Summary
Luminescent bacteria (γ-Proteobacteria: Vibrionaceae) are found in complex bilobed light organs of both sepiolid and loliginid squids (Mollusca: Cephalopoda). Despite the existence of multiple strain colonization between Vibrio bacteria and loliginid squids, specificity at the genus level still exists and may influence interactions between symbiotic and free-living stages of the symbiont. The environmentally transmitted behaviour of Vibrio symbionts bestows a certain degree of recognition that exists prior and subsequent to the colonization process. Therefore, we identified bacterial genes required for successful colonization of loliginid light organs by examining transcripts solely expressed in either the light organ or free-living stages. Selective capture of transcribed sequences (SCOTS) was used to differentiate genes expressed by the same bacterium when thriving in two different environments (i.e. loliginid light organs and seawater). Genes specific for squid light organs included vulnibactin synthetase, outer membrane protein W and dihydroxy dehydratase, which have been associated with the maintenance of bacterial host associations in other systems. In contrast, genes that were solely expressed in the free-living condition consisted of transcripts recognized as important factors for bacterial survival in the environment. These transcripts included genes for methyl accepting chemotaxis proteins, arginine decarboxylase and chitinase. These results provide valuable information regarding mechanisms determining specificity, establishment, and maintenance of bacteria–squid associations.
Introduction
Bacterial–host interaction during mutualistic or pathogenic symbioses can be a dynamic association where microorganisms use survival and reproduction strategies to fight the normal defence mechanisms of the host (Mekalanos, 1985; McFall-Ngai and Ruby, 1998). Because expression of virulence determinants in bacteria is regulated by both environmental and host factors (Heithoff et al., 1997; Jones and Nishiguchi, 2006; Soto et al., 2009), many novel genes that are not expressed during in vitro growth are known to be regulated by in vivo factors within the host (Heithoff et al., 1997) at the onset of symbiosis. For instance, Graham and Clark-Curtiss (1999) demonstrated differential gene expression of Mycobacterium tuberculosis upon interaction with cultured primary human microphages and during their free-living state. Likewise, studies on Salmonella typhimurium showed variation in expression profiles following colonization of mouse tissue when compared with growth outside of the host (Slauch et al., 1994). Both studies elucidated the importance of genes coding for membrane, stress and regulatory functions in the establishment and maintenance of these associations. Interactions between bacteria in the family Vibrionaceae and squid light organs is not an exception to this rule, with Vibrio fischeri undergoing differential gene expression upon colonization of the light organs of bobtail squids in the genus Euprymna (Mollusca: Cephalopoda) (Jones and Nishiguchi, 2006).
Associations between bobtail squids (Cephalopoda: Sepiolidae) and Vibrionaceae bacteria (γ-proteobacteria: Vibrionaceae) have been previously studied to understand the evolution and regulation of specificity of environmentally transmitted symbioses (Nishiguchi et al., 1998; Nishiguchi, 2002; Jones et al., 2006). Ruby and Asato (1993) demonstrated that luminous bacteria in squid light organs benefit from this association and exhibit higher growth rates than their free-living counterpart. Likewise, there is a benefit for the squid host, which uses the light produced by its bacterial partner for counterillumination (Jones and Nishiguchi, 2004).
Similarly, squid species in the family Loliginidae (Mollusca: Cephalopoda) are known to possess bacteriogenic light organs (Alexeyev, 1992; Anderson, 2000). The economic importance of loliginid squid fisheries (Chotiyaputta et al., 2002; Nootmorn and Chotiyaputta, 2002) has furthered scientific interest in the characterization of bacterial populations colonizing these specialized tissue complexes (Guerrero-Ferreira and Nishiguchi, 2007). Previous studies have provided evidence of an association between the marine pathogen Vibrio harveyi with light organs of loliginid squids. These findings have raised questions regarding the potential of this symbiosis as a temporary reservoir for pathogenic Vibrio species such as V. harveyi (Guerrero-Ferreira and Nishiguchi, 2007; Dunlap et al., 2008). Considering the dual life history of V. harveyi, it is important to understand the genetic factors involved in its transition from pathogenic to mutualistic lifestyles to obtain valuable clues on how these unique associations have arisen.
Research approaches to identify genes selectively expressed by bacteria during their symbiotic states within host animals or cells have revealed a great deal about how virulence factors are regulated (Slauch et al., 1994; Graham and Clark-Curtiss, 1999; Daigle et al., 2001; Somboonwiwat et al., 2006), as well as identified factors required for successful colonization and persistence (Camilli and Mekalanos, 1995; Faruque et al., 2004; Jones and Nishiguchi, 2006). SCOTS (Selective Capture Of Transcribed Sequences) has been successfully used to compare gene expression of the same bacterium existing in two different environments (Graham and Clark-Curtiss, 1999; Daigle et al., 2001; Hou et al., 2002; Jones and Nishiguchi, 2006). This technique has been recognized as a useful tool to understand selective pressures associated with persistence of bacteria in the environment as well as host colonization. Therefore, we examined differences in gene expression of environmental (seawater) and symbiotic (light organ associated) bacterial isolates from the loliginid squid Uroteuthis chinensis using SCOTS to determine bacterium responsiveness to either host or environment.
Results and discussion
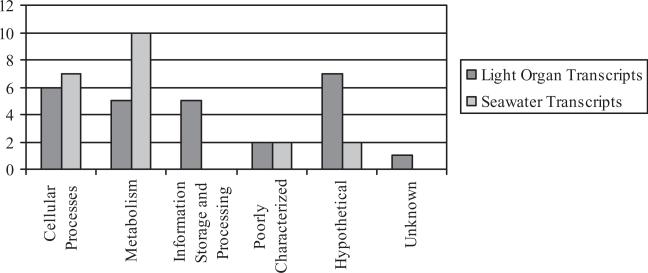
SCOTS has been proven to be a successful method for identification of genes expressed either during symbiosis (associated with a host) or in its free-living state (Graham and Clark-Curtiss, 1999; Graham et al., 2002; Dozois et al., 2003; Faucher et al., 2006). Successful use of SCOTS in mutualistic relationships such as the Euprymna–V. fischeri association has contributed to the knowledge of factors responsible for colonization and persistence of the symbiont within the light organ of the squid as well as prior to infection in the surrounding environment (Jones and Nishiguchi, 2006). We examined the utility of this method by comparing light organ-expressed genes with those solely expressed in seawater. A total of 47 genes were found, with 27 transcripts identified from light organ isolated bacteria and 20 transcripts from those isolates grown in seawater (Tables 1 and 2). Contamination by ribosomal RNA (rRNA) after capture hybridizations, which is one of the concerns during SCOTS, was ruled out by completing southern blot hybridizations during primary verification. Tables 1 and 2 also show distribution of transcripts by gene types indicating that seawater-expressed genes are most associated with cellular processes (seven transcripts) and metabolism (10 transcripts). Light organ transcripts detected by SCOTS showed a more uniform distribution among different gene categories including genes for cellular processes (six transcripts), metabolism (five transcripts), and information storage and processing (five transcripts) (Fig. 1). Lack of detection of seawater transcripts under the functional category of information storage and processing (translation, ribosomal structure and biogenesis, transcription, DNA replication, recombination and repair) after SCOTS may indicate that these genes are almost equally expressed under both conditions, therefore being blocked during enrichment of each SCOTS library. However, specific genes (shown in Table 1) are selectively expressed during the bacteria symbiotic lifestyle, which highlights their importance for symbiosis.
Table 1.
Genes expressed by vibrio isolates in the light organs of loliginid squids.
| Class | Clone name | Homology | Gene/protein coded |
|---|---|---|---|
| Cellular processes | Xba086,135 | VVA1310 | Vulnibactin synthetase, amide synthase subunit |
| Cellular processes | Xba171 | VIBHAR_06625 | Sensory histidine kinase CreC |
| Cellular processes | Xba103,014 | VIBHAR_04770 | Cell wall-associated hydrolase |
| Cellular processes | Xba064 | VF-2110 | Putative transporter YaaJ |
| Cellular processes | Xba065 | VIBHAR_06639 | ompW gene for outer membrane protein W |
| Cellular processes | Xba175 | Vibrio harveyi HY01 | Cell wall-associated hydrolase |
| Metabolism | Xba010 | VIBHAR_00348 | Gamma glutamyltransferase |
| Metabolism | Xba112 | pVHA1-VHW-1 | Quaternary ammonium compound resistance protein |
| Metabolism | Xba126 | VIBHAR_00512 | Partial ilvD gene |
| Metabolism | Xba106 | VIBHAR_03235 | Uridylate kinase (UMP kinase) PyrH |
| Metabolism | Xba008 | VIBHAR_02101 | Non-ribosomal peptide synthetase modules |
| Information storage and processing | Xba111 | VIBHAR_00057 | 30S ribosomal protein S12 |
| Information storage and processing | Xba006 | VIBHAR_06651 | lysR family transcriptional regulator |
| Information storage and processing | Xba005 | VIBHAR_06734 | Ribosomal protein S6 modification protein |
| Information storage and processing | Xba169 | Vibrio harveyi HY01-A1Q_5079 | Pseudouridine synthase, Rsu |
| Information storage and processing | Xba134 | VIBHAR_0565 | Integrase IntI |
| Poorly characterized | Xba172 | VV1_1061 | Orf122-like protein |
| Poorly characterized | Xba168 | VIBHAR_00255 | RNA-binding protein |
| Hypothetical | Xba108 | VIBHAR_00327 | Hypothetical protein |
| Hypothetical | Xba170 | VV1_0932 | Hypothetical protein |
| Hypothetical | Xba174 | V12B01_06372 | Hypothetical protein |
| Hypothetical | Xba176 | A55_B0062 | Hypothetical protein |
| Hypothetical | Xba177 | A55_B0062 | Hypothetical protein |
| Hypothetical | Xba178 | VIBHAR_01012 | Hypothetical protein |
| Hypothetical | Xba179 | VC274080_B0002 | Conserved hypothetical protein |
| Unknown | Xba173 | VV20845 | CMCP6 locus tag, product unknown |
Table 2.
Genes expressed by vibrio isolates in seawater.
| Class | Clone name | Homology | Gene/protein coded |
|---|---|---|---|
| Cellular processes | Sal105,141 | VIBHAR_01497 | Arsenate reductase (arsC) |
| Cellular processes | Sal003 | VIBHAR_05872 | ATP-dependent chaperone clpB |
| Cellular processes | Sal198 | VIBHAR_05104 | Putative arsenate reductase |
| Cellular processes | Sal077 | VIBHAR_04747 | Methyl-accepting chemotaxis protein (MTA/SAH nucleosidase) |
| Cellular processes | Sal205 | VIBHAR_05559 | Heat shock protein |
| Cellular processes | Sal209 | VIBHAR_02921 | Tyrosine-phosphatase |
| Cellular processes | Sal005 | VIBHAR_04747 | Methyl accepting chemotaxis protein |
| Metabolism | Sal203 | VIBHAR_05421 | phenylalanine monooxygenase |
| Metabolism | Sal127 | VIBHAR_06354 | d-serine deaminase |
| Metabolism | Sal142 | VIBHAR_00053 | Nitrite reductase (nirB) |
| Metabolism | Sal143,180 | VIBHAR_06737 | Arginine decarboxylase (speA) |
| Metabolism | Sal197,204 | VIBHAR_03711 | d-aspartate kinase |
| Metabolism | Sal103,163 | VIBHAR_00698 | Aminoglycoside acetyltranferase |
| Metabolism | Sal104 | VIBHAR_05656 | Chitinase |
| Metabolism | Sal107 | VIBHAR_03431 | Cellobiose phosphorylase |
| Metabolism | Sal 202 | VIBHAR_01806 | Adenosylmethionine-8amino-7-oxononanoate transaminase |
| Metabolism | Sal217 | VIBHAR_00151 | Manganese superoxide dismutase |
| Poorly characterized | Sal207 | VIBHAR_03535 | GTP-binding protein |
| Poorly characterized | Sal019 | VIBHAR_07101 | RNA binding protein |
| Hypothetical | Sal 090 | VIBHAR_00469 | Hypothetical protein |
| Hypothetical | Sal102 | VIBHAR_00194 | Hypothetical protein |
Fig. 1.
Distribution of gene categories of transcripts expressed by free-living and light organ-associated bacteria captured through SCOTS.
Gene expression of vibrio isolates in the light organ of loliginid squids
Graf and Ruby (1998) argued that the crypt epithelium within the light organ provides amino acids and potentially other nutrients to the symbiotic bacteria housed therein. This might indicate that colonization of the nutrient-rich, light organ environment provides a selective advantage to the symbiont over free-living ecological variants. However, the same experiments demonstrated that auxotrophic mutants of squid symbionts were able to grow and establish themselves within the light organ environment at lower densities than wild-type isolates. Similarly, this study demonstrated that some amino acids required for bacterial growth are found in low levels within the light organ environment. Because of this, genes required for amino acid synthesis are expected to be expressed in the light organ. In our study, we confirmed that genes required for the production of specific amino acids are being expressed during symbiosis, with ilvD (dihydroxyacid dehydratase) being solely expressed in the light organ. ilvD (Tarleton and Ely, 1991) catalyses the fourth step in the metabolic pathway leading to production of isoleucine (from pyruvate) and valine (from alpha-oxobutyrate). The product of the reaction catalysed by dihydroxyacid dehydratase is also the starting point in the alternative pathway producing leucine (Gottschalk, 1986). Production of leucine and valine is necessary within the light organ matrix where levels of these two amino acids are low, reaching only 0.07 and 0.09 mM respectively in Euprymna scolopes (Graf and Ruby, 1998). Conversely, concentrations of the same amino acids measured within symbiotic cells (including free and peptide forms) are among the highest, with 1.58 mM of leucine and 1.26 mM of valine, indicating that these molecules are being synthesized within the squid light organ. This suggests a major role for this amino acid in the proliferation of the association. Production of ilvD mutants of Vibrio bacteria and further colonization studies on squids would give insights on the role of ilvD on the persistence of the association.
An underlying role of ilvD is evident in Vibrio cholerae, the causative agent of cholera. In a human in vivo expression technology study (IVET), Lombardo and colleagues (2007) identified ilvD as one of the V. cholerae genes selectively expressed during human infection. In a different study, a large-scale signature-tagged mutagenesis screen (Merrell et al., 2002) also demonstrated the requirement of ilvD for human colonization by V. cholerae. This study showed that mutations affecting the entire ilv operon were associated with decreased virulence in the mouse model. Contribution of ilvD to the capability of Vibrio bacteria to colonize loliginid squid is yet to be characterized. However, since V. cholerae requires this gene for colonization of epithelial tissues, it may also indicate a connection between ilvD expression and colonization of eukaryotic tissue (such as squids) by Vibrio bacteria in general.
Our study also demonstrates selective expression of the gene for the amide synthetase subunit of vulnibactin synthetase, the enzyme responsible for production of vulnibactin, a siderophore first isolated from low-iron cultures of Vibrio vulnificus (Noriyuki et al., 1994). Acquisition of iron from the host is a requirement for colonization by host tissue-associated microbes. It is a common feature in pathogenic systems such as Vibrio anguillarum (Actis et al., 1986; Wolf and Crosa, 1986), V. cholerae and V. harveyi (Henderson and Payne, 1994; Owens et al., 1996). In mutualistic associations, studies with V. fischeri mutants with depleted siderophore production revealed that the ability to respond to limited iron levels may be connected to symbiosis efficiency (Graf and Ruby, 2000). Owens and colleagues (1996) also established a correlation between siderophore production and the type of bacterial isolates, and determined that free-living strains of V. harveyi produce higher amounts of siderophore compared with isolates associated with invertebrates. This difference in siderophore production was in contrast with our findings that light organ-associated vibrios selectively expressed the vulnibactin synthetase gene, necessary for bacterial siderophore production within the light organ. However, because Owens and colleagues (1996) focused on measurements of V. harveyi siderophore activity on agar plates prepared, their results do not reflect the expression of genes related with siderophore-producing pathways in vivo. Few studies have focused on iron sequestration and regulation of sidephore production of symbiotic bacteria during mutualistic associations with animal tissues (Bagg and Neilands, 1987; Aznar and Alcaide, 1992; Naidu and Yadav, 1997; Moeck and Coulton, 1998). Since bacteria harboured in the light organ are found in high concentrations (~108–1010 in an adult Euprymna), the necessity of having siderophores to capture iron resources may be fuelled by the competition among this community (i.e. limited resources in the light organ). Further examination of siderophore activity and regulation of gene expression associated with iron assimilation is necessary to fully comprehend the role of iron sequestration in colonization of loliginid light organs by Vibrionaceae bacteria.
Comparable to most siderophores, vibriobactin is a small cyclic peptide with the ability to bind environmental iron. This siderophore–iron complex is recognized by bacterial surface receptors such as ompW and transported into the cytoplasm (Moeck and Coulton, 1998). Expression of the ompW gene for outer membrane protein W was also detected in our SCOTS analysis. The importance of iron sequestration in symbiosis, specifically with regards to growth and replication of cytochrome systems of bacterial symbionts residing in eukaryotic hosts, has been previously observed (Bagg and Neilands, 1987; Henderson and Payne, 1994; Dozois et al., 2003). It is also known that production of siderophore is frequently accompanied by the synthesis of outer membrane proteins acting as receptors for the iron–siderophore complex (Bagg and Neilands, 1987; Dai et al., 1992).
In addition, outer membrane proteins have been shown to enhance adhesion and colonization when tested in animal models. However, their exact role in virulence is not completely understood (Faruque et al., 2004). In mutualistic associations, Jones and Nishiguchi (2006) reported V. fischeri expression of an integral membrane protein during colonization of E. scolopes light organs. Moreover, Aeckersberg and colleagues (2001) presented evidence connecting the presence of a specific bacteria outer membrane protein (i.e. OmpU) for the establishment of successful mutualistic associations with sepiolid squids. Results from our SCOTS screening indicate that expression of genes for membrane proteins, such as ompW in symbiotic bacteria, may be necessary for successful colonization of the loliginid light organ, because of their direct relevance to in vivo recognition of iron–siderophore complexes.
Equally important for maintenance of colonization efficiency is the expression of the pyrH gene. This gene codes for the protein uridine monophosphate kinase (UMP kinase) that participates in pyrimidine metabolism catalysing the conversion of UMP into UDP (Voet and Voet, 2004). Kim and colleagues (2003) identified pyrHas one of the genes in V. vulnificus that is expressed preferentially in vivo. Vibrio vulnificus mutants unable to produce UMP kinase exhibited retarded growth on media and a reduction in the ability to infect HeLa cells. Therefore, pyrH expression has relevance in host colonization by Vibrio bacteria by directly reducing bacterial growth and decreasing infectivity.
Gene expression of vibrio isolates in seawater
Analysis of gene expression profiles of seawater-grown bacteria yielded a set of genes dominated by those responsible for bacterial metabolism. Among transcripts identified by SCOTS, a chitinase gene was hypothesized to be present, since Vibrio species use chitin as a carbon source (Yu et al., 1991; Svitil et al., 1997). Vibrios play a critical role in transforming chitin, a highly insoluble polysaccharide, into a form usable to other organisms (Bassler et al., 1991). Moreover, it has been reported that some species in the family Vibrionaceae are able to monitor surrounding environmental conditions by recognizing and migrating towards low concentrations of chitin oligosaccharides (Yu et al., 1991). Similarly, in most cooperative interactions studied in nature, hosts provide nutrient-rich environments for bacterial growth, whereas the bacteria contribute by providing the host with products of specific bacterial processes. It might be expected that free-living vibrios would use chitin metabolic machinery to monitor host tissue until successfully detecting chitin contained in the squid light organ (McFall-Ngai, 1998). This type of mechanism would potentially be part of the interactions that account for the first encounter between bacteria and host in the squid–Vibrio symbiosis. Upon first contact, other processes such as mucus and nitric oxide production occur (Nyholm et al., 2000; Davidson et al., 2004) that allow for the successful and permanent establishment of the symbionts within the crypts of the light organ (McFall-Ngai and Ruby, 1998). Our results not only support the importance of host-produced chitin on the initiation of squid-vibrio symbiosis but also imply a more active role of the symbiont in the establishment of this association.
In addition, our screening exhibited arginine decarboxylase (ADC) expression by V. harveyi loliginid light organ isolates grown in seawater. ADC degrades arginine to produce agmatine, and increases pH by removing acidic carboxyl groups and releasing CO2 from their substrates (Stim and Bennett, 1993). A potential role of ADC in the environment is the formation of biofilm. ADC, similar to other amino acid decarboxylases, is involved in the production of polyamines (Stim and Bennett, 1993) that modulate bacterial biofilms within Vibrionaceae species (Kierek-Pearson and Karatan, 2005; Patel et al., 2006). Studies have shown that factors that modulate biofilm formation (such as ADC and uridine diphosphate dehydrogenase, UDPDH) by Vibrionaceae bacteria are important in light organ symbiosis. For example, analysis of V. fischeri UDPDH mutants demonstrated that their biofilmforming ability is reduced in vitro. In addition, these mutants were not detected in any part of the crypt region during colonization assays (Ariyakumar, 2007; Ariyakumar and Nishiguchi, 2009).
Also important for bacterial colonization of eukaryotic hosts is the expression of genetic factors responsible for DNA methylation (Mahan et al., 2000). This study provides evidence for expression in seawater of the 5′-methylthioadenosine/S-adenosylhomocysteine (MTA/SAH) nucleosidase gene. Methylation reactions depend on the appropriate methyl donor to be available when necessary. While S-adenosylmethionine (SAM) is the main donor of methyl groups in the cell, the end-product of the reaction is S-adenosylhomocysteine (SAH) that negatively regulates SAM-dependent methyltransferases. Therefore, SAH needs to be readily metabolized by MTA/SAH nucleosidase to adenine and S-ribosylhomocysteine. The activity of MTA/SAH nucleosidase is indispensable for the maintenance of methylation reactions in bacteria (Stepkowski et al., 2005). It has been demonstrated that severe defects in colonization of eukaryotic tissue occur when methylation processes are affected in methylation-defective S. typhimurium mutants (Heithoff et al., 1999). Such increases in methylation events and efficient regulation of methylation reactions may be accomplished by preferential expression of genes coding for MTA/SAH nucleosidases. Also, with methylation reactions being linked to successful eukaryotic colonization by bacteria, selective expression of methylation genes may be important for improving the likelihood of successful colonization of squid tissue by Vibrio bacteria.
Also important in metabolic processes in bacteria is the expression of chaperone molecules such as ClpB. The gene coding for this chaperone molecule was exclusively expressed during seawater growth of symbiotic bacteria. ClpB is important for the refolding of luciferase (Zavilgelsky et al., 2004), the molecule responsible for light emission and important for quorum sensing in bacteria (Bassler, 1999). Zavilgelsky and colleagues (2004) demonstrated that presence of ClpB accelerated the refolding of luciferase and increased its efficiency approximately 10-fold. Considering that luciferases of Vibrionaceae bacteria are sensitive to thermoinactivation (Zavilgelsky et al., 2004), activity of ClpB would be considerably important in locations where water temperatures can change between winter and summer months (Jones et al., 2006; Soto et al., 2009), or where temperatures can exceed 28°C (Soto et al., 2009). Since bioluminescence is an important aspect of light organ symbioses, this phenomenon is extremely important for the fitness of light-producing bacteria.
Physiological adaptation is essential to maintain homeostasis. Metabolic processes responsible for such adaptations are regulated by the activity of sensory and regulatory proteins that control gene expression and enzymatic activity. Some of these mechanisms are conserved across bacterial species, while others are the result of microbial adaptation to specific environmental niches. Our research presents evidence of gene expression profiles that may explain the ability of a bacterium to transition between a free-living to mutualistic lifestyle, given its mechanisms of transmission (environmental). This study also provides support for differential gene expression relative to the ecology of V. harveyi bacteria and their ability to occupy multiple niches when grown in seawater and in squid hosts. Examining the processes that select for the evolution of symbiosis in the ocean can be better understood by defining important genetic factors that contribute to successful initiation and persistence of symbiotic associations. Future studies will include detailed investigations of these genes and the metabolic pathways related to synthesis or degradation of their products, and will address questions concerning host specificity and the array of mechanisms necessary for beneficial associations.
Experimental procedures
Bacterial growth conditions and extraction of total RNA
Specimens of U. chinensis were obtained from squid trawls off the coast of Cairns, Australia during the 2008 summer season. Light organs were dissected from the mantle cavity and a portion of it homogenized in sterile seawater. The rest of the light organs were kept in RNA Later (Applied Biosystems/Ambion, Austin, TX) for further extraction of mRNA. Serial homogenate dilutions were incubated on sea-water tryptone agar (70% seawater v/v, 0.5% tryptone w/v, 0.3% yeast extract w/v, 0.3% glycerol v/v and 1.5% technical grade agar) at room temperature for 16 h. Individual colonies of luminous bacteria were isolated and used to inoculate 5 ml of SWT media and shaker incubated (250 r.p.m.) overnight. An aliquot (900 μl) of bacterial suspension was used for glycerol stocks. To identify transcripts selectively expressed by the U. chinensis isolates in seawater, bacteria were grown in sterile seawater (non-artificial) in a 28°C shaker incubator (250 r.p.m.) for 48 h. At this time an optical density (OD600) of about 0.5 was reached. Seawater was obtained from Santa Catalina Island, CA. Bacteria were concentrated by centrifugation and RNA extracted following the protocol described below.
For light organ-expressed transcripts, three (partial) U. chinensis light organs were homogenized in RNA Later (Applied Biosystems/Ambion, Austin, TX) and centrifuged at 12 000 g for 2 min to pellet eukaryotic tissue, separating bacterial cells from the rest of the squid light organ. RNA extraction was completed using a protocol modified from Mangan and colleagues (1997). This protocol was incorporated in the RiboPure™ kit (Applied Biosystems/Ambion, Austin, TX), and manufacturer instructions were followed. Following RNA isolation, removal of rRNA from total RNA samples was performed with MICROBExpress (Applied Biosystems/Ambion, Austin, TX). This additional step removes considerable amounts of rRNA from bacterial total RNA samples, reducing the chance of rRNA-derived complementary DNAs (cDNAs) being generated during SCOTS.
cDNA library construction
Complementary DNA libraries were constructed using the method proposed by Graham and Clark-Curtiss (1999; 2000). The first strand of cDNA was synthesized from total RNA using a Superscript II reverse transcriptase (Invitrogen Corporation, Carlsbad, CA), following manufacturer's instructions. Oligonucleotides with a random 9-mer at the 3′ end were used for this first step of cDNA library construction (Table 3). Second-strand synthesis was performed with the Klenow fragment of DNA polymerase (New England Biolabs, Beverly, MA) following manufacturer's instructions. Double-stranded cDNA was purified from remaining salts, enzymes and unincorporated nucleotides using the PCR purification kit (Qiagen, Valencia, CA).
Table 3.
Primers for SCOTS protocol and verification.
| Name | Sequence (5′-3′) | Application |
|---|---|---|
| Xba random | TGCTCTAGACGTCCTGATGGTT9(N) | cDNA library |
| Xba | TGCTCTAGACGTCCTGATGGTT | cDNA amplification |
| Sal random | ATATGTCGACTGAATTCCGTAGG9(N) | cDNA library |
| Sal | ATATGTCGACTGAATTCCGTAGG | cDNA amplification |
| 16S forward | AGAGTTTGATCMTGGCTCAG | rRNA cloning |
| 23S reverse | ATGGTTAAGCCTCACGGGCA | rRNA cloning |
| speA sense | GTTAAACCACGTCTTGGCTTGCGT | Secondary verification |
| speA antisense | ACCACCACCCACATCGAGGTATTT | Secondary verification |
| Chitinase sense | TCAGTCGGTGGATGGACGCT | Secondary verification |
| Chitinase antisense | GACGCTGATAATGGCGACAT | Secondary verification |
| ilvD sense | TCCGTCGAATACATGGTCAA | Secondary verification |
| ilvD antisense | TTTCAAACGCTGCTTTGTTG | Secondary verification |
| ompW sense | CCACCTACCTTTATGGTCC | Secondary verification |
| ompW antisense | GGTTTGTCGAATTAGCTTCACC | Secondary verification |
| vibH sense | TTGATGGCTACAGCTTGCAC | Secondary verification |
| vibH antisense | ATTGATCCACAGCGGTAAGG | Secondary verification |
Isolation of rRNA operon
In order to block cDNA derived from rRNA during SCOTS, the full ribosomal operon was PCR-amplified from total DNA using primers 16S Forward (Edwards et al., 1989) and 23S Reverse (Jones and Nishiguchi, 2006) (Table 3). PCR reactions yielded a product of approximately 5 kb in length, which was excised from a 1% agarose gel in 1× TAE buffer (40 mM Tris-Acetate, 1 mM EDTA, pH 8.0) using QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). The purified ribosomal operon was subsequently inserted into the pCR® 2.1-TOPO® cloning vector (3.9 kb) from the TOPO® TA Cloning kit (Invitrogen Corporation, Carlsbad, CA).
Selective Capture Of Transcribed Sequences (SCOTS)
Normalization of cDNA libraries was conducted by three rounds of capture hybridization following a protocol from Jones and Nishiguchi (2006), modified from Graham and Clark-Curtiss (1999). Briefly, 30 μg of sonicated, biotinylated, genomic DNA (gDNA) and 50 μg of sonicated, cloned, ribosomal operon (rRNA) were precipitated and re-suspended in 40 μl of 10 mM 3-[4-(2-Hydroxyethyl)-1-piperazinyl] propanesulfonic acid (EPPS) (Sigma-Aldrich, St. Louis, MO). Simultaneously, 125 μl of amplified cDNAs from each condition were equally precipitated and re-suspended in 40 μl of EPPS. Then, hybrids and cDNAs were denatured (2 min at 98°C), normalized (30 min at 55°C) and mixed with 10 μl of 1 M sodium chloride. After an additional incubation at 55°C for 1 h, a single gDNA–rRNA hybridization reaction was added to each cDNA library and incubated at 55°C overnight.
Following overnight hybridizations, gDNA–rRNA hybrids were recovered using Dynal streptavidin-coated magnetic beads (Dynal/Invitrogen Corporation, Carlsbad, CA) with a magnetic stand (Applied Biosystems/Ambion, Austin, TX). Biotinylated gDNA–cDNA hybrids were washed and cDNA eluted from gDNA using 100 μl of 0.5 M NaOH and 0.1 M NaCl.
Eluted cDNA was PCR-amplified with a non-random primer (Table 3) specific for each growth condition, then purified and normalized through four additional rounds of capture hybridizations. Conversely, all reactions were completed using 1/10 of the volumes used in the first round.
Enrichment of cDNA was conducted in a way similar to capture hybridizations. However, biotinylated gDNA, blocked with rRNA, was additionally blocked with 25 μl of cDNA that was amplified after the last round of capture hybridization (seawater-derived cDNA to examine genes expressed in the light organ and vice versa). Hybrids were collected using Dynal streptavidin-coated magnetic beads. cDNA was eluted, PCR-amplified and purified using the aforementioned protocols. PCR products (Xba-amplified libraries for light organ-expressed transcripts and Sal libraries for seawater-expressed transcripts) were cloned using the TOPO® TA Cloning kit (Invitrogen Corporation, Carlsbad, CA).
Southern hybridizations for primary verification
Southern hybridizations were carried out to eliminate false-positive sequences that escaped the subtraction process. Both Xba (light organ-expressed) and Sal (seawater-expressed) libraries were denatured with denaturing solution (0.5 N NaOH, 1.5 M NaCl) for 5 min and treated with neutralizing solution (1.5 M NaCl, 0.5 M Tris-HCl, pH 7.4) for 5 min and 2× SSC (0.3 M NaCl, 0.03 M sodium citrate) for 5 min. Samples were then transferred onto a positively charged nylon membrane and cross-linked utilizing a Stratalinker UV cross-linker (Stratagene, La Jolla, CA). Hybridization was then completed against DIG-labelled probes created from the fifth round captured sequences used to block the cross-linked sequences during the enrichment reactions.
Membranes were pre-hybridized in 15 ml of hybridization solution at 40°C and then hybridized at 40°C overnight. Hybridization was performed with 20 ng ml-1 of probe in DIG EasyHyb hybridization solution that was added to the membrane. After hybridization, two 2 min washes were completed using a low-stringency buffer (2× SSC, 0.1% SDS), followed by a 20 min washes at 65°C (with rotation) in high-stringency buffer (0.1× SSC, 0.1% SDS). A final wash with maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5) was executed for 5 min at room temperature.
Detection was completed by exposing the membrane to 0.5 ml of anti-DIG antibody/alkaline phosphatase enzyme conjugate for 20 min and washing twice with 4 ml of maleic acid buffer for 10 min. Detection buffer (0.1 M Tris-HCl, 0.1 M NaCl, 50 mM MgCl2) was then used for 5 min to start visualization with the enzyme substrate solution [4-nitroblue tetrazolium chloride (NBT) and 5-bromo-4chloro-3-indolyl phosphate]. No signal indicated that the clone was exclusively expressed in either the light organ or the seawater condition.
Sequence analysis
Positive clones were sequenced using an ABI 3100 capillary sequencer (Applied Biosystems, Foster City, CA), and edited using Sequencher v 4.6 (Gene Codes Corporation, Ann Arbor, MI). Sequences were then compared with the National Center for Biotechnology Information (NCBI) database using blast 2.2.11 (Basic Local Alignment Search Tool, NCBI, NLM, NIH, Bethesda, MD) for initial confirmation of sequence identity. In addition, the presence and location of transcripts in the V. harveyi genome was determined (Tables 1 and 2) by comparisons with the V. harveyi genome (Bassler et al., 2007).
Secondary verification using reverse transcriptase PCR
Four clones were picked for secondary verification using reverse transcription with the SuperScript™ One-Step RT-PCR system (Table 3). Primers were developed for each gene based on their corresponding sequence to the V. harveyi genome (Bassler et al., 2007). PCR reactions were executed as described previously by Jones and Nishiguchi (2006).
Acknowledgements
The authors thank B.W. Jones for his valuable advice on the SCOTS protocol and S. Edmands for collecting seawater from the USC Catalina Marine Station. C. Gorman assisted with sequencing of seawater and light organ transcripts. Funding was provided by NSF-IOS 0744498 and NIH-NIAID 1SC1AI081659 to M.K.N.
References
- Actis LA, Fish W, Crosa JH, Kellerman K, Ellenberger SR, Hauser FM, Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775 (pJM1). J Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeckersberg F, Lupp C, Feliciano B, Ruby EG. Vibrio fischeri outer membrane protein ompU plays a role in normal symbiotic colonization. J Bacteriol. 2001;183:6590–6597. doi: 10.1128/JB.183.22.6590-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev DO. The systematic position of bioluminescent squids of the family Loliginidae (Cephalopoda: Myopsida). Zool Zhurnal. 1992;71:12–23. [Google Scholar]
- Anderson FE. Phylogenetic relationships among loliginid squids (Cephalopoda: Myopsida) based on analyses of mulitiple data sets. Zool J Linnean Soc. 2000;130:603–633. [Google Scholar]
- Ariyakumar DS. Masters Thesis. New Mexico State University; 2007. Genetic factors regulating the symbiosis between sepiolid squids (Cephalopoda: Sepiolidae) and their Vibrio symbionts. [Google Scholar]
- Ariyakumar DS, Nishiguchi MK. Characterization of two host specific genes, mannose sensitive hemagglutinin (mshA) and uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio fischeri–Euprymna tasmanica mutualism. FEMS Microbiol Lett. 2009;299:65–73. doi: 10.1111/j.1574-6968.2009.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar R, Alcaide E. Siderophores and related outer membrane proteins produced by pseudomonads isolated from eels and freshwater. FEMS Microbiol Lett. 1992;98:269–275. doi: 10.1016/0378-1097(92)90168-n. [DOI] [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Molecular mechanisms of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B, Clifton SW, Fulton L, Delehaunty K, Fronick C, Harrison M, et al. The Vibrio harveyi Genome Sequencing Project. Genetics, Genome Sequencing Center, National Center for Biotechnology Information, NIH; Bethesda, MD, USA: 2007. [Google Scholar]
- Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Gibbons PJ, Yu C, Roseman S. Chitin utilization by marine bacteria. Chemotaxis to chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24268–24275. [PubMed] [Google Scholar]
- Camilli A, Mekalanos JJ. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiyaputta C, Nootmorn P, Jirapunpipat K. Review of cephalopod fishery production and long term changes in fish communities in the Gulf of Thailand. Bull Mar Sci. 2002;71:223–238. [Google Scholar]
- Dai J-H, Lee Y-S, Wong H-D. Effects of iron limitation of production of a siderophore, outer membrane proteins, and hemolysin and on hydrophobicity, cell adherance, and lethality for mice of Vibrio parahaemolyticus. Infect Immun. 1992;60:2952–2956. doi: 10.1128/iai.60.7.2952-2956.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle F, Graham JE, Curtiss R., III Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS). Mol Microbiol. 2001;41:1211–1222. doi: 10.1046/j.1365-2958.2001.02593.x. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid–vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Dozois CM, Daigle F, Curtiss R., 3rd Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap PV, Davis KM, Tomiyama S, Fujino M, Fukui A. Developmental and microbiological analysis of the inception of bioluminescent symbiosis in the marine fish Nuchequula nuchalis (Perciformes: Leiognathidae). Appl Environ Microbiol. 2008;74:7471–7481. doi: 10.1128/AEM.01619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Nair GB, Mekalanos JJ. Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae. DNA Cell Biol. 2004;23:723–741. doi: 10.1089/dna.2004.23.723. [DOI] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk G. Bacterial Metabolism. Springer; New York, NY, USA: 1986. [Google Scholar]
- Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Ruby EG. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNA's synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JE, Peek RMJ, Krishna U, Cover TL. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology. 2002;123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JE, Clark-Curtiss JE. Identifying Mycobacterium tuberculosis virulence determinants – new technologies for a difficult problem: response. Trends Microbiol. 2000;8:100. doi: 10.1016/s0966-842x(99)01697-2. [DOI] [PubMed] [Google Scholar]
- Guerrero-Ferreira RC, Nishiguchi MK. Biodiversity among luminescent symbionts from squid of the genera Uroteuthis, Loliolus and Euprymna (Mollusca: Cephalopoda). Cladistics. 2007;23:497–506. doi: 10.1111/j.1096-0031.2007.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Conner CP, Hanna PC, Julio SM, Hentschel U, Mahan MJ. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- Henderson DP, Payne SM. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun. 1994;62:5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JY, Graham JE, Clark-Curtiss JE. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect Immun. 2002;70:3714–3726. doi: 10.1128/IAI.70.7.3714-3726.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Jones BW, Nishiguchi MK. Differentially expressed genes reveal adaptations between free-living and symbiotic niches of Vibrio fischeri in a fully established mutualism. Can J Microbiol. 2006;52:1218–1227. doi: 10.1139/w06-088. [DOI] [PubMed] [Google Scholar]
- Jones BW, Lopez JE, Huttenburg J, Nishiguchi MK. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol Ecol. 2006;15:4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- Kierek-Pearson K, Karatan E. Biofilm development in bacteria. Adv Appl Microbiol. 2005;57:79–111. doi: 10.1016/S0065-2164(05)57003-5. [DOI] [PubMed] [Google Scholar]
- Kim YR, Lee SE, Kim CM, Kim SY, Shin EK, Shin DH, et al. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun. 2003;71:5461–5471. doi: 10.1128/IAI.71.10.5461-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M-J, Michalski J, Martinez-Wilson H, Morin C, Hilton T, Osorio CG, et al. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc Natl Acad Sci USA. 2007;104:18229–18234. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. The development of cooperative associations between animals and bacteria: establishing détente among domains. Am Zool. 1998;38:593–608. [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Sepiolids and vibrios: when first they meet. Bioscience. 1998;48:257–265. [Google Scholar]
- Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- Mangan JA, Sole KM, Mitchison DA, Butcher PD. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos JJ. Cholera toxin: genetic analysis, regulation, and the role in pathogenesis. Curr Top Microbiol Immun. 1985;118:97–118. doi: 10.1007/978-3-642-70586-1_6. [DOI] [PubMed] [Google Scholar]
- Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- Moeck GS, Coulton JW. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- Naidu AJ, Yadav M. Influence of iron, growth temperature and plasmids on siderophore production in Aeromonas hydrophila. J Med Microbiol. 1997;46:833–838. doi: 10.1099/00222615-46-10-833. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microbiol Ecol. 2002;44:10–18. doi: 10.1007/BF03036870. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid–Vibrio symbioses. Appl Environ Microbiol. 1998;64:3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nootmorn P, Chotiyaputta C. Species diversity, biomass and community structure of cephalopods off Adang-Rawi Archipelago, Thailand. Bull Mar Sci. 2002;71:591–600. [Google Scholar]
- Noriyuki O, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7:109–116. doi: 10.1007/BF00140480. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal–bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens L, Austin DA, Austin B. Effect of strain origin on siderophore production in Vibrio harveyi isolates. Dis Aquat Organ. 1996;27:157–160. [Google Scholar]
- Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. Polyamines are essential for the formation of plague biofilm. J Bacteriol. 2006;188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Slauch JM, Mahan MJ, Mekalanos JJ. In vivo expression technology for selection of bacterial genes specifically induced in host tissues. Methods Enzymol. 1994;235:481–492. doi: 10.1016/0076-6879(94)35164-3. [DOI] [PubMed] [Google Scholar]
- Somboonwiwat K, Supungul P, Rimphanitchayakit V, Aoki T, Hirono I, Tassanakajon A. Differentially expressed genes in hemocytes of Vibrio harveyi-challenged shrimp Penaeus monodon. J Biochem Mol Biol. 2006;39:26–36. doi: 10.5483/bmbrep.2006.39.1.026. [DOI] [PubMed] [Google Scholar]
- Soto W, Gutierrez J, Remmenga MD, Nishiguchi MK. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb Ecol. 2009;57:140–150. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepkowski T, Brzezinski K, Legocki AB, Jaskólski M, Béna G. Bayesian phylogenetic analysis reveals two-domain topology of S-adenosylhomocysteine hydrolase protein sequences. Mol Phylogenet Evol. 2005;34:15. doi: 10.1016/j.ympev.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Stim KP, Bennett GN. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitil AL, Chadhain S, Moore JA, Kirchman DL. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl Environ Microbiol. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton JC, Ely B. Isolation and characterization of ilvA, ilvBN, and ilvD mutants of Caulobacter crescentus. J Bacteriol. 1991;173:1259–1267. doi: 10.1128/jb.173.3.1259-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet D, Voet JG. Biochemistry. John Wiley & Sons; Hoboken, NJ, USA: 2004. [DOI] [PubMed] [Google Scholar]
- Wolf MK, Crosa JH. Evidence for the role of a siderophore in promoting Vibrio anguillarum infections. J Gen Microbiol. 1986;132:2944–2952. doi: 10.1099/00221287-132-10-2949. [DOI] [PubMed] [Google Scholar]
- Yu C, Lee AM, Bassler BL, Roseman S. Chitin utilization by marine bacteria. A physiological function for bacterial adhesion to immobilized carbohydrates. J Biol Chem. 1991;266:24260–24267. [PubMed] [Google Scholar]
- Zavilgelsky GB, Kotova VY, Mazhul MM, Manukhov IV. The effect of Clp proteins on DnaK-dependent refolding of bacterial luciferases. Mol Biol (Moscow) 2004;38:427–433. [PubMed] [Google Scholar]