Macrophages lacking SPSB2 have increased NO production and enhanced pathogen-killing capabilities due to decreased ubiquitin-mediated destruction of iNOS.
Abstract
Inducible nitric oxide (NO) synthase (iNOS; NOS2) produces NO and related reactive nitrogen species, which are critical effectors of the innate host response and are required for the intracellular killing of pathogens such as Mycobacterium tuberculosis and Leishmania major. We have identified SPRY domain–containing SOCS (suppressor of cytokine signaling) box protein 2 (SPSB2) as a novel negative regulator that recruits an E3 ubiquitin ligase complex to polyubiquitinate iNOS, resulting in its proteasomal degradation. SPSB2 interacts with the N-terminal region of iNOS via a binding interface on SPSB2 that has been mapped by nuclear magnetic resonance spectroscopy and mutational analyses. SPSB2-deficient macrophages showed prolonged iNOS expression, resulting in a corresponding increase in NO production and enhanced killing of L. major parasites. These results lay the foundation for the development of small molecule inhibitors that could disrupt the SPSB–iNOS interaction and thus prolong the intracellular lifetime of iNOS, which may be beneficial in chronic and persistent infections.
Introduction
Nitric oxide (NO) is an important effector molecule that is involved in many physiological and pathological processes (Bogdan, 2001; Ricciardolo et al., 2004). In particular, the rapid generation of NO by the inducible NO synthase (iNOS) is a source of potent reactive nitrogen species (RNS) that are an important component of host defense and play a critical role in the killing of intracellular pathogens (Kröncke et al., 1998; Bogdan et al., 2000; Chakravortty and Hensel, 2003).
iNOS (or NOS2) is one of three isoforms of the NO synthases, which are responsible for the synthesis of NO and citrulline from l-Arg, oxygen, and NADPH (nicotinamide adenine dinucleotide phosphate; Knowles and Moncada, 1994; Nathan and Xie, 1994a; MacMicking et al., 1997b; Alderton et al., 2001). Transcription of iNOS is induced across a wide range of cells and tissues by a variety of stimuli such as microbial components and cytokines. Its activity is calcium independent, in contrast to neuronal NO synthase (nNOS; NOS1) and endothelial NO synthase (eNOS; NOS3), which are constitutively expressed and calcium dependent.
A role for iNOS in antimicrobial host defense was first demonstrated by deletion of the iNOS gene in mice, which then failed to restrain replication of both the bacterium Listeria monocytogenes (MacMicking et al., 1995) and the protozoan parasite Leishmania major (Wei et al., 1995; Diefenbach et al., 1998). T helper type 1 (Th1) responses, the production of IFN-γ, IL-12 and IFN-α/β, and the subsequent induction of iNOS in macrophages are considered important factors in clearance of L. major parasites (Diefenbach et al., 1998). iNOS has since been shown to be important in other infectious diseases (Kröncke et al., 1998; Chakravortty and Hensel, 2003), with iNOS-deficient mice infected with Mycobacterium tuberculosis showing significantly higher risk of dissemination and increased mortality (MacMicking et al., 1997a; Chan et al., 2001; Chakravortty and Hensel, 2003). Consistent with this, administration of an iNOS inhibitor led to the reactivation of tuberculosis in mice (Flynn et al., 1998). A recent study has shown that a polymorphism in the NOS2A (iNOS) gene influences susceptibility to tuberculosis, suggesting a role for iNOS in the pathogenesis of human disease (Gómez et al., 2007). In support of this, alveolar macrophages from patients infected with M. tuberculosis express iNOS, and these cells lose their antimycobacterial activity when treated with iNOS inhibitors (Nicholson et al., 1996).
Over the past decade, many additional roles have been recognized for iNOS in the immune system. TNF/iNOS-producing dendritic cells not only mediate innate immune defense against L. monocytogenes infection in mice (Serbina et al., 2003) but also regulate IgA production in mucosa-associated lymphoid tissues, with impairment of IgA class switching in iNOS-deficient mice (Tezuka et al., 2007). Production of NO by iNOS in response to infection can kill invasive microbes on one hand, but cause NO-mediated cytotoxicity and tissue damage on the other (Kröncke et al., 1998; Bogdan, 2001). In some situations, the expression of iNOS is associated with inflammation and more severe disease outcomes (Bogdan, 2001). Excessive NO production triggers fatal cardiovascular collapse during septic shock, but the degree of protection from sepsis-related mortality in iNOS-deficient mice varies considerably according to the genetic background (Laubach et al., 1995; MacMicking et al., 1995; Wei et al., 1995).
The production of NO needs to be tightly regulated, and, as such, iNOS expression and activity are controlled at multiple levels, including transcriptional, translational, and posttranslational regulation (Knowles and Moncada, 1994; Nathan and Xie, 1994b; MacMicking et al., 1997b; Alderton et al., 2001). In particular, iNOS is regulated by ubiquitination and proteasomal degradation and has a relatively short half-life in primary bronchial epithelial cells and several cell lines (Musial and Eissa, 2001; Kolodziejski et al., 2002, 2004). Protein ubiquitination involves the sequential transfer of a ubiquitin molecule via an E1-E2-E3 enzyme cascade (Hershko and Ciechanover, 1998), the final step of which requires an E3 ligase that functions both as a substrate recognition module and a catalyst for the transfer of ubiquitin to a Lys in the substrate protein (Petroski and Deshaies, 2005). The SOCS (suppressor of cytokine signaling) box is a 40-residue domain, first identified in the SOCS proteins that, in the presence of elongin BC, recruits Cullin5 and Rbx2 to form an active E3 ubiquitin ligase (Kamura et al., 1998; Zhang et al., 2001). The SOCS box is present in >40 proteins encompassing at least nine different families that are defined by their protein interaction domain. The SPRY domain–containing SOCS box protein family (SPSB1–4; also known as SSB-1 to -4) is composed of a central SPRY protein interaction domain and a C-terminal SOCS box (Hilton et al., 1998; Nicholson and Hilton, 1998).
The SPSB2 SPRY domain structure consists of an N-terminal helical region packed against a distorted β-sandwich (Masters et al., 2006; Yao et al., 2006; Kuang et al., 2009). The SPRY domains of SPSB1, SPSB2, and SPSB4 but not SPSB3 interact with prostate apoptosis response protein 4 (Par-4; Masters et al., 2006), whereas the SPSB1 Drosophila melanogaster homologue, GUSTAVUS, interacts with the DEAD box RNA helicase VASA (Styhler et al., 2002). Short, contiguous sequences were identified recently in Drosophila VASA (DINNNN) and human Par-4 (ELNNNL) that interact with an extended interface on the SPSB SPRY domain formed by five variable loops (Woo et al., 2006). The presence of a similar motif in VASA and Par-4 implies a common mechanism for binding partner recognition by this subclass of SPRY domains.
Although iNOS had been shown to be ubiquitinated and degraded in cell lines (Musial and Eissa, 2001; Kolodziejski et al., 2002, 2004), the responsible E3 ubiquitin ligases were unknown. In this study, we demonstrate that SPSB2 interacts with an NNN-containing motif in the N-terminal region of iNOS. We also show that SPSB2 is the adaptor protein in the E3 ubiquitin ligase complex that ubiquitinates iNOS, targeting it for proteosomal degradation and thereby regulating NO production and macrophage killing of L. major, the intracellular parasite responsible for human cutaneous leishmaniasis.
Results
Identification of iNOS as a potential SPSB binding partner
The short sequences DINNNN in Drosophila VASA and ELNNNL in human Par-4 have been shown to mediate their interaction with the SPRY domains of GUSTAVUS and SPSB1 and SPSB2, respectively (Woo et al., 2006). However, this motif is not present in mouse or human VASA or in mouse Par-4. This lack of conservation across species suggests that neither Par-4 nor VASA is a key physiological target of the mammalian SPSB proteins. Therefore, we sought other candidate targets for SPSB proteins.
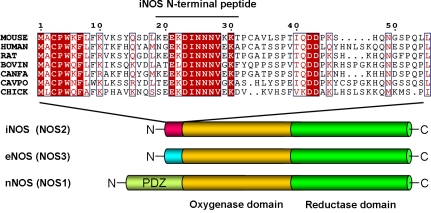
A sequence analysis using ScanProsite (de Castro et al., 2006) identified 11 mouse and 16 human proteins that contained the [DE]-[IL]-N-N-N sequence. One of these candidates was the inducible form of NO synthase (iNOS). The DINNN sequence (residues 23–27) is located in the N-terminal region of mouse iNOS, before the first structured domain (the oxygenase domain). This motif and certain flanking residues are conserved in iNOS sequences from different species, including human, mouse, bovine, chicken, and goldfish, but nNOS and eNOS do not contain this motif (Fig. 1). The N-terminal region of iNOS is predicted to be intrinsically disordered using the programs FoldIndex (Prilusky et al., 2005) and IUPred (Dosztányi et al., 2005; unpublished data), further suggesting that this region could be accessible for SPSB binding.
Figure 1.
Domain structure of NO synthases and amino acid sequence alignments of the N-terminal region of iNOS proteins. The EKDINNNVXK sequence is conserved in iNOS (NOS2) but is not present in either eNOS (NOS3) or nNOS (NOS1). The sequence of the mouse iNOS N-terminal peptide used in ITC and NMR experiments (iNOS19–31) is indicated. The amino acid numbering refers to the mouse protein (Swiss-Prot ID P29477). Outlining indicates regions of conserved amino acid sequence. Red lettering indicates amino acid similarity, and red shading indicates amino acid identity.
The SPSB2 SPRY domain interacts with the iNOS N-terminal sequence
A series of wild-type and mutant peptides corresponding to residues 19–31 of murine iNOS were synthesized, and their binding affinities for the SPSB2 SPRY domain (residues 12–224) were measured using isothermal titration calorimetry (ITC; Fig. S1 and Table I). The wild-type iNOS peptide bound SPSB2 with high affinity (Kd 13 nM). Mutation of Asp23, Asn25, and Asn27 to Ala dramatically reduced the binding affinity, indicating that these residues make a major contribution to binding, which is consistent with the structural requirements previously reported for the GUSTAVUS–VASA interaction (Woo et al., 2006). Additional conserved residues flanking the DINNN sequence (Lys22, Val28, and Lys30) also contribute to binding, as Ala substitutions of these residues produced two- to fourfold lower SPSB2 binding affinities compared with wild-type peptide. Indeed, when these three residues were mutated to Ala simultaneously, SPSB2 binding affinity was decreased by ∼24-fold, suggesting that the SPSB binding sequence in iNOS is more extensive than that reported for the GUSTAVUS–VASA or SPSB–Par-4 interactions. Binding of an SPSB2 SPRY domain in which Tyr120 had been mutated to Ala was assessed by ITC; the iNOS peptide bound Y120A-SPSB2 (12–224) with ∼5,000-fold reduced affinity, indicating that Tyr120 in SPSB2 is critical for binding to the iNOS peptide, as it is for SPSB2 binding to Par-4 (Masters et al., 2006).
Table I.
ITC analysis of interactions between the SPSB2 SPRY domain and wild-type or mutant iNOS N-terminal peptides
| Peptide | Sequence | Kd |
| nM | ||
| A | Ac-KEEKDINNNVKKT-NH2 | 13.3 ± 3.0 |
| B | Ac-KEAKDINNNVKKT-NH2 | 14.0 ± 3.0 |
| C | Ac-KEEADINNNVKKT-NH2 | 127 ± 23 |
| D | Ac-KEAADINNNVKKT-NH2 | 65.4 ± 7.4 |
| E | Ac-KEEKAINNNVKKT-NH2 | 21,600 ± 750 |
| F | Ac-KEEKDANNNVKKT-NH2 | 23.5 ± 9.9 |
| G | Ac-KEEKDIANNVKKT-NH2 | 17,200 ± 7,400 |
| H | Ac-KEEKDIQNNVKKT-NH2 | 40,500 ± 7,200 |
| I | Ac-KEEKDINANVKKT-NH2 | 826 ± 20 |
| J | Ac-KEEKDINNAVKKT-NH2 | NDa |
| K | Ac-KEEKDINNQVKKT-NH2 | NDa |
| L | Ac-KEEKDINNNAKKT-NH2 | 56.8 ± 11.2 |
| M | Ac-KEEKDINNNVKAT-NH2 | 29.8 ± 12.2 |
| N | Ac-KEEKDINNNAKAT-NH2 | 180 ± 39 |
| O | Ac-KEEADINNNAKAT-NH2 | 311 ± 37 |
Typical ITC raw data and titration curves are shown in Fig. S1. Bold letters indicate the amino acid substitution. The Kd column shows dissociation constants with errors from the Origin-calculated association constants transferred as the same fractions of primary values.
Binding affinity was too low to be determined under these conditions.
Nuclear magnetic resonance (NMR) spectroscopy was used to further characterize the SPSB2–iNOS peptide interaction. Titration of the unlabeled iNOS N-terminal peptide into a 15N-labeled SPSB2 (12–224) sample caused a gradual disappearance of the free subset of SPSB2 cross-peaks and the simultaneous appearance of a bound subset of cross-peaks in the 1H-15N heteronuclear single quantum coherence (HSQC) spectra (Fig. 2 A). The residues that exhibited chemical shift perturbations were found on a continuous surface on SPSB2 in the vicinity of Tyr120, Val206, and Trp207, forming an iNOS peptide–binding site (Fig. 2 B).
Figure 2.
SPSB2 interacts with full-length iNOS protein, and Tyr120 in the SPSB2 SPRY domain is critical for binding. (A) Overlay of the 1H-15N HSQC spectra of 0.1 mM 15N-labeled SPSB2 (12–224) in the absence (orange) and presence (green) of unlabeled iNOS peptide at SPSB2/iNOS peptide molar ratios of 1:1.5. Samples were in 95% H2O/5% 2H2O containing 10 mM Na phosphate, 50 mM Na chloride, 2 mM EDTA, 2 mM DTT, and 0.02% (wt/vol) Na azide at pH 7.0. Spectra were recorded at 500 MHz and 22°C. (B) Ribbon model of SPSB2 (12−224; PDB ID number 3EK9; Kuang et al., 2009) with residues whose 1H-15N cross-peaks experienced chemical shift perturbations upon iNOS peptide binding shown in pink. Helices, strands, and loops are colored green, gold, and purple, respectively. (C) BMDMs from C57BL/6 mice were incubated with 20 ng/ml IFN-γ and 1 µg/ml LPS overnight, lysed, and incubated with NHS-Sepharose beads coupled with recombinant SPSB2 or Y120A-SPSB2 protein or with uncoupled NHS-Sepharose beads (Bead) for 3 h at 4°C. Associated iNOS protein was detected by Western blotting (top). Equivalent amounts of SPSB2 and Y120A-SPSB2 were confirmed by reprobing with anti-SPSB2 antibodies (bottom). (D) 293T cells were transiently transfected with vector alone or cDNA encoding Flag-tagged SPSB1, SPSB2, SPSB3, or SPSB4. Cells were lysed and mixed with BMDM lysates from cells induced to express iNOS. Flag-tagged proteins were immunoprecipitated using anti-Flag antibodies, and coimmunoprecipitation of iNOS was detected by Western blotting (top). Membranes were stripped and reprobed with rat anti-Flag antibodies (middle). Comparative expression of Flag-tagged proteins in 293T lysates is shown in the bottom panel. (E) BMDMs from either wild-type (Spsb2+/+) or SPSB2-deficient mice (Spsb2−/−) were incubated with (+) or without (−) 20 ng/ml IFN-γ and 1 µg/ml LPS overnight and lysed, and endogenous SPSB2 proteins were immunoprecipitated using rabbit anti-SPSB2 antibody coupled to NHS-Sepharose. Associated iNOS protein was detected by Western blotting (top). Membranes were stripped and reprobed using biotinylated anti-SPSB2 antibody (middle). iNOS induction was confirmed by Western blotting of protein lysates (bottom). (F) 293T cells were transiently transfected with cDNA encoding Flag-tagged wild-type or mutant SPSB2 in which residues in the SPSB2 SPRY domain were mutated to Ala (and for Tyr120, also to Phe). Cells were lysed and mixed with BMDM lysates from cells induced to express iNOS. Flag-tagged proteins were immunoprecipitated using anti-Flag antibodies, and associated iNOS was detected by Western blotting. IP, immunoprecipitation.
SPSB1, SPSB2, and SPSB4 interact with full-length iNOS
iNOS protein was induced in bone marrow–derived macrophages (BMDMs) by treatment with lipopolysaccharide (LPS) and IFN-γ. Cells were lysed, and iNOS-expressing lysates were incubated with SPSB2 (12–224) protein coupled to Sepharose beads. SPSB2-associated proteins were then separated by SDS-PAGE, and iNOS was detected by Western blotting with specific antibodies. Full-length iNOS protein bound to SPSB2 (12–224), but Y120A-SPSB2 (12–224) pulled down only a minimal amount of iNOS, confirming that Tyr120 is critical for the interaction (Fig. 2 C). These results are consistent with our ITC data and indicate that the iNOS peptide–binding site on the SPSB2 SPRY domain identified by NMR is involved in binding full-length iNOS protein.
To investigate whether other SPSB family members were able to interact with iNOS, 293T cells were transiently transfected with cDNA expressing SPSB1, SPSB2, SPSB3, or SPSB4 with N-terminal Flag epitope tags. The 293T cells were lysed and mixed with iNOS-expressing lysates derived from BMDMs. SPSB proteins were then immunoprecipitated using anti-Flag antibodies coupled to Sepharose, and association with iNOS was analyzed by Western blotting. SPSB2 and SPSB4 but not SPSB3 were able to coprecipitate iNOS protein. In comparison with SPSB2 and SPSB4, SPSB1 bound iNOS relatively weakly, taking into account that slightly less SPSB1 protein was expressed, as detected by an anti-Flag blot of cell lysates. This implies that, although SPSB1, SPSB2, and SPSB4 were able to interact with iNOS, binding affinity may differ among these family members (Fig. 2 D).
To determine whether iNOS and SPSB2 formed an endogenous complex, C57BL/6 BMDMs were stimulated with LPS/IFN-γ and then lysed, and SPSB2 proteins were immunoprecipitated and analyzed by Western blotting with anti-iNOS antibodies. iNOS coimmunoprecipitated with SPSB2 from wild-type but not SPSB2-deficient macrophages (Spsb2−/−; Fig. 2 E), indicating that iNOS was likely to be a genuine physiological target of SPSB regulation. Less SPSB2 protein was immunoprecipitated in LPS/IFN-γ–activated BMDMs compared with unstimulated controls, with a faster-migrating nonspecific band evident in Spsb2−/− lanes. Probing of lysates using specific antibody confirmed iNOS expression in cells derived from both wild-type and Spsb2−/− mice (Fig. 2 E). The apparent down-regulation of SPSB2 protein occurred at the mRNA level, with a 10-fold reduction in SPSB2 mRNA levels after 4-h treatment with LPS/IFN-γ (Fig. S2).
To further interrogate the SPSB2–iNOS binding interface, 293T cells were transiently transfected with cDNA expressing either SPSB2 or various SPSB2 mutants (Masters et al., 2006; Kuang et al., 2009), and interaction with iNOS was assessed by anti-Flag immunoprecipitation and Western blotting for iNOS association. Mutation of Arg100/Gly101, Tyr120, Leu123/Leu124/Leu125, or Val206 to Ala or mutation of Tyr120 to Phe greatly abrogated SPSB2 interaction with iNOS. Mutation of Thr102/His103, Ser126/Asn127/Ser128, or Trp207 to Ala dramatically reduced SPSB2 interaction with iNOS, whereas mutation of either Asp118/His119 or Gln160/Leu161 had no effect. Reblot of the anti-Flag immunoprecipitates confirmed expression of all of the SPSB2 mutants (Fig. 2 F).
SPSB2 appeared the most likely candidate as the key physiological regulator of iNOS in macrophages, particularly as SPSB1 and SPSB4 did not appear to be expressed in BMDMs (Masters et al., 2005). Therefore, our subsequent experiments have focused on this member of the SPSB family.
SPSB2 regulates the proteasomal degradation of iNOS
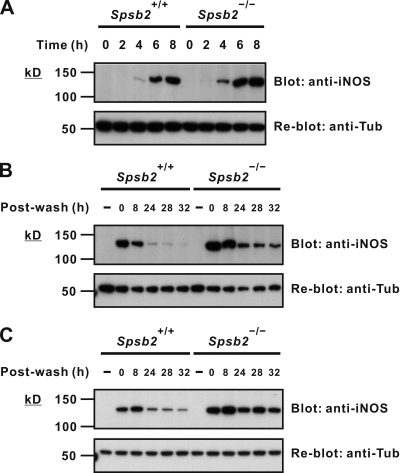
To determine whether the kinetics or magnitude of iNOS expression were altered in the absence of SPSB2, BMDMs from SPSB2-deficient mice (Spsb2−/−) or wild-type littermates were stimulated with LPS/IFN-γ for various times and then lysed, and iNOS expression was detected by Western blot. Although the initial kinetics of iNOS induction appeared to be the same in both wild-type and Spsb2−/− BMDMs, there was a modest increase in iNOS protein in Spsb2−/− BMDMs after 4-, 6-, and 8-h stimulation (Fig. 3 A). A greater difference in iNOS expression between wild-type and Spsb2−/− BMDMs was evident after the stimulus was removed and iNOS had begun to be degraded (Fig. 3 B). Indeed, clearance of iNOS in wild-type BMDMs was essentially complete at 24 h after wash, whereas iNOS was still visible after 32 h in Spsb2−/− BMDMs. This enhanced expression of iNOS was also observed when LPS was used alone (Fig. S3). The results indicate that the initial kinetics of iNOS induction are unaltered in Spsb2−/− BMDMs but the clearance of iNOS protein by the degradation machinery is impaired. SPSB2 regulation of iNOS expression was confirmed in peritoneal macrophages elicited by thioglycollate injection, with iNOS expression after wash significantly prolonged in Spsb2−/− cells (Fig. 3 C).
Figure 3.
iNOS clearance is reduced after stimulus in SPSB2-deficient macrophages. (A and B) BMDMs from either wild-type littermates (Spsb2+/+) or SPSB2-deficient mice (Spsb2−/−) were incubated with IFN-γ and LPS for the times indicated (A) or incubated with or without (−) IFN-γ and LPS overnight, washed, replenished with fresh medium, and lysed at the indicated times after wash (B). Lysates were then separated by SDS-PAGE and analyzed by Western blotting using anti-iNOS antibody (top). Equivalent protein loading was confirmed by stripping and reprobing membranes with antitubulin antibody (anti-Tub; bottom). (C) Thioglycollate-elicited peritoneal macrophages from Spsb2−/− and littermate control mice (Spsb2+/+) were cultured overnight in medium containing LPS/IFN-γ, washed, replenished with fresh medium, and lysed at the indicated times after wash. iNOS expression was detected by Western blotting (top). Equivalent protein loading was confirmed by stripping and reprobing membranes with antitubulin antibody (bottom).
As genetic deletion of SPSB2 appeared to result in an elevation of iNOS levels, we investigated whether artificially increasing SPSB2 levels could correspondingly down-regulate iNOS. BMDMs were generated from mice expressing a Flag-tagged Spsb2 transgene under a constitutive promoter (Spsb2T/+) and from wild-type littermates. LPS/IFN-γ–stimulated iNOS expression was reduced in Spsb2T/+ BMDMs after overnight stimulation (time 0) and rapidly decreased after wash in comparison with wild-type controls (Fig. 4 A), indicating that iNOS degradation was enhanced in these macrophages. iNOS degradation was not enhanced in BMDMs from mice expressing the Spsb2 transgene lacking the SOCS box (Spsb2ΔSBT/+), showing that the SPSB2 regulation of iNOS is SOCS box dependent (Fig. 4 B). Moreover, regulation of iNOS protein by SPSB2 was dependent on the proteasome, as the enhanced iNOS degradation was abrogated in Spsb2T/+ BMDMs treated with the proteasomal inhibitor MG-132 (Fig. 4 C). Macrophage expression of the SPSB2 transgenes was confirmed by anti-Flag immunoprecipitation and Western blotting (Fig. 4 D, top). Reblot of the anti-Flag immunoprecipitates with anti-iNOS antibody showed coimmunoprecipitation of iNOS with both full-length and SOCS box–deleted forms of SPSB2 (Fig. 4 D, upper middle).
Figure 4.
Expression of an SPSB2 transgene enhances iNOS degradation; this requires the SOCS box and is proteasome dependent. (A, B, and C) BMDMs from littermate controls (Spsb2+/+), SPSB2 transgenic mice (Spsb2T/+), or SPSB2 transgenic mice lacking the SOCS box (Spsb2ΔSBT/+) were incubated with or without (−) LPS/IFN-γ overnight, washed, replenished with fresh medium with (+) or without (−) 10 µM of the proteasomal inhibitor MG-132, and lysed at the indicated times after wash. Proteins were then analyzed by Western blotting using anti-iNOS antibody (top). Equivalent protein loading was confirmed by stripping and reprobing membranes with antitubulin antibody (anti-Tub; bottom). (D) Expression of Flag-tagged SPSB2 transgenes in LPS/IFN-γ–treated BMDMs from Spsb2T/+ and Spsb2ΔSBT/+ mice was confirmed by anti-Flag immunoprecipitation (IP) and Western blotting (top). Membranes were stripped and reprobed for iNOS association (upper middle). iNOS expression was confirmed by Western blotting of cell lysates (lower middle) and equivalent protein levels by Western blotting with antitubulin antibody (bottom).
SPSB2 induces iNOS ubiquitination in vitro
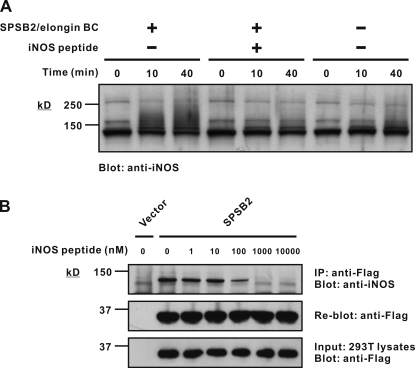
SOCS box proteins recruit an E3 ubiquitin ligase complex that, in the presence of E1 and E2 enzymes, polyubiquitinates interacting proteins, targeting them for proteasomal degradation. A cell-free ubiquitination assay was established to demonstrate SPSB2-dependent ubiquitination of iNOS. LPS/IFN-γ–stimulated macrophage lysates from Spsb2−/− mice were used as a source of iNOS and incubated with ubiquitin and a trimeric SPSB2/elongin BC complex in the presence of E1 and E2 (UbcH5a) enzymes, Rbx2, and Cullin5 for various times. The reaction mixtures were then analyzed by SDS-PAGE and Western blotting with anti-iNOS antibodies. Enhanced polyubiquitination of iNOS, as indicated by high molecular weight laddering, was observed after 10- and 40-min incubation in the presence of the SPSB2/elongin BC complex (Fig. 5 A and Fig. S4). This was completely inhibited by the addition of free iNOS peptide (Ac-KEEKDINNNVKKT-NH2). Coomassie blue staining was used to demonstrate equal input of the E1, E2, and E3 components (Fig. S3).
Figure 5.
SPSB2 induces iNOS ubiquitination in vitro. (A) An in vitro ubiquitination assay was performed using recombinant E1, E2, and E3 ligase components and macrophage lysates as a source of iNOS. Excess free iNOS peptide was added as indicated. The reaction mixture was then separated by SDS-PAGE and analyzed by Western blotting with anti-iNOS antibody. Coomassie blue stain of the gel is shown in Fig. S2. (B) 293T cells were transiently transfected with cDNA expressing Flag-tagged SPSB2, lysed, and mixed with iNOS-expressing macrophage lysates containing increasing amounts of free iNOS peptide. Anti-Flag immunoprecipitates were then assessed for iNOS interaction by SDS-PAGE and Western blotting with anti-iNOS antibody. IP, immunoprecipitation.
To confirm that the free peptide was indeed competing with full-length iNOS for interaction with SPSB2, 293T cells were transiently transfected with cDNA expressing Flag-tagged SPSB2, lysed, and mixed with iNOS-expressing macrophage lysates containing increasing amounts of free iNOS peptide. The SPSB2 interaction with iNOS was modestly inhibited by 100 nM of iNOS peptide and completely inhibited by 1 µM of peptide (Fig. 5 B).
SPSB2 regulation of iNOS results in altered NO output
BMDMs from C57BL/6, Spsb2−/−, Spsb2T/+, or Spsb2ΔSBT/+ mice were stimulated with 2 and 20 ng/ml LPS for 24 h, and the culture supernatant was assayed for NO2− (a stable breakdown product of NO) production using the Griess reaction. In both instances, Spsb2−/− BMDMs produced significantly more NO2− compared with wild-type macrophages, whereas NO2− generation by Spsb2T/+ BMDMs was reduced (Fig. 6 A). Peritoneal macrophages from Spsb2−/− mice also produced significantly more NO2− than macrophages from wild-type controls (Fig. 6 B). To determine whether SPSB2 regulates NO production in response to live bacilli, BMDMs from C57BL/6, Spsb2−/−, Spsb2T/+, and Spsb2ΔSBT/+ mice were preincubated with or without IFN-γ, washed, and infected with L. monocytogenes. Cells were then washed and cultured in DME containing 10 µg/ml gentamicin, a membrane-impermeant antibiotic, and NO2− production was measured 16 h after infection. Spsb2−/− BMDMs produced slightly more NO2− than wild-type macrophages, whereas NO2− generation by Spsb2T/+ BMDMs was comparable with wild-type in the absence of IFN-γ and reduced in the presence of IFN-γ (Fig. 6 C). Compared with that induced by LPS, the difference in NO production between Spsb2−/− and wild-type macrophages appeared to be modest in response to L. monocytogenes infection. In comparison, Spsb2−/− BMDMs infected with Mycobacterium bovis (Bacillus Calmette-Guérin [BCG]) produced more NO2− than wild-type BMDMs 24 and 48 h after infection; NO2− production was augmented in the presence of IFN-γ, and by 48 h, the amounts were similar between wild-type and Spsb2−/− cells (Fig. 6 D).
Figure 6.
SPSB2-deficient macrophages show elevated NO production. (A and B) BMDMs from C57BL/6, Spsb2−/−, Spsb2T/+, and Spsb2ΔSBT/+ mice (A) and peritoneal macrophages from Spsb2+/+ and Spsb2−/− mice (B) were cultured for 24 h in medium containing either 2 or 20 ng/ml LPS. (C) BMDMs from C57BL/6, Spsb2−/−, Spsb2T/+, and Spsb2ΔSBT/+ mice were preincubated with or without 20 ng/ml IFN-γ, washed with PBS, and infected with L. monocytogenes in DME without antibiotics for 30 min. Cells were then washed and cultured in DME containing 10 µg/ml gentamicin for 16 h. (A, B, and C) Data are shown as mean ± SD (n ≥ 3, where each replicate represents cells derived from individual mice). (D) BMDMs from Spsb2+/+, Spsb2−/−, and Spsb2T/+ mice were stimulated overnight with (+) or without (−) IFN-γ and then infected with M. bovis BCG for 2 h, extracellular bacteria were removed, and supernatants were assayed for NO production 24 and 48 h after infection. The pound signs (#) indicate that nitrite is below detectable levels. Data are shown as mean ± SD of quadruplicate cultures from one of two experiments. *, P < 0.05.
L. major killing is enhanced in Spsb2−/− macrophages
iNOS has previously been shown to be critical for the clearance of L. major parasites in mouse models of disease, with iNOS knockout mice being highly susceptible to infection (Wei et al., 1995; Diefenbach et al., 1998). This led us to investigate whether the observed changes in iNOS and NO production were sufficient to result in enhanced macrophage killing of L. major parasites. BMDMs from Spsb2+/+, Spsb2−/−, or Spsb2T/+ mice were infected with L. major parasites in the presence or absence of IFN-γ, and the culture supernatants were assayed for NO2−. In the absence of cytokine, secreted NO2− was almost undetectable. However, NO2− was readily detected in infected cultures containing IFN-γ, with SPSB2-deficient macrophages producing slightly more NO2− than wild-type macrophages (Fig. 7 A). The ability of the macrophages to kill L. major parasites was then assessed. The initial infection rates were comparable at 5 h after infection, but by 48 h after infection, macrophages from Spsb2−/− mice showed significantly enhanced killing of L. major parasites when compared with macrophages derived from Spsb2+/+ control mice (Fig. 7, B and C). The mean number of parasites per infected cell did not differ between the two genotypes (unpublished data). To confirm that the observed differences were indeed caused by enhanced iNOS expression, the experiment was repeated with the inclusion of a specific iNOS inhibitor after infection (1400W; Garvey et al., 1997). Initial 5-h infection rates were again comparable (18.6 ± 1.7% for Spsb2+/+ and 19.1 ± 2.5% for Spsb2−/−), and the addition of the iNOS inhibitor reversed the enhanced killing seen in Spsb2−/− macrophages (Fig. 7 C). Thus, the modest elevation in NO production by SPSB2-deficient macrophages correlates with increased parasite killing in vitro. However, both the low level of NO production (Fig. 7 A) and the modest increase in parasite survival in the presence of the iNOS inhibitor (Fig. 7 C) suggest that the main mechanism by which macrophages control L. major replication is likely to be independent of iNOS. Coupled with the enhanced parasite survival in iNOS-deficient mice (Wei et al., 1995) and recent studies that demonstrate an important role for iNOS-producing dendritic cells (Serbina et al., 2003; Copin et al., 2007; De Trez et al., 2009), our data would suggest that cell types other than macrophages are important for the clearance of L. major parasites in mouse models of infection.
Figure 7.
SPSB2-deficient macrophages show enhanced killing of L. major parasites. (A) BMDMs from Spsb2+/+, Spsb2−/−, and Spsb2T/+ mice were incubated in the presence of L. major promastigotes with or without 10 ng/ml IFN-γ for 48 h. Culture supernatants were then assayed for NO production. Data are shown as mean of triplicate cultures ± SD and are representative of four separate experiments. (B and C) BMDMs from Spsb2+/+ and Spsb2−/− mice were infected with L. major promastigotes. The percentage of infected cells was determined at 5 and 48 h after infection. (C) 5 µM of the iNOS inhibitor 1400W was added to cultures 5 h after infection. Data are shown as mean ± SD (n = 3, where each replicate represents cells derived from individual mice). *, P < 0.05; **, P < 0.005.
Discussion
The rapid production of NO by iNOS constitutes a critical frontline response to microbial infection. Given the potential harm caused by excessive NO production, it is paramount that the initiation and extent of iNOS expression are tightly regulated. In this study, we have identified SPSB2 as the E3 ubiquitin ligase adaptor protein responsible for regulating iNOS protein levels. In response to endotoxin and various infectious agents, iNOS and NO are enhanced in macrophages from SPSB2-deficient mice, and this change in NO production is sufficient to enhance macrophage killing of L. major parasites in vitro. iNOS and NO are correspondingly decreased in macrophages engineered to express SPSB2 under a constitutive promoter.
SPSB proteins, like other SOCS box–containing proteins, are predicted to regulate the expression of interacting protein partners through a SOCS box–associated E3 ubiquitin ligase complex, which ubiquitinates the substrate protein, targeting it for proteasomal degradation. Before our identification of iNOS, the physiological target for SPSB2 was unknown. Several SPSB-binding proteins such as Par-4, VASA, and c-Met have been identified (Styhler et al., 2002; Wang et al., 2005; Masters et al., 2006), and it seems likely that, at least for Par-4 and VASA, the underlying molecular basis for interaction with the SPRY domain is the presence of a D/E-I/L-NNN peptide motif (Woo et al., 2006). We have shown that the interaction with iNOS is mediated by a core DINNN sequence, although our NMR and ITC data suggest that the SPSB2 SPRY–peptide interface is more extensive than that described for either VASA or Par-4, with additional flanking residues in iNOS such as Lys22, Val28, and Lys30 making a substantial contribution to the interaction. Indeed, the affinity of SPSB2 for the iNOS peptide (Kd 13 nM) is higher than that reported for either SPSB1 or SPSB2 binding to Par-4 (Kd 146 nM and 4.7 µM, respectively; Woo et al., 2006). The high affinity of the peptide interaction and the ability of the synthetic iNOS peptide to competitively inhibit binding to full-length iNOS strongly imply that this motif is the critical interaction site.
Many SOCS box–containing E3 ubiquitin ligases such as SOCS1 and SOCS3 have multiple target proteins (Croker et al., 2008; Dimitriou et al., 2008). Although we have demonstrated that iNOS is a biologically relevant SPSB2 target, it is possible that there are others also containing the alNNN sequence (where a and l are acidic and aliphatic residues, respectively). Moreover, all four SPSB proteins reportedly interact with the hepatocyte growth factor receptor c-Met, and overexpression of SPSB1 but not other SPSB proteins enhanced hepatocyte growth factor signaling. The interaction with c-Met was shown to be direct using purified recombinant and in vitro transcribed proteins (Wang et al., 2005), but c-Met does not contain the NNN sequence, raising the possibility that yet another interaction site exists on the SPRY domain of the SPSB family.
Functional redundancy within the SPSB family may contribute to the modest phenotype observed in SPSB2-deficient mice, which have a decreased blood urea level and 25% reduction in circulating platelets (Masters et al., 2005). The basal level of iNOS activity appears to be continuously induced in organs such as lung, kidney, and liver (Nathan and Xie, 1994b; Ricciardolo et al., 2004), where SPSB2 is the predominant SPSB protein (Masters et al., 2005). Loss of SPSB2 in the knockout, coupled with low levels of SPSB1 and SPSB4 in these organs might lead to increased iNOS activity and thus increased Arg consumption. As Arg is also converted to urea by arginase (Wu and Morris, 1998), a reduction in available Arg may well explain the decreased blood urea levels in the SPSB2 knockout mice.
We observed enhanced NO levels in SPSB2-deficient cells after challenge with endotoxin (LPS), gram-positive L. monocytogenes and mycobacteria, and L. major parasites, all of which trigger host responses via different Toll-like receptors and signaling pathways to converge on the rapid induction of iNOS (Korbel et al., 2008; Liese et al., 2008; Corr and O’Neill, 2009). Although mycobacterial infection of BMDMs resulted in enhanced NO production in cells lacking SPSB2 (Fig. 6 D), we did not observe any significant differences in M. tuberculosis (H37Rv) survival in SPSB2-deficient macrophages compared with wild-type controls (not depicted). In this instance, either the levels of NO may not reach a level that is detrimental to mycobacterial survival or the mycobacteria may be able to adopt protective mechanisms to overcome the elevated levels of NO. The in vitro infection of macrophages does not always reflect disease progression in mouse models of infection (unpublished data), and future work will address the in vivo consequences in mice lacking SPSB2.
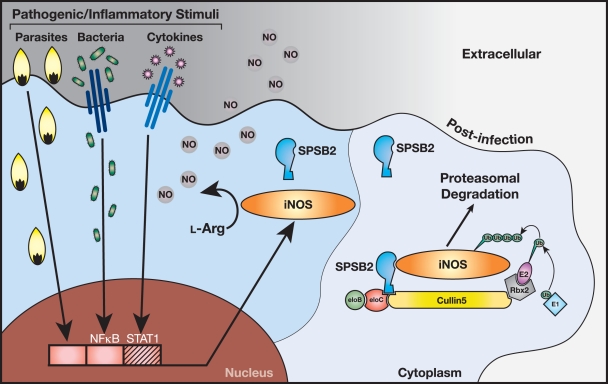
Interestingly, SPSB2 mRNA and protein were transiently down-regulated after LPS/IFN-γ treatment, the lowest level of SPSB2 mRNA corresponding to the initial induction of iNOS protein, as detected by Western blotting (Fig. 2 E and Fig. S2). This suggests another level of cellular control and could potentially provide an alternative physiological mechanism for the stimulation of iNOS activity. However, activation of Toll-like receptor pathways will induce multiple downstream effects (including down-regulation of SPSB2), whereas targeting the SPSB2–iNOS interaction would specifically enhance iNOS protein expression. The production of NO and related RNS are central to the killing of many microbial pathogens, not just the few examined in this study. SPSB2 clearly has a nonredundant role in macrophages and may, in general, function as a cellular sensor to negatively regulate NO production and limit toxicity once the initial infection has been resolved and the flood of cytokines ceases (Fig. 8). Interestingly, SPSB1 and SPSB4 are also able to bind iNOS when overexpressed, and it remains possible that all three proteins act in concert to regulate the magnitude and extent of iNOS expression in different disease/cellular contexts. Future studies will be directed at clarifying whether other SPSB proteins also have a role in regulating iNOS.
Figure 8.
SPSB2 is the adaptor protein in the E3 ubiquitin ligase complex that ubiquitinates iNOS, targeting it for proteasomal degradation. iNOS is rapidly induced in response to microbial pathogens and cytokines via the nuclear factor κB (NF-κB) and STAT1 signaling pathways. iNOS utilizes l-Arg to generate NO, a source of potent RNS that play a critical role in the killing of intracellular pathogens. SPSB2 interacts with iNOS via the SPSB2 SPRY domain and binds to an elongin BC complex via its SOCS box, recruiting Cullin5 and Rbx2 to form an active E3 ubiquitin ligase that polyubiquitinates iNOS, targeting it for proteasomal degradation.
The role of iNOS is not limited to fighting infectious diseases, with iNOS and NO implicated in a diverse range of pathologies such as inflammation and autoimmunity (Kröncke et al., 1998; Bogdan, 2001), Alzheimer’s disease (Nathan et al., 2005; Colton et al., 2006), asthma (Xiong et al., 2000; Prado et al., 2006), diabetes (Cnop et al., 2005), and cancer (Fukumura et al., 2006). Understanding the structural basis for the SPSB–iNOS interaction lays the foundation for the development of small molecule inhibitors that could disrupt the endogenous SPSB–iNOS interaction and thus prolong the intracellular lifetime of iNOS. Providing more of the stimulus systemically can often result in widespread toxic effects. One of the advantages of disrupting SPSB2-mediated degradation of iNOS is that the effects should only be observed in the target cells, specifically where and when iNOS is expressed in response to infection. Although extending the half-life of iNOS may result in cellular toxicity, this has the potential to be of particular benefit in chronic and persistent infections such as tuberculosis or leishmaniasis. In these diseases, even in individuals who appear asymptomatic, the pathogen manages to evade the host defense system (Bogdan, 2008), emerging with devastating effect as individuals age or the immune system is weakened as is the case in AIDS patients.
Materials and methods
Mice
Mice with a homozygous deletion of the Spsb2 gene (Spsb2−/−) have been described previously (Masters et al., 2005) and were maintained on a C57BL/6 background. pUBc constructs containing the SPSB2 (Spsb2) coding region with (residues 3–265; Spsb2T/+) and without the SOCS box (residues 2–224; Spsb2ΔSBT/+) were generated to express SPSB2 with an N-terminal Flag epitope under the ubiquitin C promoter. Transgenic constructs were injected into C57BL/6 blastocysts followed by implantation into pseudopregnant C57BL/6 females. Progeny were screened by Southern blotting for germline transmission of the transgene. Protein expression was confirmed by immunoprecipitation and Western blotting with anti-Flag antibodies. All mice were bred in pathogen-free facilities at the Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia. Ethics approval was obtained from the Animal Ethics Committee at the Walter and Eliza Hall Institute of Medical Research.
Cytokines and antibodies
LPS from Salmonella minnesota was obtained from Enzo Life Sciences, Inc. Murine IFN-γ was obtained from PeproTech, and murine TNF was obtained from eBioscience. Rabbit polyclonal anti-SPSB2 antibodies have been described previously (Masters et al., 2005). Affinity-purified anti-SPSB2 antibodies were either conjugated to N-hydroxysuccinimide (NHS)–Sepharose at 1.5 mg/ml or biotinylated using sulfo-NHS-biotin (Thermo Fisher Scientific) according to the manufacturer’s instructions. Mouse monoclonal anti-iNOS antibody was obtained from BD, mouse monoclonal anti–α-tubulin antibody was obtained from Sigma-Aldrich, and rat anti-Flag antibody was a gift from D. Huang and L. O’Reilly (The Walter and Eliza Hall Institute of Medical Research).
Preparation of SPSB2 protein
The construct used for expression of recombinant murine SPSB2 SPRY domain protein (residues 12–224) has been described previously (Yao et al., 2005). SPSB2 (12–224) was expressed as a GST fusion protein in BL21 (DE3) Escherichia coli. For ITC and coprecipitation experiments, bacteria were grown in Luria broth. For NMR analysis, bacteria were grown in M9 minimal media supplemented with 15N NH4Cl (99%; 1 g/liter). The GST fusion protein was purified from clarified cell lysates using glutathione–Sepharose 4B (GE Healthcare) and then cleaved in situ using thrombin (Roche). The cleaved SPSB2 (12–224) protein was then concentrated and further purified by gel filtration using a Superdex 200 column (GE Healthcare).
ITC
Wild-type and mutant iNOS N-terminal peptides, corresponding to Lys19-Thr31 of mouse iNOS, were synthesized by GL Biochem Ltd (Table I). These peptides were N-terminally acetylated and C-terminally amidated. All ITC measurements were performed at 25°C using an Omega VP-ITC (MicroCal Inc.). SPSB2 (12–224) was dialyzed against buffer (100 mM Tris-HCl and 150 mM NaCl, pH 8.0), and wild-type and mutant iNOS N-terminal peptides were prepared in the same buffer from 5-mM stocks. Solutions of 5–10 µM SPSB2 (12–224) in the cell were titrated by injection of a total of 290 µl of 50–500 µM of iNOS peptides. Data analysis was performed using the evaluation software Origin version 5.0 (MicroCal Inc.). Parameters were derived from a nonlinear least squares fit to the data, using the “one set of sites” model.
NMR spectroscopy
NMR spectra were recorded on a spectrometer (Avance 500; Bruker) equipped with a TXI cryoprobe. The 1H chemical shifts were referenced indirectly to DSS at 0 ppm via the H2O signal, and the 15N chemical shifts were referenced indirectly using the absolute frequency ratio (Wishart et al., 1995). Spectra were processed using Topspin version 1.3 (Bruker BioSpin) and analyzed using XEasy version 1.3 (Bartels et al., 1995). An 15N-labeled SPSB2 (12−224) sample for NMR analysis was prepared in H2O containing 5% 2H2O, 10 mM Na phosphate, 50 mM Na chloride, 2 mM EDTA, 2 mM DTT, and 0.02% (wt/vol) Na azide at pH 7.0. A two-dimensional 1H-15N HSQC spectrum of 0.1 mM 15N-labeled SPSB2 (12−224) was recorded at 22°C using a data matrix size of 2,048 × 256 and with 128 scans per t1 increment. The spectral widths were 13.5 ppm for 1H and 40.0 ppm for 15N; carrier frequencies were 4.7 ppm for 1H and 118 ppm for 15N. Unlabeled iNOS N-terminal peptide (wild type) was then titrated into the 15N-labeled SPSB2 (12−224) sample, and 1H-15N HSQC spectra were recorded at 15N-labeled SPSB2/iNOS peptide ratios of 1:0.5, 1:1, and 1:1.5.
Generation of macrophages
Murine BMDMs were derived by culture of whole bone marrow in DME supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, 10% FBS (Sigma-Aldrich), and 20% L-cell–conditioned medium as a source of macrophage colony-stimulating factor (Wormald et al., 2006) and maintained at 37°C in a humidified atmosphere with 10% CO2. FACS analysis confirmed that >95% of adherent cells were positive for CD11b expression (Mac-1) after 6 d in culture. To elicit macrophages, mice were given an intraperitoneal injection with 1 ml of aged 3% Brewer thioglycollate medium (Difco) and sacrificed after 3 d; peritoneal cells were then harvested by lavage of the peritoneal cavity with PBS. Before stimulation, peritoneal cells were cultured overnight in DME supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, 10% FBS, and 20% L-cell–conditioned medium. FACS analysis confirmed that adherent cells were >98% positive for CD11b (Mac-1).
cDNA constructs and transient transfection of 293T cells
pEFBOS constructs encoding murine SPSB proteins with an N-terminal Flag epitope tag were generated by PCR to give fragments with in-frame AscI and MluI restriction enzyme sites at the N and C termini, respectively, and subcloned into the mammalian expression vector pEF-Flag-I, a derivative of pEFBOS. SPSB point mutants were generated using the PCR-based technique, splicing by overlap extension (Masters et al., 2006; Kuang et al., 2009). 293T cells (DuBridge et al., 1987) were maintained in DME supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 10% FBS. Cells were transiently transfected using FuGene6 Reagent (Roche) according to the manufacturer’s instructions.
Immunoprecipitation and Western blotting
In coprecipitation experiments, bacterially expressed SPSB2 (12–224) and SPSB2 protein in which Tyr120 had been mutated to Ala (Y120A-SPSB2; 12–224) were conjugated to NHS-activated Sepharose (GE Healthcare) according to the manufacturer’s instructions. iNOS expressing BMDM lysates were precleared with Sepharose beads alone for 1 h and then incubated with SPSB2-coupled Sepharose for 3 h at 4°C. iNOS interaction was then detected by SDS-PAGE and Western blotting.
For immunoprecipitation experiments, BMDMs and peritoneal macrophages were incubated with IFN-γ and LPS overnight and lysed in kinase assay lysis buffer (Nicholson et al., 1995) containing protease inhibitors (Complete Cocktail tablets; Roche), 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 1 mM NaF. Proteins were immunoprecipitated using anti-Flag antibody conjugated to Sepharose (M2; Kodak). Proteins were separated by SDS-PAGE under reducing conditions and electrophoretically transferred to BioTrace polyvinylidene fluoride membranes (Millipore). Membranes were blocked overnight in 10% wt/vol skim milk and incubated with primary antibody for 2 h. Antibody binding was visualized with either peroxidase-conjugated sheep anti–mouse Ig antibody (GE Healthcare) or peroxidase-conjugated sheep anti–rabbit Ig antibody (Millipore) and the ECL system (GE Healthcare). To reblot, the membrane was first stripped of antibodies in 0.1 M Gly, pH 2.9.
Real-time quantitative PCR
BMDMs were generated as described in Generation of macrophages and replated at 1.0 × 106 cells/well on 6-well plates (Costar) in DME containing 10% FCS and 20% L-cell–conditioned media. The following day, triplicate cultures derived from three independent mice were incubated with 10 ng/ml murine IFN-γ and 100 ng/ml LPS. Total cellular RNA was isolated using the RNeasy kit (QIAGEN), and quantitative PCR was performed essentially as described previously (Masters et al., 2005) using the QuantiTect SYBR green PCR kit (QIAGEN).
Primer sequences used were as follows: GAPDH (glyceraldehyde 3-phosphate dehydrogenase) forward, 5′-TTGTCAAGCTCATTTCCTGGT-3′; and reverse, 5′-TTACTCCTTGGAGGCCATGTA-3′; and SPSB2 forward, 5′-AAGAAGAGTGGAGGAACCACAAT-3′; and reverse, 5′-CAAAGGCAGAGTGGATATTTGAC-3′. Relative expression was determined by normalizing the quantity of the gene of interest to the quantity of GAPDH. Each measurement was performed in duplicate.
Protein production for ubiquitination assays
Mouse Cullin5 was coexpressed as two domains, the N-terminal domain (1–384) and C-terminal domain (385–780; Zheng et al., 2002). The C-terminal domain of Cullin5 was cloned into the second multiple cloning site of pACYC-DUET (EMD), whereas mouse Rbx2 was cloned into the first multiple cloning site, resulting in a His6 tag at its N terminus. The N-terminal domain of Cullin5 was cloned as a GST fusion protein into pGEX-4T1, and the two vectors were coexpressed in BL21 (DE3) cells to yield a ternary GST-Cul5(N-terminal domain)/His6-Rbx2/Cul5(C-terminal domain) complex. Expression was performed in Luria broth media at 18°C overnight after induction using 1 mM isopropyl β-d-1-thiogalactopyranoside when the OD600 was 0.7. The cells were harvested by centrifugation and then lysed using lysozyme and sonication. The complex was bound to Ni–nitrilotriacetic acid resin, washed, and eluted in buffer containing 250 mM imidazole. The eluant was then bound to glutathione-Sepharose and washed thoroughly in PBS to remove excess Rbx2. The complex was then eluted from the resin by thrombin proteolysis of the GST fusion tag and purified by size-exclusion chromatography using a Superdex 200 16/60 column. Finally, the purified E3 ligase was concentrated to 2 mg/ml. Murine UbcH5a (E2) was expressed as a GST fusion protein by cloning into pGEX-4T and expressed in BL21 (DE3) cells at 37°C for 2 h after isopropyl β-d-1-thiogalactopyranoside induction. After cell lysis, the protein was purified using glutathione-Sepharose and eluted by thrombin digestion. Size-exclusion chromatography using a Superdex 75 16/60 column was performed as the final step in purification. Human E1 (GST tagged) was purchased from Enzo Life Sciences, Inc. Bovine ubiquitin was purchased from Sigma-Aldrich.
Ubiquitination assays
Cell-free ubiquitination reactions contained 0.1 µM E1, 2.5 µM E2, 2.5 µM E3, 50 µM ubiquitin, and 5 µM SPSB2/elongin BC and were incubated for the indicated time periods at 37°C. Cell lysate was added as substrate. Reactions were performed in 20 µl in 20 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, 2.5 mM ATP, and 0.1 mM DTT and were stopped by the addition of 2× SDS-PAGE loading buffer and heating at 95°C for 5 min. Results were visualized by Western blotting using anti-iNOS monoclonal antibody after SDS-PAGE.
NO production
L. monocytogenes strain was maintained by weekly subculture on horse blood agar. Cultures were periodically renewed from freeze-dried stock to maintain a constant level of virulence (Zhan and Cheers, 1998). BMDMs (5 × 105 cells per well) were preincubated with or without 20 ng/ml IFN-γ overnight, washed with PBS, and infected with L. monocytogenes at a ratio of 100 bacteria per cell in DME without antibiotics for 30 min. Cells were then washed with PBS and cultured in DME with 10% FBS containing 10 µg/ml gentamicin (Sigma-Aldrich) to kill residual or adherent extracellular bacteria. Culture supernatants were assayed for nitrite 16 h after infection. BMDMs were infected with M. bovis BCG at a multiplicity of 3:1 for 2 h without antibiotics in the absence or presence of 20 ng/ml IFN-γ, and extracellular bacilli were removed by washing. Culture supernatants were then assayed for nitrite 24 and 48 h after infection. L. major promastigotes (LRC-L137 V 121) were grown in M199 medium with 10% FBS and were in the stationary phase of growth. BMDMs were cultured in a U-bottom 96-well plate (105 cells per well) and preincubated with 10 ng/ml of cytokines for 2 h before infection with promastigotes at a ratio of five promastigotes per cell. Cytokines were maintained throughout. Supernatants were assayed for nitrite at 48 h.
NO production by macrophages in response to LPS, L. monocytogenes, M. bovis BCG, or L. major was determined by measuring nitrite (NO2−), a stable and nonvolatile breakdown product of NO, in culture supernatants using the Griess reaction (Stuehr and Nathan, 1989). 100 µl of culture supernatants was mixed with 10 µl of 1% sulphanilamide, incubated at room temperature for 10 min, and then mixed with 10 µl of 0.1% N-1-naphthylethylenediamine dihydrochloride in 2.5% polyphosphoric acid and further incubated at room temperature for 5 min. Absorbance was measured at 550 nm, and nitrite concentration was determined by comparison with Na nitrite standards. The statistical significance of data was determined using the Student’s t test.
In vitro killing of L. major parasites
BMDMs were plated onto glass coverslips at a density of 5 × 104 macrophages per well in 0.5 ml DME with 10% FBS and 20% L-cell–conditioned medium and allowed to adhere for 3 d. Nonadherent cells were removed by washing, and adherent cells were infected with L. major promastigotes at a ratio of 10 parasites per cell for 4 h. Free parasites were removed by vigorous washing, and cells were incubated for a further 48 h. 5 µM of iNOS inhibitor 1400W (Garvey et al., 1997) was added to cultures at 5 h after infection. The cells were fixed and stained with Giemsa, and the percentage of macrophages infected with parasites was determined for each time point with at least 300 cells counted per sample (Scott et al., 2000).
Online supplemental material
Fig. S1 shows ITC raw data and titration curves for SPSB2 and iNOS peptide interactions. Fig. S2 shows that SPSB2 mRNA is down-regulated after LPS/IFN-γ treatment. Fig. S3 shows that iNOS clearance is reduced after LPS stimulus in SPSB2-deficient macrophages. Fig. S4 shows that iNOS peptide can inhibit iNOS ubiquitination. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912087/DC1.
Acknowledgments
We thank Ms. Sarah Freeman for technical assistance and Ms. Liana Mackiewicz for excellent animal husbandry. We thank Prof. Nicos Nicola and Dr. Ben Croker for reviewing this manuscript.
This work was supported in part by the National Health and Medical Research Council, Australia (Program grant 461219 and Project grant 461233 to R.S. Norton and S.E. Nicholson, fellowships to R.S. Norton and S.E. Nicholson, and Independent Research Institutes Infrastructure Support Scheme grant 361646) as well as a Victorian State Government operational infrastructure support grant.
Footnotes
Abbreviations used in this paper:
- BCG
- Bacillus Calmette-Guérin
- BMDM
- bone marrow–derived macrophage
- eNOS
- endothelial NO synthase
- HSQC
- heteronuclear single quantum coherence
- iNOS
- inducible NO synthase
- ITC
- isothermal titration calorimetry
- LPS
- lipopolysaccharide
- NHS
- N-hydroxysuccinimide
- NMR
- nuclear magnetic resonance
- nNOS
- neuronal NO synthase
- NO
- nitric oxide
- RNS
- reactive nitrogen species
- SOCS
- suppressor of cytokine signaling
References
- Alderton W.K., Cooper C.E., Knowles R.G. 2001. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 357:593–615 10.1042/0264-6021:3570593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Xia T.H., Billeter M., Güntert P., Wüthrich K. 1995. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 6:1–10 10.1007/BF00417486 [DOI] [PubMed] [Google Scholar]
- Bogdan C. 2001. Nitric oxide and the immune response. Nat. Immunol. 2:907–916 10.1038/ni1001-907 [DOI] [PubMed] [Google Scholar]
- Bogdan C. 2008. Mechanisms and consequences of persistence of intracellular pathogens: leishmaniasis as an example. Cell. Microbiol. 10:1221–1234 10.1111/j.1462-5822.2008.01146.x [DOI] [PubMed] [Google Scholar]
- Bogdan C., Röllinghoff M., Diefenbach A. 2000. The role of nitric oxide in innate immunity. Immunol. Rev. 173:17–26 10.1034/j.1600-065X.2000.917307.x [DOI] [PubMed] [Google Scholar]
- Chakravortty D., Hensel M. 2003. Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect. 5:621–627 10.1016/S1286-4579(03)00096-0 [DOI] [PubMed] [Google Scholar]
- Chan E.D., Chan J., Schluger N.W. 2001. What is the role of nitric oxide in murine and human host defense against tuberculosis?Current knowledge. Am. J. Respir. Cell Mol. Biol. 25:606–612 [DOI] [PubMed] [Google Scholar]
- Cnop M., Welsh N., Jonas J.C., Jörns A., Lenzen S., Eizirik D.L. 2005. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 54:S97–S107 10.2337/diabetes.54.suppl_2.S97 [DOI] [PubMed] [Google Scholar]
- Colton C.A., Vitek M.P., Wink D.A., Xu Q., Cantillana V., Previti M.L., Van Nostrand W.E., Weinberg J.B., Weinberg B., Dawson H. 2006. NO synthase 2 (NOS2) deletion promotes multiple pathologies in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 103:12867–12872 10.1073/pnas.0601075103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R., De Baetselier P., Carlier Y., Letesson J.J., Muraille E. 2007. MyD88-dependent activation of B220-CD11b+LY-6C+ dendritic cells during Brucella melitensis infection. J. Immunol. 178:5182–5191 [DOI] [PubMed] [Google Scholar]
- Corr S.C., O’Neill L.A. 2009. Listeria monocytogenes infection in the face of innate immunity. Cell. Microbiol. 11:703–709 10.1111/j.1462-5822.2009.01294.x [DOI] [PubMed] [Google Scholar]
- Croker B.A., Kiu H., Nicholson S.E. 2008. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 19:414–422 10.1016/j.semcdb.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E., Sigrist C.J., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34:W362–W365 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Trez C., Magez S., Akira S., Ryffel B., Carlier Y., Muraille E. 2009. iNOS-producing inflammatory dendritic cells constitute the major infected cell type during the chronic Leishmania major infection phase of C57BL/6 resistant mice. PLoS Pathog. 5:e1000494 10.1371/journal.ppat.1000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Schindler H., Donhauser N., Lorenz E., Laskay T., MacMicking J., Röllinghoff M., Gresser I., Bogdan C. 1998. Type 1 interferon (IFNalpha/beta) and type 2 nitric oxide synthase regulate the innate immune response to a protozoan parasite. Immunity. 8:77–87 10.1016/S1074-7613(00)80460-4 [DOI] [PubMed] [Google Scholar]
- Dimitriou I.D., Clemenza L., Scotter A.J., Chen G., Guerra F.M., Rottapel R. 2008. Putting out the fire: coordinated suppression of the innate and adaptive immune systems by SOCS1 and SOCS3 proteins. Immunol. Rev. 224:265–283 10.1111/j.1600-065X.2008.00659.x [DOI] [PubMed] [Google Scholar]
- Dosztányi Z., Csizmok V., Tompa P., Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 21:3433–3434 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- DuBridge R.B., Tang P., Hsia H.C., Leong P.M., Miller J.H., Calos M.P. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.L., Scanga C.A., Tanaka K.E., Chan J. 1998. Effects of aminoguanidine on latent murine tuberculosis. J. Immunol. 160:1796–1803 [PubMed] [Google Scholar]
- Fukumura D., Kashiwagi S., Jain R.K. 2006. The role of nitric oxide in tumour progression. Nat. Rev. Cancer. 6:521–534 10.1038/nrc1910 [DOI] [PubMed] [Google Scholar]
- Garvey E.P., Oplinger J.A., Furfine E.S., Kiff R.J., Laszlo F., Whittle B.J., Knowles R.G. 1997. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J. Biol. Chem. 272:4959–4963 10.1074/jbc.272.8.4959 [DOI] [PubMed] [Google Scholar]
- Gómez L.M., Anaya J.M., Vilchez J.R., Cadena J., Hinojosa R., Vélez L., Lopez-Nevot M.A., Martín J. 2007. A polymorphism in the inducible nitric oxide synthase gene is associated with tuberculosis. Tuberculosis (Edinb.). 87:288–294 10.1016/j.tube.2007.03.002 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hilton D.J., Richardson R.T., Alexander W.S., Viney E.M., Willson T.A., Sprigg N.S., Starr R., Nicholson S.E., Metcalf D., Nicola N.A. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA. 95:114–119 10.1073/pnas.95.1.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Sato S., Haque D., Liu L., Kaelin W.G., Jr., Conaway R.C., Conaway J.W. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872–3881 10.1101/gad.12.24.3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R.G., Moncada S. 1994. Nitric oxide synthases in mammals. Biochem. J. 298:249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejski P.J., Musial A., Koo J.S., Eissa N.T. 2002. Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc. Natl. Acad. Sci. USA. 99:12315–12320 10.1073/pnas.192345199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejski P.J., Koo J.S., Eissa N.T. 2004. Regulation of inducible nitric oxide synthase by rapid cellular turnover and cotranslational down-regulation by dimerization inhibitors. Proc. Natl. Acad. Sci. USA. 101:18141–18146 10.1073/pnas.0406711102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel D.S., Schneider B.E., Schaible U.E. 2008. Innate immunity in tuberculosis: myths and truth. Microbes Infect. 10:995–1004 10.1016/j.micinf.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Kröncke K.D., Fehsel K., Kolb-Bachofen V. 1998. Inducible nitric oxide synthase in human diseases. Clin. Exp. Immunol. 113:147–156 10.1046/j.1365-2249.1998.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z., Yao S., Xu Y., Lewis R.S., Low A., Masters S.L., Willson T.A., Kolesnik T.B., Nicholson S.E., Garrett T.J., Norton R.S. 2009. SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding. J. Mol. Biol. 386:662–674 10.1016/j.jmb.2008.12.078 [DOI] [PubMed] [Google Scholar]
- Laubach V.E., Shesely E.G., Smithies O., Sherman P.A. 1995. Mice lacking inducible nitric oxide synthase are not resistant to lipopolysaccharide-induced death. Proc. Natl. Acad. Sci. USA. 92:10688–10692 10.1073/pnas.92.23.10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese J., Schleicher U., Bogdan C. 2008. The innate immune response against Leishmania parasites. Immunobiology. 213:377–387 10.1016/j.imbio.2007.12.005 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D., Nathan C., Hom G., Chartrain N., Fletcher D.S., Trumbauer M., Stevens K., Xie Q.W., Sokol K., Hutchinson N., et al. 1995. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 81:641–650 10.1016/0092-8674(95)90085-3 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D., North R.J., LaCourse R., Mudgett J.S., Shah S.K., Nathan C.F. 1997a. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA. 94:5243–5248 10.1073/pnas.94.10.5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J., Xie Q.W., Nathan C. 1997b. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350 10.1146/annurev.immunol.15.1.323 [DOI] [PubMed] [Google Scholar]
- Masters S.L., Palmer K.R., Stevenson W.S., Metcalf D., Viney E.M., Sprigg N.S., Alexander W.S., Nicola N.A., Nicholson S.E. 2005. Genetic deletion of murine SPRY domain-containing SOCS box protein 2 (SSB-2) results in very mild thrombocytopenia. Mol. Cell. Biol. 25:5639–5647 10.1128/MCB.25.13.5639-5647.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S.L., Yao S., Willson T.A., Zhang J.G., Palmer K.R., Smith B.J., Babon J.J., Nicola N.A., Norton R.S., Nicholson S.E. 2006. The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues. Nat. Struct. Mol. Biol. 13:77–84 10.1038/nsmb1034 [DOI] [PubMed] [Google Scholar]
- Musial A., Eissa N.T. 2001. Inducible nitric-oxide synthase is regulated by the proteasome degradation pathway. J. Biol. Chem. 276:24268–24273 10.1074/jbc.M100725200 [DOI] [PubMed] [Google Scholar]
- Nathan C., Xie Q.W. 1994a. Nitric oxide synthases: roles, tolls, and controls. Cell. 78:915–918 10.1016/0092-8674(94)90266-6 [DOI] [PubMed] [Google Scholar]
- Nathan C., Xie Q.W. 1994b. Regulation of biosynthesis of nitric oxide. J. Biol. Chem. 269:13725–13728 [PubMed] [Google Scholar]
- Nathan C., Calingasan N., Nezezon J., Ding A., Lucia M.S., La Perle K., Fuortes M., Lin M., Ehrt S., Kwon N.S., et al. 2005. Protection from Alzheimer’s-like disease in the mouse by genetic ablation of inducible nitric oxide synthase. J. Exp. Med. 202:1163–1169 10.1084/jem.20051529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S.E., Hilton D.J. 1998. The SOCS proteins: a new family of negative regulators of signal transduction. J. Leukoc. Biol. 63:665–668 [DOI] [PubMed] [Google Scholar]
- Nicholson S.E., Novak U., Ziegler S.F., Layton J.E. 1995. Distinct regions of the granulocyte colony-stimulating factor receptor are required for tyrosine phosphorylation of the signaling molecules JAK2, Stat3, and p42, p44MAPK. Blood. 86:3698–3704 [PubMed] [Google Scholar]
- Nicholson S., Bonecini-Almeida Mda.G., Lapa e Silva J.R., Nathan C., Xie Q.W., Mumford R., Weidner J.R., Calaycay J., Geng J., Boechat N., et al. 1996. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J. Exp. Med. 183:2293–2302 10.1084/jem.183.5.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski M.D., Deshaies R.J. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Prado C.M., Leick-Maldonado E.A., Yano L., Leme A.S., Capelozzi V.L., Martins M.A., Tibério I.F. 2006. Effects of nitric oxide synthases in chronic allergic airway inflammation and remodeling. Am. J. Respir. Cell Mol. Biol. 35:457–465 10.1165/rcmb.2005-0391OC [DOI] [PubMed] [Google Scholar]
- Prilusky J., Felder C.E., Zeev-Ben-Mordehai T., Rydberg E.H., Man O., Beckmann J.S., Silman I., Sussman J.L. 2005. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 21:3435–3438 10.1093/bioinformatics/bti537 [DOI] [PubMed] [Google Scholar]
- Ricciardolo F.L., Sterk P.J., Gaston B., Folkerts G. 2004. Nitric oxide in health and disease of the respiratory system. Physiol. Rev. 84:731–765 10.1152/physrev.00034.2003 [DOI] [PubMed] [Google Scholar]
- Scott C.L., Roe L., Curtis J., Baldwin T., Robb L., Begley C.G., Handman E. 2000. Mice unresponsive to GM-CSF are unexpectedly resistant to cutaneous Leishmania major infection. Microbes Infect. 2:1131–1138 10.1016/S1286-4579(00)01267-3 [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. 2003. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 19:59–70 10.1016/S1074-7613(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Stuehr D.J., Nathan C.F. 1989. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 169:1543–1555 10.1084/jem.169.5.1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Lasko P. 2002. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell. 3:865–876 10.1016/S1534-5807(02)00361-1 [DOI] [PubMed] [Google Scholar]
- Tezuka H., Abe Y., Iwata M., Takeuchi H., Ishikawa H., Matsushita M., Shiohara T., Akira S., Ohteki T. 2007. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 448:929–933 10.1038/nature06033 [DOI] [PubMed] [Google Scholar]
- Wang D., Li Z., Messing E.M., Wu G. 2005. The SPRY domain-containing SOCS box protein 1 (SSB-1) interacts with MET and enhances the hepatocyte growth factor-induced Erk-Elk-1-serum response element pathway. J. Biol. Chem. 280:16393–16401 10.1074/jbc.M413897200 [DOI] [PubMed] [Google Scholar]
- Wei X.Q., Charles I.G., Smith A., Ure J., Feng G.J., Huang F.P., Xu D., Muller W., Moncada S., Liew F.Y. 1995. Altered immune responses in mice lacking inducible nitric oxide synthase. Nature. 375:408–411 10.1038/375408a0 [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 6:135–140 10.1007/BF00211777 [DOI] [PubMed] [Google Scholar]
- Woo J.S., Suh H.Y., Park S.Y., Oh B.H. 2006. Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell. 24:967–976 10.1016/j.molcel.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Wormald S., Zhang J.G., Krebs D.L., Mielke L.A., Silver J., Alexander W.S., Speed T.P., Nicola N.A., Hilton D.J. 2006. The comparative roles of suppressor of cytokine signaling-1 and -3 in the inhibition and desensitization of cytokine signaling. J. Biol. Chem. 281:11135–11143 10.1074/jbc.M509595200 [DOI] [PubMed] [Google Scholar]
- Wu G., Morris S.M., Jr 1998. Arginine metabolism: nitric oxide and beyond. Biochem. J. 336:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q., Tezuka Y., Kaneko T., Li H., Tran L.Q., Hase K., Namba T., Kadota S. 2000. Inhibition of nitric oxide by phenylethanoids in activated macrophages. Eur. J. Pharmacol. 400:137–144 10.1016/S0014-2999(00)00354-X [DOI] [PubMed] [Google Scholar]
- Yao S., Masters S.L., Zhang J.G., Palmer K.R., Babon J.J., Nicola N.A., Nicholson S.E., Norton R.S. 2005. Backbone 1H, 13C and 15N assignments of the 25 kDa SPRY domain-containing SOCS box protein 2 (SSB-2). J. Biomol. NMR. 31:69–70 10.1007/s10858-004-6924-6 [DOI] [PubMed] [Google Scholar]
- Yao S., Liu M.S., Masters S.L., Zhang J.G., Babon J.J., Nicola N.A., Nicholson S.E., Norton R.S. 2006. Dynamics of the SPRY domain-containing SOCS box protein 2: flexibility of key functional loops. Protein Sci. 15:2761–2772 10.1110/ps.062477806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Cheers C. 1998. Control of IL-12 and IFN-gamma production in response to live or dead bacteria by TNF and other factors. J. Immunol. 161:1447–1453 [PubMed] [Google Scholar]
- Zhang J.G., Metcalf D., Rakar S., Asimakis M., Greenhalgh C.J., Willson T.A., Starr R., Nicholson S.E., Carter W., Alexander W.S., et al. 2001. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc. Natl. Acad. Sci. USA. 98:13261–13265 10.1073/pnas.231486498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., et al. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 416:703–709 10.1038/416703a [DOI] [PubMed] [Google Scholar]