Cdc20 and Mad2 or Bub1 don’t come together in Mps1-null cells, resulting in a dramatic acceleration of anaphase onset (see also related papers by Hewitt et al. and Santaguida et al. in this issue).
Abstract
The spindle assembly checkpoint (SAC) in mammals uses cytosolic and kinetochore-based signaling pathways to inhibit anaphase. In this study, we use chemical genetics to show that the protein kinase Mps1 regulates both aspects of the SAC. Human MPS1-null cells were generated via gene targeting and reconstituted with either the wild-type kinase (Mps1wt) or a mutant version (Mps1as) sensitized to bulky purine analogues. Mps1 inhibition sharply accelerated anaphase onset, such that cells completed mitosis in 12 min, and prevented Cdc20’s association with either Mad2 or BubR1 during interphase, i.e., before the appearance of functional kinetochores. Furthermore, intramitotic Mps1 inhibition evicted Bub1 and all other known SAC transducers from the outer kinetochore, but contrary to a recent study, did not perturb aurora B–dependent phosphorylation. We conclude that Mps1 has two complementary roles in SAC regulation: (1) initial cytoplasmic activation of Cdc20 inhibitors and (2) recruitment of factors that promote sustained anaphase inhibition and chromosome biorientation to unattached kinetochores.
Introduction
Accurate chromosome segregation is essential for cell viability, organismal development, and tumor suppression. Accordingly, eukaryotes have evolved several mechanisms to defend against chromosome segregation errors. Paramount among these is the so-called spindle assembly checkpoint (SAC), which inhibits anaphase onset until all kinetochore pairs have attached to microtubules (MTs) emanating from both spindle poles, generating a stable configuration termed chromosome biorientation (for review see Musacchio and Salmon, 2007). In biochemical terms, the SAC acts by inhibiting the Cdc20-bound form of the anaphase-promoting complex/cyclosome (APC/C), a large ubiquitin protein ligase (Peters, 2006). Repression of APC/CCdc20 activity stabilizes its downstream targets including securin and cyclin B, which directly block sister chromatid separation and mitotic exit.
Early time-lapse and laser ablation studies pointed to a central role for unattached kinetochores in checkpoint signaling (Rieder et al., 1994, 1995; Li and Nicklas, 1995). Consistent with this notion, all known SAC transducers, including the protein kinases Mps1, Bub1, and BubR1 and the nonkinase components Mad1, Mad2, and Bub3, associate with unattached kinetochores in prometaphase (for review see Musacchio and Salmon, 2007). In particular, it is thought that kinetochore-localized Mad1/Mad2 heterodimers catalyze the conversion of soluble open Mad2 (O-Mad2) to a closed conformer (C-Mad2) that stably binds to and inhibits Cdc20 (De Antoni et al., 2005). However, other compelling data argue that SAC signaling does not entirely depend on kinetochores. First, complexes of Cdc20 bound to Mad2 and/or BubR1 (sometimes referred to as the mitotic checkpoint complex) have been detected in interphase mammalian cells and yeast strains that lack functional kinetochores (Fraschini et al., 2001; Sudakin et al., 2001). Second, Mad2 and BubR1 are required not only to prolong mitosis in the presence of unattached kinetochores, but also to specify the minimum length of M phase under unperturbed conditions (Meraldi et al., 2004). In contrast, inactivation of other SAC components or factors required for kinetochore–MT binding does not accelerate M phase. Third, BubR1’s essential mitotic functions can be reconstituted with an N-terminal fragment that binds Cdc20 but cannot localize to kinetochores (Malureanu et al., 2009). Together, these observations argue that Mad2 and BubR1 are components of a cytosolic timer that actively restrains anaphase onset, affording early mitotic cells time to mature their kinetochores and (if necessary) engage the kinetochore-dependent branch of the SAC. However, the upstream factors that govern this timer remain elusive.
Although the SAC is conserved throughout Eukarya, efforts to define the order in which its components act relative to one another have yielded unexpectedly divergent results. For instance, studies in human cells have consistently positioned Mps1 near the distal end of the SAC, as depleting this kinase via RNAi results in the selective loss of Mad2 from kinetochores (Stucke et al., 2002; Liu et al., 2003, 2006; Jelluma et al., 2008; Tighe et al., 2008). In contrast, genetic analyses in yeast and immunodepletion experiments in Xenopus laevis egg extracts place Mps1 at the apex of the SAC, upstream of not only Mad2 but also Bub1, BubR1/Mad3, and Mad1 (Hardwick et al., 1996; Abrieu et al., 2001; Vigneron et al., 2004; Wong and Fang, 2005). Similarly, although human Mps1 reportedly facilitates chromosome alignment by direct phosphorylation of the aurora B kinase regulator borealin, and thus is necessary to sustain full aurora B activity in human cells (Jelluma et al., 2008), the aurora B–related kinase Ipl1 retains its normal localization and full activity in Mps1-deficient yeast strains (Maure et al., 2007). These findings have been interpreted as evidence of species-specific differences in kinetochore organization and SAC regulation, but other explanations (e.g., technical issues related to the completeness or specificity of Mps1 inactivation) have not been excluded.
To clarify these issues, we created human cells in which both copies of the MPS1 locus could be deleted via gene targeting. The resulting MPS1-null cells were complemented with versions of the kinase that differ at a single amino acid within the ATP-binding site, conferring resistance or sensitivity to bulky purine analogues. Using this chemical genetic system, we investigated the role of Mps1 in M phase progression and SAC signaling. Our experiments identify a novel interphase function for Mps1, whereby it ensures that Cdc20 binds Mad2 and BubR1 before kinetochores have matured and can generate their own anaphase inhibitory signals. Mps1 is also critical for the subsequent phase of SAC signaling, as its inhibition evicts all known SAC mediators from prometaphase kinetochores. Furthermore, we find that although human Mps1 indeed controls chromosome biorientation, it does so independently of aurora B regulation, as indicated by undiminished phosphorylation of multiple aurora B substrates in Mps1-inhibited cells. Collectively, these findings reveal new insights into the SAC response in mammalian cells and provide new tools for interrogating this response in a rapid and specific manner without collateral inhibition of aurora B.
Results
Chemical genetics reveals the M phase–timing function of Mps1
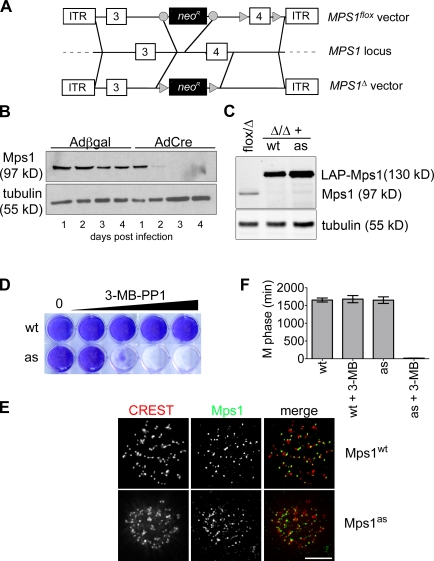
To establish a tight genetic background for functional experiments, we used adeno-associated virus (AAV)–mediated gene targeting (Berdougo et al., 2009) to conditionally delete MPS1 from the human genome. In brief, two vectors were constructed, such that exon 4 of the MPS1 locus was either flanked by loxP sites or deleted outright (Fig. 1 A). Conceptually, removal of this exon truncates the open reading frame at codon 121, upstream of sequences required for kinase activity and kinetochore localization (Stucke et al., 2004). Both vectors were used to sequentially infect human telomerase reverse transcriptase (hTERT)–immortalized retinal pigment epithelial (RPE) cells with targeting efficiencies of 6% and 3%, respectively. To initiate gene deletion, MPS1flox/Δ cells were infected with adenoviruses expressing Cre recombinase (AdCre) or β-galactosidase (Adβgal) as a negative control. Mps1 expression ceased within 48 h of AdCre infection without the appearance of any new immunoreactive species (Fig. 1 B). As anticipated, MPS1Δ/Δ clones could not be recovered by limiting dilution (unpublished data), indicating that this kinase is essential in mammals.
Figure 1.
Generation of Mps1 conditional–null and analogue-sensitive human cells. (A) Schematic of AAV vectors used to mutate the MPS1 locus. Circles and triangles denote FRT and loxP sites, respectively. ITR, inverted terminal repeat. (B) MPS1flox/Δ cells were infected with the indicated adenoviruses and analyzed for Mps1 expression by immunoblotting. (C) MPS1flox/Δ and MPS1Δ/Δ cells complemented by wild-type (wt) or analogue-sensitive (as) Mps1 transgenes were analyzed by immunoblotting. LAP, localization and affinity purification. (D) Allele-specific inhibition of Mps1as. Cells were cultured in the presence of the bulky purine analogue 3MB-PP1 (from left to right: 0, 0.078, 0.313, 1.25, and 5 µM) for 7 d and stained with crystal violet. (E) Cells were fixed and stained with antibodies to centromere autoantigens (CREST; red) and transgene-encoded Mps1wt and Mps1as (green). Bar, 10 µm. (F) Mps1as inhibition overrides the SAC. Cells were filmed at 10-min intervals during treatment with nocodazole in the presence or absence of 10 µM 3MB-PP1 to assess the length of M phase (defined as the period of cell rounding by phase-contrast microscopy). Error bars indicate SEM.
Next, MPS1flox/Δ cells were transduced with retroviruses expressing either the wild-type kinase (Mps1wt) or an analogue-sensitive mutant (M602A; hereafter Mps1as) fused to a localization and affinity purification tag (Cheeseman and Desai, 2005). In vitro Mps1as was considerably less active than Mps1wt, which is similar to other analogue-sensitive kinases (Fig. S1; Bishop et al., 2000; Jones et al., 2005; Burkard et al., 2007; Holland et al., 2010). Nevertheless, both Mps1as and Mps1wt supported the growth of MPS1-null cells (Fig. 1, C and D) and localized to kinetochores (Fig. 1 E), demonstrating their functionality in vivo. Crucially, Mps1as cells alone were susceptible to growth inhibition by the bulky purine analogue 3MB-PP1 (Fig. 1 D; Burkard et al., 2007).
To evaluate the integrity of these alleles with respect to SAC signaling, Mps1wt and Mps1as cells were challenged with spindle poisons that either globally depolymerize MTs (nocodazole) or prevent spindle bipolarization by targeting the kinesin Eg5 (monastrol and S-trityl-l-cysteine [STLC]). Afterward, mitotic indices were determined by MPM-2 staining and flow cytometry (Fig. S1). In the absence of 3MB-PP1, cells of all genotypes arrested with comparably high efficiencies. However, in the presence of 3MB-PP1, Mps1as cells demonstrated little if any increase in the mitotic index relative to unchallenged controls. To complement this endpoint assay, we measured the duration of M phase in individual nocodazole-treated cells by time-lapse phase-contrast microscopy. Strikingly, Mps1as inhibition reduced the length of M phase (defined in this assay as the period of cell rounding) from 1,651 ± 463 min to 18 ± 12 min (Fig. 1 F).
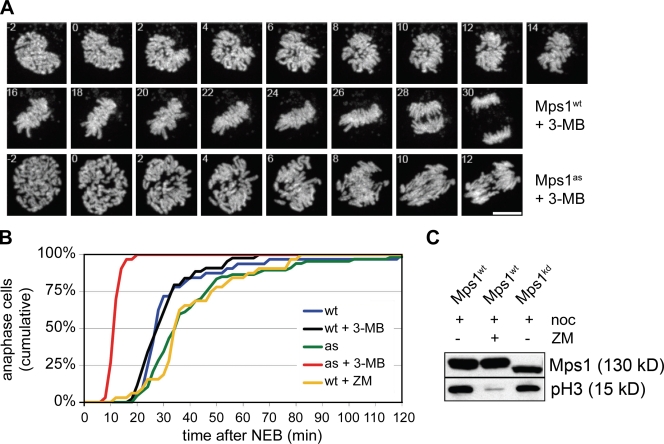
Acceleration of M phase to this degree was noteworthy and unexpected, as both wild-type cells and those lacking the kinetochore-dependent arm of the SAC require ∼30 min to complete mitosis (Meraldi et al., 2004). To score mitotic timing more precisely, we generated cells expressing a histone H2B–mCherry fusion protein and imaged them at higher temporal and spatial resolution using spinning-disk confocal microscopy (Fig. 2 A). Mps1wt cells progressed from nuclear envelope breakdown (NEB) to anaphase in 34 ± 19 min, whereas untreated Mps1as cells exhibited slightly longer kinetics of 42 ± 26 min (Fig. 2 B). However, upon 3MB-PP1 treatment, the NEB to anaphase interval in Mps1as cells fell to just 12 ± 2 min, as also occurs in Mad2- or BubR1-depleted HeLa cells (Meraldi et al., 2004). Because Mps1 is thought to regulate aurora B (Jelluma et al., 2008), we tested whether the latter is also required to sustain normal M phase timing. However, treating cells with the aurora kinase inhibitor ZM447439 (hereafter ZM; Ditchfield et al., 2003) actually delayed anaphase onset slightly rather than accelerating it (Fig. 2 B). Aurora B inhibition also failed to block Mps1’s self-catalyzed phosphorylation and consequent upshift on SDS-PAGE (Fig. 2 C). We conclude that Mps1 plays a key role in setting the basal length of mitosis but does so by a mechanism that is independent of aurora B.
Figure 2.
Mps1 is a component of the M phase timer. (A) Mps1wt and Mps1as cells stably expressing mCherry-tagged histone H2B were treated with 10 µM 3MB-PP1 and filmed at 1-min intervals by spinning-disk confocal microscopy. Maximum intensity projections are shown (Videos 1–6). Bar, 10 µm. (B) Quantification of the mitotic timing defect in Mps1-inhibited cells. Cumulative frequency of anaphase onset is plotted as a function of time after NEB. (C) Strong aurora B inhibition does not block the autophosphorylation of Mps1. HEK293 cells were transfected with GFP-tagged Mps1 expression plasmids, arrested in M phase with nocodazole (noc), and treated with or without 2 µM ZM for 2 h. Cell extracts were resolved by SDS-PAGE and probed with antibodies to serine 10–phosphorylated histone H3 (to confirm suppression of aurora B kinase activity) and GFP (to assess Mps1’s electrophoretic mobility, which becomes retarded by its mitotic activation and autophosphorylation; Stucke et al., 2004).
Mps1 is continuously required for the assembly of Cdc20 inhibitory complexes during interphase and mitosis
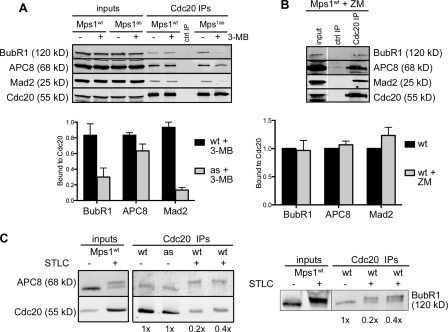
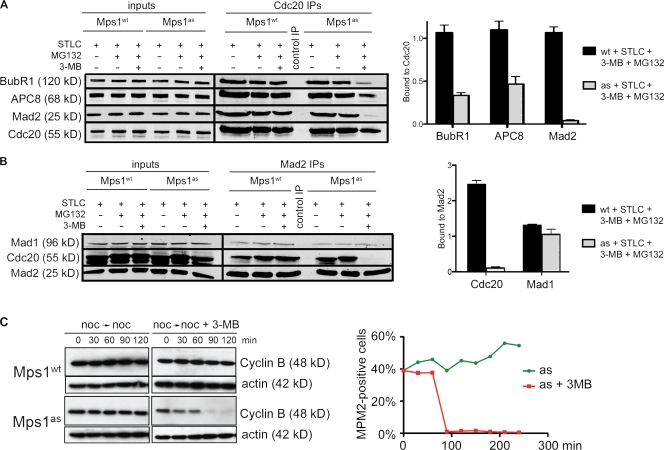
M phase timing is known to depend on Mad2 and BubR1 but not on other SAC components or mediators of kinetochore–MT attachment (Meraldi et al., 2004). Both Mad2 and BubR1 bind to Cdc20 either simultaneously (Sudakin et al., 2001; Herzog et al., 2009) or separately (Tang et al., 2001; Nilsson et al., 2008; Kulukian et al., 2009) and inhibit APC/CCdc20-mediated ubiquitylation. Notably, Cdc20 inhibitory complexes are present during interphase (Sudakin et al., 2001; Tang et al., 2001; Malureanu et al., 2009) before either MT-binding proteins or SAC transducers have been targeted to centromeres (Cheeseman and Desai, 2008). To probe Mps1’s role in the formation of such complexes, asynchronous Mps1wt and Mps1as cultures were treated with 3MB-PP1 for 2 h and depleted of mitotic cells by shake off, after which Cdc20 and associated proteins were immunoprecipitated and analyzed by quantitative immunoblotting. Mps1 inhibition caused Cdc20 to dissociate from Mad2 and BubR1, whereas its association with the APC/C was only mildly affected (Fig. 3 A). In contrast, ZM treatment failed to disrupt these interphase complexes (Fig. 3 B) in keeping with its inability to accelerate mitotic timing (Fig. 2 B). Importantly, the APC8 and BubR1 polypeptides analyzed in these experiments lacked mitotic phosphorylation-induced mobility shifts (Fig. 3 C; Kraft et al., 2003; Elowe et al., 2007; Lénárt et al., 2007), confirming that Mps1 regulates their interaction with Cdc20 specifically during interphase.
Figure 3.
Mps1 directs the assembly of Cdc20 inhibitory complexes in interphase. (A) Mps1wt and Mps1as cells were treated with 3MB-PP1 for 2 h. Extracts were immunoprecipitated with antibodies to Cdc20 and resolved by SDS-PAGE. Levels of BubR1, APC8, Mad2, and Cdc20 in each IP were determined by quantitative immunoblotting. The ratio of each protein to Cdc20 was computed and normalized to ratios obtained from untreated Mps1wt cells (= 1.0). (B) Mps1wt cells were treated with ZM for 2 h and processed as in A. White line indicates that intervening lanes have been spliced out. (C) Interphase and mitotic Cdc20 IPs were electrophoresed on 6% gels to resolve mitotically phosphorylated APC8 (left) and BubR1 (right) polypeptides as distinct species. Numbers at the bottom are loading amounts. Black lines indicate that intervening lanes have been spliced out. ctrl, control. Error bars indicate SEM.
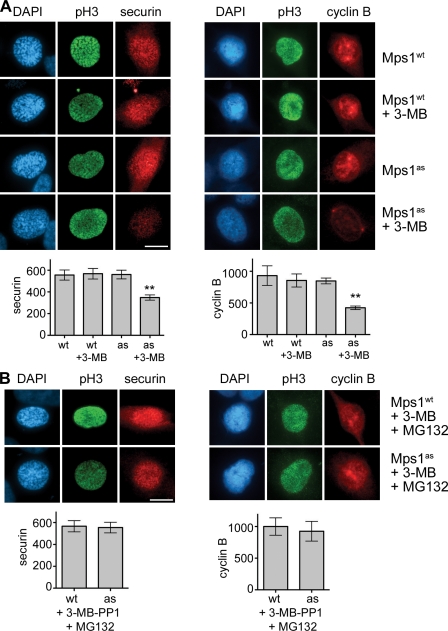
Recent evidence indicates that BubR1 binding to Cdc20 is required to prevent premature turnover of APC/CCdc20 substrates in early mitotic cells (Malureanu et al., 2009). Consistent with its effect on this interaction, Mps1 inhibition reduced the abundance of two well-characterized APC/CCdc20 substrates, securin and cyclin B, during prophase (Fig. 4 A). This decrease was attributable to proteasome-mediated turnover, as it was fully suppressed by MG132 treatment (Fig. 4 B). We conclude that by promoting the formation of Cdc20 inhibitory complexes, Mps1 protects cyclin B and securin from degradation in early mitosis and thus guards against premature anaphase onset.
Figure 4.
Mps1 protects cyclin B and securin from premature destruction. (A) Cells of the indicated genotypes were treated with or without 3MB-PP1, fixed, and stained with antibodies to serine 10–phosphorylated histone H3 (pH3) and securin (left) or cyclin B (right). Nuclei were counterstained with DAPI. Prophase cells were identified by their uniform pH3 staining (Hendzel et al., 1997), partially condensed chromatin (left), and nuclear import of cyclin B (right; Hagting et al., 1999). Note that cyclin B staining used methanol fixation, which preserves chromatin condensation less well than aldehyde fixation. (B) Cells were treated with both 3MB-PP1 and MG132 for 2 h and analyzed as in A. Asterisks indicate statistically significant deviation (P < 0.01 by one-way analysis of variance) from Mps1wt cells treated with 3MB-PP1. Error bars indicate SEM. Bars, 10 µm.
To extend these results, we tested whether Mps1 also controls the assembly of Cdc20 inhibitory complexes during mitosis. In brief, Mps1wt and Mps1as cells were treated overnight with STLC and collected by shake off. Each population of pure (>95%) mitotic cells was transferred to medium containing STLC, 3MB-PP1, and/or MG132. After 2 h, cells were analyzed by Cdc20 immunoprecipitation (IP) and quantitative immunoblotting. Intramitotic Mps1 inhibition dissociated Mad2 and BubR1 from Cdc20 (Fig. 5, A and B). In contrast, Mad2’s interaction with its kinetochore receptor Mad1 was unaffected (Fig. 5 B). In the absence of MG132, intramitotic Mps1 inhibition resulted in rapid destruction of cyclin B and M phase exit (Fig. 5 C). We conclude that Mps1 activity is continuously required for the assembly of Cdc20 inhibitory complexes during interphase and mitosis.
Figure 5.
Mps1 continuously stabilizes Cdc20 inhibitory complexes in M phase. (A) Mitotic cells were harvested by STLC treatment and shake off and incubated in medium containing STLC, 3MB-PP1, and/or MG132 for 2 h. Extracts were immunoprecipitated with antibodies to Cdc20 and resolved by SDS-PAGE. Levels of BubR1, APC8, Mad2, and Cdc20 in each IP were determined by quantitative immunoblotting. The ratio of each protein to Cdc20 was computed and normalized to ratios obtained from STLC-treated Mps1wt cells (= 1.0). (B) Extracts were analyzed as in A except that Mad2 antibodies were used for IP, and Mad1 antibodies were also used for immunoblotting. (C) Cells were arrested in M phase via overnight treatment with nocodazole. At time 0, cells were either treated with 3MB-PP1 or left untreated as a control. Samples were collected at 30-min intervals for analysis of cyclin B levels (left) and determination of mitotic indices (right). Black lines indicate that intervening lanes have been spliced out. Error bars indicate SEM.
Mps1 promotes chromosome alignment independently of aurora B
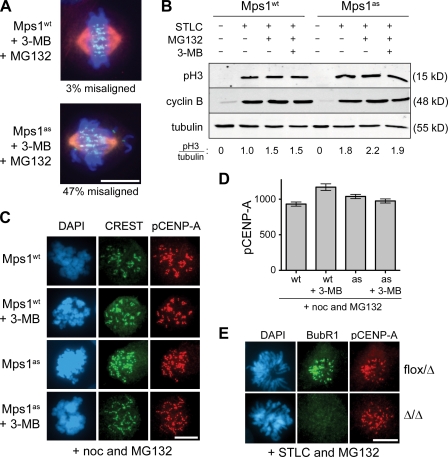
In addition to restraining anaphase onset, Mps1 is necessary for alignment of sister chromatids at the metaphase plate, a function that has been attributed to Mps1-dependent activation of aurora B (Jelluma et al., 2008). In an attempt to confirm this finding, we treated Mps1wt and Mps1as cells with 3MB-PP1 and MG132 to analyze chromosome alignment while blocking anaphase onset. Although almost all cells with active Mps1 had well-formed metaphase plates, 47% of Mps1-inhibited cells had misaligned chromosomes near one or both spindle poles as well as broader than normal metaphase plates (Fig. 6 A). In parallel, we measured levels of phosphorylated histone H3 and centromere protein A (CENP-A) via quantitative immunoblotting and fluorescence microscopy with phosphospecific antibodies (Fig. 6, B–D). Surprisingly, allele-specific Mps1 inhibition failed to suppress the phosphorylation of either aurora B substrate. To corroborate these results, we examined CENP-A phosphorylation levels in MPS1-null cells and again detected no significant loss of phosphorylation (Fig. 6 E). These results have two important implications: (1) Mps1 promotes metaphase chromosome alignment independently of aurora B, and (2) Mps1 inhibition can be used to interrogate the SAC without collateral suppression of aurora B–dependent phosphorylation.
Figure 6.
Mps1 promotes chromosome biorientation independently of aurora B. (A) Cells of the indicated genotypes were treated with 3MB-PP1 and MG132 for 1 h, fixed, and stained to detect centromeres (CREST; green), spindle MTs (α-tubulin; red), and chromosomes (DAPI; blue). (B) Mitotic Mps1wt and Mps1as cells were collected via STLC treatment and shake off and maintained in the presence of STLC, MG132, and/or 3MB-PP1 for 2 h. Lysates were resolved by SDS-PAGE and quantitative immunoblotting. The ratio of phosphorylated histone H3 (pH3) to tubulin was computed for each lane and normalized to the ratio in STLC-treated Mps1wt cells. (C) Cells were treated with nocodazole (noc) for 4 h to activate the SAC and treated for an additional 4 h with nocodazole, MG132, and/or 3MB-PP1. Centromeres (CREST) and serine 7–phosphorylated CENP-A (pCENP-A) were detected by immunofluorescence microscopy. (D) Quantification of results in C is shown. At least 100 centromeres in at least five cells were scored per sample. Error bars indicate SD. (E) MPS1flox/Δ cells were infected with Adβgal (top) or AdCre (bottom). 3 d later, both populations were treated with STLC and MG132 for 30 min, fixed, and stained with the indicated antibodies. BubR1 loss from kinetochores was used as a functional marker of Mps1 inactivation (Fig. 7). Bars, 10 µm.
Mps1 recruits Bub1 and all other SAC transducers to the outer kinetochore and is necessary for centromeric targeting of shugoshin
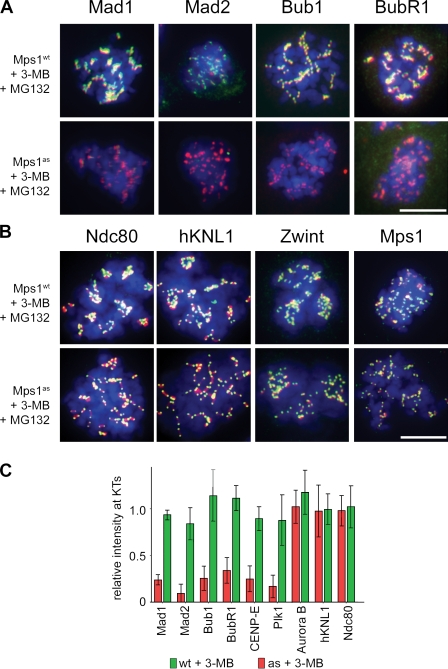
To investigate why chromosomes misalign despite normal phosphorylation of aurora B substrates, we examined known regulators of SAC signaling, kinetochore–MT attachment, and error correction in Mps1-inhibited cells using quantitative microscopy (Fig. 7). Remarkably, all SAC transducers tested, including Bub1, BubR1, Mad1, Mad2, and Zw10, were evicted after Mps1 inhibition (Fig. 7 and Fig. S2). Also lost were CENP-E and Plk1, which are important stabilizers of kinetochore–MT attachment (Fig. S2). In contrast, core MT-binding factors such as Ndc80 and human KNL1 (also called AF15q14, blinkin, or CASC5) remained tightly bound at the kinetochore, as did Mps1 itself (Fig. 7). Similarly, aurora B and INCENP were properly localized at the inner centromere (Fig. S2), which is consistent with the intact phosphorylation of CENP-A in these cells (Fig. 6).
Figure 7.
Mps1 kinase activity is continuously required to maintain Bub1 and all other SAC effectors at unattached kinetochores. (A and B) Mps1wt and Mps1as cells were treated with nocodazole for 4 h to activate the SAC and treated with nocodazole, MG132, and/or 3MB-PP1 for an additional 2 h. Cells were fixed and stained with antibodies against the indicated SAC or kinetochore components (green) and CREST antiserum (red). Note that cells not treated with 3MB-PP1 retained normal kinetochore localization patterns and, thus, have been omitted from these montages for clarity. Additional kinetochore/centromere proteins analyzed in this assay are shown in Fig. S2. (C) Kinetochore (KT)-specific signal intensities were determined in 3MB-PP1–treated Mps1as and Mps1wt cells (>100 kinetochores in more than five cells per sample) and normalized to the equivalent values in untreated Mps1wt cells. Error bars indicate SEM. Bars, 10 µm.
This spectrum of kinetochore targeting defects parallels the known consequences of Bub1 inactivation in both mammalian cells and frog egg extracts (Sharp-Baker and Chen, 2001; Johnson et al., 2004; Perera et al., 2007). In addition to regulating the SAC, Bub1 facilitates pericentromeric cohesion by phosphorylating histone H2A on threonine 120, which in turn recruits the cohesin protector Sgo1 (Kawashima et al., 2010). Interestingly, inactivating Mps1 hindered T120 phosphorylation at centromeres, causing Sgo1 to spread out onto chromosome arms (Fig. S3). Together, these findings demonstrate that Mps1 regulates Bub1 spatially and functionally, providing a simple explanation for the chromosome biorientation defects of Mps1-inhibited cells, which occurred despite normal levels of aurora B–catalyzed phosphorylation.
Cytosol-specific rescue of Mps1 inhibition restores mitotic timing and SAC proficiency to human cells
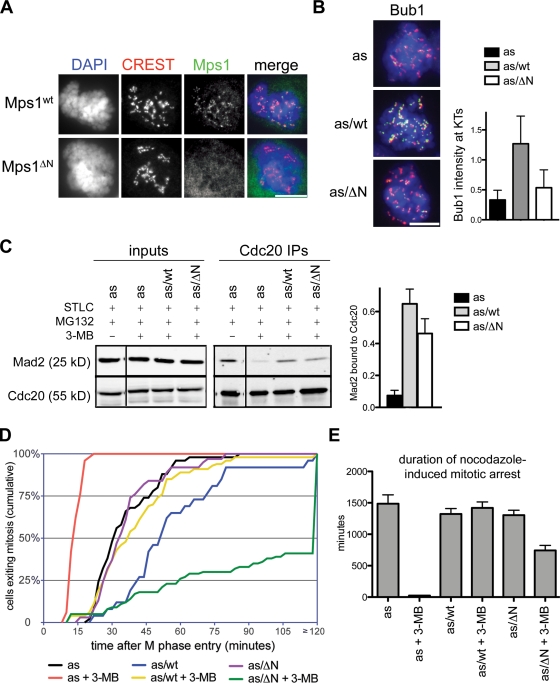
Both Mad2 and BubR1 can inhibit APC/CCdc20 and regulate the timing of anaphase onset independently of kinetochores (Meraldi et al., 2004; Malureanu et al., 2009). To determine whether the same is true for Mps1, we exploited the fact that Mps1’s association with kinetochores depends on its N terminus (Liu et al., 2003; Stucke et al., 2004). In brief, we constructed a mutant allele lacking the first 100 amino acids (hereafter referred to as Mps1ΔN) that indeed fails to localize to kinetochores (Fig. 8 A). Both Mps1ΔN and Mps1wt were introduced into Mps1as cells as mCherry fusions, generating Mps1as/ΔN and Mps1as/wt cells. These cells were treated with 3MB-PP1 to inactivate Mps1as and probe the functionality of the remaining allele. This assay revealed that Mps1 localization is crucial for targeting Bub1 to kinetochores (Fig. 8 B) but not for assembling Cdc20–Mad2 complexes (Fig. 8 C). Interestingly, Mps1ΔN not only rescued the accelerated mitosis seen in Mps1as cells, but actually prolonged it in a 3MB-PP1–dependent manner (Fig. 8 D), suggesting that this cytosolic kinase can respond to (but not correct) biorientation defects caused by inhibition of kinetochore-bound Mps1as. As a direct test of SAC proficiency, we quantified the duration of M phase in each cell line upon treatment with nocodazole (Fig. 8 E). Although Mps1as cells completed mitosis in 23 ± 5 min, Mps1as/ΔN cells remained in M phase for 742 ± 80 min or roughly half as long as Mps1as/wt cells (1,419 ± 95 min; Fig. 8 E). These data establish that Mps1 can indeed delay anaphase without being targeted to kinetochores. Nevertheless, its targeting substantially increases the perdurance of this delay. Thus, Mps1 uses both soluble and kinetochore-dependent mechanisms to generate and maintain the wait anaphase signal.
Figure 8.
Cytosolic-specific rescue of Mps1 inhibition restores mitotic timing and SAC proficiency to human cells. (A) Cells were fixed and stained with antibodies to centromere autoantigens (CREST; red) and transgene-encoded Mps1wt and Mps1ΔN (green). (B) Mps1as, Mps1as/wt, and Mps1as/ΔN cells were treated with nocodazole for 4 h to activate the SAC and treated with nocodazole, MG132, and/or 3MB-PP1 for an additional 2 h. Cells were fixed and stained with antibodies against Bub1 (green) and CREST antiserum (red). Kinetochore (KT)-specific signal intensities were determined in 3MB-PP1–treated Mps1as, Mps1as/wt, and Mps1as/ΔN cells (>100 kinetochores in more than five cells per sample) and normalized to values in untreated Mps1as cells. Bar, 10 µm. (C) Mitotic cells were harvested by overnight STLC treatment and shake off and incubated in medium containing STLC, 3MB-PP1, and/or MG132 for 2 h. Extracts and Cdc20 immunoprecipitates were analyzed as in Fig. 5. Recovery of Mad2 relative to Cdc20 was determined and normalized to its recovery from STLC- and MG132-treated Mps1as cells (= 1.0). Black lines indicate that intervening lanes have been spliced out. (D) Quantification of mitotic timing is shown. Cumulative frequency of anaphase onset is plotted as a function of time after NEB. (E) Mps1ΔN rescues the SAC. Cells were traced by phase-contrast microscopy during treatment with nocodazole ± 3MB-PP1 (compare with Fig. 1 F). Error bars indicate SEM.
Discussion
Classically, the SAC has been conceptualized as a sensor of kinetochore–MT attachment defects. However, recent studies have painted a more complex picture, as the terminal anaphase inhibitors Mad2 and BubR1 (but not other canonical SAC components or kinetochores themselves) are also required to sustain basal M phase timing. These and related observations have led to newer models in which Mad2 and/or BubR1 can be activated away from kinetochores. Indeed, Mad2 and BubR1 can spontaneously form a potent APC/CCdc20 inhibitor in vitro from purified proteins (Kulukian et al., 2009), which could, in theory, account for the cytosolic assembly of Cdc20 inhibitory complexes during interphase (Sudakin et al., 2001; Malureanu et al., 2009). However, our data reveal that such complexes cannot be formed or maintained in the absence of Mps1 kinase activity. Consequently, Mps1-inhibited cells progressed from NEB to anaphase onset in just 12 min, a time frame similar to the delay between Cdk1 and APC/CCdc20 activation in cycling Xenopus egg extracts (Pomerening et al., 2005).
How might Mps1 promote the cytosolic formation of Cdc20 inhibitory complexes? Quantitative models of the SAC indicate that the rate at which a single unattached kinetochore generates Mad2–Cdc20 heterodimers is too slow to account for the global suppression of APC/CCdc20 activity under these conditions (Ciliberto and Shah, 2009). Rather, this high degree of sensitivity requires further rounds of Mad2–Cdc20 complex assembly in the cytoplasm (De Antoni et al., 2005). We speculate that Mps1 activates this cytosolic amplification mechanism either by phosphorylating soluble Mad2 or Mad2–Cdc20 complexes directly (Wassmann et al., 2003) or by suppressing p31comet, a structural mimic of Mad2 that competitively destabilizes Mad2–Cdc20 complexes (Xia et al., 2004; Vink et al., 2006; Yang et al., 2007). This would also explain why a cytosolic form of the kinase (Mps1ΔN) was able to restore Mad2–Cdc20 binding and inhibit anaphase onset in Mps1as cells (Fig. 8). Nevertheless, long-term maintenance of this inhibition (e.g., during chronic treatment with spindle poisons) depends on Mps1’s targeting to kinetochores, which presumably aids Mps1’s phosphorylation of docking partners and/or activators of Bub1 and other SAC mediators. In support of this view, cytosolic versions of Bub1 only partially rescue the SAC deficiency and chromosome misalignment phenotypes of Bub1 RNAi cells (Kiyomitsu et al., 2007; Klebig et al., 2009).
Many of the functions of Mps1 elucidated in this study were inapparent when this kinase was strongly (>90%) depleted from human cells using RNAi (Stucke et al., 2002; Liu et al., 2003, 2006; Jelluma et al., 2008; Tighe et al., 2008). Two observations argue that this difference reflects more complete inactivation of Mps1 using gene deletion and chemical genetics, rather than an off-target effect on other kinases or ATPases. First and foremost, Mps1wt cells were treated in the same manner as Mps1as cells and proved to be completely resistant to 3MB-PP1 in all assays. Second, the epistasis pattern exposed by Mps1 inhibition in human cells mirrors those defined by orthogonal methods in other model systems (i.e., immunodepletion in Xenopus egg extracts and strong Mps1 overproduction in budding yeast; Hardwick et al., 1996; Vigneron et al., 2004; Wong and Fang, 2005). This concurrence suggests that Mps1’s proximal targets and their mode of regulation by phosphorylation are likely to be conserved among all eukaryotes.
It was recently reported that Mps1 is needed to sustain normal levels of aurora B kinase activity and, thus, is required for proper alignment of chromosomes at the metaphase plate (Jelluma et al., 2008). Although maloriented chromosomes were frequently observed in Mps1-inhibited cells, we failed to detect any significant decrease in the phosphorylation of histone H3 or CENP-A, two well-known in vivo substrates of aurora B. One potentially relevant difference is that our analysis used human RPE cells (a nontransformed and chromosomally stable cell type), whereas the earlier study used U2OS cells, a cancer cell line that harbors multiple genetic alterations. Such alterations might have disabled other regulatory networks that normally sustain aurora B activity independently of Mps1. Alternatively, because the RNAi machinery regulates centromeric heterochromatin (Fukagawa, 2004), it is conceivable that high levels of some Mps1-directed shRNAs interfere with the processing of endogenous centromere-derived transcripts to produce subtle anomalies in inner centromere structure and function that manifest as a synthetic defect in aurora B regulation. Consistent with our findings, two orthogonal Mps1 kinase inhibitors, AZ3146 and reversine, also induce SAC override and chromosome biorientation defects without dampening aurora B activity in vivo (in this issue, see Hewitt et al., 2010; Santaguida et al., 2010).
In mammals, the SAC plays an essential role in suppressing aneuploidy, developmental anomalies, and malignancy (Weaver and Cleveland, 2009). Moreover, SAC signaling can either enhance or attenuate the ability of spindle poisons to kill cancer cells (Swanton et al., 2007; Gascoigne and Taylor, 2008; Janssen et al., 2009). Thus, understanding how the SAC operates in molecular terms remains an important objective with broad scientific and medical relevance. Because the SAC itself is required for most methods of synchronizing cells in mitosis, biochemical analyses of SAC signaling require the development of specific and fast-acting inhibitors that can be applied to homogenous populations of mitotic cells. Although aurora B inhibitors can sometimes be used for this purpose, whether these compounds actually inactivate the SAC or merely lead to its satisfaction via stabilization of kinetochore–MT attachments remains controversial (Ditchfield et al., 2003; Hauf et al., 2003; Pinsky and Biggins, 2005; Vanoosthuyse and Hardwick, 2009; Yang et al., 2009). In contrast, Mps1 inhibition was effective in overriding the SAC even when MTs were depolymerized yet did not perturb phosphorylation of endogenous aurora B substrates. By enabling more surgical manipulation of the SAC, the tools developed in this study should clarify how this pathway and others interact to achieve high fidelity chromosome segregation in human cells.
Materials and methods
Cell culture and chemicals
All cell lines were maintained in the following media supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin: HEK293 and Phoenix retroviral packaging lines, DME, hTERT-RPE1 cells, and a 1:1 mixture of DME and Ham’s F-12 medium supplemented with 2.5 mM L-glutamine. Where indicated, 200 ng/ml nocodazole (Sigma-Aldrich), 100 µM monastrol (EMD), 5 µM STLC (Acros Organics), 10 µM 3MB-PP1 (Burkard et al., 2007), 10 µM MG132 (Enzo Life Sciences, Inc.), and/or 2 µM ZM (Tocris Bioscience) were added.
Molecular biology and retroviral transgenesis
To generate the MPS1flox targeting construct, PfuTurbo polymerase (Agilent Technologies) was used to amplify left and right homology arms from bacteria artificial chromosome clone RP11-472L12. Both homology arms were cloned into pNY (Burkard et al., 2007; Terret et al., 2009), and a BglII-marked loxP site was added via linker ligation. The entire targeting construct was transferred to pAAV as an NotI fragment. All manipulated regions were checked by sequencing to ensure their integrity. A similar strategy was used to create the pAAV-MPS1Δ construct used to delete the second allele in MPS1flox/+ cells. Procedures for preparation of infectious AAV particles, transduction of hTERT-RPE1 cells, and isolation of properly targeted clones have been described previously (Berdougo et al., 2009).
For retroviral transduction, inserts were cloned into pQCXIN and pQCXIB (Takara Bio Inc.), and the resulting plasmids were cotransfected with a vesicular stomatitis virus glycoprotein envelope expression construct into Phoenix cells. Infectious supernatants were collected, diluted 1:1 with complete medium containing 20 µg/ml Polybrene, and applied to target cells for 12 h. Selection with 0.4 mg/ml G418 or 5 µg/ml blasticidin was initiated 48 h later.
Antibodies, flow cytometry, immunofluorescence microscopy, and time-lapse imaging
A list of all antibodies used in this study is shown in Table S1. For flow cytometry, cells were fixed in 70% ethanol at −20°C for 24 h. Afterward, cells were rehydrated, blocked in 1% fetal calf serum in 0.1% Triton X-100 and PBS, and stained with a mouse anti–MPM-2 antibody and an Alexa Fluor 633–conjugated anti–mouse secondary antibody (Invitrogen). For immunofluorescence microscopy, cells were plated onto chamber slides (LabTek; Sigma-Aldrich) and grown to 70% confluence. For all antigens except cyclin B, cells were fixed in 4% PFA for 15 min and permeabilized in 0.5% Triton X-100 for 5 min (protocol 1) or simultaneously fixed and permeabilized in 4% PFA and 0.2% Triton X-100 for 20 min (protocol 2), blocked in 3% BSA for 30 min, and incubated in primary antibody for 2 h at room temperature. For cyclin B (Fig. 4), cells were fixed in −20°C methanol for 20 min and rehydrated in PBS for 5 min before blocking in 3% for 30 min and incubation with primary antibody for 2 h at room temperature. Alexa Fluor 488–, 594–, and 647–conjugated goat antibodies were used for secondary detection. Cells were counterstained with DAPI before mounting in Prolong Plus (Invitrogen). For time-lapse experiments, cells were grown on 6-well plates (for phase-contrast imaging) or 35-mm glass-bottomed dishes (for confocal imaging).
Widefield and phase-contrast images were acquired on an inverted microscope (TE2000; Nikon) equipped with 10× and 40× long–working distance and 100× oil objectives, single-bandpass excitation and emission filters, a camera (ORCA ER; Hamamatsu Photonics), a temperature-controlled stage enclosure with CO2 support (Solent Scientific), and NIS Elements software (Nikon). Spinning-disk microscopy (Fig. 2) was performed on a microscope (Axiovert 200; Carl Zeiss, Inc.) fitted with a confocal head (UltraView; PerkinElmer), a 512 × 512 EM charge-coupled device camera (iXon; Andor), XYZ piezo stage (Prior Scientific), and temperature-controlled enclosure with CO2 support (Solent Scientific). Imaging was performed in normal growth media at 37°C. Acquisition was performed with MetaMorph (MDS Analytical Technologies). For deconvolution microscopy (Fig. 1 E and Fig. S2), an image restoration system (DeltaVision; Applied Precision) based on a microscope (IX-70; Olympus) with a 100× oil objective and a cooled charge-coupled device camera (CoolSnap QE; Photometrics) was used. Calculations were performed using measured point-spread functions. Individual images were cropped and assembled into figures using PhotoShop (CS4; Adobe).
Extract preparation and IP
For analysis of mitotic Cdc20 inhibitory complexes, cells were arrested in STLC for 16 h, isolated by shake off, and plated in medium containing STLC, 3MB-PP1, and/or MG132 for 2 h. Cells were harvested, washed twice in ice-cold PBS, and snap frozen in a dry ice/methanol bath. For analysis of interphase complexes, cells were treated with 3MB-PP1 or ZM447439 for 2 h, depleted of the mitotic fraction by shake off, and collected by trypsin/EDTA treatment before snap freezing. Cell pellets were thawed on ice before resuspension in buffer B (140 mM NaCl, 30 mM Hepes, pH 7.8, 5% glycerol, 1 mM DTT, 0.2 µM microcystin, and 1× protease inhibitor cocktail [Sigma-Aldrich]) and disruption by nitrogen cavitation (1,250 psi for 45 min; Parr Instruments). Extracts were clarified by centrifugation at 20,000 g for 30 min and quantified by assay (Bio-Rad Laboratories). 2 mg mitotic extracts or 1 mg interphase extracts were used for each IP. In brief, extracts were incubated with mouse monoclonal antibody to Cdc20 cross-linked to Dynabeads using BS3 (Thermo Fisher Scientific). After 2 h, beads were washed four times in buffer B, resuspended in 1× Laemmli buffer, and boiled for 5 min to elute Cdc20 and any associated proteins.
Quantitative immunoblotting
SDS-PAGE resolved proteins were transferred to PVDF membranes and incubated with primary antibodies as indicated in Table S1. Secondary antibodies (goat anti–rabbit IRDye 680 and goat anti–mouse IRDye 800CW) were used at 1:5,000. Fluorescence was measured using an infrared imaging system (Odyssey; LI-COR Biosciences) according to the manufacturer’s instructions.
Online supplemental material
Fig. S1 shows Mps1wt and Mps1as autophosphorylation assays and that Mps1 kinase activity is required for mitotic arrest in response to several spindle poisons. Fig. S2 shows the effect of Mps1 inhibition on kinetochore targeting of mitotic proteins referenced in Fig. 7 C. Fig. S3 shows that Mps1 promotes Bub1-catalyzed histone H2A phosphorylation and targeting of Sgo1 to centromeres. Videos 1–6 show Mps1wt and Mps1as cells progressing through mitosis in the presence of 10 µM 3MB-PP1. Table S1 lists all of the antibodies used in this study, their working dilution, and application. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001050/DC1.
Acknowledgments
We thank I. Cheeseman, D. Cleveland, G. Gorbsky, and A. Musacchio for sharing reagents, and A. North at the Rockefeller University Bio-Imaging Core Facility for expert assistance with spinning-disk confocal and Deltavision microscopy.
This study was supported by the National Institutes of Health (grants CA107342 and GM094972 to P.V. Jallepalli).
Footnotes
Abbreviations used in this paper:
- AAV
- adeno-associated virus
- APC/C
- anaphase-promoting complex/cyclosome
- CENP
- centromere protein
- hTERT
- human telomerase reverse transcriptase
- IP
- immunoprecipitation
- MT
- microtubule
- NEB
- nuclear envelope breakdown
- RPE
- retinal pigment epithelial
- SAC
- spindle assembly checkpoint
- STLC
- S-trityl-l-cysteine
References
- Abrieu A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., Labbé J.C. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93 10.1016/S0092-8674(01)00410-X [DOI] [PubMed] [Google Scholar]
- Berdougo E., Terret M.E., Jallepalli P.V. 2009. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol. Biol. 545:21–37 10.1007/978-1-60327-993-2_2 [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 407:395–401 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Burkard M.E., Randall C.L., Larochelle S., Zhang C., Shokat K.M., Fisher R.P., Jallepalli P.V. 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 104:4383–4388 10.1073/pnas.0701140104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Ciliberto A., Shah J.V. 2009. A quantitative systems view of the spindle assembly checkpoint. EMBO J. 28:2162–2173 10.1038/emboj.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Musacchio A. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E.A. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21:2205–2219 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20:6648–6659 10.1093/emboj/20.23.6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T. 2004. Assembly of kinetochores in vertebrate cells. Exp. Cell Res. 296:21–27 10.1016/j.yexcr.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Taylor S.S. 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 14:111–122 10.1016/j.ccr.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Hagting A., Jackman M., Simpson K., Pines J. 1999. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 9:680–689 10.1016/S0960-9822(99)80308-X [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 273:953–956 10.1126/science.273.5277.953 [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J.M. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 323:1477–1481 10.1126/science.1163300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt L., Tighe A., Santaguida S., White A.M., Jones C.D., Musacchio A., Green S., Taylor S.S. 2010. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol. 190:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., Cleveland D.W. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Kops G.J., Medema R.H. 2009. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. USA. 106:19108–19113 10.1073/pnas.0904343106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., van den Broek N.J., Cruijsen C.W., van Osch M.H., Lens S.M., Medema R.H., Kops G.J. 2008. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 132:233–246 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- Jones M.H., Huneycutt B.J., Pearson C.G., Zhang C., Morgan G., Shokat K., Bloom K., Winey M. 2005. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 15:160–165 10.1016/j.cub.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.M. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609 10.1093/emboj/cdg627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.M. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632 10.1038/373630a0 [DOI] [PubMed] [Google Scholar]
- Liu S.T., Chan G.K., Hittle J.C., Fujii G., Lees E., Yen T.J. 2003. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 14:1638–1651 10.1091/mbc.02-05-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure J.F., Kitamura E., Tanaka T.U. 2007. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 17:2175–2182 10.1016/j.cub.2007.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 13:566–579 10.1016/j.devcel.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493 10.1016/j.tcb.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Pomerening J.R., Kim S.Y., Ferrell J.E., Jr 2005. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 122:565–578 10.1016/j.cell.2005.06.016 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Schultz A., Cole R., Sluder G. 1994. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127:1301–1310 10.1083/jcb.127.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R.H. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250 10.1083/jcb.153.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke V.M., Silljé H.H., Arnaud L., Nigg E.A. 2002. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 21:1723–1732 10.1093/emboj/21.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke V.M., Baumann C., Nigg E.A. 2004. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 113:1–15 10.1007/s00412-004-0288-2 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C., Marani M., Pardo O., Warne P.H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A.A., et al. 2007. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 11:498–512 10.1016/j.ccr.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H. 2001. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237 10.1016/S1534-5807(01)00019-3 [DOI] [PubMed] [Google Scholar]
- Terret M.-E., Sherwood R., Rahman S., Qin J., Jallepalli P.V. 2009. Cohesin acetylation speeds the replication fork. Nature. 462:231–234 10.1038/nature08550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbé J.C., Castro A., Lorca T. 2004. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 15:4584–4596 10.1091/mbc.E04-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., Musacchio A. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Wassmann K., Liberal V., Benezra R. 2003. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 22:797–806 10.1093/emboj/cdg071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Cleveland D.W. 2009. The role of aneuploidy in promoting and suppressing tumors. J. Cell Biol. 185:935–937 10.1083/jcb.200905098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong O.K., Fang G. 2005. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J. Cell Biol. 170:709–719 10.1083/jcb.200502163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., Yu H. 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23:3133–3143 10.1038/sj.emboj.7600322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D.R., Machius M., Rizo J., Yu H., Luo X. 2007. p31comet blocks Mad2 activation through structural mimicry. Cell. 131:744–755 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Kenny A.E., Brito D.A., Rieder C.L. 2009. Cells satisfy the mitotic checkpoint in taxol, and do so faster in concentrations that stabilize syntelic attachments. J. Cell Biol. 186:675–684 10.1083/jcb.200906150 [DOI] [PMC free article] [PubMed] [Google Scholar]