Abstract
The HMG (high-mobility group)-box-containing chromatin-remodelling factor SRY (sex-determining region on the Y chromosome) plays a key role in sex determination. Its role in the nucleus is critically dependent on two NLSs (nuclear localization signals) that flank its HMG domain: the C-terminally located ‘β-NLS’ that mediates nuclear transport through Impβ1 (importin β1) and the N-terminally located ‘CaM-NLS’ which is known to recognize the calcium-binding protein CaM (calmodulin). In the present study, we examined a number of missense mutations in the SRY CaM-NLS from human XY sex-reversed females for the first time, showing that they result in significantly reduced nuclear localization of GFP (green fluorescent protein)–SRY fusion proteins in transfected cells compared with wild-type. The CaM antagonist CDZ (calmidazolium chloride) was found to significantly reduce wild-type SRY nuclear accumulation, indicating dependence of SRY nuclear import on CaM. Intriguingly, the CaM-NLS mutants were all resistant to CDZ's effects, implying a loss of interaction with CaM, which was confirmed by direct binding experiments. CaM-binding/resultant nuclear accumulation was the only property of SRY found to be impaired by two of the CaM-NLS mutations, implying that inhibition of CaM-dependent nuclear import is the basis of sex reversal in these cases. Importantly, the CaM-NLS is conserved in other HMG-box-domain-containing proteins such as SOX-2, -9, -10 and HMGN1, all of which were found for the first time to rely on CaM for optimal nuclear localization. CaM-dependent nuclear translocation is thus a common mechanism for this family of important transcription factors.
Keywords: calmodulin, high-mobility group box (HMG box), nuclear transport, sex-determining region on the Y chromosome (SRY), SRY-related HMG box (SOX), XY sex reversal
Abbreviations: CaM, calmodulin; CDZ, calmidazolium chloride; CLSM, confocal laser-scanning microscopy; CREB, cAMP-response-element-binding protein; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; FL, full-length; GST, glutathione transferase; HMG, high-mobility group; HRP, horseradish peroxidase; HTC, hepatoma tissue culture; IB, intracellular buffer; Imp, importin; NLS, nuclear localization signal; p.t., post-transfection; SRY, sex-determining region on the Y chromosome; SOX, SRY-related HMG-box; T-ag, SV40 (simian virus 40) large tumour antigen
INTRODUCTION
HMG (high-mobility group) non-histone chromosomal proteins include the AT-hook, HMGN and HMG domain families [1]. Members of the SOX [SRY (sex-determining region on the Y chromosome)-related HMG-box] family of chromatin-remodelling factors play important developmental roles, SRY itself playing a key role in mammalian sex determination, as indicated by the fact that 15% of all XY sex-reversed individuals carry mutations in SRY [2–4]. Expressed in the indifferent gonad 7 weeks post-fertilization in humans, SRY's specific role is in the nucleus, activating/co-ordinating the expression of genes such as the related protein SOX9 that result in the differentiation of the pre-Sertoli cells to produce a testis, and suppress genes favouring formation of the female gonad [3–6]. In the case of XY sex reversal due to impaired action of SRY (Swyer syndrome), patients present with CGD (complete gonadal dysgenesis), female external genitalia, and one in two develop gonadal tumours [2–4,7].
Like other HMG proteins, SRY is able to bind and bend specific DNA targets through its HMG-box domain [3,8]. The majority of sex-reversing mutations in SRY result in impaired DNA binding/bending, but a number, several of which do not affect DNA binding, map to one of SRY's two independently functioning NLSs (nuclear localization signals) that flank the HMG-box domain (Figure 1) [3,4,7,9–11]. Little is known regarding the mechanism by which the N-terminal ‘CaM-NLS’ (amino acids 59–77) functions, apart from the fact that it can bind the Ca2+-binding protein CaM (calmodulin) in vitro [12], whereas the C-terminal ‘β-NLS’ (amino acids 128–134) appears to be conventional, mediating nuclear import dependent on the Impβ1 (importin β1) member of the Imp superfamily and the guanine-nucleotide-binding protein Ran [13].
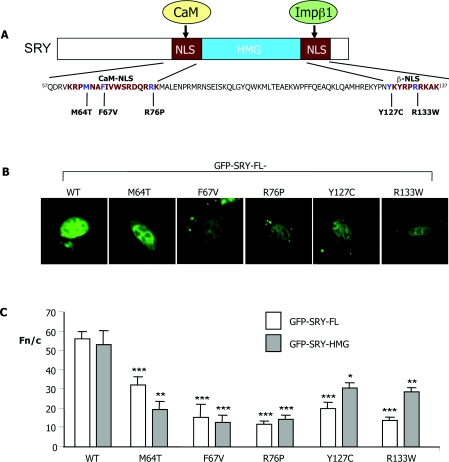
Figure 1. Sex-reversing mutations in the CaM-NLS reduce SRY nuclear accumulation.
(A) The SRY protein is shown diagrammatically, with the CaM-NLS and β-NLSs flanking the HMG-box (single-letter amino acid code) highlighted together with the sex-reversing point mutations analysed in the present study (see the Materials and methods section for patient details). (B) TM4 cells were transiently transfected to express the indicated GFP–SRY-HMG/FL fusion proteins, and imaged live 24 h later by CLSM using a 40× water-immersion objective. (C) Results of quantitative analysis, whereby CLSM images such as those in (B) were analysed using ImageJ software for the extent of nuclear accumulation measured as the Fn/c (see the Materials and methods section). Results are means±S.E.M. (n>7); ***P<0.0001; **P<0.005; *P<0.01 for Fn/c values for the NLS mutant constructs significantly different from those of WT.
We previously described a sex-reversing NLS mutation R133W in the β-NLS that impairs Impβ1 binding and SRY nuclear localization specifically [10], the results indicating that nuclear import mediated by the β-NLS/Impβ1 is essential for sex determination. We also found that two sex-reversing mutations in the CaM-recognized CaM-NLS did not affect Impβ1 binding [10], implying effects on nuclear import distinct from those involving Impβ1. CaM is a ubiquitous Ca2+-binding protein, highly conserved in eukaryotes [14,15], which regulates a plethora of Ca2+-responsive cellular processes. A role for CaM in nuclear import is implied by the fact that the potent CaM antagonist CDZ (calmidazolium chloride) reduces nuclear accumulation of SOX9 [16,17], whereas a CaM-dependent, Ran-independent nuclear import pathway has been described for the yeast HMG-box protein Nhp6Ap [18].
In the present study, we analysed a number of sex-reversing mutations from patients in terms of their CaM/Impβ1-binding and nuclear transport properties for the first time. We found that CDZ can inhibit SRY nuclear transport and, importantly, that sex-reversing CaM-NLS mutations result in loss of sensitivity to CDZ. Of several CaM-NLS mutants examined for the first time, we show that all have reduced nuclear import, and that several are impaired directly in CaM binding. Of these, the M64T and R76P CaM-NLS mutants, which possess normal DNA-binding/bending activity [3,10], have normal Impβ1 binding, implying that impaired nuclear import through reduced ability to bind CaM is the basis of sex reversal. Importantly, we also show for the first time that other HMG-box domain proteins, along with a non-histone chromosomal protein, HMGN1, show similar properties to SRY in terms of a CaM-facilitated nuclear import pathway, implying a general role for CaM in the nuclear transport of HMG proteins.
MATERIALS AND METHODS
SRY patient alleles
All sex-reversed patients examined in the present study have 46 chromosomes, XY gonadal dysgenesis and single nucleotide transversions that result in substitution of a single amino acid. The detailed clinical profiles for patients carrying the R76P, R133W, F67V, M64T and Y127C alleles have been described previously [3,4,9].
Plasmid construction
Plasmids encoding GFP (green fluorescent protein)–SRY-HMG or GFP–SRY-FL (full-length) proteins for mammalian cell expression, and His6-tagged GFP–SRY-HMG for bacterial expression were generated using Gateway™ technology (Invitrogen) [19]. SRY-HMG (residues 55–141) and full-length constructs were generated by PCR as described previously [13,20], using primers containing attB sites to enable the derivation of expression plasmids via recombination. The control constructs encoding GFP–CREB (cAMP-response-element-binding protein)-(252–341) and GFP–T-ag [SV40 (simian virus 40) large tumour antigen]-(111–135) [19] were derived similarly. Point mutations in SRY-FL- and SRY-HMG-box-encoding plasmids were introduced using the QuikChange™ site-directed mutagenesis system (Stratagene). Subsequent to confirmation by DNA sequencing, the mutated sequences were recombined into the various expression constructs as described above. In the case of plasmids encoding the pEGFP-SOX-HMG domains of the SOX2 [21], SOX9 [22] and SOX10 [23] proteins for mammalian cell expression, the HMG domains were amplified by PCR using appropriate primers and cloned into the BamHI/BglII sites of pEGFP-C1. The HMGN1-pEGFP plasmid was kindly supplied by Michael Bustin [24].
Cell culture
COS-7 African green monkey kidney, GC-2 mouse germ, TM4 mouse Sertoli, P19 mouse embryonic carcinoma, HeLa cervical cancer and rat HTC (hepatoma tissue culture) cell lines were all cultured in DMEM (Dulbecco's modified Eagle's medium) (ICN), supplemented with 10% (v/v) heat inactivated FBS (fetal bovine serum) (CSL), 1 mM L-glutamine, 1 mM penicillin/streptomycin and 20 mM Hepes, with the exception of TM4 cells which were cultured in 1:1 Ham's F12/DMEM, supplemented with 1 mM L-glutamine, 1 mM penicillin/streptomycin, 20 mM Hepes and 5% (v/v) FBS in a 5% CO2 humidified atmosphere at 37 °C.
Transfection/CLSM (confocal laser-scanning microscopy)
COS-7, GC-2 and HeLa cells were transfected using Lipofectamine™ (Invitrogen) 16 h after plating according to the manufacturer's instructions, whereas TM4 and P19 cell lines were transfected using Mirus LT-1 (Mirus TransIT®). Cells were imaged live by CLSM (Bio-Rad Laboratories MRC-500) using a 40× water-immersion objective 24 h p.t. (post-transfection), and image analysis of digitized confocal images was performed using the ImageJ (NIH) public domain software. Nuclear (Fn) and cytoplasmic (Fc) fluorescence was determined subsequent to the subtraction of autofluorescence (also quantified), and used to derive the nuclear to cytoplasmic ratio (Fn/c), a parameter that enables differences in expression levels between individual transfected cells to be standardized. Where indicated, cells were treated with 5, 2 or 1.2 μM CDZ (COS-7/GC-2/HeLa, TM4 and P19 cells respectively) 12 h p.t. or 8 h p.t. for HeLa cells before CLSM imaging/image analysis as above.
Subcellular fractionation and immunoblotting
COS-7 cells were transfected to express GFP–SRY constructs and subcellular fractionation as described previously [25]. Briefly, intact cells were harvested 24 h p.t. using 0.02% EDTA and washed twice with PBS, and nuclei were extracted upon incubation in nuclei isolation buffer {320 mM sucrose, 0.1% Triton X-100 and Complete™ EDTA-free protease inhibitor cocktail tablets (Roche) in IB [intracellular buffer: 110 mM KCl, 5 mM NaHCO3, 5 mM MgCl2, 1 mM EGTA, 0.1 mM CaCl2 and 20 mM Hepes (pH 7.4)]} for 5 min on ice. Nuclei were pelleted by centrifugation at 2000 g for 5 min at 4 °C, the cytoplasmic fraction (supernatant) was collected, and intact nuclei were washed twice and lysed with 1% CHAPS and Complete™ protease inhibitor cocktail for 5 min on ice. Insoluble material was removed by centrifugation at 20000 g at 4 °C for 5 min [25]. Nuclei and cytoplasmic extracts were then subjected to SDS/PAGE (12% gel) and transferred on to a PVDF membrane that was probed with anti-GFP (1:1000 dilution; Roche), anti-(histone 2B) (1:500 dilution; Millipore) or anti-α/β-tubulin (1:500 dilution; Cell Signaling Technology) antibodies followed by HRP (horseradish peroxidase)-conjugated goat anti-(mouse IgG) (1:10000 dilution; Millipore) and HRP-conjugated goat anti-(rabbit IgG) (1:10000 dilution; Millipore) antibodies before development using the Western Lighting Chemiluminescence reagent (PerkinElmer). Densitometric analysis was performed on digitized images of immunoblots using ImageJ software. Nuclear (N) and cytoplasmic (C) density was determined and used to derive the nuclear to cytoplasmic ratio (N/C) to assess the presence of GFP–SRY in the nucleus compared with cytoplasm. Immunoblot analysis of whole cell lysates indicated that all constructs were expressed to comparable levels and in intact form (see Supplementary Figure S1 at http://BiochemJ.org/bj/430/bj4300039add.htm).
Bacterial expression and biotinylation of recombinant proteins
WT (wild-type) and mutant SRY-HMG domain variants were expressed in Escherichia coli strain BL21-Rosetta2 (Novagen) as His6-tagged GFP-fusion proteins [13,19], with expression induced for 4 h using 1 mM IPTG (isopropyl β-D-thiogalactoside) at 28 °C at a D595 of 0.6, followed by purification under denaturing conditions. Impβ1 was purified from bacteria as a GST (glutathione transferase)-fusion protein under native conditions [13], and biotinylated using the sulfo-NHS-biotin (sulfo-N-hydroxysuccinimide biotin) reagent (Pierce) [26].
In vitro nuclear transport assays
Mechanically perforated rat HTC cells were used to assess nuclear import of WT GFP–SRY-HMG in conjunction with CLSM, as described previously [13,27] using a 60× oil-immersion lens, with image analysis performed as above. Experiments were carried out in 5 μl volumes, containing 30 μM WT GFP–SRY-HMG, a 70 kDa Texas Red–dextran to assess nuclear integrity, untreated reticulocyte lysate (45 μg/μl) and an ATP-regenerating system (0.125 g/ml creatine kinase, 30 mM creatine phosphate and 2 mM ATP) with and without CDZ, as indicated.
Native PAGE
Native PAGE assays were performed to assess binding [28] of SRY to GST–Impβ1 or CaM (Calbiochem); 1 μM WT His6–GFP–SRY-HMG was incubated in the absence or presence of 1.5 μM GST–Impβ1 and/or 6 μM CaM (as indicated), in IB supplemented with 2 mM Ca2+ for 20 min at room temperature (22 °C), loaded with 40% sucrose and electrophoresed on a pre-run 8% native polyacrylamide gel for ~6 h at 80 V at 4 °C in TBE (0.9 M Tris, 1.125 M boric acid and 20 mM EDTA, pH 7.5). Fluoroimaging was then performed using the Wallach Arthur 1422 Multiwavelength Fluorimager to enable visualization of fluorescent protein–protein complexes.
AlphaScreen® assays
AlphaScreen® (amplified luminescence proximity homogeneous assay screen) determinations of binding constants were performed as described previously [26] in triplicate in a 384-well white opaque plate (PerkinElmer): 30 nM (2 μl of 375 nM) of His6-tagged proteins were added to each well, followed by 20 μl of the appropriate concentration of 0–15 nM biotinylated Impβ1 (see above) or biotin-labelled CaM (Calbiochem), prepared by serial dilution in IB (110 mM KCl, 5 mM NaHCO3, 5 mM MgCl2 and 20 mM Hepes, pH 7.4, with Ca2+ added as indicated). Impβ1/CaM binding to proteins was allowed to proceed for 30 min at room temperature. All subsequent additions and incubations were made in subdued lighting conditions due to photosensitivity of the beads; 1 μl of a 1:10 dilution of acceptor beads and 1 μl of 2.5% BSA were added simultaneously and incubated for 90 min at room temperature. A 1 μl sample of a 1:10 dilution of donor beads was then added to give a final sample volume of 25 μl and incubated at room temperature for 2 h. The assay was read on a PerkinElmer FusionAlpha plate reader according to the manufacturer's recommendations. Triplicate values were averaged and curve fitting (three-parameter sigmoidal) performed using the Sigmaplot software and the equation y=a/{1+exp−[(x−x0)/b]} [13,26]; where a represents maximal binding, and x represents the apparent dissociation constant.
RESULTS
CaM-NLS mutations impair SRY nuclear accumulation
A number of sex-reversing mutations have been described in SRY (e.g. see Table 1), of particular interest being those that do not have any impact on any known activities of SRY, including DNA binding and bending [10]. To test whether these affect SRY's nuclear import capabilities directly, plasmids encoding GFP–SRY fusion proteins (both SRY-FL and SRY-HMG) with or without the M64T, F67V, R76P, Y127C and R133W point mutations (Figure 1A) were transiently expressed in the testicular TM4 Sertoli cell line, and cells were subjected to CLSM imaging (Figure 1B). Exclusively nuclear localization was observed for the WT SRY protein, with clear cytoplasmic localization in addition to nuclear accumulation observed for all of the NLS mutant derivatives (Figure 1B). This result was confirmed by quantifying the extent of nuclear accumulation through image analysis (see Figure 1C), whereby the nuclear to cytoplasmic ratio (Fn/c) was determined as described previously [29,30]. The extent of nuclear accumulation of SRY appeared to be similar for full-length and HMG-derivative constructs, with the WT constructs showing Fn/c values of ~60 (indicative of 60-fold higher levels in the nucleus than in the cytoplasm). In comparison, constructs containing the β-NLS and CaM-NLS mutations showed significantly (P<0.01) reduced (~40–80%) levels of nuclear accumulation (Fn/c<30; see Figure 1C). Essentially identical results were observed for the constructs expressed in the GC-2 testicular (see Figures 4C and 4D and Table 1) and P19 embryonic carcinoma (results not shown) cell lines, as well as the COS-7 cell line from kidney (see below), suggesting that the mechanisms determining nuclear import of SRY are not specific to testicular cells.
Table 1. Summary of properties of the sex-reversing NLS-mutation-containing SRY-HMG proteins used in the present study.
ND, not determined.
| Effect on DNA* | Ability to bind‡ | |||||
|---|---|---|---|---|---|---|
| SRY mutant | Binding | Bending | Effect on nuclear accumulation† | CDZ sensitivity† | CaM | Impβ1 |
| WT | High affinity | Bends DNA 66 ° | Accumulates strongly in the nucleus | Reduced 2-fold | High affinity | High affinity |
| M64T | WT activity | WT activity | Reduced 2–3-fold | None | Reduced ~5-fold | High affinity |
| F67V | Reduced >99.9% | ND | Reduced ~3-fold | None | Reduced 3-fold | High affinity |
| R76P | WT activity | WT activity | Reduced ~1.5-fold | None | Reduced ~3-fold | High affinity |
| Y127C | No activity | ND | Reduced ~2-fold | Reduced 2-fold | High affinity | Reduced 4-fold |
| R133W | WT activity | WT activity | Reduced ~2-fold | Reduced ~2-fold | ND | Reduced 2-fold |
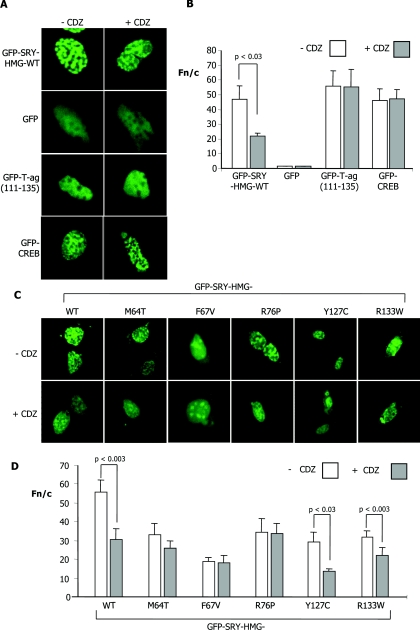
Figure 4. Specific inhibition of SRY nuclear accumulation by the CaM antagonist CDZ; CaM-NLS mutations prevent inhibition by CDZ.
GC-2 cells were transiently transfected to express the indicated GFP-fusion proteins, treated with or without 5 μM CDZ 12 h p.t., and then imaged live 12 h later by CLSM using a 40× water-immersion objective. (A) Confocal images indicate the localization patterns of WT GFP–SRY-HMG and the control proteins GFP, GFP–T-ag-(111–135) and GFP–CREB-(252–341). (B) Results of quantitative analysis, whereby CLSM images such as those in (A) were analysed as described in Figure 1(C). Results are means±S.E.M. (n>13) with significant differences between values in the presence and absence of CDZ denoted by the P values. (C) Confocal images of typical cells indicating localization patterns of the GFP–SRY-HMG WT and NLS mutant proteins, in the absence and presence of CDZ. (D) Results for quantitative analysis (mean±S.E.M.; n>22), from a single typical experiment, from a series of at least three similar experiments; P values are indicated where the Fn/c values are significantly different for constructs in the presence of CDZ.
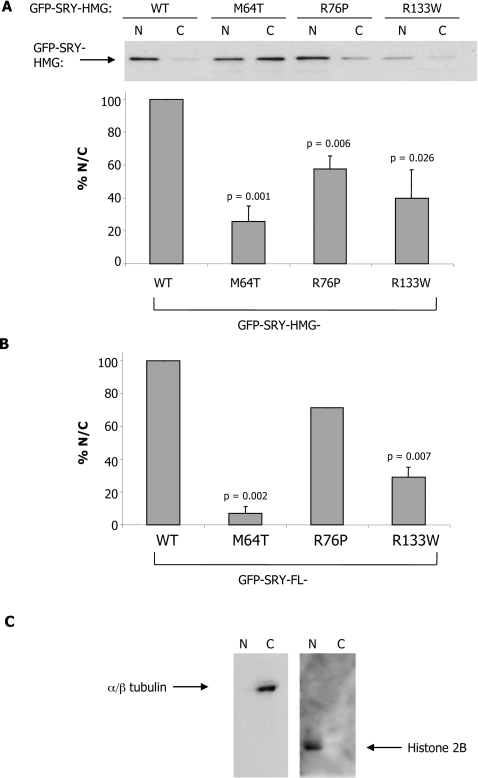
These results were confirmed by immunoblot/densitometric analysis on separated nuclear and cytoplasmic fractions for COS-7 cells transiently transfected to express the indicated GFP–SRY proteins (Figures 2A and 2B); probing the nuclear and cytoplasmic fractions using anti-α/β-tubulin and anti-(histone 2B) antibodies (Figure 2C) confirmed the integrity of the fractionation approach, with only α/β-tubulin detected in the cytoplasmic fraction and only histone 2B identified in the nuclear fraction. The analysis shown in Figure 2A (upper panel) indicates increased amounts of WT GFP–SRY-HMG in the nuclear fraction compared with the cytoplasmic fraction, with clearly increased amounts of GFP–SRY in the cytoplasmic fractions for all of the mutant proteins. Densitometric analysis (Figure 2A, lower panel) to determine the relative nuclear to cytoplasmic levels (N/C, subsequent to subtraction for background signal) indicated significantly (P<0.026) reduced N/Cs for the M64T, R76P and R133W mutants (71, 44 and 60% respectively), compared with WT. Similar results were observed for analysis of GFP–SRY-FL proteins (Figure 2B), with significantly (P<0.007) higher levels of WT GFP–SRY-FL protein in the nuclear fraction compared with the cytoplasmic fraction, compared with the M64T, R76P and R133W (93, 29 and 71% reduced N/Cs respectively compared with WT) NLS mutant derivatives. These results clearly support the idea that sex-reversing mutations in both the CaM- and β-NLS impair SRY nuclear accumulation in living cells, completely consistent with the results generated by CLSM analysis (Figure 1); the results strongly imply that CaM plays an important role in SRY nuclear import.
Figure 2. Sex-reversing mutations in CaM-NLS reduce SRY nuclear accumulation, as shown using immunoblot analysis.
(A) COS-7 cells were transiently transfected to express GFP–SRY-HMG WT and mutant proteins, and nuclear (N) and cytoplasmic (C) fractionation was performed 24 h p.t. with immunoblots probed with an anti-GFP antibody (see the Materials and methods section). Densitometric analysis (lower panel) was performed on images such as those in the upper panel, as described in the Materials and methods section, to calculate the relative amount of GFP-fusion protein in the nucleus and cytoplasmic fraction (% N/C). Results are means±S.E.M. for three similar experiments, with P values indicated for significant differences compared with WT. (B) COS-7 cells were transfected to express GFP–SRY-FL WT and mutant proteins and densitometric analysis was performed to quantify the relative amount of nuclear to cytoplasmic protein. Results are means±S.E.M. for three similar experiments, with P values indicated for significant differences compared with WT. (C) Integrity of the subcellular fractionation method as demonstrated by immunoblots for anti-α/β-tubulin and anti-(histone 2B) antibodies.
CaM antagonists inhibit SRY nuclear accumulation
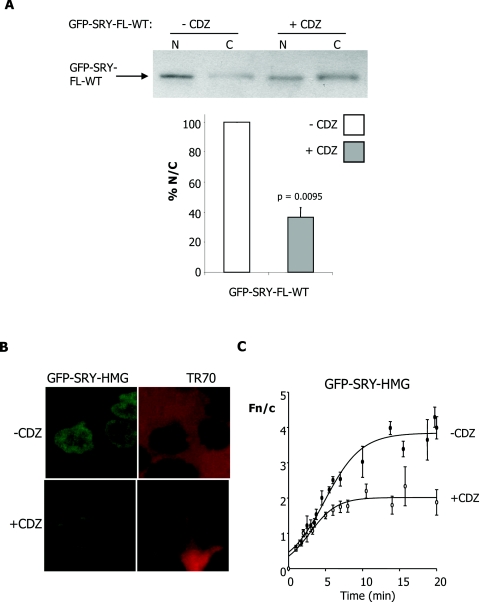
To establish the dependence on CaM for SRY nuclear import, cells were initially transfected to express WT GFP–SRY-FL protein and treated 12 h p.t. with the potent and specific CaM antagonist CDZ, a widely used imidazole compound which binds directly to CaM to prevent binding to and modulation of the activity of CaM's various specific binding partners [16,31]. Cells were subjected to subcellular fractionation, as above, 24 h p.t. and immunoblot analysis performed to demonstrate higher levels of GFP–SRY-FL in the cytoplasmic fraction upon CDZ treatment (Figure 3A, upper panel). This was confirmed by densitometric analysis (Figure 3A, lower panel), indicating significantly (P=0.0095) lower (approx. 63% reduced) levels of GFP–SRY-FL in the nuclear fraction compared with the cytoplasmic fraction in the presence of CDZ.
Figure 3. Specific inhibition of SRY nuclear accumulation by the CaM antagonist CDZ.
(A) Immunoblot analysis for the nuclear and cytoplasmic fractions of COS-7 cells transfected with WT GFP–SRY-FL, treated with 5 μM CDZ 12 h p.t. and probed with anti-GFP antibody. Densitometric analysis (lower panel) of images such as those in the upper panel, were performed as described in Figure 2. Results are means±S.E.M. for two similar experiments, with P values indicated for significant differences compared with in the absence of CDZ. (B) Nuclear import of WT GFP–SRY-HMG was reconstituted in vitro using mechanically perforated HTC cells, in the presence of exogenously added cytosol, an ATP-regenerating system and a 70 kDa Texas Red–dextran (TR70), in the absence and presence of CDZ (70 μM), as indicated. CLSM images of cells after 18–20 min of incubation are shown on the left in each case, with images for nuclear exclusion of TR70 to demonstrate nuclear integrity on the right. (C) Quantitative analysis is shown for nuclear uptake over time, whereby CLSM images such as those on the left were analysed as described in Figure 1(C). Results are means±S.E.M. for a single typical experiment, from a series of three similar experiments (see Table 2 for pooled data), with each data point representing more than three separate measurements of Fn and Fc above background. Curves were fitted (three-parameter sigmoidal) using SigmaPlot software.
In a second approach, nuclear transport of WT GFP–SRY-HMG protein was reconstituted in vitro using mechanically perforated HTC cells as described previously [10,13,32] and the transport kinetics determined using quantitative CLSM. WT SRY was seen to accumulate strongly in intact nuclei (Figure 3B, left-hand panel); quantitative analysis of the extent of nuclear accumulation confirming this observation (Figure 3B, right-hand panel), with a maximal Fn/c of 4 observed after a period of 15 min in the presence of exogenously added cytosol. Addition of 70 μM CDZ resulted in significantly (P=0.0006) reduced (42%) maximal nuclear accumulation of WT SRY (Table 2). Similar results were observed for SRY in the absence of exogenously added cytosol, with significantly reduced nuclear accumulation observed in the presence of CDZ (Table 2).
Table 2. Pooled data for the in vitro nuclear kinetics of WT GFP–SRY-HMG.
Results are means±S.E.M. for n replicates (where n is indicated in parentheses). Fn/cmax is the maximal nuclear accumulation. P values were calculated using Student's t test with GraphPad Prism 5.0, relative to in the absence of CDZ. NS, not significant.
| Conditions | Fn/cmax (%) | P value |
|---|---|---|
| +Cytosol (3) | 100±0.01 | |
| +Cytosol+70 μM CDZ (3) | 58±4.3 | 0.0006 |
| −Cytosol (5) | 100±0.01 | |
| −Cytosol+35 μM DMSO (3) | 91±33 | NS |
| −Cytosol+35 μM CDZ (3) | 56±19 | 0.0199 |
| −Cytosol+70 μM CDZ (2) | 35±3.9 | <0.0001 |
In a third approach, cells transfected to express the indicated GFP–SRY-HMG protein were treated with CDZ 12 h p.t. and imaged with CLSM. Results indicated that the WT protein showed an increase in cytoplasmic fluorescence (Figure 4A), with quantitative analysis confirming significantly (P<0.03) ~45% reduced levels (Table 1) of nuclear accumulation in the presence of CDZ (Figure 4B) compared with in its absence, consistent with a dependence on CaM for SRY nuclear accumulation. Similar results (not shown) were observed using the CaM antagonist TFP (trifluoroperazine) in place of CDZ. Control molecules were analysed in the same way including GFP alone, GFP T-ag-(111–135), containing the Impα/β-recognized NLS of T-ag, and GFP-CREB-(252–341), containing the Impβ1-recognized NLS of CREB [33] (Figures 4A and 4B). In all cases, no effect of CDZ treatment was evident, with no increase in cytoplasmic fluorescence observed (Figure 4A), and quantitative analysis supported this idea (Figure 4B). Clearly, the effects observed for SRY appear to be specific and not the result of general effects of CaM antagonism upon the nuclear transport machinery/Imps/Ran etc.
Similar experiments were performed for the constructs carrying the β-NLS and CaM-NLS mutations (Figures 4C and 4D). Although already showing increased cytoplasmic fluorescence in the case of the β-NLS and CaM-NLS sex-reversing mutations compared with WT, a further significant (P<0.02) ~30–50% reduction in nuclear accumulation in the presence of CDZ was observed for proteins carrying the R133W and Y127C mutations in the β-NLS (Figures 4C and 4D). Intriguingly, the proteins carrying the M64T, F67V and R76P mutations in the CaM-NLS did not show a further reduction (Figures 4C and 4D) in nuclear accumulation upon CDZ treatment beyond the lower level of accumulation already observed in its absence. The clear implication was that the CaM-NLS mutations impair SRY's CaM-dependent nuclear import pathway.
CaM-NLS mutations impair CaM binding to SRY
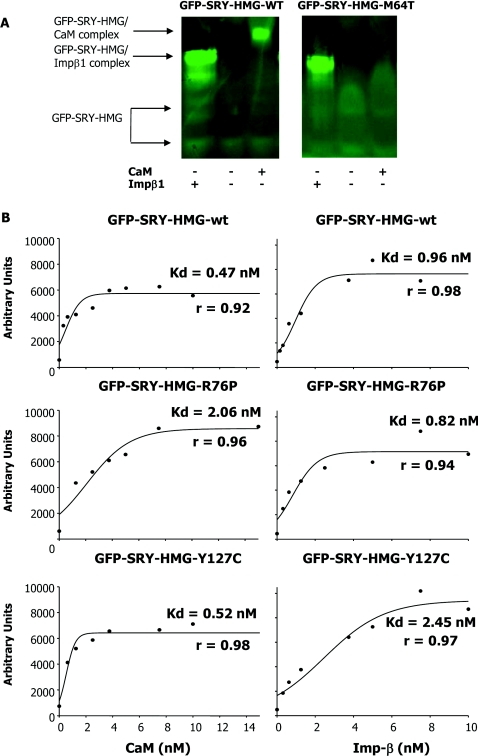
To test whether the CaM-NLS mutations affect SRY binding to CaM directly, bacterially expressed His6-tagged GFP–SRY-HMG derivatives were initially analysed by native PAGE, whereby SRY proteins were incubated with either CaM (in the presence of Ca2+) or Impβ1 and then subjected to electrophoresis; SRY alone shows a diffuse band in the absence of a binding partner as observed previously [13], with a focused band indicative of complex formation [13,26,30,31]. Figure 5(A) shows typical contrasting results for the comparison of WT with the M64T CaM-NLS mutant derivative, where the latter shows a clear lack of complex formation with CaM, but essentially normal binding to Impβ1. Table 3 summarizes the data for all of the mutants analysed, where the CaM-NLS mutants in all cases showed reduced CaM binding, but normal Impβ1 binding, whereas the β-NLS Y127C mutation-carrying SRY fusion protein bound CaM in similar fashion to the WT SRY protein (but showed impaired Impβ1 binding).
Figure 5. Sex-reversing mutations in the CaM-NLS specifically impair binding to CaM, but not Impβ1, as shown using native PAGE and AlphaScreen® assays.
(A) A native PAGE gel mobility-shift assay (see the Materials and methods section) for Impβ1 and CaM binding to GFP–SRY-HMG WT and M64T mutant was performed, where GFP–SRY-HMG (1 μM) was incubated in the absence and presence of Impβ1 (1.5 μM) and CaM (6 μM) in the presence of Ca2+ (2 mM) for 20 min at room temperature, and then fluoroimaging was performed subsequent to PAGE (see the Materials and methods section). Results are shown for a single typical assay from a series of two similar experiments. (B) AlphaScreen® assays were performed as described in the Materials and methods section. Results are shown for a single typical experiment (pooled data are shown in Table 2), with the apparent dissociation constants (Kd) indicated for binding of biotinylated CaM (left) or biotinylated Impβ1 (right) to SRY-HMG (WT and mutants). Results for regression analysis (r value) for the accuracy of the curve fitting are presented in all cases.
Table 3. Pooled data for the binding affinities of GFP–SRY proteins to CaM and Impβ1, as determined using native PAGE and AlphaScreen® assays.
Results in native PAGE experiments such as those in Figure 5(A), were scored according to binding strength: +++, strong binding; +, weak binding; –, no binding. Results are means±S.E.M. for n (in parentheses) experiments for the AlphaScreen® binding experiments (see Figure 5B); where significant differences in binding affinities of the mutant derivatives compared with that of the WT were observed, P values were calculated using Student's t test with InStat2.01. NS, not significant.
| CaM binding | Impβ1 binding | |||
|---|---|---|---|---|
| SRY derivative | Native PAGE | AlphaScreen® Kd (nM) | Native PAGE | AlphaScreen® Kd (nM) |
| WT HMG | +++ (6) | 0.43±0.07 (13) | +++ (6) | 0.94±0.14 (13) |
| HMG-M64T | – (2) | 2.05±0.31 (7) P<0.0001 | +++ (2) | 1.42±0.20 (3) NS |
| HMG-F67V | – (1) | 1.25±0.27 (4) P=0.0004 | +++ (1) | 1.09±0.05 (5) NS |
| HMG-R76P | – (3) | 1.18±0.20 (3) P=0.0004 | +++ (3) | 0.89±0.22 (5) NS |
| HMG-Y127C | +++ (3) | 0.62±0.05 (5) NS | + (3) | 3.60±0.72 (4) P<0.0001 |
Although the native PAGE indicated markedly reduced CaM and normal Impβ1 binding for CaM-NLS mutants, a more sensitive direct protein binding assay, an AlphaScreen® assay, was used to confirm these results, as well as to provide an estimate of the binding affinities of the respective SRY proteins for CaM and Impβ1. His6–GFP–SRY-HMG showed high-affinity binding to both CaM (dependent on Ca2+, results not shown) and Impβ1, with apparent dissociation constants (Kd values) of ~0.43 and 0.94 nM respectively (Table 3). SRY fusion proteins carrying sex-reversing CaM-NLS mutations all showed significantly (P<0.004) reduced CaM binding (Table 3); the R76P variant, for example, showed a 4-fold higher Kd for CaM (2.05 nM; Table 3 and Figure 5B, middle panels). In contrast, the β-NLS Y127C-carrying SRY fusion protein showed normal CaM binding (Figure 5B, bottom panels). Consistent with the results for native PAGE, the CaM-NLS mutants all bound to Impβ1 with high affinity comparable with WT, in contrast with the Y127C mutant, which showed ~4-fold higher Kd and significantly (P<0.0001) impaired Impβ1 binding (Table 3 and Figure 5B, bottom panels). Both native PAGE and AlphaScreen® assay results clearly imply that the CaM-NLS mutations directly affect CaM, but not Impβ1 binding; this presumably represents the basis of the reduced nuclear localization (Figures 1, 2 and 4) and lack of CDZ sensitivity of nuclear accumulation (Figure 3) of SRY carrying CaM-NLS mutations compared with WT.
CaM-dependent nuclear transport is conserved in HMG proteins
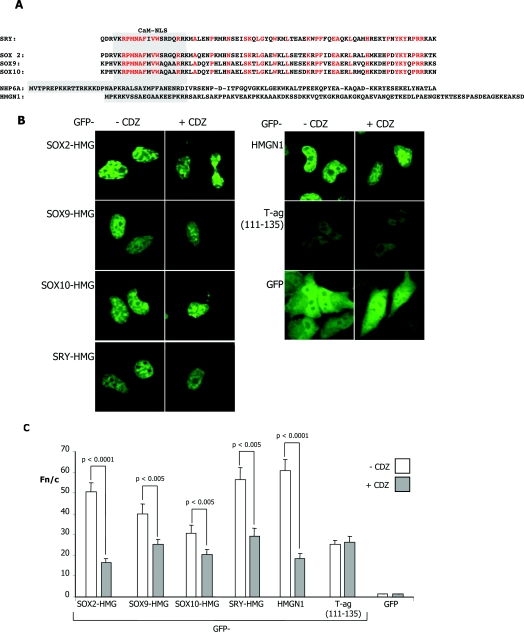
The results above strongly implicate CaM as playing a critical role in SRY nuclear import in fetal development and sex determination. Intriguingly, the HMG box of SRY is >50% conserved in other SOX family members which play key roles in development [34], with striking homology evident within the CaM-NLS (Figure 6A), implying conservation of function. To test whether these sequences may be functional in conferring CaM-dependent nuclear localization, HeLa cells were transiently transfected to express various GFP–SOX-HMG (SOX2-HMG, SOX9-HMG and SOX10-HMG) domains as well as the more distantly related HMGN1 nucleosomal binding protein, with CDZ added 16 h p.t, followed by CLSM imaging 8 h later. All GFP–SOX-HMG proteins, as well as GFP–HMGN1, showed exclusively nuclear accumulation (Figure 6B), with >30-fold higher levels of protein in the nucleus compared with the cytoplasm (Fn/c ~30; Figures 6B and 6C). Significantly, in the presence of CDZ, increased cytoplasmic fluorescence was evident (Figure 6B), which was confirmed by quantitative analysis, which indicated significantly (P<0.005) reduced (30–70%) nuclear accumulation for all of the HMG domain-containing proteins in the presence of CDZ (Figure 6C). This was in contrast with the control proteins GFP and T-ag, which showed no effect of CDZ (Figures 6B and 6C). The results clearly imply that SOX proteins similar to SRY as well as the more distantly related HMGN1 protein possess a functional CaM-dependent nuclear import pathway. CaM-dependent nuclear import thus appears to be an intrinsic property of the HMG domain of SOX proteins, and HMG proteins generally.
Figure 6. Specific inhibition of nuclear accumulation by the specific CaM antagonist CDZ of HMG-containing proteins.
(A) Alignment of SRY, SOX2, SOX9, SOX10 and Nhp6Ap (NHP6A) HMG-box domains, along with the HMGN1 nucleosomal binding protein. The text in red represents conserved residues in the HMG domains of the SOX proteins and the residues in the grey box are the potential CaM-binding domains. (B) HeLa cells were transiently transfected to express the indicated GFP-fusion proteins, treated with or without 5 μM CDZ 16 h p.t. and imaged 8 h later by CLSM using a 40× water-immersion objective. (C) Results of quantitative analysis, whereby images such as those in (B) were analysed as described in Figure 1(C). Results are mean±S.E.M. (n>30) from a single typical experiment, from a series of three similar experiments; P values are indicated for Fn/c values that were significantly different in the presence of CDZ.
DISCUSSION
The present study demonstrates for the first time a specific role for CaM in nuclear import of HMG-box family nuclear factors, as well as the central importance thereof for SRY's role in sex determination, in that CaM-NLS mutations specifically impairing CaM binding/SRY nuclear import result in sex reversal. Since the M64T and R76P CaM-NLS mutants show essentially normal DNA-binding and -bending activities [9,10], the only detectable effect of the mutations, as demonstrated for the first time, is impaired CaM binding and consequently impaired nuclear import. Significantly, there is a clear correlation between the extent of the reduction in CaM binding and in nuclear accumulation for these mutants; the M64T mutation results in more severely impaired CaM binding (~5-fold higher Kd; see Tables 1 and 3), and a 2–3-fold reduction in nuclear accumulation as a consequence (see Table 1 and Figures 2A and 2B), whereas the R76P mutant shows a smaller effect in terms of the reduction in CaM-binding affinity (~3-fold higher Kd), and a consequently lower (~1.5-fold) reduction in nuclear accumulation. Clearly, the affinity of CaM binding is critical to nuclear import efficiency, with even a ~3-fold reduction in binding affinity able to have a significant effect on nuclear accumulation, and in turn likely to be the basis of sex reversal in vivo.
The present study clearly implies that the CaM-dependent CaM-NLS-mediated nuclear import pathway of SRY is essential for sex determination. That CaM's role is almost certainly in nuclear entry rather than in facilitating SRY nuclear retention is consistent with the fact that SRY nuclear accumulation does not appear to involve binding to nuclear components [13] and that the CaM-NLS alone is sufficient to target even large carrier proteins to the nucleus [10,35]. CaM is known to be able to localize in either nucleus or cytoplasm in various tissues including the testis [15]; the exact mechanism by which it mediates/contributes to SRY nuclear import remains to be determined, but one possibility is that CaM facilitates nuclear import through mediating direct interaction of SRY with the NPC. Subsequent to this, CaM release from SRY is likely to be effected by DNA binding in the nucleus, which has been shown in vitro [12]. Since CaM is ubiquitous in essentially all cell types, this mechanism appears to be unlikely to relate uniquely to the testis, but rather to essentially all tissues/cell types, consistent with the effects of CaM antagonism on HMG-family nuclear factor transport shown in the present study for the widely used COS-7 and HeLa cell lines (Figures 2, 3 and 6).
The results of the present study and others indicate that SRY has distinct dual nuclear import pathways mediated by the CaM-NLS through CaM and β-NLS through Impβ1. Mutation in either NLS can lead to sex reversal, despite the fact that nuclear import through the other NLS is functional. The most informative mutations in this respect are the M64T/R76P CaM-NLS and R133W β-NLS (see [10]) mutations, none of which markedly affects DNA binding and bending, all only specifically impairing the nuclear import pathways mediated by the NLS in which they are mutated. The fact that all of these mutations cause sex reversal indicates clearly that both nuclear import pathways are essential to SRY's function in sex determination; perturbing either pathway is sufficient to result in a failure to form a testis. The implication of this is that the level of SRY in the nucleus is critical to sex determination, consistent with its potential role as a repressor of genes responsible for embryonic female development [3,4].
Importantly, the results of the present study have wider implications in that they indicate for the first time that the nuclear accumulation of SOX proteins distinct from SRY also appear to be dependent on CaM, along with the HMGN1, consistent with the homology in their respective HMG domains. The clear implication is that there is a general role for CaM in mediating the nuclear transport of HMG proteins, consistent with the recent observation that the Nhp6Ap yeast HMG box protein may have a CaM-dependent Ran-independent nuclear import pathway [18]. The present study and that for Nhp6Ap [18] thus establish a role for CaM in mediating nuclear transport, highlighting yet another important function for this versatile protein. Detailed examination of the molecular basis of this is the focus of future work in this laboratory.
Online data
AUTHOR CONTRIBUTION
The majority of the work in terms of experimental work and manuscript preparation was performed by Gurpreet Kaur. Aurelie Delluc-Clavieres commenced the project, generating the plasmid for bacterial expression of the WT SRY protein, while Ivan Poon generated GFP-tagged WT constructs for mammalian expression, for both the SRY-FL and SRY-HMG derivatives. Jade Forwood assisted with manuscript preparation. Dominic Glover generated the GFP-tagged CREB construct and assisted with subcellular fractionation/immunoblot methodology/analysis. David Jans supervised the project, helping to develop the project and preparing the manuscript.
FUNDING
This work was supported by the National Health and Medical Research Council, Australia [fellowship grant number 334013 and project grant number 143710) and the Australian Research Council (Centre of Excellence CE348239).
References
- 1.Lefebvre V., Dumitriu B., Penzo-Mendez A., Han Y., Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veitia R. A., Salas-Cortes L., Ottolenghi C., Pailhoux E., Cotinot C., Fellous M. Testis determination in mammals: more questions than answers. Mol. Cell. Endocrinol. 2001;179:3–16. doi: 10.1016/s0303-7207(01)00460-9. [DOI] [PubMed] [Google Scholar]
- 3.Harley V. R., Clarkson M. J., Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) box 9] Endocr. Rev. 2003;24:466–487. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- 4.Knower K. C., Kelly S., Harley V. R. Turning on the male: SRY, SOX9 and sex determination in mammals. Cytogenet. Genome Res. 2003;101:185–198. doi: 10.1159/000074336. [DOI] [PubMed] [Google Scholar]
- 5.Swain A., Lovell-Badge R. Mammalian sex determination: a molecular drama. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- 6.Gasca S., Canizares J., De Santa Barbara P., Mejean C., Poulat F., Berta P., Boizet-Bonhoure B. A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11199–11204. doi: 10.1073/pnas.172383099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uehara S., Hashiyada M., Sato K., Nata M., Funato T., Okamura K. Complete XY gonadal dysgenesis and aspects of the SRY genotype and gonadal tumor formation. J. Hum. Genet. 2002;47:279–284. doi: 10.1007/s100380200040. [DOI] [PubMed] [Google Scholar]
- 8.Werner M. H., Bianchi M. E., Gronenborn A. M., Clore G. M. NMR spectroscopic analysis of the DNA conformation induced by the human testis determining factor SRY. Biochemistry. 1995;34:11998–12004. doi: 10.1021/bi00037a042. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell C. L., Harley V. R. Biochemical defects in eight SRY missense mutations causing XY gonadal dysgenesis. Mol. Genet. Metab. 2002;77:217–225. doi: 10.1016/s1096-7192(02)00165-8. [DOI] [PubMed] [Google Scholar]
- 10.Harley V. R., Layfield S., Mitchell C. L., Forwood J. K., John A. P., Briggs L. J., McDowall S. G., Jans D. A. Defective importin β recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7045–7050. doi: 10.1073/pnas.1137864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith J. M., Koopman P. A. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 2004;20:4–8. doi: 10.1016/j.tig.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Harley V. R., Lovell-Badge R., Goodfellow P. N., Hextall P. J. The HMG box of SRY is a calmodulin binding domain. FEBS Lett. 1996;391:24–28. doi: 10.1016/0014-5793(96)00694-1. [DOI] [PubMed] [Google Scholar]
- 13.Forwood J. K., Harley V., Jans D. A. The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin β1. J. Biol. Chem. 2001;276:46575–46582. doi: 10.1074/jbc.M101668200. [DOI] [PubMed] [Google Scholar]
- 14.Vetter S. W., Leclerc E. Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 15.Kortvely E., Gulya K. Calmodulin, and various ways to regulate its activity. Life Sci. 2004;74:1065–1070. doi: 10.1016/j.lfs.2003.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Argentaro A., Sim H., Kelly S., Preiss S., Clayton A., Jans D. A., Harley V. R. A SOX9 defect of calmodulin-dependent nuclear import in campomelic dysplasia/autosomal sex reversal. J. Biol. Chem. 2003;278:33839–33847. doi: 10.1074/jbc.M302078200. [DOI] [PubMed] [Google Scholar]
- 17.Preiss S., Argentaro A., Clayton A., John A., Jans D. A., Ogata T., Nagai T., Barroso I., Schafer A. J., Harley V. R. Compound effects of point mutations causing campomelic dysplasia/autosomal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. J. Biol. Chem. 2001;276:27864–27872. doi: 10.1074/jbc.M101278200. [DOI] [PubMed] [Google Scholar]
- 18.Hanover J. A., Love D. C., DeAngelis N., O'Kane M. E., Lima-Miranda R., Schulz T., Yen Y. M., Johnson R. C., Prinz W. A. The high mobility group box transcription factor Nhp6Ap enters the nucleus by a calmodulin-dependent, Ran-independent pathway. J. Biol. Chem. 2007;282:33743–33751. doi: 10.1074/jbc.M705875200. [DOI] [PubMed] [Google Scholar]
- 19.Baliga B. C., Colussi P. A., Read S. H., Dias M. M., Jans D. A., Kumar S. Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 2003;278:4899–4905. doi: 10.1074/jbc.M211512200. [DOI] [PubMed] [Google Scholar]
- 20.Cohen D. R., Sinclair A. H., McGovern J. D. SRY protein enhances transcription of Fos-related antigen 1 promoter constructs. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4372–4376. doi: 10.1073/pnas.91.10.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelberman D., Rizzoti K., Avilion A., Bitner-Glindzicz M., Cianfarani S., Collins J., Chong W. K., Kirk J. M., Achermann J. C., Ross R., et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo–pituitary–gonadal axis in mice and humans. J. Clin. Invest. 2006;116:2442–2455. doi: 10.1172/JCI28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudbeck P., Schmitz M. L., Baeuerle P. A., Scherer G. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat. Genet. 1996;13:230–232. doi: 10.1038/ng0696-230. [DOI] [PubMed] [Google Scholar]
- 23.Rehberg S., Lischka P., Glaser G., Stamminger T., Wegner M., Rosorius O. Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol. Cell. Biol. 2002;22:5826–5834. doi: 10.1128/MCB.22.16.5826-5834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda T., Catez F., Gerlitz G., Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol. Cell. Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glover D. J., Leyton D. L., Moseley G. W., Jans D. A. The efficiency of nuclear plasmid DNA delivery is a critical determinant of transgene expression at the single cell level. J. Gene Med. 2010;12:77–85. doi: 10.1002/jgm.1406. [DOI] [PubMed] [Google Scholar]
- 26.Wagstaff K. M., Jans D. A. Intramolecular masking of nuclear localization signals: analysis of importin binding using a novel AlphaScreen-based method. Anal. Biochem. 2006;348:49–56. doi: 10.1016/j.ab.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Forwood J. K., Kaur G., Jans D. A. Nuclear import properties of the sex-determining factor SRY. Methods Mol. Biol. 2007;390:83–97. doi: 10.1007/978-1-59745-466-7_6. [DOI] [PubMed] [Google Scholar]
- 28.Wagstaff K. M., Dias M. M., Alvisi G., Jans D. A. Quantitative analysis of protein–protein interactions by native page/fluorimaging. J. Fluoresc. 2005;15:469–473. doi: 10.1007/s10895-005-2819-5. [DOI] [PubMed] [Google Scholar]
- 29.Poon I. K., Oro C., Dias M. M., Zhang J. P., Jans D. A. A tumor cell-specific nuclear targeting signal within chicken anemia virus VP3/apoptin. J. Virol. 2005;79:1339–1341. doi: 10.1128/JVI.79.2.1339-1341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005;65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- 31.Sunagawa M., Kosugi T., Nakamura M., Sperelakis N. Pharmacological actions of calmidazolium, a calmodulin antagonist in cardiovascular system. Cardiovasc. Drug Rev. 2000;18:211–221. [Google Scholar]
- 32.Ghildyal R., Ho A., Wagstaff K. M., Dias M. M., Barton C. L., Jans P., Bardin P., Jans D. A. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin β1 independent of importin α. Biochemistry. 2005;44:12887–12895. doi: 10.1021/bi050701e. [DOI] [PubMed] [Google Scholar]
- 33.Forwood J. K., Lam M. H., Jans D. A. Nuclear import of CREB and AP-1 transcription factors requires importin-β1 and Ran but is independent of importin-α. Biochemistry. 2001;40:5208–5217. doi: 10.1021/bi002732+. [DOI] [PubMed] [Google Scholar]
- 34.Guth S. I., Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell. Mol. Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudbeck P., Scherer G. Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J. Biol. Chem. 1997;272:27848–27852. doi: 10.1074/jbc.272.44.27848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.