Abstract
MUC7 12-mer is a cationic antimicrobial peptide derived from the N-terminal region of human low molecular weight salivary mucin. In order to gain new insights into the modes of action of the 12-mer against opportunistic fungal pathogen Candida albicans, we examined changes in the gene expression profile of C. albicans upon exposure to this peptide. Cells at an early logarithmic phase were exposed to 6 μM peptide and grown until an OD600 of approximately 0.4 was reached. Changes in gene expression were determined by microarray analysis and showed that 19 out of the total of 531 genes, whose expression was elevated in response to the peptide, are regulated by the calcium/calcineurin signalling pathway. Inactivation of this pathway by deletions, or by FK506, caused hypersensitivity to the peptide, demonstrating the importance of this pathway to the defence of C. albicans against the MUC7 peptide. Other differentially expressed genes that were detected include those encoding subunits of proteasome, and genes involved in cell stress, iron metabolism, cell wall maintenance, and small molecule transport. The presented results suggest that the calcium/calcineurin signalling pathway plays a role in the adaptation of C. albicans to the MUC7 antimicrobial peptide.
Keywords: Candida albicans, antimicrobial peptide, calcium signalling, transcription profiling
Introduction
C. albicans, in healthy people, resides as a commensal microorganism. However, in immunocompromized patients it becomes an opportunistic fungal pathogen, capable of causing mucosal, cutaneous, and systemic infections including oropharyngeal candidiasis (OPC). OPC is the most frequent opportunistic infection in patients with HIV and AIDS (Egusa et al., 2008), but it also manifests in other immunocompromized patients, such as oncology patients with neutropenia, bone marrow and organ transplant recipients, and patients receiving immunosuppressive drugs (Ostrosky-Zeichner et al., 2002). OPC remains a significant cause of morbidity despite the use of effective antiretroviral therapy (Egusa et al., 2008).
Most of the currently used antifungal agents target the fungal cell membrane sterol, ergosterol and the cell wall. Ergosterol function is impaired by either its direct inhibition by polyene antifungals (amphotericin-B, nystatin) or by inhibition of its synthesis by azoles (fluconazole, itraconazole, voriconazole) or allylamines. Azoles are fungistatic and vulnerable to resistance, whereas polyenes are fungicidal but cause serious host toxicity. A newer class of antifungal drugs, the echinocandins, damage the fungal cell wall by blocking the synthesis of beta-(1,3) glucan. Resistance to the echinocandins, although rare, is also emerging (Baixench et al., 2007). Due to the problems of toxicity and resistance associated with the current antifungal agents, the treatment of fungal infections has become an increasing medical problem (Rex et al., 1995; Sims et al., 2005; Kanafani & Perfect, 2008).
Among other approaches for the development of new antifungal agents, antimicrobial peptides are receiving increasing attention (Zanetti et al., 2002). Unlike many currently used antimicrobial compounds, they show little or no toxicity toward mammalian cells and are believed to have low tendency to elicit resistance (Zanetti et al., 2002). Cationic antimicrobial peptides (CAMPs) are small, ribosomally synthesized peptides grouped together by two major common characteristics: an overall positive net charge, and their specificity toward bacterial, fungal and other microbial cells. It is widely believed that the cationic nature is responsible for an initial interaction with usually negatively charged surfaces of microorganisms, effectively raising local concentration and facilitating interaction with the plasma membrane. This second stage, resulting in formation of pores and/or translocation of peptides into the cytoplasm, appears to be associated with their amphipathic nature.
MUC7 12-mer is a cationic peptide derived from the N-terminal region of the human low-molecular-weight salivary mucin, MUC7. It possesses potent bacteriocidal and fungicidal activity against many microorganisms including Streptococcus mutans, and C. albicans, (Satyanarayana et al., 2000; Situ et al., 2003; Wei & Bobek, 2004; Wei et al., 2006), and thus it has emerged as a potential candidate for antifungal drug development. To better understand the mechanism of action of the MUC7 12-mer peptide, we have, and are continuing to employ multiple approaches, such as the development and characterization of a C. albicans mutant strain that is resistant to this peptide (Lis & Bobek, 2008), and a fitness screen of Saccharomyces cerevisiae gene deletion mutant pool grown in the presence of the MUC7 12-mer peptide (Lis et al., 2009).
Genome-wide expression profiling has been extensively used to determine the inhibitory effects of antifungals, including the response of C. albicans to representatives of the four classes of antifungal agents, namely ketoconazole, amphotericin B, caspofungin and flucytosine (Liu et al., 2005). More recently, gene expression profiling in combination with genome-wide phenotypic screen, using S. cerevisiae, was used to identify both common and unique effects of two amphibian-derived cationic antimicrobial peptides: dermaseptin and magainin (Morton et al., 2007). In this study, we analyzed the changes in the transcriptional profile of C. albicans upon exposure to the MUC7 12-mer.
Materials and Methods
Strains and growth conditions
Transcription profiling was performed on C. albicans clinical isolate DIS (denture-induced stomatitis, provided by M. Edgerton, University at Buffalo). C. albicans mutant strains (cmp1Δ/Δ, crz1Δ/Δ, crz2Δ/Δ and rim101Δ/Δ) and the reference strains (DAY286, DAY185) were generously provided by Aaron Mitchell (Carnegie Mellon University). Yeasts were grown on Sabouraud Dextrose Broth (SDB). For experiments involving treatment with the peptide, an eight-fold diluted SDB (sometimes referred to as 1/8 SDB) was used, since, like many CAMPs, the MUC7 peptide is ineffective in the full strength medium (Wei et al., 2007). Diluted medium allows for the peptide to be fully active without significantly affecting the growth of all the C. albicans strains used for this study. MUC7 peptide (RKSYKCLHKRCR) was synthesized and tested for purity by Bio-Synthesis (Lewisville, TX).
Treatment of C. albicans with the peptide for transcription profiling
Two independent, 250 ml liquid cultures of C. albicans (in eight-fold diluted SDB medium) were started by inoculation with cells grown overnight on SD agar plate, to an OD600 nm of 0.08 at 30° C. When cultures reached early log phase (OD600 of 0.1), they were split into two 125 ml cultures. One of the 125 ml cultures was exposed to MUC7 12-mer at a final concentration of 6 μM while the other 125 ml culture remained untreated. At this concentration the growth rate is reduced to approximately one half of the untreated cells. Cultures were grown until they reached OD600 nm of approximately 0.4 (3 h and 5 h for untreated and treated cells, respectively). The cells were then collected by centrifugation and stored at −80° C. Each pair of samples (untreated and MUC7-treated cells) from the same original 250 ml culture constitutes a single experiment.
RNA preparation
RNA was isolated using the hot phenol method (Liu et al., 2005; Schmitt et al., 1990). Quantity and quality of RNA were determined spectrophotometrically and on a 1% denaturing agarose gel.
Microarray design and hybridization
The nucleotide sequences corresponding to 6165 open reading frames of C. albicans were downloaded from the Galar Fungail European Consortium (Assembly 19, http://www.pasteur.fr/Galar_Fungail/CandidaDB/). Following the Affymetrix Design Guide, we designed two separate probe sets for each ORF, each consisting of 13 perfectly matched and 13 mismatched overlapping 25 bp oligonucleotides, to the 3′ 600 bp region. Microarrays were constructed by NimbleGen Systems (Madison, WI) in conjunction with Affymetrix. RNA from each sample in each of the two independent experiments was prepared and labelled for hybridization, with each sample hybridized on a separate array. Hybridizations were performed as per the manufacturer’s protocol.
Microarray data analysis
Microarray data were analyzed using GeneSpring software. Normalized microarray data from the MUC7-treated sample were compared to the normalized microarray data from the corresponding untreated sample for each experiment. Genes were considered to be differentially expressed if their expression changed 2-fold in two independent experiments.
Growth rate measurements
C. albicans parental and mutant strains grown overnight on SD agar were suspended in 1/8 SDB and the cell density (measured on spectrophotometer) was adjusted to an OD 600 equivalent of 0.005. Each suspension was divided into two aliquots, one was supplemented with 3 μM MUC7 peptide and 200 μl of each suspension was transferred into 96-well microtiter plate wells in triplicates. Plates were incubated at 30°C and the optical densities of cultures were measured at different time points in a microtiter plate reader. OD values of three wells were averaged. Plotted OD 595 values for each time-point are the means of three separate experiments. (Note that OD values measured by the microtiter plate reader do not correspond to those measured by the spectrophotometer).
MIC and MFC
Minimal Inhibitory Concentration (MIC) and Minimal Fungicidal Concentration (MFC) assays were performed in 1/8 SDB as described previously (Wei & Bobek, 2004). The reported values represent the majority of at least three independent experiments.
Time-kill experiments
For evaluation of killing efficiency, C. albicans parental and mutant strains grown overnight on SD agar were suspended in 1/8 SDB to a density of approximately 0.3 at OD 600, and were incubated with 5 μM peptide at 37°C. At time intervals samples were removed and mixed with NaCl and propidium iodide (PI) at final concentrations of 0.1 M and 25 μM, respectively. Percentages of PI-stained, killed cells were determined by fluorescence microscopy.
Chequerboard microdilution assays
The assays were essentially performed as previously described (Wei & Bobek, 2004). Briefly, two-fold serial dilutions of the MUC7 12-mer and FK506 were prepared in 1/8 SDB. Each dilution of one agent was mixed with all dilutions of the other in a 96-well microtiter plate. Suspension of freshly grown C. albicans cells was added to each well to a final density of 104 cells per ml. The plate was then incubated at 37°C for 48 h. Optical density at 595 nm was measured on a microtiter plate reader. The fractional inhibitory concentration (FIC) was calculated as a ratio between the MIC of an agent used in combination and the MIC of the agent tested alone. The FIC index (the sum of the FICs of each agent) was calculated for the most equally effective concentration of drugs.
Results
We aimed at monitoring the response of fungus C. albicans to treatment with sub-lethal concentrations of MUC7 12-mer during early logarithmic growth, by detecting changes of transcription activities of genes induced by the peptide.
By a criterion of a reproducible change of at least two-fold, we have identified a total of 531 genes that were up-regulated and 468 genes that were down-regulated in response to treatment with the peptide. The distribution of the MUC7 12-mer responsive genes that were up-regulated showed that the category with the largest number of responses was that of unknown function (27%), followed by “other” (17 %). Responses by genes that were of particular interest were in categories of Pol II transcription (7%), protein modification (4%), and protein degradation (3%). The up-regulated genes included 19 genes regulated by the Ca2+/calcineurin signalling pathway (Table 1), and 8 genes encoding subunits of 20S and 26S proteasome (Table 2). The distribution of the down-regulated genes showed that the largest number of responses was again in the genes of unknown function (34%), followed by small molecule transport (14%). Among them were 14 genes involved in iron metabolism (Table 3). This data suggests that, in C. albicans, the MUC7 12-mer peptide activates the calcineurin pathway and the 20S and 26S proteasome, and down-regulates the expression of genes involved in iron metabolism. Thus we focused our attention on these three groups of genes. Other differentially expressed genes included those involved in cell stress (YHB1, SSA1, TRX2, GTT1, CTA1, HSP12, HYR1), cell wall maintenance (SWI6, STE11, MNN2, MNN3, CHS4, CHS5, KRE5, ALS2, ALS12, GSL23) and small molecule transport (QDR1, QDR2, ERG6, VMA2, VMA4, VMA8, SNQ2, MDR1, CDR3).
Table 1.
List of C. albicans genes up-regulated in response to treatment with MUC7 12-mer associated with calcium/calcineurin signalling pathway
| CandidaDB namea | Common name (CGD)b | Systematic name | Fold changec | Description |
|---|---|---|---|---|
| ECM331 | ECM331 | orf19.4255 | 4.4 | involved in cell wall biogenesis and architecture (by homology) |
| IFO2 | orf19.1765 | 7.1 | unknown function | |
| IML2 | orf19.7229 | 3.0 | unknown function | |
| IPF10262 | orf19.2726 | 2.8 | unknown function | |
| IPF11548 | orf19.3854 | 4.0 | serine threonine protein kinase (by homology) | |
| IPF11900 | orf19.4180 | 3.2 | unknown function | |
| MNT4 | KTR4 | orf19.4475 | 3.0 | putative mannosyltransferase |
| IPF13407 | orf19.753 | 7.9 | unknown function | |
| IPF11667 | orf19.4771 | 2.7 | unknown function | |
| IPF14030 | orf19.851 | 14.2 | unknown function | |
| MNN4 | orf19.2957 | 9.3 | regulates the mannosylphosphorylation (by homology) | |
| PDI1 | PDI1 | orf19.5130 | 4.9 | protein disulfide-isomerase precursor (by homology) |
| PLC3 | FGR22 | orf19.1586 | 2.3 | phosphatidylinositol phospholipase C |
| RNH1 | RNH1 | orf19.5563 | 3.7 | ribonuclease H (by homology) |
| RTA4 | RTA4 | orf19.6595 | 3.9 | protein involved in 7-aminocholesterol resistance (by homology) |
| SAC6.3f | SAC6 | orf19.5544 | 3.7 | actin filament bundling protein, fimbrin, 3-prime end (by homology) |
| SAC6.5f | SAC6 | orf19.5544 | 3.2 | actin filament bundling protein, fimbrin, 5-prime end (by homology) |
| VPS1 | VPS1 | orf19.1949 | 5.2 | member of the dynamin family of GTPases (by homology) |
| CRZ2 | CRZ2 | orf19.2356 | 4.9 | putative transcription factor (by homology) |
| CRZ1 | CRZ1 | orf19.7359 | 3.4 | putative transcription factor (by homology) |
= Name as assigned by CandidaDB (http://genolist.pasteur.fr/CandidaDB/)
= Name as assigned by the Candida Genome Database (CGD) (http://www.candidagenome.org/)
= Fold change compared to untreated control sample, as described in Materials and Methods)
= Description as provided by CandidaDB
Table 2.
List of C. albicans genes up-regulated in response to treatment with MUC7 12-mer associated with 20S and 26S proteasomes
| CandidaDB namea | Common name (CGD)b | Systematic name | Fold changec | Description |
|---|---|---|---|---|
| PRE10 | PRE10 | orf19.6582 | 2.8 | 20S proteasome subunit C1 (by homology) |
| PRE5 | PRE5 | orf19.7178 | 3.5 | 20S proteasome subunit alpha6 (by homology) |
| PRE6 | PRE6 | orf19.544.1 | 2.5 | 20S proteasome subunit |
| RPT2 | RPT2 | orf19.5440 | 3.5 | 26S proteasome regulatory subunit (by homology) |
| RPT4 | RPT4 | orf19.482 | 2.6 | 26S proteasome regulatory subunit (by homology) |
| RPN5.3f | RPN5 | orf19.4032 | 3.1 | subunit of the regulatory particle of the proteasome, 3' end |
| RPN6 | RPN6 | orf19.1299 | 3.2 | subunit of the regulatory particle of the proteasome (by homology) |
| RPN9 | orf19.1993 | 2.4 | 26S proteasome regulatory particle (by homology) |
= Name as assigned by CandidaDB (http://genolist.pasteur.fr/CandidaDB/)
= Name as assigned by the Candida Genome Database (CGD) (http://www.candidagenome.org/)
= Fold change compared to untreated control sample, as described in Materials and Methods)
= Description as provided by CandidaDB
Table 3.
List of C. albicans genes down-regulated in response to treatment with MUC7 12-mer associated with iron metabolism.
| CandidaDB namea | Common name (CGD)b | Systematic name | Fold changec | Descriptiond |
|---|---|---|---|---|
| YFH1 | YFH1 | orf19.1413 | 0.5 | regulates mitochondrial iron accumulation (by homology) |
| ISU1 | ISU1 | orf19.6548 | 0.4 | unknown function |
| ISA2 | orf19.6811 | 0.4 | mitochondrial protein required for iron metabolism (by homology) | |
| FTR2 | FTR2 | orf19.7231 | 0.3 | high affinity iron permease |
| FET5 | FET34 | orf19.4215 | 0.3 | multicopper oxidase (by homology) |
| FET33 | FET3 | orf19.4211 | 0.1 | cell surface ferroxidase (by homology) |
| FET34.3eoc | FET34 | orf19.4215 | 0.4 | iron transport multicopper oxidase, 3-prime end (by homology) |
| SIT1 | SIT1 | orf19.2179 | 0.2 | ferrioxamine B permease by homology |
| CFL2 | CFL2 | orf19.1264 | 0.4 | ferric reductase (by homology) |
| FRE32 | CFL4 | orf19.1932 | 0.3 | ferric reductase (by homology) |
| FRE30.5eoc | FRE30 | orf19.6140 | 0.3 | ferric reductase-like, 5-prime end (by homology) |
| FRE31 | CFL5 | orf19.1930 | 0.4 | ferric reductase (by homology) |
| IPF1537 | YAH1 | orf19.336 | 0.4 | putative adrenodoxin and ferrodoxin (by homolgy) |
| IPF3174 | orf19.3167 | 0.5 | heme A:Farnesyl transferase (by homology) |
= Name as assigned by CandidaDB (http://genolist.pasteur.fr/CandidaDB/)
= Name as assigned by the Candida Genome Database (CGD) (http://www.candidagenome.org/)
= Fold change compared to untreated control sample, as described in Materials and Methods)
= Description as provided by CandidaDB
Upregulation of a number of genes regulated by calcineurin detected in this screen suggested that this environmental signalling pathway may play an important role in protecting cells from the effects of the MUC7 peptide under the employed growth condition. To explore this hypothesis, we individually tested C. albicans strains lacking both alleles of genes encoding catalytic subunit of calcineurin (cmp1Δ/Δ) or of calcineurin-regulated transcription factor Crz1p (crz1Δ/Δ), along with the reference strains DAY185 and DAY286, for susceptibilities to the MUC7 peptide. We also tested two additional deletion strains, rim101Δ/Δ and crz2Δ/Δ. Transcription factor Rim101p regulates response to alkaline pH in many fungal species, including C. albicans. We have recently shown that the signalling pathway it controls is necessary for the survival and growth of S. cerevisiae treated with MUC7 12-mer (Lis et al., 2009). Moreover, some of the genes associated with induction of this pathway in C. albicans are also up-regulated following treatment with the peptide (not shown). CRZ2 also plays a role in response of C. albicans to changes in external pH, along with calcineurin and Rim101p (Kullas et al., 2007) and exhibited elevated transcription in our microarray screening (4.9 fold).
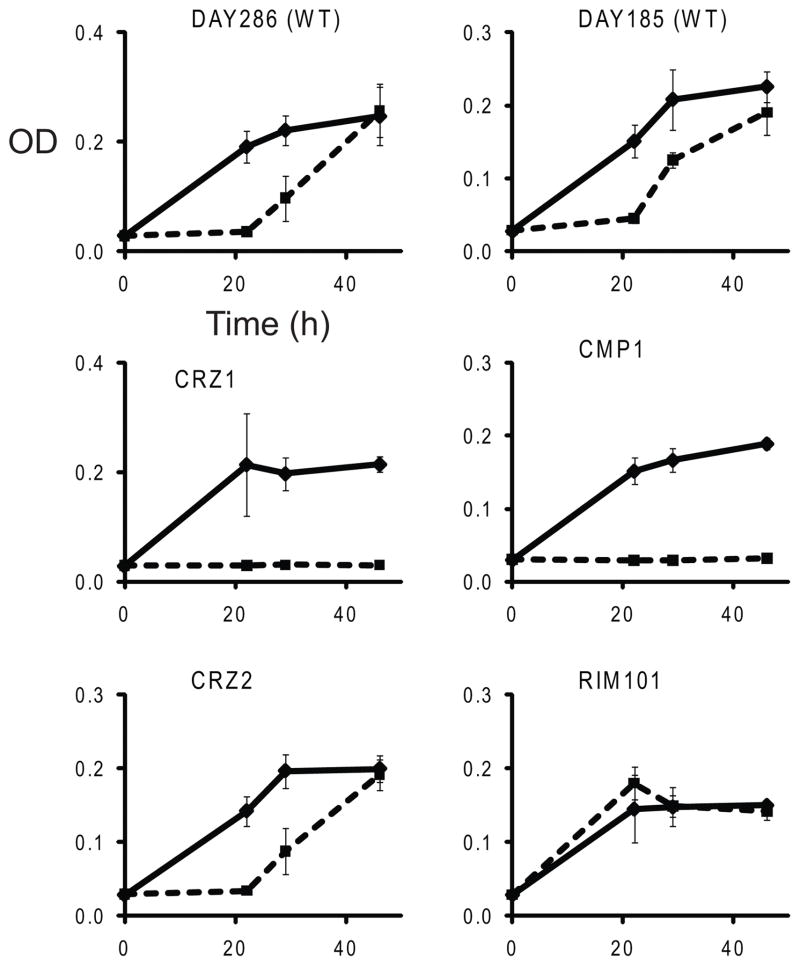
First we determined the effect of MUC7 peptide on the strains’ growth rates. During continuous culture supplemented with 3 μ of the peptide (a concentration at which the differences between strains were the most pronounced), the growth rates of the two wild type strains were reduced, whereas the growth of crz1Δ/Δ and cmp1Δ/Δ mutants was completely inhibited (Fig. 1). The crz2Δ/Δ strain was affected by the peptide only to the same extent as the wild type strains. Interestingly, MUC7 peptide, at the concentration used for this experiment, had almost no effect on growth of the rim101Δ/Δ mutant (Fig. 1).
Fig. 1.
Growth rates of the wild type (WT) C. albicans and selected deletion mutants in 1/8 SDB at 30°C in the presence (squares) or absence (diamonds) of 3 μM MUC7 12-mer peptide. Time point values are means of three independent experiments. Error bars represent standard deviation.
In addition to growth rate experiments, we also determined MIC and MFC of the 12-mer against selected WT and mutant strains (Table 4). The obtained MIC values are consistent with the growth rate experiments described above. Moreover, MFC values were, in all cases, identical to the MIC values, which confirmed our earlier finding that MUC7 12-mer is a fungicidal peptide (Wei & Bobek, 2004).
Table 4.
MIC and MFC of MUC7 12-mer peptide against selected C. albicans strains.
| Strain | MIC | MFC |
|---|---|---|
| DAY286 | 2.5 | 2.5 |
| crz1Δ/Δ | 1.25 | 1.25 |
| cmp1Δ/Δ | 1.25 | 1.25 |
| rim101Δ/Δ | 5 | 5 |
MIC and MFC (in μM) were determined in 1/8 SDB as described earlier (Wei & Bobek, 2004). The values represent majority of at least three independent tests.
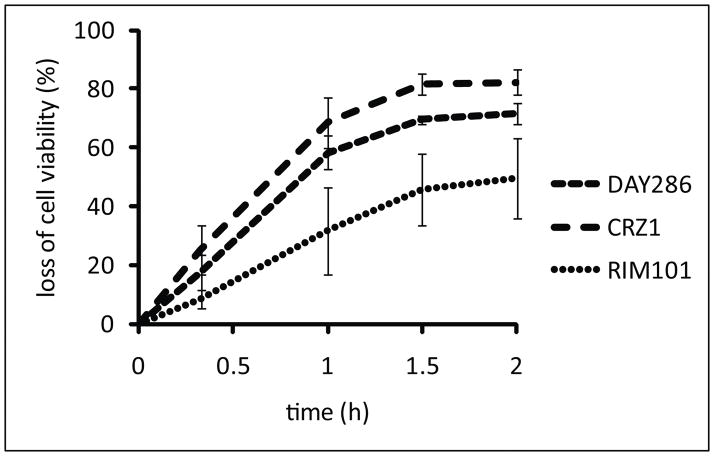
In order to compare the effect of MUC7 peptide on viabilities of the WT and selected mutant strains, we performed a time course killing experiments within the first two hours of incubation in 1/8 SDB medium (Fig. 2). Efficiency of killing by the peptide was highest for the crz1Δ/Δ mutant followed by that of the WT strain DAY286, and then by the mutant rim101Δ/Δ. These results were consistent with the results obtained by growth rate experiments in Fig. 1.
Fig. 2.
Killing rates of C. albicans strains by the MUC7 12- mer. Cells were incubated in 1/8 SDB at 37°C with 5 μM peptide. At indicated time points loss of cell viability was determined by PI staining and fluorescence microscopy. Values represent the means of four independent experiments. Error bars represent standard deviation.
The importance of calcineurin in defence against the MUC7 12-mer peptide was further confirmed by examining the effect of its inhibitor FK506 on growth of yeasts in the presence of the peptide. The results of several chequerboard microdilution assays of combinations of the MUC7 peptide with FK506 showed a moderate synergistic effect between the two agents, with FIC index values between 0.25 and 0.53 (average=0.39, n=5).
Discussion
We are reporting on the transcription profiling of C. albicans exposed to the antimicrobial peptide MUC7 12-mer during growth in liquid medium. Among the differentially expressed genes with known function, the largest group of the up-regulated genes is associated with the calcium/calcineurin signal transduction pathway. The identified set consists of 19 genes out of 60, which were previously determined to be regulated by calcineurin via Crz1p (Karababa et al., 2006). This pathway constitutes a common and conserved regulatory mechanism among all eukaryotic organisms. The external signal triggers influx of calcium into cytoplasm. Calcium-bound calmodulin activates the major regulatory protein calcineurin, a serine/threonine phosphatase composed of two subunits, Cna1p (catalytic) and Cnb1p (regulatory). One of the protein substrates of activated calcineurin is a transcription factor Crz1p. Crz1p, which in a phosphorylated form localizes in the cytoplasm, is translocated to the nucleus upon dephosphorylation by calcineurin, where it activates an array of genes. In fungi, calcium signalling appears to regulate responses to a variety of environmental conditions, each specific to a particular species. In C. albicans, calcineurin regulates a response to neutral and alkaline pH (Kullas et al., 2007). Calcineurin has also been shown to be essential for survival of treatment with fluconazole, a membrane-destabilizing drug targeting ergosterol metabolism. Inactivation of the pathway by cyclosporin A or FK506, or by deletion of the gene encoding one of the calcineurin subunits, in combination with fluconazole resulted in the killing of cells by this otherwise fungistatic drug. Similar results were obtained for several other membrane-destabilizing agents, including other azoles, SDS and metal ions (Cruz et al., 2002; Sanglard et al., 2003). The authors have also determined that calcineurin was essential specifically for protection against membrane perturbations but not against cell wall stress (Cruz 2002).
Our results presented here show that, as in the examples described above, calcium/calcineurin signalling affords partial protection for C. albicans from the MUC7 12-mer peptide. This is demonstrated by stronger growth inhibition of the mutant strains harbouring deletions of genes encoding either catalytic domain of calcineurin (cmp1Δ/Δ) or transcription regulator Crz1p (crz1Δ/Δ) (Fig. 1), and by synergism between MUC7 12-mer and FK506. Unlike azoles, which are fungistatic, MUC7 peptide is fungicidal. The inactivation of calcium signalling results in a higher efficiency of killing by the peptide, as demonstrated by lower MFC of crz1Δ/Δ and cmp1Δ/Δ mutants, and by higher killing rate of crz1Δ/Δ mutant, compared to the wild type strain (Table 4 and Fig. 2).
In contrast to the effect of mutations described above, deletion of gene encoding transcription regulator, Rim101p, reduced sensitivity to the peptide. This finding is somewhat surprising, as our recent fitness profiling of genome-wide deletion pool of S. cerevisiae revealed hypersensitivity of the rim101Δ mutant (and of several other null mutants defective in activation of the RIM101 pathway) to the 12-mer peptide (Lis et al., 2009). In S. cerevisiae, the Rim101p-controlled pathway is known to primarily regulate response to neutral and alkaline pH, and to certain ions (Lamb & Mitchell, 2003). In contrast, in C. albicans, regulations of response to low as well as high pH appear to be intertwined and more complex. According to a model proposed by Kullas et al. (2007), calcineurin (via Crz1p) and Rim101p regulate growth at alkaline pH, while adaptation to acidic environment requires Crz1p and Crz2p and is also indirectly controlled by Rim101p. Thus, the striking difference in the susceptibilities of RIM101 null mutants to the peptide between S. cerevisiae and C. albicans (hypersensitivity vs. resistance) probably reflects notable differences in regulatory mechanisms governed by Rim101p in these two yeast species (Bensen et al., 2004). Similar difference between the two species in the susceptibilities of RIM101 mutants have been revealed in a recent phenotypic screen of transcription regulators in C. albicans (Homann et al., 2009). RIM101 deletion mutants of C. albicans and S. cerevisiae exhibited, respectively, weak resistance and weak sensitivity to fenpropimorph, an antifungal drug targeting ergosterol metabolism and acting synergistically with FK506 (Onyewu et al., 2003). Despite the differences noted above, however, in both fungi the lack of the Rim101p transcription factor renders cells sensitive to alkaline pH and lithium ions (Bensen et al., 2004). Therefore, the response to the MUC7 peptide, while probably overlapping with responses to changes in external pH, is a distinct phenomenon.
Our earlier results have shown that MUC7 12-mer binds to the surface of C. albicans cells where it accumulates and then, in a concentration-dependent, all-or-none manner breaks membrane integrity leading to an influx of peptide molecules and immediate cell death (Lis and Bobek 2008). One can envision a scenario where the peptide interacting with the surface of plasma membrane imposes a stress perceived by the cell as similar to other environmental cues triggering calcium response. In consequence, some properties of the membrane are adjusted resulting in lessened sensitivity of cells to the peptide. According to the presented view, deletion of the RIM101 gene may constitutively trigger similar, perhaps stronger, alterations of membrane properties leading to an even lower sensitivity. These hypothetical alterations or adjustments of membrane properties may result in diminished peptide binding or, alternatively, in a lower peptide concentration required for the transition from surface-bound state to crossing of the membrane and concomitant cell death.
Another group of genes induced by the MUC7 peptide, encodes subunits of proteasome. This complex structure consists of the 20S core particle and 19S regulatory particle, together constituting the 26S proteasome involved in ATP-dependent degradation of ubiquitinated proteins (Wolf & Hilt, 2004). Proteasome plays a role in a variety of normal cellular functions via the turnover of proteins and in response to stress by degrading damaged, misfolded and oxidized proteins. Activation of genes encoding proteasome subunits has been detected in S. cerevisiae and C. albicans in response to oxidative stress (Wang et al., 2006; Costa et al., 2007). The induction of proteasome genes observed here may reflect a response to a stress to which yeasts are subjected during their growth in the presence of MUC7 peptide.
Among the genes whose expressions were diminished by the MUC7 peptide, several encode proteins involved in iron transport and metabolism (Table 3). Some of these genes are regulated by the availability of iron (Lan et al., 2004). Similar repression of a number of iron transport genes has been observed following the treatment of C. albicans with various antifungal agents (Liu et al., 2005). The significance of these changes in iron metabolism, induced by the MUC7 peptide, is currently unclear and will require further studies.
In summary, the obtained data revealed the importance of calcium signalling as a mechanism of adaptation of C. albicans yeasts to the action of MUC7 12-mer, and changes in gene expression reflecting common responses to cellular damage inflicted by the peptide.
Acknowledgments
This work was supported by NIH/NIDCR RO1 grant DE009820 (LAB). We thank Dr. Aaron Mitchell (Carnegie Mellon University) for providing the C. albicans deletion mutants, and acknowledge Sanjay Bhatt for technical assistance and proofreading of the manuscript.
References
- Baixench MT, Aoun N, Desnos-Ollivier M, Garcia-Hermoso D, Bretagne S, Ramires S, Piketty C, Dannaoui E. Acquired resistance to echinocandins in Candida albicans: case report and review. J Antimicrob Chemother. 2007;59:1076–1083. doi: 10.1093/jac/dkm095. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- Costa V, Quintanilha A, Moradas-Ferreira P. Protein oxidation, repair mechanisms and proteolysis in Saccharomyces cerevisiae. IUBMB Life. 2007;59:293–298. doi: 10.1080/15216540701225958. [DOI] [PubMed] [Google Scholar]
- Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. Embo J. 2002;21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egusa H, Soysa NS, Ellepola AN, Yatani H, Samaranayake LP. Oral candidosis in HIV- infected patients. Curr HIV Res. 2008;6:485–499. doi: 10.2174/157016208786501445. [DOI] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanafani ZA, Perfect JR. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis. 2008;46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Kullas AL, Martin SJ, Davis D. Adaptation to environmental pH: integrating the Rim101 and calcineurin signal transduction pathways. Mol Microbiol. 2007;66:858–871. doi: 10.1111/j.1365-2958.2007.05929.x. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53 :1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- Lis M, Bobek LA. Proteomic and metabolic characterization of a Candida albicans mutant resistant to the antimicrobial peptide MUC7 12-mer. FEMS Immunol Med Microbiol. 2008;54:80–91. doi: 10.1111/j.1574-695X.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- Lis M, Fuss JR, Bobek LA. Exploring the Mode of Action of Antimicrobial Peptide MUC7 12-mer by Fitness Profiling of Saccharomyces cerevisiae Genome-Wide Mutant Collection. Antimicrob Agents Chemother. 2009;53:3762–3769. doi: 10.1128/AAC.00668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother. 2005;49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CO, Hayes A, Wilson M, Rash BM, Oliver SG, Coote P. Global phenotype screening and transcript analysis outlines the inhibitory mode(s) of action of two amphibian-derived, alpha-helical, cationic peptides on Saccharomyces cerevisiae. Antimicrob Agents Chemother. 2007;51:3948–3959. doi: 10.1128/AAC.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyewu C, Blankenship JR, Del Poeta M, Heitman J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob Agents Chemother. 2003;47:956–964. doi: 10.1128/AAC.47.3.956-964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrosky-Zeichner L, Rex JH, Bennett J, Kullberg BJ. Deeply invasive candidiasis. Infect Dis Clin North Am. 2002;16:821–835. doi: 10.1016/s0891-5520(02)00034-x. [DOI] [PubMed] [Google Scholar]
- Rex JH, Rinaldi MG, Pfaller MA. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48 :959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Satyanarayana J, Situ H, Narasimhamurthy S, Bhayani N, Bobek LA, Levine MJ. Divergent solid-phase synthesis and candidacidal activity of MUC7 D1, a 51-residue histidine-rich N-terminal domain of human salivary mucin MUC7. J Pept Res. 2000;56:275–282. doi: 10.1034/j.1399-3011.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims CR, Ostrosky-Zeichner L, Rex JH. Invasive candidiasis in immunocompromised hospitalized patients. Arch Med Res. 2005;36:660–671. doi: 10.1016/j.arcmed.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Situ H, Wei G, Smith CJ, Mashhoon S, Bobek LA. Human salivary MUC7 mucin peptides: effect of size, charge and cysteine residues on antifungal activity. Biochem J. 2003;375:175–182. doi: 10.1042/BJ20030779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, Jiang YY. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med. 2006;40:1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Wei GX, Bobek LA. In vitro synergic antifungal effect of MUC7 12-mer with histatin-5 12-mer or miconazole. J Antimicrob Chemother. 2004;53:750–758. doi: 10.1093/jac/dkh181. [DOI] [PubMed] [Google Scholar]
- Wei GX, Campagna AN, Bobek LA. Effect of MUC7 peptides on the growth of bacteria and on Streptococcus mutans biofilm. J Antimicrob Chemother. 2006;57:1100–1109. doi: 10.1093/jac/dkl120. [DOI] [PubMed] [Google Scholar]
- Wei GX, Campagna AN, Bobek LA. Factors affecting antimicrobial activity of MUC7 12-mer, a human salivary mucin-derived peptide. Ann Clin Microbiol Antimicrob. 2007;6:14. doi: 10.1186/1476-0711-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim Biophys Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Zanetti M, Gennaro R, Skerlavaj B, Tomasinsig L, Circo R. Cathelicidin peptides as candidates for a novel class of antimicrobials. Curr Pharm Des. 2002;8:779–793. doi: 10.2174/1381612023395457. [DOI] [PubMed] [Google Scholar]