Abstract
This article expands upon recent interest in Bayesian hierarchical models in quantitative genetics by developing spatial process models for inference on additive and dominance genetic variance within the context of large spatially referenced trial datasets of multiple traits of interest. Direct application of such multivariate models to large spatial datasets is often computationally infeasible because of cubic order matrix algorithms involved in estimation. The situation is even worse in Markov chain Monte Carlo (MCMC) contexts where such computations are performed for several thousand iterations. Here, we discuss approaches that help obviate these hurdles without sacrificing the richness in modeling. For genetic effects, we demonstrate how an initial spectral decomposition of the relationship matrices negates the expensive matrix inversions required in previously proposed MCMC methods. For spatial effects we discuss a multivariate predictive process that reduces the computational burden by projecting the original process onto a subspace generated by realizations of the original process at a specified set of locations (or knots). We illustrate the proposed methods using a synthetic dataset with multivariate additive and dominant genetic effects and anisotropic spatial residuals, and a large dataset from a scots pine (Pinus sylvestris L.) progeny study conducted in northern Sweden. Our approaches enable us to provide a comprehensive analysis of this large trial which amply demonstrates that, in addition to violating basic assumptions of the linear model, ignoring spatial effects can result in downwardly biased measures of heritability.
Keywords: Bayesian inference, Cross-covariance functions, Genetic trait models, Heredity, Hierarchical spatial models, Markov chain Monte Carlo, Multivariate spatial process, Spatial predictive process
1. INTRODUCTION
Increasing international carbon credit markets and demand for wood fiber to supply burgeoning bioeconomies has spurred interest in improving forest ecosystem services. Much of this demand will be met by large plantation forests and, as a result, foresters seek methods to improve the productivity of these plantations. Tree breeding is one of the most important methods to improve productivity. Here, the focus is on increasing the genetic gain in future generations of certain traits by identifying and mating superior individuals that have high breeding values. In plantation settings, foresters are typically interested in improving traits which increase wood production (e.g., increasing both stem height and diameter). Depending on the anticipated use of the wood, there are other traits considered such as those that determine stem quality (e.g., the number and form of knots produced by branches from the stem). The principal objective is to reveal whether interrelationship among these various traits of primary or secondary interest are favorable or unfavorable for future selection (i.e., evaluating if traits are positively or negatively correlated depending on the objectives of the study). Henderson and Quaas (1976) introduced the multitrait animal (or tree or individual) model in quantitative genetics. Efficient breeding requires accurate estimates of the genetic variance and covariance parameters within the multitrait animal model and individual breeding values (Lynch and Walsh 1998).
Quantitative genetics studies the inheritance of polygenic traits, focusing upon estimation of additive genetic variance and heritability often estimated as the proportion of additiuve variance out of the total genetic and unexplained variation. A high heritability should result in a larger selection response, that is, a higher probability for genetic gain in future generations. A feature common to genetic trials in forestry and agriculture is the presence of systematic heterogeneity among observational units. If this spatial heterogeneity is neglected in the model, estimates of the genetic parameters might be biased (Dutkowski et al. 2002; Cappa and Cantet 2007). For spatially arranged observational units, heterogeneity typically results from small-scale environmental conditions (e.g., soil characteristics, microclimates, light availability, etc.) that might cause correlation among units as a function of distance and/or direction. While traditional randomized designs sought to negate any confounding spatial correlation, recent interest has shifted to models accounting for spatial correlation that, most often, incorporate spatial correlation through differencing or by constructing covariates using residuals from neighboring units (see, e.g., Zimmerman and Harville 1991; Cullis et al. 1998).
Hierarchical random effects models implemented through Markov chain Monte Carlo (MCMC) methods have recently gained popularity within quantitative genetics (see, e.g., Sorensen and Gianola 2002; Waldmann and Ericsson 2006; and references therein). Both single-component and blocked Gibbs samplers have been proposed. However, the former suffer from slow mixing and the latter often involve matrix decompositions whose complexity increases as O(n3) in the number of observations, n, at every iteration of the MCMC algorithm. These computationally intensive matrix calculations limit inference to small datasets insufficient for breeding purposes. Recently, Finley et al. (2009a) proposed a class of hierarchical models for a single outcome variable with genetic and spatial random effects that are suited for large datasets. A regular spectral decomposition of genetic covariance matrices together with a reduced-rank spatial process enable full Bayesian inference using MCMC methods.
The current article extends the work of Finley et al. (2009a) to a multivariate spatial setting, where georeferenced trees with known pedigrees have been observed. Multiple traits of breeding interest are measured on each tree. We posit that the variation within and between these traits can be explained by heredity, spatial variation (e.g., soil characteristics or small scale topographic attributes), and some unstructured measurement error. Here we focus upon three traits that are vitally important in the production of trees for lumber and biomass: height (H), diameter at 1.4 meters from the ground (D), and branch angle (B). The first two traits describe the amount of wood produced by a tree (i.e., the wood volume) and the third describes both the wood quality (i.e., due to knots resulting from branches connecting to the tree bole) and structural integrity of the tree (i.e., susceptibility to breakage due to heavy snow loadings). By estimating the variability of each trait’s additive and, to some extent, nonadditive effects while adjusting for their associations within a location, along with the breeding values, plantation tree nurseries are better able to choose among breeding stock to produce seed with the greatest genetic potential for the desired traits.
One approach builds hierarchical multivariate spatial models by exploiting conditional relationships (spatial regressions) between the variables (Royle and Berliner 1999). This can accrue some computational benefits (see, e.g., Banerjee, Carlin, and Gelfand 2004, section 7.2.2) and can be useful in situations where there is a natural chronology, or causality, for the conditioning. For genetic traits, however, such chronology is often absent or, at best, controversial and one seeks joint multivariate models (e.g., Gelfand et al. 2004; Jin, Banerjee, and Carlin 2007).
For the genetic effects we make use of separable association structures and derive a multivariate eigen-decomposition that separates heredity from association among the traits and produces significant computational benefits. For multivariate spatial dependence, a computationally feasible choice is “separable” or “intrinsic” association structures (see, e.g., Banerjee, Carlin, and Gelfand 2004, chapter 7). Here each trait is assigned the same spatial correlation function. It is unlikely that the strength and nature of association will be the same for the different traits and we seek more flexible correlation structures that accommodate a different spatial correlation function for each trait.
A “linear model of coregionalization” (Gelfand et al. 2004) yields arbitrarily rich multivariate correlation models, but its estimation for large datasets is onerous. A reduced-rank multivariate “kriging” model can help achieve dimension reduction. One possibility is to use low-rank smoothing splines that have been fairly conspicuous in univariate settings (Kamman and Wand 2003; Ruppert, Wand, and Carroll 2003; Crainiceanu, Diggle, and Rowlingson 2008). With multiple traits we require multivariate splines whose specifications may be awkward. A reduced-rank multivariate spatial process is arguably more natural and at least as flexible here with the nice interpretability of correlation functions (see Banerjee et al. 2008; Cressie and Johannesson 2008).
A key point here is to address the issue of systematic biases in the estimates of variance components that arise in low-rank kriging or spline models. Since most existing low-rank methods focus upon prediction and smoothing, this has remained largely unaddressed in the aforementioned literature. Here we offer further insight into this systematic bias and improve the predictive process model with a bias-adjustment that easily applies to multivariate applications. Again, the spatial process approach that we adopt here facilitates in understanding and rectifying these biases that, arguably, would have been more difficult to resolve with multivariate splines.
In Section 2, we develop the multivariate spatial models for the data. Section 3 discusses the dimension-reducing predictive process models, systematic biases in estimating variance components from such models and their remedy. Section 4 outlines the implementation details for the genetic and spatial effects. A synthetic data analysis and the scots pine analyses are presented in Section 5. Finally, Section 6 concludes the article with a brief discussion.
2. MULTIVARIATE SPATIAL MODELS FOR PROGENY TRIALS
For our subsequent development we consider a finite collection of spatial locations, say 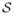 = {s1, …, sn}, referenced by Easting–Northing, where m phenotypic traits of a tree have been observed. Let Yj(si) denote the outcome variable corresponding to the jth trait at location si and Y(si) = (Y1(si), …, Ym(si))′ is the collection of the outcome measures in site si. Models for trait outcome measures, that is, the Yj(si)’s, should account for inherent association between the traits within each location and between observations across locations.
= {s1, …, sn}, referenced by Easting–Northing, where m phenotypic traits of a tree have been observed. Let Yj(si) denote the outcome variable corresponding to the jth trait at location si and Y(si) = (Y1(si), …, Ym(si))′ is the collection of the outcome measures in site si. Models for trait outcome measures, that is, the Yj(si)’s, should account for inherent association between the traits within each location and between observations across locations.
2.1 Multivariate Trait Model With Structured Random Effects
Finley et al. (2009a) (also see references therein) discussed structured random effect models for a single trait. Here we extend their work to multivariate traits over spatial domains. The following multivariate spatially referenced trait model
| (1) |
Here X(si)′ is an m × p block-diagonal matrix ( ), with lth diagonal element being the 1 × pl vector xl(si)′ and is a p × 1 vector of regression coefficients, with each βl a pl × 1 vector of regression coefficients corresponding to xl(si)′. This specifies the mean structure that accounts for large-scale variation in the outcome. Finer-scale variation in the dependent variable at each location si is modeled using three structured random effects. The first is an m × 1 vector of additive effects ai = (ai1, …, aim)′, the second is an m× 1 vector of dominance effects di, defined analogously to ai, and the third is an m × 1 vector of spatial processes w(si). Finally, we account for the unstructured residuals ε(si) that follow a zero-centered multivariate normal distribution with zero mean and an m × m diagonal matrix, denoted , as its dispersion matrix. Therefore, each outcome variable accommodates its respective variability in measurement—a phenomenon not uncommon in progeny trial datasets.
While ai and di play the role of correlated random effects to capture extraneous variation in the outcome attributable to additive and dominant effects, the spatial effects are assumed to arise from realizations of a spatial stochastic process. Conceptually, w(s) is an unknown function of space and exists at every every location s residing in our spatial domain 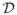 . The realizations of this process over a finite set of locations, such as
. The realizations of this process over a finite set of locations, such as 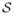 , yields spatially correlated random effects, but w(s), being an unknown, perhaps arbitrarily complex, function of space can also act as a surrogate for missing spatially informative predictors in the model. A challenge is to specify computationally feasible multivariate spatial processes. We discuss this in Section 2.3.
, yields spatially correlated random effects, but w(s), being an unknown, perhaps arbitrarily complex, function of space can also act as a surrogate for missing spatially informative predictors in the model. A challenge is to specify computationally feasible multivariate spatial processes. We discuss this in Section 2.3.
In modeling these multivariate effects, we need to model associations between the outcome variables within a site as well as association across sites. The m × 1 additive and dominance vector effects, ai and di, are assumed to have the m × m dispersion matrices Σa and Σd to model within-site association between the different types. These associations are assumed to remain invariant across the locations. One approach is to “separate” the correlation between the different trees from the association between the traits within a location. For example, we can specify cov{ai, aj} = ãijΣa, where Σa is a m × m dispersion matrix capturing the association attributable to additive effects between the traits. This specification produces valid probability models whenever Σa is positive definite and the A = {ãij} is a positive-definite matrix. The dominance effects di is modeled analogously as cov{di, dj} = d̃ijΣd where Σd is the dispersion matrix modeling association between the traits attributable to dominance effects and D = {d̃ij} is positive definite. Lynch and Walsh (1998) detail calculation of A and D from pedigree information.
Estimating (1) from large datasets can be a computationally onerous task as it involves the inverses of the dispersion matrices of , d defined analogously and w = (w(s1)′, …, w(sn)′)′. For the genetic effects, separability simplifies computations through the Kronecker products var{a} = A ⊗ Σa and var{d} = D ⊗ Σd. Modeling the multivariate spatial effects flexibly in a computationally feasible manner is, however, more challenging and we explore this in the subsequent sections.
2.2 Spatial Cross-Covariance Functions for Multivariate Processes
Spatially structured dependence is introduced in (1) through a multivariate (m × 1) spatial process w(s) ~ GP(0, Cw(·, ·; θ)) (e.g., Cressie 1993), where the cross-covariance function Cw(s1, s2) is defined to be the m × m matrix with (i, j)th entry cov{wi(s1), wj(s2)}. The cross-covariance function completely determines the joint dispersion structure implied by the spatial process. Specifically, for any n and any arbitrary collection of sites 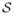 = {s1, …, sn} the nm × 1 vector of realizations w = (w(s1)′, …, w(sn)′)′ will have the variance–covariance matrix given by Σw, an nm × nm block matrix whose (i, j)th block is the cross-covariance matrix Cw(si, sj). Since Σw must be symmetric and positive definite, it is immediate that the cross-covariance function must satisfy (i) Cw(si, sj) = Cw(sj, si)′ for any pair of locations and (ii)
for all xi, xj ∈ ℜm\{0}.
= {s1, …, sn} the nm × 1 vector of realizations w = (w(s1)′, …, w(sn)′)′ will have the variance–covariance matrix given by Σw, an nm × nm block matrix whose (i, j)th block is the cross-covariance matrix Cw(si, sj). Since Σw must be symmetric and positive definite, it is immediate that the cross-covariance function must satisfy (i) Cw(si, sj) = Cw(sj, si)′ for any pair of locations and (ii)
for all xi, xj ∈ ℜm\{0}.
The first condition asserts that Σw be symmetric, although the cross-covariance function itself need not be. The second ensures the positive definiteness of Σw and is in fact quite stringent: this condition must hold for any arbitrary set of distinct locations. The above definition implies that Cw(s, s) is symmetric and positive definite. In fact, it is precisely the variance–covariance matrix for the elements of w(s) within site s. We say that w(s) is stationary if Cw(si, sj) = Cw(hij), where hij = si − sj, while w(s) is isotropic if Cw(si, sj) = Cw(||hij||), that is, the cross-covariance function depends only upon the distance between the sites. For stationary processes, condition (i) above implies that Cw(−h) = Cw(h)′, so that symmetric cross-covariance functions are even, that is, Cw(−h) = Cw(h). For isotropic processes, the cross-covariance function will be even and the matrix is necessarily symmetric.
One possibility for Cw(s1, s2) is a separable specification, ρ(s1, s2; φ)Σw, where Σw is an m × m covariance matrix between the traits and ρ(s1, s2; φ) is a spatial correlation function (e.g., Zimmerman and Harville 1991; Cressie 1993; Stein 1999). This implies var(w) = R(φ) ⊗ Σw, where R(φ) = {ρ(si, sj; φ)}. Correlation functions must be positive definite, that is, the matrix R(φ) must be positive definite for any finite collection of sites. When this function depends only upon the separation (difference) between the coordinates, we say the function, or the spatial process, is (weakly) stationary. Moreover, if the function depends only upon the distance between the locations, then we say that the process is isotropic. A well-known result due to Bochner (e.g., Cressie 1993; Stein 1999) characterizes all real-valued positive-definite functions as characteristic functions of symmetric random variables. A separable spatial association structure is rather restrictive here, assigning a single set of spatial correlation parameters φ to be shared by all the traits. This can be inappropriate: why should all the traits have, for example, the same strength of spatial association?
More general cross-covariance functions are not routine to specify since they demand that for any number of locations and any choice of these locations the resulting covariance matrix for the associated data be positive definite. Various constructions are possible. One possibility is the moving average approach of Ver Hoef, Cressie, and Barry (2004). The technique is also called kernel convolution and is a well-known approach for creating rich classes of stationary and nonstationary spatial processes (e.g., Higdon 2002). Yet another approach could attempt a multivariate version of local stationarity, extending ideas in Fuentes (2002). The primary characterization theorem for cross-covariance functions (Yaglom 1987) says that real-valued functions, say Cij(h), will form the elements of a valid cross-covariance matrix if and only if each Cij(h) has the cross-spectral representation Cij(h) = ∫ exp(2πιt′h) d(Cij(t)), where , with respect to a positive definite measure C(·), that is, the cross-spectral matrix is positive definite for any Borel subset B ⊆ ℜd. The cross-spectral representation provides a very general representation for cross-covariance functions. Matters simplify when Cij(t) is assumed to be square-integrable ensuring that a spectral density function cij(t) exists such that d(Cij(t)) = cij(t) dt. Now one simply needs to ensure that are positive definite for all t ∈ ℜd. Corollaries of the above representation lead to the approaches proposed by Gaspari and Cohn (1999) and Majumdar and Gelfand (2007) for constructing valid cross-covariance functions as convolutions of covariance functions of stationary random fields. For isotropic settings the development is very similar; customary notations use Cw(hij) where hij = ||si − sj|| is the distance between any two sites.
2.3 A Simple Construction of Valid Cross-Covariance Functions
Cross-spectral forms are often computationally infeasible resulting in integral representations with embedded parameters that are difficult to estimate. We motivate an alternative approach as follows. For the separable model, notice that we can write Cw(s1, s2; θ) = LΘ(s1, s2; φ)L′ where Σw = LL′ is a Cholesky factorization and Θ(s1, s2; φ) = ρ(s1, s2; φ)Im. More generally, any valid cross-covariance matrix can be expressed as Cw(s1, s2; θ) = L(s1)Θ(s1, s2; φ)L(s2)′. We call Θ(s1, s2) the cross-correlation function which must satisfy the two conditions for a cross-covariance function and, in addition, must satisfy Θ(s, s) = Im. This implies that Cw(s, s) = L(s)L(s)′ and L(s) identifies with a matrix square root (e.g., Cholesky) of Cw(s, s). A flexible and computationally feasible approach generalizes the spatial correlation function Θ(s1, s2; φ) as a diagonal matrix with ρk(s1, s2; φk), k = 1, …, m, as its diagonal elements. This incorporates a set of m correlation parameters, or even m different correlation functions, offering an attractive, easily interpretable and flexible approach. This approach resembles the linear model of coregionalization (LMC) as in, for example, Wackernagel (2006), Gelfand et al. (2004), and Zhang (2007). See, also, Reich and Fuentes (2007) for a Bayesian non-parametric adaptation.
For modeling L(s), without loss of generality one can assume that is a lower-triangular square root; the one-to-one correspondence between the elements of L(s) and Cw(s, s) is well known (see, e.g., Harville 1997, p. 229). Therefore, L(s) determines the association between the elements of w(s) within s. Choices for modeling L(s) include an inverse spatial-Wishart process for L(s)L(s)′ (Gelfand et al. 2004), element-wise modeling with Gaussian and log-Gaussian processes, or using parametric association structures suggested by the design under consideration. When stationarity is assumed, so that Cw(s, s) = Cw(0) and L(s) = L, we could assign a prior, for example, inverse Wishart, to LL′. Alternatively, instead of a lower-triangular Cholesky factor, we could opt for a spectral square root, setting L(s) = P(s) Λ (s)1/2, where Cw(s, s) = P(s) ΛP(s)′ is the spectral decomposition for Cw(s, s). We can further parameterize the orthogonal m × m orthogonal matrix function P(s) in terms of the p(p − 1)/2 Givens angles θij(s) for i = 1, …, p − 1 and j = i + 1, …, p (Daniels and Kass 1999). Specifically, where i and j are distinct and Gij(θij(s)) is the p × p identity matrix with the ith and jth diagonal elements replaced by cos(θij(s)), and the (i, j)th and (j, i)th elements replaced by ± sin(θij(s)), respectively. Given P(s) for any s, the θij(s)’s are unique within range (−π/2, π/2). These may be further modeled by means of Gaussian processes on a suitably transformed function, say .
2.4 Choice of Correlation Function
Finally, we turn to the choice of the spatial correlation functions. The nonstationary Matérn correlation function (Paciorek and Schervish 2006),
offers a very flexible choice for ρ(si, sj; φ(si, sj)). Here φ(si, sj) = (ν, Σ(si), Σ(sj)) varies across the domain (nonstationary), Γ(ν) is the Gamma function, κν is a Bessel function of the second kind and is the Mahalanobis distance between si and sj. When Σ(s) = Σ is constant over space, we have , where d(si, sj) = (si − sj)TΣ−1(si − sj). This yields a stationary but anisotropic process which can be parameterized as Σ = G(ψ) × Λ2GT(ψ) where G(ψ) is a rotation matrix with angle ψ and Λ is the diagonal matrix with positive diagonal elements, the λ’s. Thus, φ = (ν, ψ, Λ). The parameter ν controls the smoothness while the rate of spatial decay is controlled by the λ2’s. Finally, a stationary and isotropic process results with Σ = I, whence with Σ−1 = I and where φ = (ν, λ) with λ controlling spatial decay. This is further simplified by fixing ν = 0.5, yielding the familiar exponential correlation ρ(si, sj; λ) = exp(−λd(si, sj)).
Once a valid cross-covariance function is specified for a multivariate Gaussian process, the realizations of w(s) over the set of observed locations 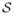 are given by N(0, Σw(θ)), where Σw(θ) is an mn × mn block matrix whose (i, j)th block is the m × m cross-covariance Cw(si, sj; θ), i, j = 1, …, n. Without further specifications, estimating (1) will involve factorizations that yield the inverse and determinant of the mn × mn matrix Σw(θ). Such computations invoke linear solvers or Cholesky decompositions of complexity O(n3m3), once every iteration, to produce estimates of θ. With large n, this is computationally infeasible and we resort to a class of dimension-reducing models in the next section.
are given by N(0, Σw(θ)), where Σw(θ) is an mn × mn block matrix whose (i, j)th block is the m × m cross-covariance Cw(si, sj; θ), i, j = 1, …, n. Without further specifications, estimating (1) will involve factorizations that yield the inverse and determinant of the mn × mn matrix Σw(θ). Such computations invoke linear solvers or Cholesky decompositions of complexity O(n3m3), once every iteration, to produce estimates of θ. With large n, this is computationally infeasible and we resort to a class of dimension-reducing models in the next section.
3. PREDICTIVE PROCESS MODELS FOR LARGE SPATIAL DATASETS
3.1 Approaches for Modeling Large Spatial Datasets: A Brief Review
Modeling large spatial datasets have received much attention in the recent past. Vecchia (1988) proposed approximating the likelihood with a product of appropriate conditional distributions to obtain maximum-likelihood estimates. Stein, Chi, and Welty (2004) adapt this effectively for restricted maximum likelihood estimation. Another possibility is to approximate the likelihood using spectral representations of the spatial process (Fuentes 2007). These likelihood approximations yield a joint distribution, but not a process that facilitate spatial interpolation. Furthermore, these approaches do not seem to extend easily to multivariate settings, especially in the presence of other structured dependencies such as genetic traits. Yet another approach considers compactly supported correlation functions (Furrer, Genton, and Nychka 2006; Kaufman, Schervish, and Nychka 2008; Du, Zhang, and Mandrekar 2009) that yield sparse correlation structures. More efficient sparse solvers can then be employed for kriging and variance estimation, but the tapered structures may limit modeling flexibility. Also, full likelihood-based inference still requires determinant computations that may be problematic.
In recent work Rue, Martino, and Chopin (2009) propose a promising Integrated Nested Laplace Approximation (INLA) algorithm as an alternative to MCMC that utilizes the sparser matrix structures to deliver fast and accurate posterior approximations. This uses conditional independence to achieve sparse spatial precision matrices that considerably accelerate computations, but relaxing this assumption would significantly detract from the computational benefits of the INLA and the process needs to be approximated by a Gaussian Markov Random Field (GMRF) (Rue and Held 2006). Furthermore, the method involves a mode-finding exercise for hyper-parameters that may be problematic when the number of hyperparameters is more than 10. Briefly, its effectiveness is unclear for our multivariate genetic models with different structured random effects and unknown covariance matrices as hyperparameters.
We opt for a class of models that emerge from representations of the spatial process in a lower-dimensional subspace and that easily adapt to multivariate processes. These are often referred to as low-rank or reduced-rank spatial models and have been explored in different contexts (Kamman and Wand 2003; Stein 2007, 2008; Banerjee et al. 2008; Crainiceanu, Diggle, and Rowlingson 2008; Cressie and Johannesson 2008). Many of these methods are variants of the so-called “subset of regressors” methods used in Gaussian process regressions for large datasets in machine learning (e.g., Rasmussen and Williams 2006). The idea here is to consider a smaller set of locations, or “knots,” say
, where the number of knots, n*, is much smaller than the number of observed sites, and to express the spatial process realizations over 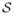 in terms of its realizations over the smaller set of knots. It is reasonable to assume that the spatial information available from the n sites could be summarized in terms of a smaller, but representative, set of locations—the knots. This is achieved by an optimal projection of w(s) using the information available from the knots.
in terms of its realizations over the smaller set of knots. It is reasonable to assume that the spatial information available from the n sites could be summarized in terms of a smaller, but representative, set of locations—the knots. This is achieved by an optimal projection of w(s) using the information available from the knots.
3.2 Predictive Process Models
A process-based reduced-rank formulation has recently been explored by Banerjee et al. (2008). We call w(s) the parent process and consider its realizations over an arbitrary set of n* locations
with n* ≪ n. Letting
be the realizations of the parent process over 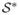 , we formulate a multivariate predictive process defined as
, we formulate a multivariate predictive process defined as
| (2) |
where 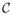 (s; θ)′ is a 1× n* block matrix, each block being m× m, Cw(s, s*; θ) is the ith block, and Σ*(θ) is the n*m × n*m dispersion matrix of w*, that is, with
as its (i, j)th block. Equation (2) reveals w̃(s) to be a zero mean m × 1 spatial process with cross-covariance matrix given by Cw̃(si, sj; θ) =
(s; θ)′ is a 1× n* block matrix, each block being m× m, Cw(s, s*; θ) is the ith block, and Σ*(θ) is the n*m × n*m dispersion matrix of w*, that is, with
as its (i, j)th block. Equation (2) reveals w̃(s) to be a zero mean m × 1 spatial process with cross-covariance matrix given by Cw̃(si, sj; θ) = 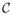 (si; θ)′Σ*(θ)−1
(si; θ)′Σ*(θ)−1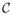 (sj; θ). The appeal here is that every spatial process (parent) model produces a corresponding predictive process version. Thus, the predictive process counterpart of the parent model in (1) is given by
(sj; θ). The appeal here is that every spatial process (parent) model produces a corresponding predictive process version. Thus, the predictive process counterpart of the parent model in (1) is given by
| (3) |
No new parameters are introduced and the cross-covariance function of the predictive process derives directly from that of the parent process.
Unlike many other knot-based methods, the predictive process model does not introduce additional parameters or in having to project the data onto a grid. Also, some approaches (e.g., Cressie and Johannesson 2008) require obtaining an empirical estimate of the covariance function. This may be challenging for the hierarchical models we envision here as the variability in the “data” is assumed to be a sum of different structured effects. Hence we cannot use variograms on the data to isolate the empirical estimates of the covariance function from the other random effects. These effects are identified through the second-stage specifications of their dispersion structures. In addition, the predictive process enjoys several desirable theoretical properties in terms of approximating the parent process. For instance, w̃(s), being a conditional expectation, can be regarded as an orthogonal projection of w(s) on an appropriate linear subspace (e.g., Stein 1999). Other properties are discussed in Banerjee et al. (2008).
3.3 Biased Estimates and the Rectified Predictive Process
For multivariate spatial processes, we have the following inequality:
| (4) |
where var{·} denotes the variance–covariance matrix and ≥ 0 indicates nonnegative definiteness. Equality holds only when s ∈ 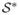 and thus the predictive process coincides with the parent process.
and thus the predictive process coincides with the parent process.
To understand the implications of the inequality in (4), let us momentarily focus upon a univariate predictive process. For the univariate process (i.e., with m = 1), let w(s) follow a spatial process with covariance function parameterized as
. The corresponding covariance function for w̃(s) is given by
, where
now represents the variance of the spatial process, r(s; φ) is the n* × 1 vector with the spatial correlation between s and
, that is,
, as its ith element and R*(φ) is the n* × n* correlation matrix of the realizations of the parent process over 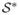 . In particular, the variance of the predictive process at any location s is given by
. The inequality in (4) implies
at every location s.
. In particular, the variance of the predictive process at any location s is given by
. The inequality in (4) implies
at every location s.
The above argument reveals the lower variability of the predictive process which leads to oversmoothing by predictive process models. This, in turn, would lead to overestimation by the predictive process model of the residual (unstructured) variance, τ2, as it attempts to absorb the remaining variability. Furthermore, note that the predictive process is inherently nonstationary. This calls for care in interpreting the parameter in predictive process models. In fact, it may be tempting to conclude that the oversmoothing caused by the predictive process would lead to underestimation of . The matter, however, is somewhat more subtle and in practice we tend to see an upward bias in the estimation of both the spatial and unstructured residual variance components (see Section 5.1).
To explain this, consider a single trait Y(s) in a univariate version (i.e., m = 1) of the model in (1). Let 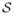 = {s1, …, sn} denote the set of observed data locations and assume (for convenience) that
= {s1, …, sn} denote the set of observed data locations and assume (for convenience) that 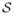 ∩
∩ 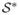 is empty, that is, the knots are completely exclusive of the data locations. Then, standard multivariate normal calculations reveal
is empty, that is, the knots are completely exclusive of the data locations. Then, standard multivariate normal calculations reveal
where w = (w(s1), …, w(sn))′,
is the n × n* cross-correlation matrix between the realizations over the locations and over the knots, and > 0 denotes positive definiteness. This implies that R(φ) + γ2 In > 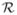 (φ)′R*(φ)−1
(φ)′R*(φ)−1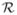 (φ) + γ2In (A > B denotes A − B is positive definite), where γ2 = τ2/σ2, or the noise–signal ratio. Since A > B implies B−1 > A−1 (Harville 1997, theorem 18.3.4, p. 434), we have (
(φ) + γ2In (A > B denotes A − B is positive definite), where γ2 = τ2/σ2, or the noise–signal ratio. Since A > B implies B−1 > A−1 (Harville 1997, theorem 18.3.4, p. 434), we have (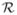 (φ)′R*(φ)−1
(φ)′R*(φ)−1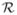 (φ) + γ2In)−1 > (R(φ) + γ2In)−1. Therefore,
(φ) + γ2In)−1 > (R(φ) + γ2In)−1. Therefore,
| (5) |
where u = y − Xβ − a − d. Here , and are n × 1 vectors of the observed outcomes, the additive and dominant effects respectively. Conditional upon the genetic effects and the regression parameters, Equation (5) reveals a systematic upward bias in likelihood-based estimates of the spatial variance parameter from the predictive process model as compared to the parent model. An analogous argument holds for estimates of τ2.
If and denote the likelihood-based estimates from the predictive process model (3) and the parent model (1), respectively, then using standard eigen-analysis (Appendix A) we obtain , where λmax(A) denotes the maximum eigen-value of the matrix A. Arguments analogous to the above can be used to demonstrate biases in variance component estimation in other low-rank spline models as well.
Using equivalence of Gaussian measures and fixed domain asymptotics Zhang (2004) proved that the spatial covariance parameters cannot all be estimated consistently in classical geostatistical settings with the isotropic Matérn correlation function (see Section 2.4), although the function σ2(λ2ν) is consistently estimable. However, assigning priors to such reparametrizations may not always be straightforward. From a Bayesian perspective, this implies that the effect of the priors on the inference of the variance components may not be easily overwhelmed by the data—even when the datasets are large.
In lieu of the above, the biases we point out are derived holding the parameters in φ as constant. In such settings, our simulation experiments reveal that both the parent model and the predictive process model often tend to estimate the regression coefficients β and γ2 = τ2/σ2 in a stable manner. However, while γ2 is estimated in a stable manner both τ2 and σ2 are systematically over-estimated. We point out that we regard the predictive process as a model in its own right, specifically suited for large spatial datasets, and not as an approximation to some model. In other words, its parameters should be interpreted with respect to (3) and not (1).
The key point is that the predictive process, univariate or multivariate, introduces nonstationarity in the spatial variance component. The variance component parameters, therefore, have different interpretations for stationary parent process models and their nonstationary predictive process counterparts. To adjust for the nonstationary spatial variance component, we need to introduce a nonstationary unstructured variance while making sure not to incur computational expenses.
Returning to the more general m× 1 multivariate processes, a rectification is obtained by adding a spatially adaptive “nugget” process to the predictive process w̃(s). More precisely, we introduce w̃ε̃(s) = w̃(s) + ε̃(s), where . We call w̃ε̃(s) the bias-adjusted predictive process and the model in (3) is now modified to
| (6) |
Notice that var{w̃ε̃(s)} = Cw(s, s; θ) = var{w(s)} and ε̃(s) also acts as a nonstationary adjustment to the residual process eliminating the systematic bias in variance components. Section 5.1 demonstrates these biases and their rectification.
We provide some brief remarks on knot selection (see Finley et al. 2009b, for details). With a fairly even distribution of data locations, one possibility is to select knots on a uniform grid overlaid on the domain. Alternatively, selection can be achieved through a formal design-based approach based upon minimization of a spatially averaged predictive variance criterion (e.g., Diggle and Lophaven 2006). However, in general, the locations are highly irregular, generating substantial areas of sparse observations where we wish to avoid placing knots, since they would be “wasted” and possibly lead to inflated predictive process variances and slower convergence. Here, more practical space-covering designs (e.g., Royle and Nychka 1998) can yield a representative collection of knots that better cover the domain. Another alternative is to apply popular clustering algorithms such as k-means or more robust median-based partitioning around medoids algorithms (e.g., Kaufman and Rousseeuw 1990). User-friendly implementations of these algorithms are available in R packages such as fields and cluster and have been used in spline-based low-rank kriging models (Ruppert, Wand, and Caroll 2003; Crainiceanu, Diggle, and Rowlingson 2008).
4. HIERARCHICAL MODEL SPECIFICATION AND IMPLEMENTATION
Let Y = (Y(s1)′, …, Y(sn)′)′ be mn×1 and X = (X(s1), …, X(sn))′ be mn × p. We assume that a and d follow zero-centered multivariate normal distributions with dispersion matrices A ⊗ Σa and D ⊗ Σd, respectively, where A = {aij} and D = {dij} and let F(θ) = 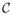 (θ)′Σ*(θ)−1, where
(θ)′Σ*(θ)−1, where 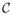 (θ)′ is the n × n* block-matrix whose (i, j)th block is the m × m matrix
with i = 1, …, n and j = 1, …, n*. Also, let Σε̃ denote the n × n block-diagonal matrix with ith block being the m × m matrix Cw(si, si; θ) −
(θ)′ is the n × n* block-matrix whose (i, j)th block is the m × m matrix
with i = 1, …, n and j = 1, …, n*. Also, let Σε̃ denote the n × n block-diagonal matrix with ith block being the m × m matrix Cw(si, si; θ) − 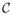 (si; θ)′Σ*(θ)−1
(si; θ)′Σ*(θ)−1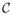 (si; θ). The hierarchical model corresponding to (6) can be written in terms of the joint density
(si; θ). The hierarchical model corresponding to (6) can be written in terms of the joint density
| (7) |
The matrices A and D are fixed, while Σa and Σd are assigned inverse-Wishart priors. The prior on β is N(β|μβ, Σβ) and is often taken as “flat,” setting to be the null matrix. One could either assume an inverse-Wishart prior for Ψ or take with , where IG is the inverse-Gamma distribution [mean = sεj/(rεj − 1)]. The spatial covariance parameters are collected into θ.
Assuming stationary cross-covariances [i.e., L(s) = L in Section 2.3], we have θ = {L, φ} and the cross-covariance matrix of the predictive process can be written as (In ⊗ L)Θ̃(θ)Σ*(θ)−1Θ̃(θ)′(In* ⊗ L′), where Θ̃(θ) is a block matrix with Θ(si, sj) as the (i, j)th block. We induce a prior on L by assigning p(LL′) = IW(η, SL), while all the elements in φ = (ν, ψ, Λ) are assigned weakly informative uniform priors (see Section 5 for details). Therefore, we write p(θ) in (7) as . Using matrix differentials, we can show that the induced prior on L is given by , where L = {lij}, and is the required jacobian of the transformation.
Estimation proceeds using MCMC employing a Gibbs sampler with Metropolis–Hastings (MH) steps (Gelman et al. 2004); see Appendix B for details. Briefly, the full conditional distributions for each of the parameters’ regression coefficients, the additive, dominant and spatial effects are all normal, while those for the matrices Σa, Σd, and Ψ are conjugate inverse-Wishart. The spatial variance covariance parameters θ = {L, φ} do not have closed-form full conditionals and are updated using MH steps. Observe that the likelihood [i.e., the last term in (7)] does not involve L or any of the spatial covariance parameters in θ and need not be evaluated for the Metropolis steps. These parameters are updated using Metropolis steps, possibly with block-updates (e.g., all the parameters in φ in one block and those in L in another). Typically, a random walk MH algorithm with (multivariate) normal proposals is adopted.
Gibbs updates still requires computing for updating a and a similar matrix for d, defined analogously, that are both nm × nm. A brute-force approach will not be computationally feasible, so we derive the following multivariate eigen-decomposition (Appendix C) to obtain
| (8) |
Here and are the spectral decompositions of A−1 and , and denotes the eigenvalues of A. Massive computational gains are achieved since the matrix A is fixed, so the spectral decomposition, which delivers the eigenvalues, needs to be evaluated only once, preceding the Gibbs sampler. All the other matrix inversions on the right hand expression in (8) involve the much smaller m × m matrices. The setting for Σd|· is exactly analogous.
A useful strategy for estimating spatial hierarchical models is to replace the conditionally independent likelihood in (7) with the marginalized likelihood obtained after integrating out the spatial effects (e.g., Banerjee et al. 2008; Finley et al. 2009b). However, this becomes problematic in the presence of other structured random effects as we are unable to arrive at forms such as (8). Instead, we integrate out the w* from (7) and design the Gibbs sampler to obtain the realizations of the predictive process w̃ε̃. More specifically, this leads to replacing N(w*|0, Σ*(θ)) × N(w̃ε̃|F(θ)w*, Σε̃) in (7) with N(w̃ε̃|0, Σw̃ε̃(θ)), where Σ w̃ε̃(θ) = Σε̃+ F(θ)Σ*(θ)F(θ)′.
Updating w̃ε̃ requires the inverse of Σw̃ε̃(θ)−1+ In ⊗ Ψ−1, an nm × nm matrix. Substantial computational benefits accrue from using the Sherman–Woodbury–Morrison (SWM) matrix identities (Henderson and Searle 1981). We first write , where is n*m × nm and M(θ) = Σ*(θ)−1 + V(θ)Σε̃ V(θ)′ is n*m × n*m. After some algebra, we obtain
where H(θ) is block-diagonal with [Ψ−1 + (Cw(si, si; θ) − 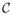 (si; θ)′Σ*(θ)−1
(si; θ)′Σ*(θ)−1 (si; θ))−1] as its m × m diagonal blocks, and J(θ) = M(θ) − V(θ)H(θ)−1V(θ)′. The determinant of Σw̃ε̃(θ) is required for updating the spatial covariance parameters in the MH steps. This can be evaluated avoiding nm × nm matrices using the identity (derived similarly to the SWM identity)
(si; θ))−1] as its m × m diagonal blocks, and J(θ) = M(θ) − V(θ)H(θ)−1V(θ)′. The determinant of Σw̃ε̃(θ) is required for updating the spatial covariance parameters in the MH steps. This can be evaluated avoiding nm × nm matrices using the identity (derived similarly to the SWM identity)
This setup still allows us to recover full posterior inference on the two components of w̃ε̃, viz. w* and ε̃. Specifically, given each posterior sample and θ(k), k = 1, …, K, we draw w*(l) from a multivariate normal distribution with mean and variance–covariance matrix Σw*|·(θ(k)) = Σ*(θ(k))−1 + F(θ(k))′Σε̃(θ(k))−1 × F(θ(k)))−1. Next, we can obtain w̃(k) = F(θ(k))w*(k). Finally, if desired, gives the posterior samples of ε̃.
5. ANALYSIS OF DATA
5.1 Synthetic Illustration of Bias-Adjustment
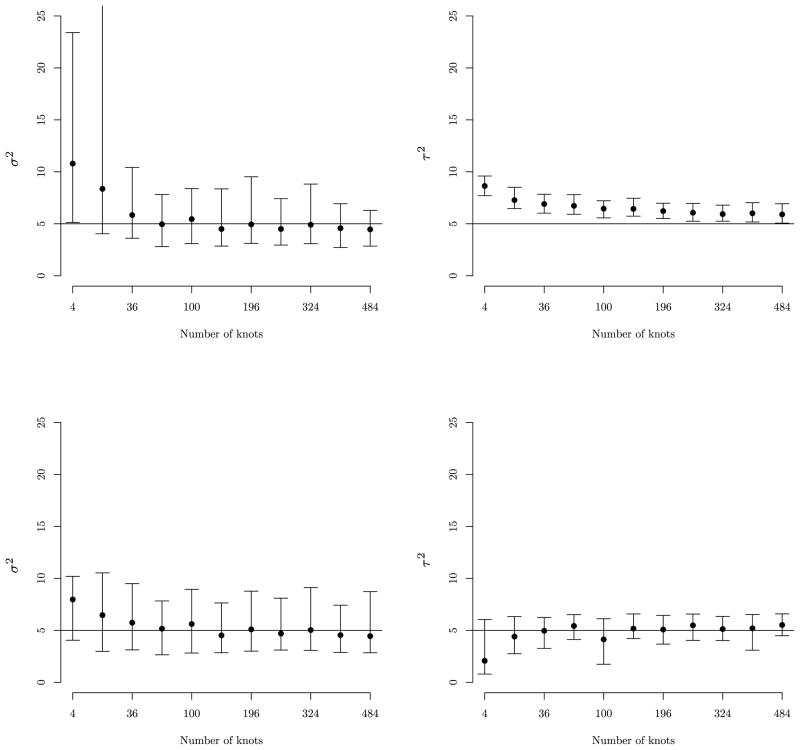
To supplement the discussion on the predictive process bias-adjustment in Section 3.3, we offer a brief analysis of a synthetic univariate dataset. This dataset comprises 500 locations distributed randomly within a unit square domain. The univariate outcome Y(s) is generated over these 500 locations from a zero-centered isotropic Gaussian process, with covariance , where , τ2 = 5, and φ = 12 [i.e., the spatial correlation drops to 0.05 at a distance of 0.25 units]. To illustrate the relationship between bias and knot intensity over the domain, we fit regular (unadjusted) and bias-adjusted predictive process models to these data based on 11 knot intensities between 0 and 500. For each model, , τ2, φ, and the model intercept β0 were estimated using three MCMC chains of post burn-in 25,000 iterations.
For brevity, we present only the results for the variance parameters of interest. Results for the unadjusted models are presented in the top subfigures in Figure 1 which illustrates the upward bias in and τ2 estimates as a function of knot intensity. Here, we see a large bias in and τ2 estimates when knot intensity is low. Increasing knot intensity does reduce the bias, but only marginally for τ2 which fails to recover the true parameter value even as the number of knots approaches the number of observations. In comparison, the bias-adjusted predictive process recovers the true and τ2 values even at low knot intensities, as illustrated in the bottom subfigures in Figure 1. For the analyses offered in Sections 5.2 and 5.3 only the bias-adjusted models are considered.
Figure 1.
The top row shows estimates of univariate model’s spatial variance σ2 (left) and residual variance τ2 (right) given increasing number of knots. The second row provides corresponding estimates from the bias-adjusted predictive process model.
5.2 Synthetic Data Analysis
One replicate of full-sib families (800 offspring in total) were constructed by crossing 50 unrelated parents according to a partial diallel design (see Kempthorne and Curnow 1961, for design details). The 850 individuals were assigned randomly to intersections of a 34 × 25 unit grid. The values of three traits of interest were generated for each individual. The synthetic population mean of each trait was described with an intercept and single slope coefficient [with corresponding variable drawn from N(0, 1)] and the multitrait random polygenic effects a and d were drawn from MVN(0, A ⊗ Σa) and MVN(0, D ⊗ Σd), respectively.
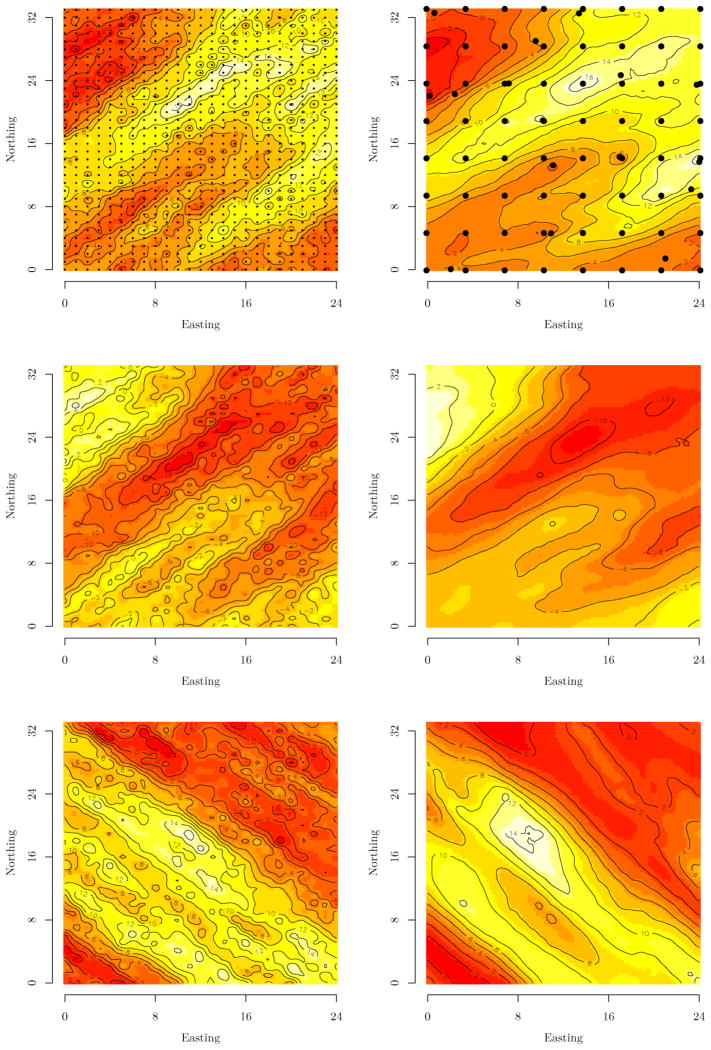
We estimate the hierarchical model in (7) using a stationary but anisotropic Matérn correlation function as described in Section 2.4. The mean fixed effects and lower-triangles of Σa and Σd are provided in the column labeled True within Table 1. In addition to the genetic effects, random spatial effects w were also added. Here, w was drawn from MVN(0, Σw), where Σw is the multivariate dispersion matrix defined in Section 2. The spatial processes associated with each response variable arise from an anisotropic Matérn correlation function. The estimates of the stationary process cross-covariance matrix (see Section 2.3), LL′ = Cw(0) ≡ Cw, along with the correlation functions’ parameters are provided in Table 2. These correlation parameters define the processes’ primary axis with an effective spatial range of ~21, that are oriented to ±45° and a second axis, perpendicular to the first, with an effective spatial range of ~9.5. Here, effective range is defined as the distance at which the correlation drops to 0.05 [i.e., when ν = 0.5 the effective range is − log(0.05)λ]. Further, the off-diagonal values in Cw define a strong negative correlation between the first and second process and zero correlation between the first two and the third process. The measurement error was drawn from MVN(0, In ⊗ Ψ), with Ψ = 5.0I3. The multitrait phenotypic values, Y, for each of the 800 individuals were then generated by model (1). The top left image in Figure 2 illustrates the planting grid and the interpolated surfaces of the synthetic spatial random effects follow down the left column. The phenotypic values of the 50 unrelated parents are assumed unknown and are therefore treated as missing data in the subsequent analysis.
Table 1.
Parameter credible intervals, 50% (2.5%, 97.5%) for the synthetic dataset nonspatial and spatial predictive process models. Credible intervals that do not include the parameter’s true value are bolded
| Parameter | True | Nonspatial | Anisotropic predictive process |
|
|---|---|---|---|---|
| 80 knots | 125 knots | |||
| β1,0 | 1 | 7.76 (6.77, 8.87) | 2.04 (0.93, 4.69) | 1.40 (−0.39, 3.75) |
| β1,1 | 5 | 5.32 (4.72, 5.89) | 5.28 (4.78, 5.74) | 5.22 (4.83, 5.66) |
| β2,0 | 10 | 3.72 (2.10, 5.09) | 10.32 (8.41, 11.90) | 13.20 (8.92, 16.60) |
| β2,1 | 15 | 14.99 (14.20, 15.64) | 14.99 (14.44, 15.51) | 15.01 (14.42, 15.60) |
| β3,0 | 20 | 27.61 (25.86, 29.38) | 20.48 (18.34, 22.85) | 22.18 (19.69, 30.61) |
| β3,1 | 25 | 25.07 (24.49, 25.64) | 24.95 (24.43, 25.49) | 24.91 (24.48, 25.39) |
| Σa;1,1 | 25 | 31.26 (22.35, 40.24) | 21.58 (15.39, 29.48) | 21.20 (13.35, 30.07) |
| Σa;1,2 | 25 | 26.61 (17.98, 35.77) | 25.60 (17.85, 35.20) | 22.17 (15.31, 30.08) |
| Σa;1,3 | 25 | 27.88 (19.37, 36.34) | 25.02 (17.39, 33.95) | 23.74 (15.45, 35.87) |
| Σa;2,2 | 50 | 55.65 (41.04, 73.43) | 50.37 (33.40, 69.94) | 42.03 (30.37, 56.79) |
| Σa;2,3 | 50 | 56.96 (40.23, 70.59) | 51.29 (33.84, 63.76) | 47.48 (35.00, 62.20) |
| Σa;3,3 | 75 | 75.43 (55.17, 93.81) | 77.19 (50.74, 94.29) | 84.04 (58.76, 99.72) |
| Σd;1,1 | 25 | 27.54 (19.90, 37.89) | 26.70 (20.56, 35.14) | 27.29 (20.68, 35.83) |
| Σd;1,2 | 25 | 19.51 (13.84, 28.78) | 22.39 (16.04, 28.31) | 24.27 (17.76, 31.56) |
| Σd;1,3 | 0 | −2.39 (−16.28, 8.38) | −2.15 (−5.87, 3.32) | 2.87 (−2.23, 6.62) |
| Σd;2,2 | 50 | 25.12 (7.69, 46.12) | 50.26 (36.58, 60.02) | 51.55 (41.26, 62.59) |
| Σd;2,3 | 0 | 0.59 (−8.71, 9.79) | −1.31 (−5.80, 8.63) | −1.32 (−8.38, 4.53) |
| Σd;3,3 | 4 | 1.79 (0.28, 9.57) | 0.93 (0.19, 2.70) | 1.31 (0.16, 4.69) |
Table 2.
Parameter credible intervals, 50% (2.5%, 97.5%) for the synthetic dataset nonspatial and spatial predictive process models (continuation of Table 1). Credible intervals that do not include the parameter’s true value are bolded
| Parameter | True | Nonspatial | Anisotropic predictive process |
||
|---|---|---|---|---|---|
| 80 knots | 125 knots | ||||
| Cw; 1,1 | 25 | – | 36.49 (15.51, 54.58) | 32.66 (4.89, 89.72) | |
| Cw; 1,2 | −25 | – | −31.98 (−55.09, −27.19) | −30.19 (−75.42, −16.18) | |
| Cw; 1,3 | 0 | – | −0.57 (−6.85, 1.37) | −1.16 (−12.03, 10.18) | |
| Cw; 2,2 | 26 | – | 31.11 (25.44, 34.09) | 30.92 (18.71, 40.23) | |
| Cw; 2,3 | 0 | – | 0.26 (−4.31, 2.21) | 1.42 (−9.23, 6.03) | |
| Cw; 3,3 | 25 | – | 55.79 (13.12, 108.21) | 38.26 (3.98, 68.04) | |
| Ψ1,1 | 5 | 13.58 (3.52, 24.65) | 1.46 (0.17, 5.93) | 1.54 (0.14, 6.89) | |
| Ψ2,2 | 5 | 43.64 (21.27, 67.24) | 0.85 (0.15, 7.22) | 4.15 (0.80, 11.31) | |
| Ψ3,3 | 5 | 31.02 (22.80, 41.83) | 1.56 (0.40, 7.07) | 5.40 (1.55, 19.22) | |
| ψw1 | 45° | – | 38.39 (22.06, 81.36) | 42.23 (8.08, 58.96) | |
| ψw2 | 45° | – | 57.30 (9.13, 84.63) | 47.21 (13.52, 86.00) | |
| ψw3 | 45° | – | 33.17 (1.03, 78.04) | 48.07 (4.87,76.89) | |
| Λw1,1 | 50 | – | 49.35 (47.94, 50.41) | 50.31 (49.17, 52.96) | |
| Λw1,2 | 10 | – | 9.79 (8.22, 11.03) | 10.05 (8.28, 11.72) | |
| Λw2,1 | 50 | – | 50.31 (49.00, 51.04) | 51.19 (49.67, 52.09) | |
| Λw2,2 | 10 | – | 9.67 (8.93, 10.32) | 9.81 (7.78, 10.68) | |
| Λw3,1 | 10 | – | 10.58 (9.84, 11.46) | 10.82 (9.49, 12.79) | |
| Λw3,2 | 50 | – | 49.60 (48.60, 50.38) | 49.51 (48.27, 53.06) | |
| νw1 | 0.5 | – | 0.57 (0.27, 0.92) | 0.62 (0.09, 1.00) | |
| νw2 | 0.5 | – | 0.68 (0.40, 0.90) | 0.60 (0.29, 0.84) | |
| νw3 | 0.5 | – | 0.36 (0.15, 0.55) | 0.68 (0.39, 1.16) | |
|
|
0.45 | 0.43 (0.32, 0.51) | 0.43 (0.31, 0.54) | 0.42 (0.27, 0.54) | |
|
|
0.48 | 0.45 (0.35, 0.55) | 0.49 (0.36, 0.63) | 0.43 (0.33, 0.53) | |
|
|
0.9 | 0.69 (0.55, 0.78) | 0.97 (0.88, 0.99) | 0.92 (0.76, 0.97) | |
Figure 2.
The left column shows the interpolated surfaces of the three response variables over the 850 synthetic data points (overlaid on top left surface). The right column shows the associated estimates of the random spatial effects based on 80 knots (overlaid on the top right surface).
The synthetic dataset is used to illustrate the extent to which the proposed methods can recover genetic parameters of interest. Assuming ignorance about the partitioning of the variance in Y, we assign an IW(m + 1, Im) prior to each genetic dispersion matrix, while each diagonal element of Ψ received an IG(2, ·) with the scale hyperparameters being set to yield a mean comparable with the residual variances from fixed effects only linear model. The maximum distance across the synthetic domain is about 40; hence, for the anisotropic Matérn models, we assign λi ~ U(0.5, 20) for i = 1, 2, 3, which corresponds to an effective range between 1.5 to 60 distance units (when ν = 0.5). Each smoothness and rotation parameter in ν and ψ were assigned U(0, 2) and U[π/2, −π/2) priors, respectively.
Four candidate models were considered: the nonspatial model with only genetic effects and three models with genetic effects and predictive process based random spatial effects. For the spatial models, we considered knot intensities of 80, 101, and 125. Knot configuration followed a lattice plus close pairs design (see Diggle and Lophaven 2006). This design consists of a regular k × k lattice of knots intensified by a set of m′ knots placed at random adjacent to the lattice knots—say within a circle having the knot point as center and a radius that is some fraction of the spacing on the lattice. This design is illustrated in the top right panel in Figure 2.
For each model, three MCMC chains were run for 25,000 iterations. The sampler was coded in C++ and leveraged threaded BLAS and LAPACK routines for matrix computations. The code was run on an Altix 350 SGI with two 64-bit 1.6 GHz Intel Itanium processors. Each chain of 25,000 iterations took less than 2 hours to complete. The CODA package in R (http://cran.r-project.org/web/packages/coda/) was used to diagnose convergence by monitoring mixing using Gelman–Rubin diagnostics and autocorrelations (Gelman et al. 2004, section 11.6). Acceptable convergence was diagnosed within 10,000 iterations and therefore 45,000 samples (15,000 × 3) were retained for posterior analysis.
Tables 1 and 2 offer parameter estimates and associated credible intervals for the nonspatial and predictive process models based on 80 and 125 knot intensities. The nonspatial model is not able to estimate correctly three of the six fixed effects (as identified in bold text in Table 1). Interestingly, despite the misspecification of the model (i.e., missing spatial effects), the additive and dominance dispersion matrices are estimated correctly, but for Σd;2,2. However, apportioning of spatial variation to pure error causes the 95% credible intervals of two diagonal elements in Ψ to not cover their true values, Table 2. This can cause a downward bias in the measures of heritability. The true heritability for the three traits are Σa;1,1/(Σa;1,1 + Σd;1,1 + Ψ1,1) = 25/(25 + 25 + 5) = 0.45, Σa;2,2/(Σa;2,2 + Σd;2,2 + Ψ2,2) = 0.48, and Σa;3,3/(Σa;3,3 + Σd;3,3 + Ψ3,3) = 0.9, and the nonspatial model estimates with 95% credible intervals are 0.43 (0.32, 0.51), 0.45 (0.35, 0.55), and 0.69 (0.55, 0.78), as provided in the Table 2. A similar downward bias is seen in the dominance ratio when calculated.
The spatial models provide improved estimates of the fixed and genetic random effects as well as estimates of heritability and dominance ratio. For example, at the 80 knot intensity the heritability estimates for the tree traits are 0.43 (0.31, 0.54), 0.49 (0.36, 0.63), and 0.97 (0.88, 0.99). We note that two parameters fall outside of the 95% credible interval in the 80 knot model (i.e., Σd;3,3 and Cw;1,2 in Tables 1 and 2, respectively). However, beyond the 80 knot intensity, all parameters are estimated correctly. The discrepancies in estimates for the lowest knot intensity are likely due to excessive smoothing of the random effects surfaces, as seen in the right column of Figure 2. Here, the response-specific anisotropic rotation and range of dependence are clearly matched between the true (left column) and estimated (right column); however, the 80 knot intensity is forced to smooth over some local variation. Again, more knots allow the model to better conform to the residual surface.
5.3 Scots Pine Data Analysis
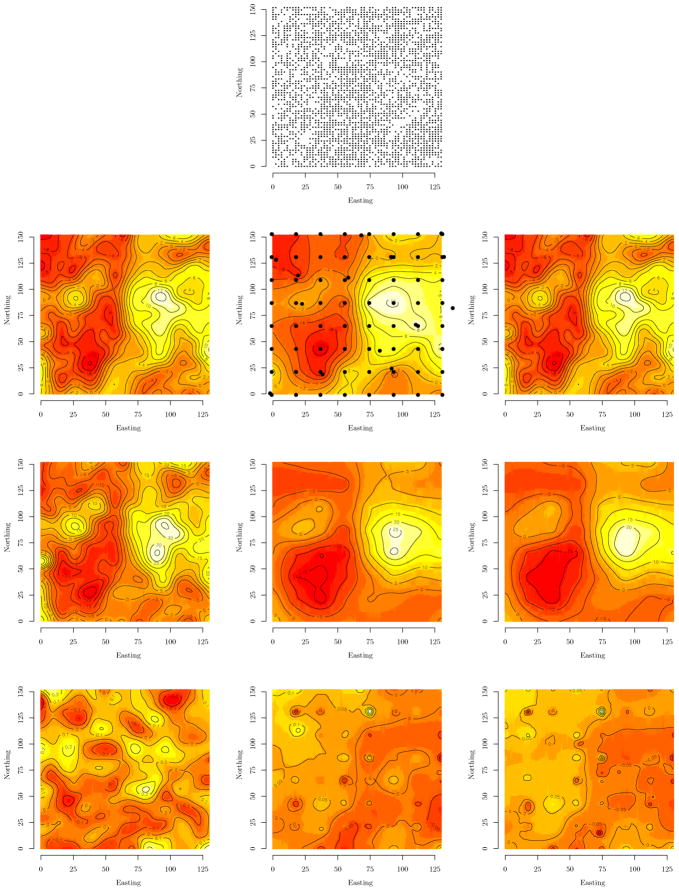
Field measurements from a 26-year-old scots pine (Pinus sylvestris L.) progeny study in northern Sweden serve as a trial dataset for our proposed methods. The original design established in 1971 (by Skogforsk; trial identification S23F7110264 Vindeln) consisted of 8160 seedlings divided into two spatially disjoint trial sites (i.e., denoted as northern and southern trial site). The analysis presented here focuses on the northern planting site. At planting, this site was subdivided into 105 8.8 m × 22 m blocks each containing 40 seedlings placed on a 2.2 m × 2.2 m grid. Parent stock was crossed according to a partial diallel design of 52 parent trees assumed unrelated and seedlings were planted out on the grids with unrestricteded randomization. In 1997, the 2598 surviving trees were measured for various traits of breeding interest, including diameter (D) (measured at 130 cm above ground), total tree height (H) and branch angle (B). The branch angle was scored such that an average tree was given score 5, and the range 1–9 reflect desirable to undesirable branch angle (i.e., horizontal branches were given low scores and upwards vertical branches high scores). The top panel in Figure 3 shows the location of the surviving trees use in the subsequent analysis.
Figure 3.
First row: Each point represents a scots pine tree location. The left column shows interpolated surface of nonspatial model residuals for stem height (H), diameter (D), and branch angle (A), respectively from top to bottom. The middle and right columns show associated estimated surfaces of spatial random effects for the 78 and 160 knot intensities modeled with the stationary isotropic Matérn correlation function. Note that to reduce clutter, the knots are overlaid on only the 78 knot H surface.
As in Section 5.2, we fit both nonspatial and spatial models with random spatial effects estimated with varying knot intensities. Specifically, we fit a nonspatial model with a single intercept and additive and dominance effects for each trait. Then random spatial effects were added with knot intensities of 78, 124, and 160. An initial exploratory analysis with an omnidirectional semivariogram of the nonspatial additive and dominance model residuals showed no evidence of an anisotropic process for any of the response variables; therefore, we consider only the spatial isotropic models here.
The left column of Figure 3 shows clear spatial association among the residuals, which is further affirmed by the empirical semivariogram for the trait of interest (not shown). Specifically, in the left column of Figure 3, we see that D and H are strongly associated and have a relatively longer spatial range compared to the surface for B. Inspection of the semivariogram supports this observation, showing an effective range of ~50 for D and H, while B’s spatial range is less than 10. This disparity in effective range suggests that a separable model, with a single spatial range parameter, would be inappropriate. Further, the relatively short spatial range of B, will require a knot configuration where the distance between knots is less than the residual spatial range. We again use the lattice plus close pairs design to help estimate small scale variability and local dependence. Using the isotropic Matérn correlation function, the decay parameters were set to φ ~ U(0.03, 3) and smoothness ν fixed at 0.05. This implies an effective spatial range of about 1 to 100 meters. Again, assuming ignorance about the partitioning of the variance in Y, we assign an IW(m + 1, Im) prior to each dispersion matrix with the exception of Ψ which received IG(2, ·) on the diagonal elements. The respective IG scale hyperparameters were set to the residual variance of the multivariate intercept-only model.
For each model, three MCMC chains were run for 25,000 iterations and 45,000 post burn-in samples (15,000 × 3) were retained for posterior analysis. The parameter estimates for the nonspatial additive and dominance model as well as the spatial models based on 78 and 160 knots are provided in Table 3. Here, overlapping 95% credible intervals suggest agreement among the nonspatial and spatial models’ fixed and genetic dispersion parameters (i.e., β, Σa, and Σd). However, the 160 knot model does generally estimate greater additive variance for several diagonal and off-diagonal elements in Σa.
Table 3.
Parameter credible intervals, 50% (2.5%, 97.5%) for the scots pine dataset nonspatial and spatial predictive process models
| Parameter | Nonspatial | Isotropic predicted process |
||
|---|---|---|---|---|
| 78 knots | 160 knots | |||
| βH,0 | 72.56 (70.45, 74.68) | 73.68 (70.70, 76.78) | 70.78 (68.39, 73.28) | |
| βD,0 | 119.64 (117.01, 122.63) | 119.19 (112.35, 126.14) | 120.31 (113.96, 126.22) | |
| βB,0 | 4.89 (4.75, 5.05) | 4.78 (4.59, 4.95) | 4.93 (4.66, 5.22) | |
| Σa;H | 29.38 (19.43, 38.19) | 21.25 (15.80, 30.79) | 33.12 (21.41, 46.57) | |
| Σa;H,D | 17.73 (6.76, 29.45) | 13.21 (3.79, 30.36) | 30.64 (10.81, 58.05) | |
| Σa;H,B | −0.99 (−1.79, −0.50) | −1.04 (−1.57, −0.66) | −1.05 (−2.09, −0.24 ) | |
| Σa;D | 43.30 (14.85, 67.31) | 52.94 (25.47, 119.27) | 89.23 (26.80, 129.97) | |
| Σa;D,B | −0.55 (−1.24, 0.12) | −0.18 (−1.14, 0.77) | −0.38 (−2.02, 1.57) | |
| Σa;B | 0.22 (0.17, 0.31) | 0.23 (0.16, 0.33) | 0.21 (0.15, 0.30) | |
| Σd;H | 1.22 (0.80, 2.46) | 7.91 (1.40, 15.15) | 2.56 (0.14, 9.29) | |
| Σd;H,D | 0.21 (−1.66, 1.09) | 0.37 (−1.35, 4.29) | −2.25 (−4.53, 5.84) | |
| Σd;H,B | −0.08 (−0.36, 0.08) | −0.13 (−0.64, 0.10 ) | −0.12 (−0.52, 0.02) | |
| Σd;D | 2.50 (1.48, 3.88) | 2.76 (1.70, 8.10) | 5.27 (2.35, 12.46) | |
| Σd;D,B | 0.05 (−0.22, 0.25) | −0.01 (−0.21, 0.38) | 0.17 (−0.23, 0.59) | |
| Σd;B | 0.19 (0.13, 0.29) | 0.18 (0.13, 0.26) | 0.17 (0.12, 0.25) | |
| Cw;H | – | 28.41 (19.88, 52.32) | 22.83 (19.69, 28.32) | |
| Cw;H,D | – | 0.56 (−0.12, 1.91) | 0.40 (−0.64, 2.24) | |
| Cw;H,B | – | −0.47 (−0.88, −0.01) | −0.28 (−0.80, 0.15) | |
| Cw;D | – | 64.29 (49.62, 86.89) | 62.39 (43.32, 163.98) | |
| Cw;D,B | – | 0.41 (−0.01, 1.05) | 0.23 (−0.46, 1.08) | |
| Cw;B | – | 0.16 (0.11, 0.19) | 0.16 (0.14, 0.24) | |
| ΨH | 134.37 (125.55, 142.99) | 87.93 (79.68, 96.13) | 86.08 (75.62, 94.43) | |
| ΨD | 954.38 (901.96, 1011.65) | 790.73 (725.24, 854.88) | 777.79 (716.86, 847.03) | |
| ΨB | 0.41 (0.32, 0.48) | 0.26 (0.19, 0.35) | 0.28 (0.20, 0.34) | |
| φwH | – | 0.05 (0.03, 0.06) | 0.07 (0.05, 0.09) | |
| φwD | – | 0.06 (0.03, 0.08) | 0.06 (0.03, 0.07) | |
| φwB | – | 0.59 (0.57, 0.61) | 0.60 (0.58, 0.62) | |
|
|
0.18 (0.12, 0.22) | 0.18 (0.14, 0.26) | 0.27 (0.18, 0.37) | |
|
|
0.04 (0.02, 0.07) | 0.06 (0.03, 0.14) | 0.10 (0.03, 0.15) | |
|
|
0.27 (0.21, 0.36) | 0.34 (0.26, 0.45) | 0.32 (0.24, 0.45) | |
We compare the candidate models using Deviance Information Criterion (DIC) (Spiegelhalter et al. 2002). With Ω denoting the generic set of parameters being estimated for each model, we compute , the expected posterior deviance, where L(Data|Ω) is the first stage likelihood from the respective model and the effective number of parameters (penalty) is , where Ω̄ is the posterior mean of the model parameters. The DIC is then computed easily from the posterior samples as with lower values indicating better models.
Table 4 provides DIC scores and associated pD for the non-spatial and predictive process models. Here, we see a large decrease in DIC moving from the nonspatial to the spatial models. As the knot intensity increases the models become more responsive to local variation in the residual surface and, as a result, we see the DIC scores incrementally decrease.
Table 4.
Model comparisons using the DIC criterion for the scots pine dataset
| Model | pD | DIC |
|---|---|---|
| Non-spatial | 1862.1 | 34,121.5 |
| Spatial bias-adjusted predictive process | ||
| 78 knots | 3574.6 | 31,682.1 |
| 124 knots | 3614.8 | 31,195.7 |
| 160 knots | 3685.7 | 31,098.4 |
From a breeding point of view, it is encouraging that the additive genetic correlations between branch angle (B) and growth traits (H and D) are small or negative, which means that faster growing trees develop more desirable branching characteristics (lower B scores denote more favorable branch angles). For instance the 160 knot model estimates the median and 95% credible intervals ρa;H,B = −0.40 (−1.00, −0.06) and ρa;D,B = −0.09 (−1.01, 0.25). For all models, we see a significant positive correlation between H and D, for example, the 160 knot model estimates ρa;H,D = 0.56 (0.45,0.75). Again, when the nonspatial model is forced to bring the spatial variation into the Ψ we see lower estimates of heritiability result, compared to those of the spatial models, specifically the 160 knot model. The panels in the middle and right columns of Figure 3 depict the estimated spatial effects surfaces for the 78 and 160 knot models. Comparing these columns to the left-most column, we see that global and local trends in the residual surfaces are captured even when the dimensionality of the problem is reduced from 2598 to 78 knots. Finally, the large disparity in the spatial range estimates, given in Table 3, supports the use of the nonseparable model.
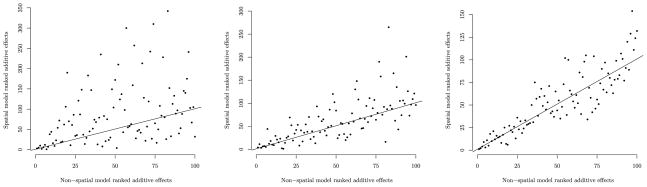
Ultimately, interest turns to estimation and subsequent ranking of the breeding values (i.e., additive effects) and specifically disparities in the ranking order given by the different models. Figure 4 offers a trait specific comparison between the ranking of the top 100 breeding values from the nonspatial and 160 knot predictive process models. Here, departures from the 1: 1 line indicate different ranking. These dispersed scatters suggest the addition of the spatial effect substantially influences breeding preference.
Figure 4.
Comparative ranking of the additive genetic effects for scots pine height (left), diameter (middle), and branch angle (right) from the nonspatial and 160 knot predictive process models. The x-axis plots the nonspatial model estimated position of the 100 highest ranked individuals and the y-axis plots the corresponding ranking from the spatial model. The straight line indicates no discrepancy in rank between the two models.
This analysis supports the idea that ignoring spatial variability in these models can lead to biased estimates of genetic variance parameters and ranking of breeding values, which could result in nonoptimal selection response and lower genetic gain (Magnussen 1993). Analysis of the scots pine data reveals that tree height and diameter are highly genetically correlated, have relatively low heritability, and display similar long distance effective spatial range. In contrast, branch angle seems to be more influenced by genetic components, as indicated by a higher heritability and shorter effective spatial range. Comparison between the nonspatial and spatial predictive process models also shows that the nonspatial model estimates lower heritability for all three traits, although the effect is less pronounced for height. Similar results have been found in other spatial genetic studies of trees. Zas (2006) reported that application of an iterative kriging method increased heritability in Pinus pinaster from a 95% confidence interval of 0.04–0.11 to 0.10–0.17 for diameter, and from 0.13–0.18 to 0.17–0.33 for height growth. In Cappa and Cantet (2007), the heritability of diameter at breast height in Eucalyptus globulus increased from 0.08 to 0.27 when a spatial spline component was included in the model. Moreover, both Zas (2006) and Cappa and Cantet (2007) showed a dramatic increase in accuracy of breeding values for spatial models, a trend that is likely also seen in the scots pine analysis presented here.
6. SUMMARY
While modeling of large spatial datasets have received much attention in the literature, estimation of variance components in reduced-rank kriging models have been explored less. Here we proposed computationally feasible multivariate spatial genetic trait models that robustly estimate variance components in datasets of sizes that normally are out of range because of prohibitive matrix computations. The process-based approach laid out here not only helps us identify a bias in the estimates of variance components that plagues most reduced-rank spline models but also leads to a remedy that neither involves additional computational burden, nor introduces any additional parameters. Future work may include extending predictive process models to space-time dynamic models and exploration of variance components in temporal contexts (see, e.g., Pourahmadi 1999; Ghosh et al. 2009).
Our simulated and field analysis reveals the spatial component to demonstrably improve the model’s ability to recapture correct estimates of fixed and genetic random effects as well as estimates of heritability and dominance ratios. The spBayes package in R (http://cran.r-project.org/web/packages/spBayes/) now include functions for these bias-adjusted multivariate predictive process models. Functions specifically geared for spatial quantitative genetics are being planned.
Finally, selecting the “knots” (Section 3) is an integral part of any reduced-rank spatial methodology. Since our field trials are conducted over a fairly regular configuration of plots, fixed knots on a regular grid provided robust inference. An arguably better strategy is to allow the knots to vary in their positions and/or their number. In particular, the knots can learn from the realizations and while they will change with each MCMC iteration, the number of adaptive knots required to deliver robust inference will likely be far smaller. These will form a part of future explorations.
Acknowledgments
The authors thank the editor, the associate editor and two anonymous referees for valuable suggestions that greatly helped to improve the manuscript. The work by the first two authors was supported in part by National Science Foundation grant DMS-0706870. The first author’s work was also supported in part by NIH grant 1-R01-CA95995.
APPENDIX A: BOUND FOR VARIANCE COMPONENT ESTIMATES
That the ratio
is bounded can be seen as follows. Let A = (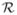 (φ)′R*(φ)−1
(φ)′R*(φ)−1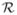 (φ) + γ2In)−1 and B = (R(φ) + γ2In)−1; thus, both are symmetric and positive-definite matrices. Then, there exists a nonsingular matrix C such that B = C′C. Writing Cu = z, where u = y − Xβ − a − d, we have
(φ) + γ2In)−1 and B = (R(φ) + γ2In)−1; thus, both are symmetric and positive-definite matrices. Then, there exists a nonsingular matrix C such that B = C′C. Writing Cu = z, where u = y − Xβ − a − d, we have
Note that the eigenvalues of C−1′AC−1 are all real and are the same as those of C−1C−1′A = B−1A (applying Theorem 21.10.1, pp. 545–546, Harville 1997) This shows that .
APPENDIX B: FULL CONDITIONAL DISTRIBUTIONS
The full conditional distributions are listed below:
For the full conditional distribution of the dispersion matrices, we have conjugate Wishart distributions. Assuming that Σa ~ IW(ra, ϒa) is the Inverse-Wishart prior, that is,
and Σd ~ IW(rd, ϒd), we obtain the full conditionals,
where A−1 = {ãij} and D−1 = {d̃ij}. Finally, with , with each , with mean rεj/sεj, we obtain the full conditionals
for j = 1, …, m, where aij, dij, and w̃ε̃j(si) are the jth elements of ai, di, and w̃ε̃(si) respectively.
APPENDIX C: DERIVATION OF (8)
In the above expressions, we can simplify as
where is the ith diagonal element of ΛA. Note that PA is the only n × n matrix here but it needs to be computed only once. Instead of having to invert a big mn × mn matrix, we now require n inversions of m × m matrices. Note that both Σa and Ψ will need to be updated in each iteration of the Gibbs.
Contributor Information
Sudipto Banerjee, Email: sudiptob@biostat.umn.edu, School of Public Health, University of Minnesota, Minneapolis, MN 55455.
Andrew O. Finley, Email: finleya@msu.edu, Departments of Forestry and Geography, Michigan State University, East Lansing, MI 48824.
Patrik Waldmann, Email: Patrik.Waldmann@genfys.slu.se, Thetastats, Uar-davgen, 91, SE-224 71 Lund, Sweden.
Tore Ericsson, Email: tore.ericsson@skogforsk.se, Skogforsk (the Forestry Research Institute of Sweden) and Associate Professor, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, Skogforsk, Box 3 918 21 Svar, Sweden.
References
- Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. Boca Raton, FL: Chapman & Hall/CRC Press; 2004. [Google Scholar]
- Banerjee S, Gelfand AE, Finley AO, Sang H. Gaussian Predictive Process Models for Large Spatial Datasets. Journal of the Royal Statistical Society, Ser B. 2008;70:825–848. doi: 10.1111/j.1467-9868.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappa EP, Cantet RJC. Bayesian Estimation of a Surface to Account for a Spatial Trend Using Penalized Splines in an Individual-Tree Mixed Model. Canadian Journal of Forest Research. 2007;37:2677–2688. [Google Scholar]
- Crainiceanu CM, Diggle PJ, Rowlingson B. Bivariate Binomial Spatial Modeling of Loa Loa Prevalence in Tropical Africa” (with discussion) Journal of the American Statistical Association. 2008;103:21–37. [Google Scholar]
- Cressie N. Statistics for Spatial Data. 2. New York: Wiley; 1993. [Google Scholar]
- Cressie N, Johannesson G. Fixed Rank Kriging for Very Large Spatial Data Sets. Journal of the Royal Statistical Society, Ser B. 2008;70:209–226. doi: 10.1111/j.1467-9868.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis BR, Gogel B, Verbyla A, Thompson R. Spatial Analysis of Multi-Environment Early Generation Variety Trials. Biometrics. 1998;54:1–18. [Google Scholar]
- Daniels MJ, Kass RE. Nonconjugate Bayesian Estimation of Covariance Matrices and Its Use in Hierarchical Models. Journal of the American Statistical Association. 1999;94:1254–1263. [Google Scholar]
- Diggle P, Lophaven S. Bayesian Geostatistical Design. Scandinavian Journal of Statistics. 2006;33:53–64. [Google Scholar]
- Du J, Zhang H, Mandrekar VS. Fixed-Domain Asymptotic Properties of Tapered Maximum Likelihood Estimators. The Annals of Statistics. 2009;37:3330–3361. [Google Scholar]
- Dutkowski GW, Silva JC, Gilmour AR, Lopez GA. Spatial Analysis Methods for Forest Genetic Trials. Canadian Journal of Forest Research. 2002;32:2201–2214. [Google Scholar]
- Finley AO, Banerjee S, Waldmann P, Ericsonn T. Hierarchical Spatial Modeling of Additive and Dominance Genetic Variance for Large Spatial Trial Datasets. Biometrics. 2009a;61:441–451. doi: 10.1111/j.1541-0420.2008.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley AO, Sang H, Banerjee S, Gelfand AE. Improving the Performance of Predictive Process Modeling for Large Datasets. Computational Statistics and Data Analysis. 2009b;53:2873–2884. doi: 10.1016/j.csda.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes M. Periodogram and Other Spectral Methods for Nonstationary Spatial Processes. Biometrika. 2002;89:197–210. [Google Scholar]
- Fuentes M. Approximate Likelihood for Large Irregularly Spaced Spatial Data. Journal of the American Statistical Association. 2007;102:321–331. doi: 10.1198/016214506000000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer R, Genton MG, Nychka D. Covariance Tapering for Interpolation of Large Spatial Datasets. Journal of Computational and Graphical Statistics. 2006;15:502–523. [Google Scholar]
- Gaspari G, Cohn SE. Construction of Correlation Functions in Two and Three Dimensions. The Quarterly Journal of the Royal Meteorological Society. 1999;125:723–757. [Google Scholar]
- Gelfand AE, Schmidt AM, Banerjee S, Sirmans CF. Nonstationary Multivariate Process Modeling Through Spatially Varying Coregionalization” (with discussion) Test. 2004;13:263–312. [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis. 2. Boca Raton, FL: Chapman & Hall/CRC Press; 2004. [Google Scholar]
- Ghosh SK, Lee H, Davis J, Bhave P. technical report. North Carolina State Univesity; 2009. Spatio-Temporal Analysis of Total Nitrate Concentrations Using Dynamic Statistical Models. [Google Scholar]
- Harville DA. Matrix Algebra From a Statistician’s Perspective. New York: Springer; 1997. [Google Scholar]
- Henderson CR, Quaas RL. Multiple Trait Evaluation Using Relatives Records. Journal of Animal Science. 1976;43:1188–1197. [Google Scholar]
- Henderson HV, Searle SR. On Deriving the Inverse of a Sum of Matrices. SIAM Review. 1981;23:53–60. [Google Scholar]
- Higdon D. Space and Space-Time Modeling Using Process Convolutions. In: Anderson C, Barnett V, Chatwin PC, El-Shaarawi AH, editors. Quantitative Methods for Current Environmental Issues. New York: Springer-Verlag; 2002. pp. 37–56. [Google Scholar]
- Jin X, Banerjee S, Carlin BP. Order-Free Coregionalized Lattice Models With Application to Multiple Disease Mapping. Journal of the Royal Statistical Society, Ser B. 2007;69:817–838. doi: 10.1111/j.1467-9868.2007.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamman EE, Wand MP. Geoadditive Models. Applied Statistics. 2003;52:1–18. [Google Scholar]
- Kaufman CG, Schervish MJ, Nychka DW. Covariance Tapering for Likelihood-Based Estimation in Large Spatial Data Sets. Journal of the American Statistical Association. 2008;103:1545–1555. [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- Kempthorne O, Curnow RN. The Partial Diallel Cross. Biometrics. 1961;17:229–250. [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Assoc; 1998. [Google Scholar]
- Magnussen S. Bias in Genetic Variance Estimates Due to Spatial Autocorrelation. Theoretical and Applied Genetics. 1993;86:349–355. doi: 10.1007/BF00222101. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Gelfand AE. Multivariate Spatial Modeling for Geostatistical Data Using Convolved Covariance Functions. Mathematical Geology. 2007;39:225–245. [Google Scholar]
- Paciorek CJ, Schervish MJ. Spatial Modelling Using a New Class of Nonstationary Covariance Functions. Environmetrics. 2006;17:483–506. doi: 10.1002/env.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourahmadi M. Joint Mean-Covariance Model With Applications to Longitudinal Data: Unconstrained Parameterisation. Biometrika. 1999;86:677–690. [Google Scholar]
- Rasmussen CE, Williams CKI. Gaussian Processes for Machine Learning. Cambridge, MA: The MIT Press; 2006. [Google Scholar]
- Reich BJ, Fuentes M. A Multivariate Nonparametric Bayesian Spatial Framework for Hurricane Surface Wind Fields. The Annals of Applied Statistics. 2007;1:249–264. [Google Scholar]
- Royle JA, Berliner LM. A Hierarchical Approach to Multivariate Spatial Modeling and Prediction. Journal of Agricultural Biological and Environmental Statistics. 1999;4:29–56. [Google Scholar]
- Royle JA, Nychka D. An Algorithm for the Construction of Spatial Coverage Designs With Implementation in SPLUS. Computers and Geosciences. 1998;24:479–488. [Google Scholar]
- Rue H, Held L. Gaussian Markov Random Fields: Theory and Applications. Boca Raton, FL: Chapman & Hall/CRC Press; 2006. [Google Scholar]
- Rue H, Martino S, Chopin N. Approximate Bayesian Inference for Latent Gaussian Models by Using Integrated Nested Laplace Approximations” (with discussion) Journal of the Royal Statistical Society, Ser B. 2009;71:1–35. [Google Scholar]
- Ruppert D, Wand MP, Caroll RJ. Semiparametric Regression. Cambridge, U.K: Cambridge University Press; 2003. [Google Scholar]
- Sorensen D, Gianola D. Likelihood, Bayesian and MCMC Methods in Quantitative Genetics. New York: Springer-Verlag; 2002. [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian Measures of Model Complexity and Fit” (with discussion) Journal of the Royal Statistical Society, Ser B. 2002;64:583–639. [Google Scholar]
- Stein ML. Interpolation of Spatial Data: Some Theory of Kriging. New York: Springer; 1999. [Google Scholar]
- Stein ML. Spatial Variation of Total Column Ozone on a Global Scale. The Annals of Applied Statistics. 2007;1:191–210. [Google Scholar]
- Stein ML. A Modeling Approach for Large Spatial Datasets. Journal of the Korean Statistical Society. 2008;37:3–10. [Google Scholar]
- Stein ML, Chi Z, Welty LJ. Approximating Likelihoods for Large Spatial Datasets. Journal of the Royal Statistical Society, Ser B. 2004;66:275–296. [Google Scholar]
- Vecchia AV. Estimation and Model Identification for Continuous Spatial Processes. Journal of the Royal Statistical Society, Ser B. 1988;50:297–312. [Google Scholar]
- Ver Hoef JM, Cressie NAC, Barry RP. Flexible Spatial Models Based on the Fast Fourier Transform (FFT) for Cokriging. Journal of Computational and Graphical Statistics. 2004;13:265–282. [Google Scholar]
- Wackernagel H. Multivariate Geostatistics: An Introduction With Applications. 3. New York: Springer-Verlag; 2006. [Google Scholar]
- Waldmann P, Ericsson T. Comparison of REML and Gibbs Sampling Estimates of Multi-Trait Genetic Parameters in Scots Pine. Theoretical and Applied Genetics. 2006;112:1441–1451. doi: 10.1007/s00122-006-0246-x. [DOI] [PubMed] [Google Scholar]
- Yaglom AM. Correlation Theory of Stationary and Related Random Functions. I. New York: Springer-Verlag; 1987. [Google Scholar]
- Zas R. Iterative Kriging for Removing Spatial Autocorrelation in Analysis of Forest Genetic Trials. Tree Genetics and Genomes. 2006;2:177–185. [Google Scholar]
- Zhang H. Inconsistent Estimation and Asymptotically Equal Interpolations in Model-Based Geostatistics. Journal of the American Statistical Association. 2004;99:250–261. [Google Scholar]
- Zhang H. Maximum-Likelihood Estimation for Multivariate Spatial Linear Coregionalization Models. Environmetrics. 2007;18:125–139. [Google Scholar]
- Zimmerman DL, Harville DA. A Random Field Approach to the Analysis of Field-Plot Experiments and Other Spatial Experiments. Biometrics. 1991;47:223–239. [Google Scholar]