B). Quantification of Western blotting data revealed that SHC1 phosphorylation was significantly increased during infections with C. trachomatis but not C. pneumoniae (Fig. 4 C). Interestingly, the SHC1p52 isoform was most rapidly and most strongly phosphorylated by C. trachomatis (Fig. 4 A), which is consistent with the observed binding preference of Tarp L2 and D (Fig. 3 B). Furthermore, immunofluorescence staining experiments showed that SHC1 activation was accompanied by recruitment of the protein to bacteria during host cell entry (Fig. S2, B and C).
Figure 4.
SHC1, MEK, and ERK are activated during early chlamydial infection. Western blots showing SHC1 phosphorylation on tyrosine residues Y239/240 (p-SHC1Y239/240, white arrowheads) and Tarp phosphorylation (p-Tarp, black arrowheads) during a time-course infection experiment (C. trachomatis L2, MOI 200; HeLa cells) (A) and a control experiment using C. pneumoniae TWAR (MOI 200, HeLa cells) showing only marginal SHC1 phosphorylation on tyrosine residues Y239/240 (p-SHC1Y239/240, white arrowheads) (B). Tarp phosphorylation was determined through use of a tyrosine phosphorylation–specific antibody. Hsp60 was used as an infection control (gray arrowhead). (C) Densitometric quantification of SHC1 p52 Y239/240 phosphorylation. Values were calculated from A and B and normalized to total SHC1 p52 (n = 2, error bars indicate standard error [SE]). (D) MEK (white arrowheads) and ERK (black arrowheads) phosphorylation during a time-course infection experiment (C. trachomatis L2, MOI 200; HeLa cells). MEK and ERK show a phosphorylation peak at ∼5 min. Densitometric quantification of MEK phosphorylation (E) and ERK phosphorylation (F). Values were calculated from D and normalized to total MEK and ERK, respectively (n = 2, error bars indicate SE). All values are normalized to the “5 minutes” infection time point. (G and H) Western blots showing MEK (G) and ERK phosphorylation (H) during an infection time course (C. trachomatis L2, MOI 200; HeLa cells). Cells were either treated with siRNA against Luciferase (control) or SHC1. MEK and ERK activation were reduced upon infection. (A, B, G, and H) Controls (N) were mock infected with SPG for 60 min.
Both SHC1 phosphorylation (van der Geer et al., 1996) and C. trachomatis infection (Su et al., 2004) have been shown to activate the MEK/ERK cascade; however, C. trachomatis–induced MEK/ERK activation has mainly been demonstrated at later infection time points, e.g., from 12 h pi (Paland et al., 2008). Because Tarp is secreted within minutes after attachment, we investigated MEK and ERK activation during this early phase of infection. Both MEK and ERK phosphorylation increased sharply 5–10 min pi in HeLa (Fig. 4, D–F) and End1/E6E7 cells (primary immortalized endocervical cells; unpublished data), then decreased to ∼20% and ∼10% above basal levels for MEK and ERK, respectively, after 1 h. MEK/ERK activation was dependent on MOI (unpublished data).
The role of SHC1 in infection and C. trachomatis–induced early MEK/ERK activation was further examined by siRNA-mediated knockdown of SHC1. Upon C. trachomatis infection, SHC1 knockdown reduced MEK and ERK phosphorylation (Fig. 4, G and H). A similar degree of reduction occurred after stimulation of noninfected SHC1 knockdown cells with inducers of MEK/ERK phosphorylation such as TNF and EGF (Fig. S2, D and E). SHC1 knockdown resulted in a 90–95% reduction at the protein level (Fig. S2 F) and did not affect adhesion, invasion, inclusion formation, or numbers of progeny (Fig. S2 G). Our results show that SHC1 phosphorylation during early infection is Tarp dependent. Furthermore, SHC1 is recruited immediately after infection, where it is not directly needed for adhesion, invasion, inclusion formation, or bacterial propagation. Rather, SHC1 plays a prominent role in early C. trachomatis–induced MEK and ERK activation.
SHC1 activation and its transcriptional regulation during infection
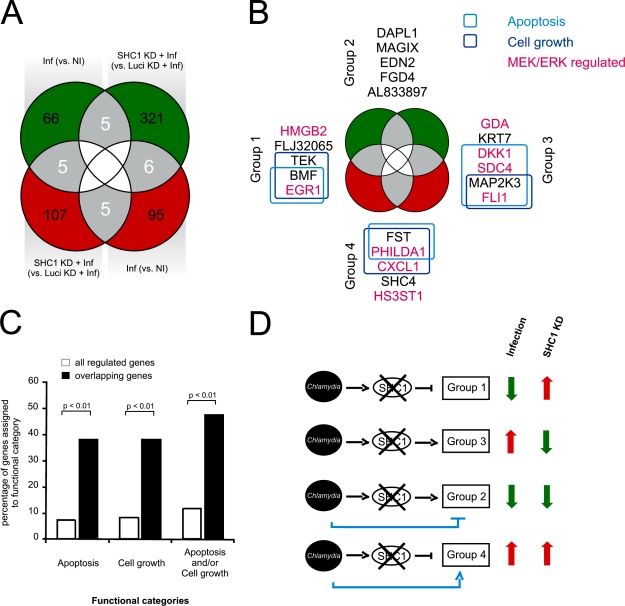
SHC1 activation is known to modulate survival, proliferation, and migration by transcriptional regulation, depending on the stimulus (Guo and Giancotti, 2004). Thus, we decided to investigate the influence of SHC1 activation on transcription upon C. trachomatis cell entry. Because of the expected delay between MEK/ERK activation and its transcriptional response, experiments were performed at 4 h pi (Fig. S2 H), a time point at which the host cell exhibits a strong transcriptional response but bacteria are still largely metabolically inactive (EB to RB conversion takes place at 8–10 h; Mathews et al., 1999). Using human DNA microarrays, we determined genes that were differentially expressed during infection of either control (without knockdown) or SHC1 knockdown cells. First, we analyzed infected versus noninfected HeLa cells at 4 h pi, revealing 182 differentially regulated genes: 76 genes were down-regulated (Fig. 5 A, top left green circle) and 106 genes were up-regulated (Fig. 5 A, bottom right red circle) in response to infection. Second, we compared infected SHC1 knockdown and infected Luciferase (control) knockdown HeLa cells, which indicates regulation of 449 SHC1-dependent genes: 332 genes were down-regulated (Fig. 5 A, top right green circle), whereas 117 genes were up-regulated (Fig. 5 A, bottom left red circle). The observed approximately threefold increase of down-regulated genes as compared with up-regulated genes upon infection of SHC1 knockdown indicates a strong activating potential of SHC1 on gene expression. Interestingly, infection of SHC1 knockdown cells down-regulated approximately fourfold more genes than infection alone (Fig. 5 A, green circles), most likely due to more genes being SHC1 rather than infection dependent. A comparison of genes regulated between all conditions (gray areas) identified a total of 21 SHC1- and infection-dependent genes (Fig. 5 A, gray overlapping areas; and Table S2).
Figure 5.
Apoptosis and cell growth genes are regulated in an SHC1-dependent manner. (A) HeLa cells were infected for 4 h (C. trachomatis L2, MOI 20) before RNA isolation and array hybridization. Gene expression profiles of infected versus noninfected cells (Inf (vs. NI)) were compared with infected SHC1 knockdown versus infected Luciferase knockdown cells (SHC1 KD + Inf (vs. Luci KD + Inf)) to determine infection and SHC1-dependently regulated genes (n = 2, each with dye swap). In total, 21 genes are regulated in both a SHC1- and infection-dependent manner (up-regulated genes are shown in red, down-regulated genes in green). (B) The 21 genes that correlate with both infection and SHC1 signaling were further analyzed using the IPA software. Of these, eight genes are grouped into the functional category “apoptosis” and another eight genes are assigned to the category “cell growth.” (C) Gene enrichment analysis of the overlapping genes (21) compared with all regulated genes (610). Both functional categories “apoptosis” and “cell growth” are significantly enriched in the 21 overlapping genes (as determined by Fisher’s exact test). (D) Model of the role of SHC1 during early infection-mediated gene regulation. C. trachomatis activates SHC1 leading to four groups of regulated genes. Expression of group 1 genes is likely inhibited by SHC1 activation, explaining their up-regulation after SHC1 knockdown in infected cells. In contrast, gene expression in group 3 is likely enhanced by SHC1 activation, causing a down-regulation of group 3 genes after SHC1 knockdown in infected cells. Genes of groups 2 and 4 show an additional regulation by C. trachomatis infection independent of SHC1, which remains after SHC1 knockdown in infected cells.
Functional analysis of the 21 differentially regulated genes using IPA revealed genes associated with apoptosis and cell growth regulation, with eight genes assigned to each category (Fig. 5 B and Table S2). Indeed, these functional gene types were significantly enriched (P < 0.01) in this gene subset in comparison to the extended gene set, i.e., ∼10% of the 610 genes versus ∼40% of the 21 genes were apoptosis related (Fig. 5 C). Surprisingly, IPA analysis also indicated that, among these 21 genes, nine genes are associated with MEK/ERK signaling, whereas 12 genes are controlled by other pathways (Fig. 5 B, Fig. S3, and Table S2). Regulation of the majority of these genes was confirmed using real-time quantitative RT-PCR (qRT-PCR; Fig. S4 and Table S3). Thus, our transcriptional data suggest that SHC1 activation is involved in regulation of apoptosis and cell growth–related genes.
Further analysis of the 21-gene subset revealed four distinct groups (Fig. 5 B and Table S2): group 1 comprises transcripts less abundant upon infection and more abundant in infected SHC1 knockdown cells (Fig. 5 D). Therefore, expression of the genes in group 1 is likely inhibited by SHC1 activation. In contrast, group 3 comprises transcripts more abundant upon infection and less abundant in infected SHC1 knockdown cells (Fig. 5 D). Consequently, expression of genes in group 3 is likely enhanced by SHC1 activation. Groups 2 and 4 include transcripts either less abundant (group 2) or more abundant (group 4) after both infection and SHC1 knockdown in infected cells (Fig. 5 D). This can be explained by infection-dependent regulation of these genes in a SHC1-independent manner, counteracted by a SHC1-dependent regulation. Consequently, knockdown of SHC1 in infected cells leads to an enforced regulation due to the remaining SHC1-independent effects. Thus, our data has identified opposing gene regulatory circuits during C. trachomatis infection: being dependent or independent of SHC1.
SHC1 activation is a survival stimulus
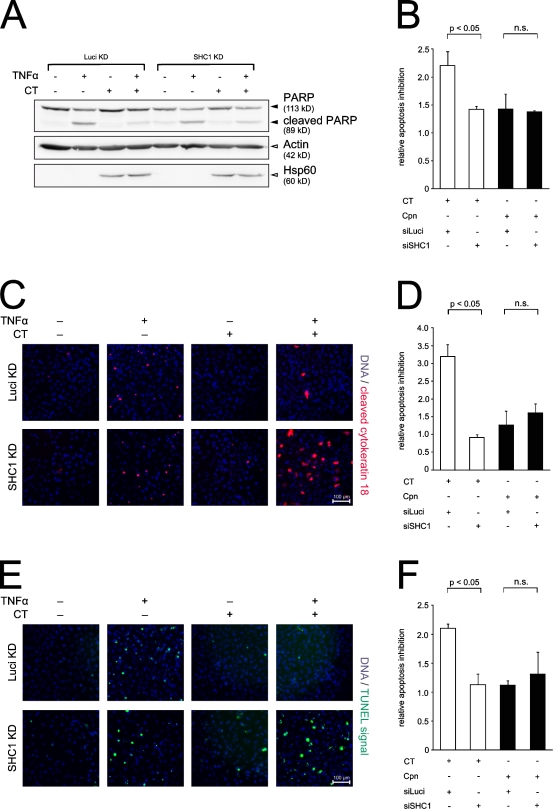
To further examine the role of SHC1 in early C. trachomatis–induced cell survival, we investigated the effect of SHC1 knockdown on host apoptosis after C. trachomatis entry (Fig. 6). For this purpose, TNF was added at 6 h pi for a duration of 4 h, i.e., 2 h after the observed transcriptional regulation changes (Fig. S2 H). Apoptosis induction levels in a combination of SHC1 or Luciferase (control) knockdown cells infected with either C. trachomatis or C. pneumoniae were determined using three different readouts: poly (ADP-ribose) polymerase (PARP) cleavage (Fig. 6, A and B), cytokeratin 18 cleavage (Fig. 6, C and D), and terminal deoxynucleotidyl transferase activity using the TUNEL assay (Fig. 6, E and F). C. trachomatis–infected cells display the highest degree of apoptosis resistance under Luciferase (control) knockdown conditions. This inhibitory phenotype was significantly reduced under SHC1 knockdown conditions using all three assays (Fig. 6, B, D, and F). Although SHC1 dependence was apparent for C. trachomatis at the early stages of infection, evidence was lacking for C. pneumoniae, which exhibited a comparably diminished degree of apoptosis resistance (Fig. 6, B, D, and F) and, notably, expresses a Tarp protein devoid of SHC1 phospho-binding sites (Clifton et al., 2005). Thus, our data demonstrate that C. trachomatis, but not C. pneumoniae, is able to confer early apoptosis resistance to infected host cells in an SHC1-dependent manner.
Figure 6.
SHC1 knockdown sensitizes early infected cells to apoptosis. (A) Western blots showing TNF-induced PARP cleavage (black arrowheads) in HeLa cells with Luciferase or SHC1 knockdown after C. trachomatis infection (MOI 50) for 6 h and an additional induction of apoptosis for 4 h (25 ng/ml TNF and 10 µg/ml cycloheximide). Actin and Hsp60 were used as loading control (white arrowheads). Infection with C. trachomatis L2 blocked PARP cleavage in infected Luciferase-transfected HeLa cells, whereas SHC1 knockdown sensitizes infected cells to apoptosis as indicated by PARP cleavage. (B) Quantification of PARP cleavage in C. trachomatis and C. pneumoniae infection from chemiluminescence imager recorded blots (data from A and not depicted, n = 3, error bars indicate SE). Band signals from PARP and cleaved PARP were measured using AIDA software, and the ratio was calculated. The ratio of apoptosis inhibition in uninfected TNF-stimulated versus infected TNF-stimulated cells is depicted. C. trachomatis–infected control cells show a high degree of apoptosis resistance, which is significantly reduced upon SHC1 knockdown. In contrast, C. pneumoniae–infected cells exhibit a diminished apoptosis resistance, independent of SHC1. (C and E) Immunostainings showing TNF-induced apoptosis in HeLa cells with Luciferase or SHC1 knockdown after infection with C. trachomatis and apoptosis induction (identical conditions to A). SHC1 knockdown sensitizes infected cells to apoptosis, whereas apoptosis is blocked in infected control cells. Apoptosis induction was assessed by cytokeratin 18 cleavage (red) and TUNEL assay (green), respectively. Nuclei are stained blue with Hoechst 33342. Image area: 450 µm × 450 µm. (D and F) Quantification of cells positive for cytokeratin 18 cleavage (data from B and not shown; n = 2, error bars indicate SE) and TUNEL (data from D and not depicted; n = 2, error bars indicate SE), respectively. A mean of 10,000 cells per condition were analyzed. Apoptosis inhibition is depicted as in B. C. trachomatis infection leads to apoptosis inhibition in control cells, but not in SHC1 knockdown cells. In contrast, C. pneumoniae fails to block apoptosis in both control and SHC1 knockdown cells.
Discussion
Here, we describe the use of protein microarrays comprising nearly every human SH2 and PTB domain to quantitatively assess Tarp-mediated interactions. Tarp was found to have the capacity to interact with many different SH2 domain–containing proteins, which suggests that Tarp contributes substantially to phosphorylation-dependent signaling. In total, we discovered 23 novel in vitro binding partners of Tarp and confirmed two (VAV2 and PI3K) that had previously been identified. High-affinity interactions with two proteins, the serovariant L2-specific protein NCK2 and the serovariant D and L2 common interaction partner SHC1, were experimentally validated, and the interaction of Tarp with SHC1 was studied in greater detail. We found that the Tarp–SHC1 interaction plays a direct role in conferring resistance to apoptosis in infected cells. Together, these data extend our understanding of how Tarp modulates host cell function.
In the nonphosphorylated state, Tarp showed serovariant-dependent affinity for host SH2 domain–containing proteins. ABL2 (ARG) was found to be the strongest interaction partner shared by both serovariants (D and L2). The ABL kinases can be activated through interaction of their SH2 domains with cellular substrates (Lewis and Schwartz, 1998; Barilá et al., 2000). ABL1 and ABL2 have been shown to be among the human kinases that phosphorylate Tarp (Elwell et al., 2008; Jewett et al., 2008; Mehlitz et al., 2008). Consequently, the interaction between nonphosphorylated Tarp and ABL, leading to subsequent activation of ABL, may constitute an initial trigger for Tarp phosphorylation (see model in Fig. S5; Hantschel et al., 2003; Nagar et al., 2003). It has previously been reported that infection of ABL/ARG−/− knockout mouse fibroblasts leads to a ∼50% reduction of Tarp phosphorylation, which suggests that ABL could be the initial Tarp kinase (Elwell et al., 2008). Interestingly, phosphorylation of the H. pylori protein CagA and the EPEC effector Tir was also shown to involve ABL (Swimm et al., 2004; Poppe et al., 2007). Furthermore, CagA, ABL, and CRKII have been shown to physically interact (Tammer et al., 2007).
The nonphosphorylated state of Tarp is short-lived, as Tarp is phosphorylated immediately after translocation (Clifton et al., 2004). Here, we demonstrate that this phosphorylation leads to a dramatic increase of SH2 domain-mediated interactions. Accordingly, the Tarp phospho-interactome has a degree of complexity similar to that of the ERBB family of receptor tyrosine kinases (Jones et al., 2006). For example, Tarp D and L2 show similar numbers of interaction partners as ERBB4 and ERBB3, respectively.
Previously, only VAV2 and the regulatory subunit of PI3K have been identified as interaction partners of phosphorylated Tarp (Lane et al., 2008). Here, we confirmed these interactions using our SH2/PTB domain microarrays. Moreover, we found that Tarp L2 interacts with several host kinases previously implicated in Tarp phosphorylation, corroborating Tarp L2 as a promiscuous phosphorylation substrate (Elwell et al., 2008; Jewett et al., 2008).
Comparison of Tarp D and L2 interactions revealed SHC1 as one of the strongest partners for both serovariants, which suggests a general role for SHC1 in C. trachomatis infections. SHC1, which has not previously been shown to interact with Tarp, is also involved in EGFR signaling, leading to MEK/ERK activation upon stimulation with EGF or TNF. MEK/ERK activation, an emerging theme in the chlamydial infection process, was first shown to be involved in glycerophospholipid acquisition by C. trachomatis (Su et al., 2004) and, more recently, as an important anti-apoptotic mechanism (Paland et al., 2008). We found pronounced activation of MEK and ERK shortly after infection coincident with phosphorylation of Tarp and SHC1. Previous work suggested that rapid phosphorylation of ERK during chlamydial infection is linked to up-regulation of the anti-apoptotic protein MCL-1 (Rajalingam et al., 2008), which we found to be independent of SHC1 (unpublished data). Moreover, in our previous work, activation of both MEK and ERK was shown to be necessary for C. trachomatis at an advanced stage of infection; however, this activation was uncoupled from upstream signaling by RAS and RAF (Gurumurthy et al., 2010). MEK activation is thought to be crucial during infections with H. pylori (Hatakeyama, 2008) and has been shown to be initiated through interaction of CagA with SHP-2 (Higashi et al., 2002), CRK (Suzuki et al., 2005), GRB2 (Mimuro et al., 2002), and c-MET (Churin et al., 2003).
Here, we describe the direct recruitment of SHC1 and the subsequent activation of MAP kinase family members MEK and ERK in the early stages of a C. trachomatis infection. However, C. trachomatis Tarp may very well trigger additional pathways causing MEK/ERK activation (Fig. S3). Candidate molecules are the adaptor protein CRKL, which is capable of activating ERK through b-RAF upon integrin signaling (Guo and Giancotti, 2004); the GTPase RASA1, directly activating b-RAF (Schubbert et al., 2007); and p56LCK, which bypasses RAS/RAF/MEK and activates ERK in a PKCε-dependent manner during Mycobacterium leprae infection (Tapinos and Rambukkana, 2005). All three factors were found to recognize phospho-Tarp in this study (Fig. 1, Table S1, and model in Fig. S5). SHC1 knockdown led to significant changes in the expression of anti-apoptotic and growth-related genes in the context of C. trachomatis infection. As deduced from knowledge-based pathway analysis, SHC1 signaling depends only partially on the MEK/ERK cascade, whereas the remaining genes are regulated by additional signaling pathways (Figs. S3 and S5). This is consistent with our observation of a significant reduction in C. trachomatis–induced cell survival upon SHC1 knockdown despite a residual activation of MEK and ERK. It is tempting to argue that this SHC1-mediated gene regulation is a MEK/ERK-independent branch of the EGFR-mediated signaling cascade. In H. pylori infection, a major outcome of CagA translocation appears to be regulation of growth-dependent genes (Keates et al., 1999; Mimuro et al., 2002). Thus, induction of growth signaling seems to be an important function of tyrosine-phosphorylated effectors. Future investigations are required to clarify whether this also holds true for other effectors like EPEC Tir, Anaplasma phagocytophilum AnkA, or Vaccinia virus A36R (Backert et al., 2008). Collectively, our observations support a model in which activation of both MEK/ERK-dependent and independent growth signaling cascades plays a central role in C. trachomatis–induced cell survival. This assigns an important anti-apoptotic function to the interaction between phospho-Tarp and SHC1 during the early stage of infection.
An alternative approach to assess protein interactions with tyrosine-phosphorylated peptides from CagA, Tir, Tarp, and BepD-F used differential mass-spectroscopy (SILAC; Selbach et al., 2009). This study revealed only a few of the Tarp interaction partners reported here, notably the previously reported interaction with PI3K. Differences in methodological approach are most likely responsible for this small degree of overlap. Our array analysis provides a list of all possible direct interaction partners. In contrast, immunoprecipitation coupled with mass spectrometry does not discriminate between direct and indirect interactions, owing to secondary protein contacts in signaling complexes. In addition, our approach provides a global view of biophysical interactions, whereas the approach of (Selbach et al., 2009) focused on a specific cell type and hence was dependent on the proteins being expressed in their cell of interest. Nonetheless, both approaches provide important and complementary information on the complexity of host–pathogen interactomes.
Here, we have shown that global interactome studies provide a powerful approach to uncover host cell counterparts of bacterial effector proteins, helping to increase our understanding of the mechanisms of host–microbe interactions. To our knowledge, this paper presents the first comprehensive interactome study of a phosphorylated bacterial effector protein using SH2/PTB domain microarrays. Our study not only confirmed several known interactions but also revealed an extensive repertoire of novel high- and low-affinity interactions with phosphorylated and nonphosphorylated versions of C. trachomatis Tarp. In addition, these interactions varied substantially between serovars. We focused here on just one of the many possible Tarp–cellular protein interactions to substantiate our hypothesis. The remaining data reported here provide a wealth of additional hypotheses to test regarding Tarp-mediated cellular signaling induced by chlamydial infection.
Materials and methods
Cell lines and bacteria
HeLa (American Type Culture Collection [ATCC] No. CRM-CCL-2) and End1/E6E7 (ATCC No. CRL-2615) cervical epithelial cells were grown in DME (Invitrogen) supplemented with 10% fetal calf serum, 5 mM l-glutamine, and 1 mM sodium pyruvate (Invitrogen). C. trachomatis Lymphogranuloma venereum (LGV) biovar strain L2/434/Bu (ATCC No. VR-902B), C. trachomatis trachoma type D strain UW-3/Cx (ATCC No. VR-885), and C. pneumoniae TWAR strain CDC/CWL-029 (ATCC No. VR-1310) were purified from confluent HeLa monolayers. In brief, HeLa cells were grown to 80% confluency and were inoculated for 2 h with the respective C. trachomatis strain. For infections with C. pneumoniae, inoculated cells were incubated at 22°C for 1 h on a shaker, centrifuged (500 rpm at 35°C) for 1 h, and then incubated for 2 h at 35°C. The medium was replaced by infection medium (DME; 5% fetal calf serum, 5 mM l-glutamine, 1 mM sodium pyruvate, and 1 µg/ml cycloheximide), and growth was allowed for 48 h. Cells were mechanically detached, and bacteria were released using ∼2–5 mm glass beads (Carl Roth GmbH + Co. KG). Low-speed supernatant (5 min at 4,000 g and 4°C) was subjected to high-speed centrifugation (25 min at 40,000 g and 4°C) to pellet the bacteria. Bacteria were washed twice with 10 ml SPG (220 mM sucrose, 50 mM sodium phosphate, and 5 mM glutamate, pH 7.4), aliquoted, and stored at −80°C in SPG.
Reagents, antibodies, and constructs
Chemicals were obtained from Sigma-Aldrich. Recombinant human TNF was obtained from BD. Antibodies against β-actin, ERK2, p-ERK1/2, MEK1/2, p-MEK1/2, PARP-1/2, SHC, p-SHC pY239/240, p-Tyr PY99, NCK1/2, and GST were acquired from Santa Cruz BioTechnology, Inc., or Cell Signaling Technology. Antibodies against the M30 epitope of cleaved cytokeratin 18 and against bacterial Hsp60 were obtained from Enzo Life Sciences, Inc. Secondary Cy2-conjugated antibodies against rabbit IgG, mouse IgG, and Cy5-conjugated antibodies against rabbit IgG and mouse IgG were obtained from Jackson ImmunoResearch Laboratories, Inc. Alexa Fluor 532 phalloidin for staining of actin was obtained from Invitrogen. Antiserum against Tarp was raised by immunization of rats with GST-Tarp fusion protein (Biogenes). Monoclonal anti–C. trachomatis OMP1 was obtained from the University of Washington. GST-Tarp was constructed by PCR amplification of nucleotides 6–818 of C. trachomatis LGV L2 Tarp or 1–939 of C. trachomatis D Tarp and ligation of the product into pGEX4T-3 (GE Healthcare) using dinucleotide sticky-end cloning. Purification of GST-Tarp was performed according to manufacturer’s instructions using glutathione sepharose (GE Healthcare).
Peptide synthesis and protein microarrays
Peptides were synthesized as described previously (Jones et al., 2006), purified to >95% by preparative reverse phase HPLC, quality controlled via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Thermo Fisher Scientific), and labeled on their N termini with 5- (and 6)-carboxytetramethylrhodamine (5(6)-TAMRA) from AnaSpec.
We have previously described cloning, expressing, and purifying virtually every human SH2 and PTB domain, as well as preparing microarrays of these domains on chemically derivatized glass surfaces (MacBeath and Schreiber, 2000; Jones et al., 2006). In brief, the coding regions for each domain were cloned from human cDNA, and the corresponding proteins were produced recombinantly in E. coli using the T7 expression system. Each domain features an N-terminal His6 tag, as well as a thioredoxin tag to facilitate the high-level production of soluble protein. After purifying each domain from large-scale bacterial culture (0.5 liter), we assessed its purity by SDS-PAGE and its aggregation state by size exclusion column chromatography. In the current version of our arrays, we eliminated domains that were impure or did not contain soluble, monomeric protein. Notably, SH2 domains derived from the signal transducers and activator of transcription (STAT) and suppressors of cytokine signalling (SOCS) families of proteins did not behave well. By cloning larger portions of STAT1 and STAT2 that included their entire SH2 domain–containing cores (Mao and Chen, 2005), we obtained soluble, monomeric material for these two proteins. In addition, we cloned, expressed, and purified the N-terminal domain of CBL (Meng et al., 1999), which contains a noncanonical SH2 domain. In total, 133 domains representing 103 proteins were used in these studies.
To facilitate the rapid and automated processing of protein microarrays, we used a NanoPrint microarrayer (TeleChem International, Inc.) to spot our proteins in quadruplicate on aldehyde displaying glass substrates, cut to the size of a microtiter plate (112.5 mm × 74.5 mm × 1 mm; Erie Scientific). 96 separate arrays were prepared on each glass substrate, and the glass was attached to the bottom of a bottomless microtiter plate using an intervening silicone gasket (Grace Bio-Labs). Two 16 × 17 microarrays were required to accommodate all 133 domains, as well as the appropriate controls (His6-tagged thioredoxin and buffer). Proteins were spotted in quadruplicate at a high concentration (40–200 µM), and a low concentration (200 nM) of cyanine 5–labeled bovine serum albumin was included in each sample to facilitate image analysis. Arrays were stored before use at −80°C.
Immediately before use, the plates were quenched with buffer A (20 mM Hepes, 100 mM KCl, and 0.1% Tween-20, pH 7.8) containing 1% BSA (wt/vol) for 30 min at room temperature, followed by several rinses with buffer A. Arrays were probed with eight different concentrations of labeled peptides (5 µM, 3 µM, 2 µM, 1 µM, 500 nM, 200 nM, 100 nM, and 10 nM) dissolved in buffer A. Peptide solution was removed after a 1-h incubation at room temperature, and arrays were washed with 150 ml of buffer A for 10 s. Arrays were rinsed briefly with double-distilled H2O and spun upside down in a centrifuge for 1 min to remove residual water.
Protein microarrays were scanned at 10-µm resolution using a scanner (LS400; Tecan). Spots were defined using the Cy5 image, and the mean fluorescence of each spot was calculated from the 5(6)-TAMRA image. We have previously shown that probing a protein microarray with a single concentration of a labeled probe can produce very misleading results (Gordus and MacBeath, 2006). We therefore probed our arrays with eight concentrations of each peptide and fit the resulting spot intensities, Fobs, to the following equation:
| (1) |
where F0 is the background fluorescence, Fmax is the maximum fluorescence at saturation, [pep] is the total peptide concentration, and KD is the equilibrium dissociation constant. For each peptide, we fit all 133 curves, one for each domain. Interactions were considered specific if the data fit well to Eq. 1 (R2 > 0.9), with KD < 2 µM and Fmax at least twofold higher than the mean fluorescence of control spots (His6-tagged thioredoxin). The resulting data were displayed graphically using Cytoscape 2.1 (http://www.cytoscape.org/).
In vitro phosphorylation, precipitation, immunoprecipitation, and immunoblotting
GST-Tarp was phosphorylated while the beads coupled to glutathione sepharose (GE Healthcare). In brief, GST-Tarp beads were incubated for 30 min at 30°C with recombinant human c-SRC (Cell Signaling Technology) in kinase buffer (25 mM Hepes, pH 7.0, 150 mM NaCl, 10 mM MgCl2, 1% Nonident P40, 5 mM dithiothreitol, 1 mM Na3VO4, 1× Complete protease inhibitors, and 6 mM ATP). GST-Tarp beads were washed three times with PBS and incubated with lysate from 107 cells (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Nonident P40, 0.5% sodium deoxycholate, and 1× Complete protease inhibitors) for 2 h at 4°C. Proteins were eluted by incubation with 20 mM of reduced glutathione in 50 mM Tris, pH 8.0. Immunoprecipitation and Western blotting were performed according to standard procedures (Sambrook and Russel, 2001). Signal detection was done using ECL/Hyperfilm (GE Healthcare). Quantification was performed using ImageJ (National Institutes of Health) and Excel (Microsoft).
siRNA transfections
HeLa cells were transfected with SHC1 SMARTpool (Thermo Fisher Scientific) or Luciferase siRNA (QIAGEN) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Knockdown was confirmed via Western blotting 72 h after transfection.
Infection time courses
HeLa or End1/E6E7 cells were seeded into 12-well cluster plates at a density of 60–70%. Cells were serum starved for 24 h in serum-free growth medium before performing infection time-course experiments. Infection was performed at indicated MOIs with either bacteria or control mock-infected cells (i.e., infected with C. trachomatis storage buffer, SPG). C. pneumoniae infections were synchronized by centrifugation for 1 h at 500 g and 4°C, and shifting to 35°C. Infection was stopped at the indicated times by removing the medium, rinsing quickly with prewarmed PBS, and immediately lysing in SDS loading buffer at 94°C for 5 min.
Immunofluorescence staining, invasion, inclusion formation, and progeny assays
For immunofluorescence, cells were either seeded in 24- or 12-well cluster plates with or without coverslips, and were infected in a humidified incubator at 35°C and 5% CO2 with MOIs indicated in the respective experiments. Cells were fixed at indicated time points with 4% PFA, washed once with PBS, and stained. In brief, cells were permeabilized with 0.2% Triton X-100 or 0.1% saponin in PBS for 30 and 10 min, respectively, and were washed 3× with PBS for 5 min at RT. Cells were blocked with 2% FCS in PBS for 45 min and were stained with primary antibodies diluted in 2% FCS in PBS for 1 h at RT. After 3× washing with PBS, samples were incubated with secondary antibodies for 1 h in blocking solution at RT in the dark. After one short wash with ddH2O, samples were mounted with Mowiol 4–88 (Carl Roth GmbH + Co. KG) and visualized at RT on a confocal microscope (photomultiplier equipped; TCS SPE; Leica) at 63× magnification (HCX Plan-Apochromat with a 63×/1.40–0.60 oil objective lens; Leica) using acquisition software (LAS AF TCS SPE; Leica). Data were processed with Photoshop (adjustment of brightness and contrast identical for all images; Adobe).
For measurement of invasion efficiency, inclusion formation, and progeny, HeLa cells were seeded in 12-well cluster plates and were siRNA transfected as described in “siRNA transfections.” On day 2, cells were split into three separate wells and then infected on day 4 at MOI 50 for invasion assays or MOI 1 for inclusion formation and progeny counts. Invasion assays were stopped at 1 and 10 h pi; cells were fixed with ice-cold methanol overnight at −20°C, stained for OMP1 and Hoechst 33342, and analyzed by automated microscopy (Scan^R system; Olympus) at 10× magnification. Data were processed using ImageJ and Excel.
Inclusion formation was quantified 24 h pi by manually counting the inclusion forming units (IFU) as inclusions per 40× field on a cell culture microscope (DM-IL; Leica). Progeny infections were grown for 48 h before glass bead lysis of host cells to release infectious particles and infection of fresh HeLa monolayers. After another 24 h of infection, cells were processed as described for inclusion formation. Data were processed with Excel.
RNA preparation, DNA microarrays, qRT-PCR, and experimental design and analysis
RNA from infected, uninfected, or transfected (and infected) HeLa cells was isolated with the RNAeasy kit (QIAGEN) according to the manufacturer’s instructions. RNA integrity was analyzed using a Bioanalyzer (2100; Agilent Technologies). DNA microarray experiments were performed as two-color dye-reversal ratio hybridizations on arrays containing 44,000 human genes (AMADID 010646; Agilent Technologies) in biological duplicates. RNA labeling was performed with a Fluorescent Linear Amplification kit (Agilent Technologies). The labeling efficiency was verified at A552nm for Cy3-CTP and A650nm for Cy5-CTP with a Nanodrop photometer (Kisker-Bioteck). Before hybridization, complimentary RNA of each product was fragmented and mixed with control targets and hybridization buffer according to the manufacturer’s instructions (Agilent Technologies). Hybridizations were performed overnight (∼17 h) at 60°C. The slides were washed according to the manufacturer’s instructions, and scanning of microarrays was performed at 5-µm resolution using a microarray laser scanner (Agilent Technologies).
Data analysis was performed on the Rosetta Resolver system 7.2 (Rosetta Biosoftware). Ratio profiles were generated from raw scan data by a processing pipeline, which includes preprocessing (feature extraction) and postprocessing (Rosetta Resolver) of data and error model adjustments to the raw scan data. Ratio profiles were combined in an error-weighted fashion (Rosetta Resolver) to create ratio experiments. Expression patterns were identified using stringent analysis criteria of 1.6-fold expression cutoffs of the ratio experiments and an anti-correlation of the dye reversal ratio profiles. Anti-correlation was determined by using the compare function to match two different hybridizations pairs and to decide how similar or dissimilar they were. By combining the first and the second criteria of analysis, data points with a low p-value (P < 0.01) were filtered out, making the analysis robust and reproducible. Additionally, by using this strategy, the data selection was independent of error models implemented in the Rosetta Resolver system.
For qRT-PCR, mRNA was isolated as described above. 10 µl mRNA was DNaseI-digested with RNase-free DNaseI (Fermentas) according to the manufacturer’s instructions. Digested mRNA was phenol/chloroform purified. In brief, 180 µl of RNase-free water (Millipore/Synergy) and 200 µl of phenol/chloroform (Roth) were added to the 20-µl digestion reaction and vortexed. Samples were phase separated at 12,000 g (4°C for 10 min), and the supernatant was combined with 200 µl of chloroform followed by vortexing. After repeated phase separation, mRNA was precipitated using 1/10 volume of 3 M sodium acetate and 2 volumes of 80% ethanol followed by centrifugation at 14,000 g (4°C for 15 min). The supernatant was removed, and the pellet was air dried in a clean bench environment and resuspended in 50 µl of RNase-free water. mRNA was reverse transcribed using the Revert Aid First Strand Synthesis kit (Fermentas) according to the manufacturer’s instructions and was diluted 1:10 with RNase free water. qRT-PCR was set up with Absolute QPCR SYBR Green Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. qRT-PCR was performed on a Step One Plus device (Applied Biosystems), and data were analyzed using the ΔΔCt method, Step One Plus software package (Applied Biosystems), and Excel. Endogenous controls were GAPDH and L13a. Splice variant-specific primers were designed using National Center for Biotechnology Information Primer Blast or Primer 3 (Rozen and Skaletsky, 2000; Table S4).
Apoptosis induction and detection, data acquisition and analysis
HeLa cells were seeded in 12-well plates and infected with C. trachomatis L2 at MOI 50 for 6 h, then apoptosis was induced by the addition of TNF (25 ng/ml) and cycloheximide (10 µg/ml) for an additional 4 h (Fig. S2 H). Infection with C. pneumoniae TWAR (MOI 50) was assisted by centrifugation (1 h at 500 g and 35°C), followed by 5 h of incubation before apoptosis induction as described for C. trachomatis. For the PARP cleavage assay, cells were directly lysed in 2× sample buffer (Laemmli) and heated at 95°C for 10 min. For quantification, Western blots were imaged using a bioluminescence reader (LAS-3000; Fujifilm) and the Image Reader LAS-3000 software (Fujifilm). Respective bands and their lane backgrounds were measured using the Advanced Image Data Analyzer (AIDA) software, and the ratio of cleaved PARP to the total PARP amount was calculated. For the cytokeratin 18, cleavage and the TUNEL assays cells were spun down before staining. Cytokeratin 18 staining was done as described in the immunofluorescence section, and TUNEL staining was performed according to manufacturer’s instructions (DeadEnd Fluorometric TUNEL System; Promega) with green fluorescence of apoptotic cells due to fluorescein-12-dUTP labeling. Nuclei were stained blue with Hoechst 33342. Images of stained PBS stored cells were acquired at RT using an automated microscope (Scan^R system [Olympus] consisting of an inverted microscope [IX81; Olympus], charge-coupled device camera [C4742-80-12AG; Hamamatsu] and the Scan^R acquisition software) at 10× magnification (UPlanS Apo 10×/0.40 NA lens; Olympus). Cells positive for cleaved cytokeratin 18, TUNEL, and nuclei were counted using ImageJ, and the ratio (cleaved cytokeratin 18 or TUNEL-positive cells/total nuclei) was calculated. A mean of 10,000 cells per condition was analyzed. Apoptosis inhibition was depicted as ratios from uninfected TNF-stimulated and infected TNF-stimulated cells.
Modeling of Tarp and SHC1 signaling
Interaction partners identified using the SH2/PTB arrays were analyzed for their participation in signaling pathways using IPA software (Ingenuity Systems) and the KEGG. Only signaling pathways associated with at least two Tarp interaction partners were considered in the model. Pathways involved in SHC1-dependent regulation of the 21 apoptosis- and cell growth–related genes were obtained by using IPA. In brief, all known transcriptional regulators upstream of the SHC1 dependently regulated genes were introduced into the model and then linked to SHC1 using the “Path Explorer” tool of IPA. Pathways were grouped for MEK/ERK dependency.
Online supplemental material
Fig. S1 summarizes Tarp interactions and putative signaling. Fig. S2 displays Western blots and images showing SHC1 phosphorylation and subcellular localization upon infection, as well as MEK/ERK signaling. Additional graphs detailing chlamydial invasion, inclusion formation and progeny after SHC1 knockdown, and an experimental time scale are also shown. Fig. S3 depicts signaling interactions between SHC1 and its downstream genes. Fig. S4 shows qRT-PCR confirmation of SHC1-dependent regulation of apoptosis and cell growth genes. Fig. S5 provides a model of Tarp in host cell signaling. Table S1 gives an overview of Tarp peptides and their dissociation constants. Table S2 lists the 21 regulated genes and their function. Table S3 provides qRT-PCR analysis of selected genes. Table S4 lists qRT-PCR primers used. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909095/DC1.
Acknowledgments
We thank L. Ann Ogilvie for editing the manuscript and H.-J. Mollenkopf and I. Wagner for help with microarray preparation, hybridization, and feature extraction.
This work was supported by grants from the National Institutes of Health (NIH; 1 R33 CA128726 to G. MacBeath) and the Bundesministerium für Bildung und Forschung through ERA-NET PathoGenoMics (RNAi-Net 0313938Ar). A. Kaushansky was supported in part by the NIH Molecular, Cellular, and Chemical Biology Training Grant (5 T32 GM07598). A.G. Gordus is the recipient of an National Science Foundation Graduate Research Fellowship, S. Banhart is a fellow the International Max Planck Research School for Infectious Diseases and Immunology (IMPRS-IDI), and J. Zielecki is supported by the German Science Foundation (GRK 1121).
Footnotes
Abbreviations used in this paper:
- CagA
- cytotoxicity-associated gene A
- EB
- elementary body
- EGFR
- EGF receptor
- EPEC
- enteropathogenic Escherichia coli
- ERK
- extracellular signal-regulated kinase
- IPA
- Ingenuity Pathway Analysis
- MEK
- MAPK/ERK kinase
- PARP
- poly (ADP-ribose) polymerase
- pi
- postinfection
- PI3K
- phosphatidylinositol 3-kinase
- PTB
- phosphotyrosine binding
- qRT-PCR
- quantitative RT-PCR
- RB
- reticulate body
- SE
- standard error
- SH2
- SRC homology 2
- Tarp
- translocated actin-recruiting phosphoprotein
- Tir
- translocated intimin receptor
References
- Arora S., Wang Y., Jia Z., Vardar-Sengul S., Munawar A., Doctor K.S., Birrer M., McClelland M., Adamson E., Mercola D. 2008. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP-on-chip. Genome Biol. 9:R166 10.1186/gb-2008-9-11-r166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahi M., Azuma T., Ito S., Ito Y., Suto H., Nagai Y., Tsubokawa M., Tohyama Y., Maeda S., Omata M., et al. 2000. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191:593–602 10.1084/jem.191.4.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Feller S.M., Wessler S. 2008. Emerging roles of Abl family tyrosine kinases in microbial pathogenesis. Trends Biochem. Sci. 33:80–90 [DOI] [PubMed] [Google Scholar]
- Barilá D., Mangano R., Gonfloni S., Kretzschmar J., Moro M., Bohmann D., Superti-Furga G. 2000. A nuclear tyrosine phosphorylation circuit: c-Jun as an activator and substrate of c-Abl and JNK. EMBO J. 19:273–281 10.1093/emboj/19.2.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham R.C., Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5:149–161 10.1038/nri1551 [DOI] [PubMed] [Google Scholar]
- Buchholz K.R., Stephens R.S. 2008. The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect. Immun. 76:3150–3155 10.1128/IAI.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.M., Splinter P.L., Tietz P.S., Huang B.Q., Billadeau D.D., LaRusso N.F. 2004. Phosphatidylinositol 3-kinase and frabin mediate Cryptosporidium parvum cellular invasion via activation of Cdc42. J. Biol. Chem. 279:31671–31678 10.1074/jbc.M401592200 [DOI] [PubMed] [Google Scholar]
- Christie P.J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451–485 10.1146/annurev.micro.58.030603.123630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churin Y., Al-Ghoul L., Kepp O., Meyer T.F., Birchmeier W., Naumann M. 2003. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161:249–255 10.1083/jcb.200208039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D.R., Fields K.A., Grieshaber S.S., Dooley C.A., Fischer E.R., Mead D.J., Carabeo R.A., Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA. 101:10166–10171 10.1073/pnas.0402829101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D.R., Dooley C.A., Grieshaber S.S., Carabeo R.A., Fields K.A., Hackstadt T. 2005. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 73:3860–3868 10.1128/IAI.73.7.3860-3868.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell C.A., Ceesay A., Kim J.H., Kalman D., Engel J.N. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 4:e1000021 10.1371/journal.ppat.1000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiani E., Giardina G., Luzi L., Cesaroni M., Quarto M., Capra M., Germano G., Bono M., Capillo M., Pelicci P., Lanfrancone L. 2007. RaLP, a new member of the Src homology and collagen family, regulates cell migration and tumor growth of metastatic melanomas. Cancer Res. 67:3064–3073 10.1158/0008-5472.CAN-06-2301 [DOI] [PubMed] [Google Scholar]
- Fan T., Lu H., Hu H., Shi L., McClarty G.A., Nance D.M., Greenberg A.H., Zhong G. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487–496 10.1084/jem.187.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S.F., Vier J., Kirschnek S., Klos A., Hess S., Ying S., Häcker G. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 200:905–916 10.1084/jem.20040402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraca A. 2002. New views on the role of endothelin (minireview). Endocr. Regul. 36:161–167 [PubMed] [Google Scholar]
- Gordus A., MacBeath G. 2006. Circumventing the problems caused by protein diversity in microarrays: implications for protein interaction networks. J. Am. Chem. Soc. 128:13668–13669 10.1021/ja065381g [DOI] [PubMed] [Google Scholar]
- Gregorc U., Ivanova S., Thomas M., Guccione E., Glaunsinger B., Javier R., Turk V., Banks L., Turk B. 2007. Cleavage of MAGI-1, a tight junction PDZ protein, by caspases is an important step for cell-cell detachment in apoptosis. Apoptosis. 12:343–354 10.1007/s10495-006-0579-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Giancotti F.G. 2004. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5:816–826 10.1038/nrm1490 [DOI] [PubMed] [Google Scholar]
- Gurumurthy R.K., Mäurer A.P., Machuy N., Hess S., Pleissner K.-P., Schuchhardt J., Rudel T., Meyer T.F. 2010. A loss-of-function screen reveals Ras- and Raf-independent MEK-ERK signaling during Chlamydia trachomatis infection. Sci. Signal. 3:ra21 10.1126/scisignal.2000651 [DOI] [PubMed] [Google Scholar]
- Hantschel O., Nagar B., Guettler S., Kretzschmar J., Dorey K., Kuriyan J., Superti-Furga G. 2003. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 112:845–857 10.1016/S0092-8674(03)00191-0 [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. 2008. SagA of CagA in Helicobacter pylori pathogenesis. Curr. Opin. Microbiol. 11:30–37 10.1016/j.mib.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Heuer D., Brinkmann V., Meyer T.F., Szczepek A.J. 2003. Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell. Microbiol. 5:315–322 10.1046/j.1462-5822.2003.00278.x [DOI] [PubMed] [Google Scholar]
- Higashi H., Tsutsumi R., Fujita A., Yamazaki S., Asaka M., Azuma T., Hatakeyama M. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA. 99:14428–14433 10.1073/pnas.222375399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck C.J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett T.J., Fischer E.R., Mead D.J., Hackstadt T. 2006. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. USA. 103:15599–15604 10.1073/pnas.0603044103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett T.J., Dooley C.A., Mead D.J., Hackstadt T. 2008. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371:339–344 10.1016/j.bbrc.2008.04.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N., Dumont D.J. 2000. Tek/Tie2 signaling: new and old partners. Cancer Metastasis Rev. 19:13–17 10.1023/A:1026555121511 [DOI] [PubMed] [Google Scholar]
- Jones R.B., Gordus A., Krall J.A., MacBeath G. 2006. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 439:168–174 10.1038/nature04177 [DOI] [PubMed] [Google Scholar]
- Juban G., Giraud G., Guyot B., Belin S., Diaz J.J., Starck J., Guillouf C., Moreau-Gachelin F., Morlé F. 2009. Spi-1 and Fli-1 directly activate common target genes involved in ribosome biogenesis in Friend erythroleukemic cells. Mol. Cell. Biol. 29:2852–2864 10.1128/MCB.01435-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keates S., Keates A.C., Warny M., Peek R.M., Jr., Murray P.G., Kelly C.P. 1999. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag- Helicobacter pylori. J. Immunol. 163:5552–5559 [PubMed] [Google Scholar]
- Kenny B., DeVinney R., Stein M., Reinscheid D.J., Frey E.A., Finlay B.B. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 91:511–520 10.1016/S0092-8674(00)80437-7 [DOI] [PubMed] [Google Scholar]
- Lane B.J., Mutchler C., Al Khodor S., Grieshaber S.S., Carabeo R.A. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 4:e1000014 10.1371/journal.ppat.1000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.M., Schwartz M.A. 1998. Integrins regulate the association and phosphorylation of paxillin by c-Abl. J. Biol. Chem. 273:14225–14230 10.1074/jbc.273.23.14225 [DOI] [PubMed] [Google Scholar]
- Luzi L., Confalonieri S., Di Fiore P.P., Pelicci P.G. 2000. Evolution of Shc functions from nematode to human. Curr. Opin. Genet. Dev. 10:668–674 10.1016/S0959-437X(00)00146-5 [DOI] [PubMed] [Google Scholar]
- MacBeath G., Schreiber S.L. 2000. Printing proteins as microarrays for high-throughput function determination. Science. 289:1760–1763 [DOI] [PubMed] [Google Scholar]
- Mao X., Chen X. 2005. Crystallization and X-ray crystallographic analysis of human STAT1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:666–668 10.1107/S1744309105017392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S.A., Volp K.M., Timms P. 1999. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Lett. 458:354–358 10.1016/S0014-5793(99)01182-5 [DOI] [PubMed] [Google Scholar]
- McDowall M., Edwards N.M., Jahoda C.A., Hynd P.I. 2008. The role of activins and follistatins in skin and hair follicle development and function. Cytokine Growth Factor Rev. 19:415–426 10.1016/j.cytogfr.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Mehlitz A., Banhart S., Hess S., Selbach M., Meyer T.F. 2008. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol. Lett. 289:233–240 10.1111/j.1574-6968.2008.01390.x [DOI] [PubMed] [Google Scholar]
- Meng W., Sawasdikosol S., Burakoff S.J., Eck M.J. 1999. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 398:84–90 10.1038/18050 [DOI] [PubMed] [Google Scholar]
- Mimuro H., Suzuki T., Tanaka J., Asahi M., Haas R., Sasakawa C. 2002. Grb2 is a key mediator of helicobacter pylori CagA protein activities. Mol. Cell. 10:745–755 10.1016/S1097-2765(02)00681-0 [DOI] [PubMed] [Google Scholar]
- Muñoz E., Xu D., Kemp M., Zhang F., Liu J., Linhardt R.J. 2006. Affinity, kinetic, and structural study of the interaction of 3-O-sulfotransferase isoform 1 with heparan sulfate. Biochemistry. 45:5122–5128 10.1021/bi052403n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B., Hantschel O., Young M.A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. 2003. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 112:859–871 10.1016/S0092-8674(03)00194-6 [DOI] [PubMed] [Google Scholar]
- Neef R., Kuske M.A., Pröls E., Johnson J.P. 2002. Identification of the human PHLDA1/TDAG51 gene: down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 62:5920–5929 [PubMed] [Google Scholar]
- Oh E.S., Couchman J.R. 2004. Syndecans-2 and -4; close cousins, but not identical twins. Mol. Cells. 17:181–187 [PubMed] [Google Scholar]
- Paland N., Böhme L., Gurumurthy R.K., Mäurer A., Szczepek A.J., Rudel T. 2008. Reduced display of tumor necrosis factor receptor I at the host cell surface supports infection with Chlamydia trachomatis. J. Biol. Chem. 283:6438–6448 10.1074/jbc.M708422200 [DOI] [PubMed] [Google Scholar]
- Pelicci G., Lanfrancone L., Grignani F., McGlade J., Cavallo F., Forni G., Nicoletti I., Grignani F., Pawson T., Pelicci P.G. 1992. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 70:93–104 10.1016/0092-8674(92)90536-L [DOI] [PubMed] [Google Scholar]
- Perfettini J.L., Gissot M., Souque P., Ojcius D.M. 2002. Modulation of apoptosis during infection with Chlamydia. Methods Enzymol. 358:334–344 10.1016/S0076-6879(02)58099-X [DOI] [PubMed] [Google Scholar]
- Poppe M., Feller S.M., Römer G., Wessler S. 2007. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene. 26:3462–3472 10.1038/sj.onc.1210139 [DOI] [PubMed] [Google Scholar]
- Rajalingam K., Sharma M., Lohmann C., Oswald M., Thieck O., Froelich C.J., Rudel T. 2008. Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS One. 3:e3102 10.1371/journal.pone.0003102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran K.S. 2001. Signaling via Shc family adapter proteins. Oncogene. 20:6322–6330 10.1038/sj.onc.1204776 [DOI] [PubMed] [Google Scholar]
- Reutershan J., Ley K. 2004. Bench-to-bedside review: acute respiratory distress syndrome - how neutrophils migrate into the lung. Crit. Care. 8:453–461 10.1186/cc2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaltesky H.J. 2000. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S., Misener S., Humana Press, Totowa, NJ: 365–386 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. 2001. Molecular Cloning: A Laboratory Manual. Third edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 2,334 pp. [Google Scholar]
- Schlessinger J., Lemmon M.A. 2003. SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE. 2003:RE12 10.1126/stke.2003.191.re12 [DOI] [PubMed] [Google Scholar]
- Schubbert S., Shannon K., Bollag G. 2007. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 7:295–308 10.1038/nrc2109 [DOI] [PubMed] [Google Scholar]
- Selbach M., Paul F.E., Brandt S., Guye P., Daumke O., Backert S., Dehio C., Mann M. 2009. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 5:397–403 10.1016/j.chom.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Su H., McClarty G., Dong F., Hatch G.M., Pan Z.K., Zhong G. 2004. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 279:9409–9416 10.1074/jbc.M312008200 [DOI] [PubMed] [Google Scholar]
- Sudol M. 1998. From Src Homology domains to other signaling modules: proposal of the ‘protein recognition code’. Oncogene. 17:1469–1474 10.1038/sj.onc.1202182 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Mimuro H., Suzuki T., Park M., Yamamoto T., Sasakawa C. 2005. Interaction of CagA with Crk plays an important role in Helicobacter pylori-induced loss of gastric epithelial cell adhesion. J. Exp. Med. 202:1235–1247 10.1084/jem.20051027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swimm A., Bommarius B., Li Y., Cheng D., Reeves P., Sherman M., Veach D., Bornmann W., Kalman D. 2004. Enteropathogenic Escherichia coli use redundant tyrosine kinases to form actin pedestals. Mol. Biol. Cell. 15:3520–3529 10.1091/mbc.E04-02-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammer I., Brandt S., Hartig R., König W., Backert S. 2007. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 132:1309–1319 10.1053/j.gastro.2007.01.050 [DOI] [PubMed] [Google Scholar]
- Tapinos N., Rambukkana A. 2005. Insights into regulation of human Schwann cell proliferation by Erk1/2 via a MEK-independent and p56Lck-dependent pathway from leprosy bacilli. Proc. Natl. Acad. Sci. USA. 102:9188–9193 10.1073/pnas.0501196102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.O., Travers A.A. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167–174 10.1016/S0968-0004(01)01801-1 [DOI] [PubMed] [Google Scholar]
- Tse S.M., Mason D., Botelho R.J., Chiu B., Reyland M., Hanada K., Inman R.D., Grinstein S. 2005. Accumulation of diacylglycerol in the Chlamydia inclusion vacuole: possible role in the inhibition of host cell apoptosis. J. Biol. Chem. 280:25210–25215 10.1074/jbc.M501980200 [DOI] [PubMed] [Google Scholar]
- Valdivia R.H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 10.1016/j.mib.2008.01.003 [DOI] [PubMed] [Google Scholar]
- van der Geer P., Wiley S., Gish G.D., Pawson T. 1996. The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions. Curr. Biol. 6:1435–1444 10.1016/S0960-9822(96)00748-8 [DOI] [PubMed] [Google Scholar]
- Willis S.N., Adams J.M. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17:617–625 10.1016/j.ceb.2005.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright H.R., Turner A., Taylor H.R. 2008. Trachoma. Lancet. 371:1945–1954 10.1016/S0140-6736(08)60836-3 [DOI] [PubMed] [Google Scholar]
- Zhao J., Kim K.A., Abo A. 2009. Tipping the balance: modulating the Wnt pathway for tissue repair. Trends Biotechnol. 27:131–136 10.1016/j.tibtech.2008.11.007 [DOI] [PubMed] [Google Scholar]