Abstract
In vitro chemical modifications in proteins, introduced during sample preparation, can complicate mass spectra and increase the potential for false-positive identifications. While several in vitro protein modifications have been described previously, additional types of such modifications may exist. Here, we report discovery of four types of in vitro protein modifications, identified by HPLC/ MS/MS analysis and nonrestrictive protein sequence alignment by PTMap, an algorithm recently developed in our laboratory. These novel in vitro modifications included ethylation of aspartate and glutamate (+28 Da), esterification of aspartate and glutamate by glycerol (+74 Da), loss of 19 Da from lysine, and addition of 108 Da to cysteine. We confirmed that these modifications occurred in vitro and not in vivo in control experiments designed to avoid conditions likely to induce the modifications. We propose a plausible molecular mechanism for the −19 Da modification of lysine. Our study therefore conclusively identifies several novel in vitro protein modifications, suggests ways to avoid these modifications, and highlights the possibility of misidentification of peptides because of in vitro modifications.
Keywords: protein modifications, PTMap, automated database searching
Introduction
Over the past two decades, mass spectrometry technologies have advanced such that protein modifications of low stoichiometry, produced in vitro or in vivo, can now be detected. Analysis of proteolytic peptides by mass spectrometers with high resolution and high mass accuracy makes it possible to correlate detected peptide masses to known protein modifications.1-3 In addition, the recent parallel development of protein sequence alignment algorithms for tandem mass spectrometry (MS/MS) data allows researchers to identify all possible protein modifications, whether known or previously undescribed.4-6 At present, more than 300 types of protein modifications have been described,2 some of which are generated in vitro.
Several types of protein modifications are known to be induced in vitro under specific protein environments:7
(1) Acrylamide adduct. SDS-PAGE, a popular method for resolving proteins, induces formation of covalent protein adducts between proteins and chemicals in the gel. The cysteine side chain can react with unpolymerized free acrylamide in the matrix to form cysteinyl-S-β-propionamide, with a mass increase of 71 Da.8-10 Acrylamide adduction at the N-terminal amine has also been reported.10
(2) β-mercaptoethanol adduct. The cysteine side chain can also be modified in vitro by β-mercaptoethanol, leading to a mass increase of 76 Da.11
(3) Oxidation. Some amino acid residues, such as methionine, tryptophan, and cysteine, are easily oxidized in vitro.12
(4) Methylation. When methanol is used in staining and destaining buffers following SDS-PAGE, methylation can be induced at the side chains of aspartate and glutamate.13
(5) Deamidation. Deamidation can happen at the side chains of asparagine and glutamine, leading to the formation of aspartate and glutamate, respectively. The rate of this reaction is dramatically increased at high pH (pH > 10).14
Here, we report systematic analysis of protein modifications in bovine serum albumin (BSA) and histone H4 by HPLC/MS/ MS in combination with protein sequence alignment by PTMap, a recently developed program for comprehensive identification of protein modifications with high accuracy. Our analysis discovered four previously unreported types of in vitro protein modifications: ethylation of aspartate and glutamate, esterification of aspartate and glutamate by glycerol, loss of 19 Da from lysine, and addition of 108 Da to cysteine. Using various control experiments, we confirmed that the four modifications originated in vitro. Our study enriches our understanding of protein in vitro modifications, provides a procedure to avoid such modifications during protein handling, and provides a caution against assuming that protein modifications matching these mass shifts originate in vivo.
Material and Methods
Proteins
BSA was purchased from Sigma (St. Louis, MO) and calf thymus core histones were purified according to a procedure described previously.15 Briefly, cold, fat-free calf thymus (200 g) was sliced into 1-2 cm3 cubes, soaked in 160 mL of 0.5 M sucrose solution for 3 min, and then mixed with 1.44 L of homogenizing buffer (0.25 M sucrose and 3.3 mM CaCl2 solution). This solution was homogenized in 250 mL portions for 30 s twice in an Oster 12-speed blender at the speed of “easy clean”. The homogenate was filtered through two layers of cheesecloth. The filtrate was centrifuged at 1000g for 10 min to obtain a wet cell pellet. The pellet was resus-pended in 4 vol of hypotonic buffer (50 mM Tris-Cl, pH 7.9, 2.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.5 mM PMSF) with slow stirring for 30 min. The suspension was centrifuged at 1600g for 10 min to collect a pellet containing nuclei. The core histones were extracted from the nuclear pellet overnight using two washes of ~4 vol of 0.4 N H2SO4. The extract was dialysed sequentially against H2O and 50 mM Tris buffer (pH 7.3) for 8 h each. The core histones were resolved using 15% SDS-PAGE and visualized by colloidal Coommasie blue staining. To isolate histone H4 protein, the core histone preparation was subjected to HPLC separation using a C4 column. The histone H4 peak was collected, dried in a SpeedVac, and resolubilized in water.
Protein Tryptic Digestion
Three sets of experiments were designed to compare the effects of various reagents used in protein storage and in-gel digestion solutions on in vitro protein modifications. (1) BSA was incubated in 50 mM ammonium bicarbonate, pH 8, with or without 20% glycerol at 4 °C for 7 days, followed by in-solution digestion using porcine trypsin (Promega, Madison, WI) at a 1:50 enzyme/ substrate ratio. (2) BSA or calf thymus core histone H4 was digested either in solution or in gel. (3) BSA was separated by SDS-PAGE and in-gel digested following a standard protocol with varied washing solutions. SDS-PAGE gels were stained overnight with a colloidal Coomassie staining solution composed of 9 vol of G-250 stain solution (ProtoBlue, National Diagnostics, Atlanta, GA) and 1 vol of ethanol. Before in-gel digestion, gels were destained with water.
For protein in-gel digestion, the protein bands of interest were washed for 8 h, with 3 buffer exchanges, with a destaining solution in 1.5 mL microcentrifuge tubes on a Tomy MT-360 microtube mixer (Tomy Digital Biology, Japan) at medium speed. In our series of experiments, four different destaining buffers were used: destaining buffer I contained ethanol/water (50:50); destaining buffer II contained acetic acid/ethanol/water (10:50:40); destaining buffer III contained methanol/water (50:50); and destaining buffer IV contained acetic acid/methanol/ water (10:50:40).16 After destaining, protein bands were first rehydrated with a 20-min wash with 1 mL of water, then cut into 1 mm3 cubes, dehydrated with acetonitrile, and dried in a SpeedVac for 20 min. The dried gel pieces were swelled by covering with 50 mM ammonium bicarbonate containing 10 ng/μL trypsin and subjected to overnight digestion at 37 °C. The resulting tryptic peptides were extracted, dried, and cleaned in a C18 Zip-tip (Millipore, Bedford, MA) as previously described.16 Protein in-solution digestion was carried out by adding trypsin stock solution into a protein solution (in 50 mM ammonium bicarbonate, pH 8.0) at a 1:50 enzyme/substrate ratio and incubating overnight at 37 °C.
Nano-HPLC/MS/MS Analysis
Tryptic peptides cleaned by a C18 Zip-tip (Millipore, Bedford, MA) were reconstituted in buffer A solution (0.1% acetic acid, 2% acetonitrile, 97.9% H2O, v/v/v). Mass analyses were performed in an LTQ-2D ion trap mass spectrometer (Thermo Scientific, San Jose, CA) equipped with a nanoelectrospray ionization source, coupled with an Agilent 1100 nanoflow HPLC system. Two microliters of a peptide sample in buffer A was manually loaded onto a capillary column (10 cm length × 75 μm i.d.) home-packed with Jupiter C12 resin (4 μm particle size, 90 Å pore diameter; Phenomenex, Torrance, CA). Peptides were eluted from the column using a 100-min gradient from 8% to 90% buffer B (0.1% acetic acid, 90% acetonitrile, 9.9% H2O, v/v/v). Eluted peptides were directly electrosprayed into the LTQ mass spectrometer with spectra acquired in a data-dependent mode that cycled between MS and MS/MS of the 10 strongest precursor ions of the previous MS spectrum.
Protein Sequence Alignment and Manual Validation of Peptide Hits
Each LC/MS data set was searched against the corresponding protein sequence with PTMap, software developed in-house to identify all possible protein modifications, including previously undescribed PTMs.6 PTMap allows confident identification of protein modifications with mass shifts ranging from −100 Da to +200 Da in 1-Da increments. When searching, trypsin was specified as the proteolytic enzyme and 3 missed cleavages were allowed. Mass errors of precursor and product ions were set at (4 Da and (0.6 Da, respectively. All peptide identifications were manually validated with high stringency according to previously published criteria, and each modification site was exclusively localized in the peptide sequence by PTMap.17
Results
We report four types of protein modifications that occur in vitro during staining and destaining gels, in-gel digestion, or protein storage. These modifications were identified by HPLC/ MS/MS analysis of tryptic peptides using the PTMap algorithm, recently developed software that enables nonrestrictive sequence alignment for identifying all possible PTMs.6 Thus, the algorithm enables us to identify previously undescribed protein modifications in addition to known ones. Comparing protein modifications under different digestion methods (in-gel or in-solution digestion) or using different sample solutions (e.g., various buffers for gel staining and destaining, and protein storage solutions) enables us to distinguish in vitro protein modifications from ones originating in vivo.
To study possible in vitro protein modifications, we resolved BSA and histone H4 by SDS-PAGE. The gel was stained and destained with buffers containing ethanol to avoid possible in vitro methylation. The resulting tryptic peptides were analyzed by nano-HPLC/MS/MS followed by nonrestrictive sequence alignment by PTMap. Here, we highlight four types of novel in vitro modifications identifieds in BSA and histone H4 during the course of this study.
Ethylation of Aspartate and Glutamate Residues
Methanol is usually included in staining buffer for visualization of proteins on SDS-PAGE gels and in destaining buffer before ingel digestion. Methanol can cause methylation of aspartate and glutamate residues.13 To distinguish between in vitro methylation and in vivo methylation of acidic residues, ethanol was used to replace methanol in the staining and destaining buffers.13,18 However, this change induces in vitro ethylation.
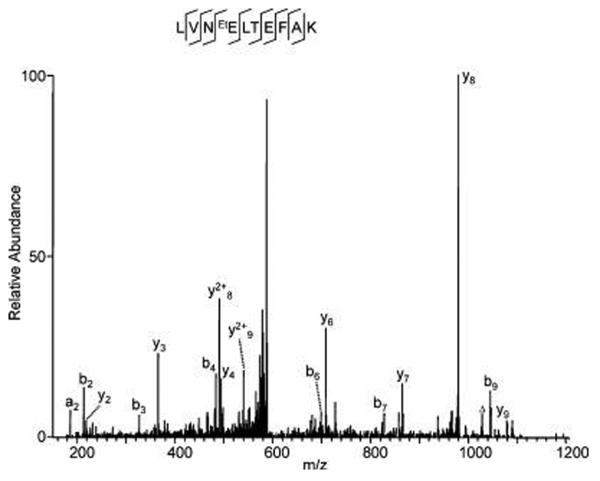
As an example, Figure 1 shows the MS/MS spectrum of a tryptic peptide from BSA that was ethylated in vitro. The ethylated peptide was identified in a sample that was destained with buffers containing ethanol, but not in samples destained without ethanol (Supporting Information T1), suggesting that ethylation happens in vitro instead of in vivo. The presence of acetic acid in the destaining buffer increases the number of ethylated tryptic peptides observed (Supporting Information T1), consistent with the ability of acidic conditions to induce esterification between alcohols and carboxylic acid moieties. We believe that the detected ethylation in tryptic peptides of BSA was produced in vitro because no ethylated peptides were present in samples that were destained with buffers devoid of ethanol (Supporting Information T1).
Figure 1.
Identification of ethylated glutamate in BSA. BSA (~300 ng) was resolved by SDS-PAGE and stained with colloidal Coomassie staining solution composed of 9 vol of G−250 stain solution (ProtoBlue, National Diagnostics, Atlanta, GA) and 1 vol of ethanol. The BSA protein band was destained with water and washed with a buffer containing acetic acid/ethanol/water (10: 50:40, v/v/v) before in-gel digestion and analysis by HPLC/MS/ MS. The labels “b” and “y” designate the N- and C-terminal fragment ions, respectively, of the peptide produced by breakage at the peptide bond in the mass spectrometer. The label “a” designates N-terminal fragments produced by breakage at the C-C bond adjacent to the peptide bond. The number represents the number of N- or C-terminal residues present in the peptide fragment. The label “Δ” designates “b”, “y” or “a” ions with water and/or ammonia loss. The same nomenclature system is used for all subsequent figures.
Likewise, the presence of acetic acid in destaining buffer containing methanol increases the number of tryptic peptides of BSA that were methylated (Supporting Information T1).
Esterification of Aspartate and Glutamate Residues by Glycerol
Glycerol is a solvent commonly used in buffers for purification and storage of proteins. Like methanol and ethanol, glycerol contains hydroxyl groups that may participate in esterification reactions with the side chains of aspartate and glutamate residues, leading to a mass shift of 74 Da. As expected, glycerol-modified tryptic peptides were identified when BSA was incubated with a buffer containing 20% glycerol (for examples, see Figure 2). The detected glycerol modifications must have occurred in vitro and not in vivo, because glycerol-modified peptides were not detected in BSA samples that were not treated with glycerol-containing buffer (data not shown).
Figure 2.
Modification of acidic residues by glycerol. BSA was incubated in 50 mM ammonium bicarbonate, pH 8, containing 20% glycerol at 4 °C for 7 days before in-solution digestion by trypsin. The tryptic peptides were analyzed by HPLC/MS/MS in an LTQ mass spectrometer. (A) A peptide modified by glycerol at a glutamate residue. (B) A peptide modified by glycerol at an aspartate residue. (C) The number of glycerol-modified peptides from BSA identified in our analysis, in the presence or absence of glycerol incubation.
Addition of 108 Da to Cysteine Residues
Cysteine residues are susceptible to diverse in vitro modifications, such as oxidation, or addition of molecules such as iodoacetamide, acrylamide, dithiothreitol, and β-mercaptoethanol.19,20 It remains unknown whether additional, undescribed modifications can occur at cysteine residues. Availability of the PTMap algorithm in our laboratory enables us to identify all possible modifications at cysteine residues, produced in vivo or in vitro.
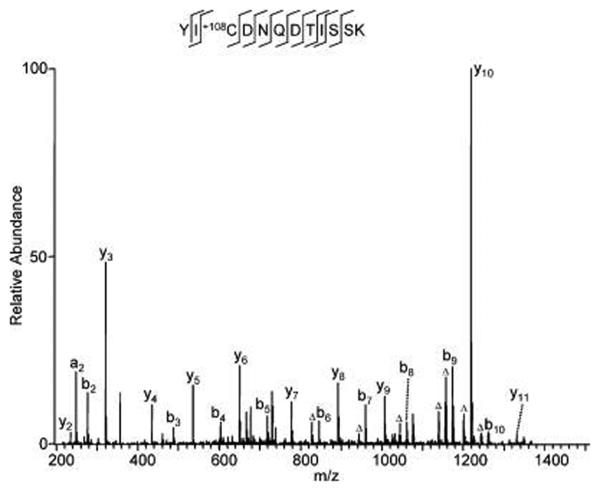
Our mass spectrometric analysis and subsequent nonrestrictive sequence alignment identified 6 cysteine residues within peptides of histone H4 that were modified by a chemical that leads to a mass shift of 108 Da (Figure 3, Supporting Information T2). To our knowledge, such a modification has not been described at cysteine before. Because the modification was not detected in tryptic peptides produced by in-solution digestion, the modification was considered to be an in vitro modification that happens during SDS-PAGE and/or gel-staining/destaining steps. The mass shift was determined with low mass accuracy that did not allow elemental composition to be determined. The structure of the modification moiety remains unknown.
Figure 3.
An in vitro-modified peptide with a mass shift of +108 at cysteine. BSA (300 ng) was resolved by SDS-PAGE and stained with colloidal Coomassie staining solution composed of 9 vol of G−250 stain solution (ProtoBlue, National Diagnostics, Atlanta, GA) and 1 vol of ethanol. The gel band was destained with buffer composed of ethanol/water (50:50, v/v) before in-gel digestion and HPLC/MS/MS analysis.
Loss of 19 Da from Lysine Residues
The side chain of lysine is subject to several in vivo post-translational modifications, including methylation, acetylation, biotinylation, ubiquitination, and sumoylation, that have pivotal roles in cell physiology and pathology. Our study of tryptic peptides from histone H4 identified 8 lysine residues that were modified by loss of 19 Da (Figure 4, Supporting Information F1). Because the modification was not identified in peptides that were produced by in-solution digestion, we believe the modification happens in vitro. To the best of our knowledge, an in vitro modification with this mass shift has not been reported before.
Figure 4.
Mechanistic study of the in vitro modification with a mass shift of −19 at lysine. (A) MS/MS of the unmodified histone H4 peptide, DNIQGITKPAIR. (B) MS/MS spectrum of the modified histone H4 peptide with loss of 19 Da at lysine, DNIQGIT−19KPAIR. (C) A possible molecular mechanism of in vitro protein modification resulting in a mass shift of −19 Da at lysine.
To explore the mechanism of this modification, we compared the peptide fragmentation pattern and retention time for the peptide of interest with and without the modification. The MS/MS spectra of the histone H4 peptide “DNIQITK*PAIR” with or without K7 modification (Figure 4A,B) showed changes in intensity among a series of y ions containing the modified lysine residue. The ion intensities of y6, y8, y92+, Y102+, Y112+ are much higher for the unmodified peptide (Figure 4A) than for the K-19 Da modified peptide (Figure 4B). In addition, the y5 and b7 ions are the most intense peaks in the spectrum of the modified peptide. Next, we noticed that the very strong y4 ion in the unmodified peptide's spectrum, produced by breakage at the K–P peptide bond, became much weaker in the modified peptide's spectrum, suggesting some significant structural change of the K–P residues. On the other hand, y5 became the strongest ion in the MS/MS spectrum of the modified peptide (Figure 4B). This change suggests that the most fragile peptide bond in the sequence is no longer the K–P bond but is the T–K bond, implying an increase in steric hindrance of the T–K bond upon modification. The HPLC retention time of the modified peptide was about 49.4 min, which is significantly delayed relative to that of the unmodified peptide (~39.5 min), suggesting an increase in hydrophobicity. Similar fragmentation pattern changes and a retention time delay were also observed for other histone H4 peptides with the Lys −19 Da modification.
Taken together, our results suggest several changes in the properties of peptides affected by this modification. First, the decrease in intensity of the y ions containing the unknown modification on the lysine side chain suggests that the modification causes the loss of basicity of the ε-amine group. Second, the change in identity of the most fragile peptide bond indicates that this modification probably introduced strong steric hindrance on the T–K peptide bond, which may lead to the same effect as proline. Third, the increase in retention time shows that the modified Lys side chain is more hydrophobic. On the basis of these observations, the known mass shift (−19 Da), and the cellular Lys metabolism pathway, we propose a reaction mechanism for the Lys −19 Da modification (Figure 4C). In the first step, the ε-amine group of lysine is oxidized by a free-radical reaction,12 which may be initiated by radicals in the SDS-PAGE gel. The reactive ε-aldehyde group would then be nucleophilically attacked by the R-amine group of the peptide backbone in a mechanism similar to glutamine or asparagine deamidation through a succinimide intermediate.21 The resulting 2-hydroxyl piperidine homologue would lose one molecule of water to form a more stable structure with a double bond. The −19 Da mass shift is the net effect of the loss of one nitrogen and five hydrogen atoms. While this model remains to be confirmed, it can explain four experimental observations arising from the modification: (1) the mass of the peptide is decreased by 19 Da; (2) the peptide becomes more hydrophobic; (3) the modified lysine residue introduces an additional steric constraint in a fashion similar to proline, thereby facilitating fragmentation; and (4) due to facile fragmentation N-terminal to the modified lysine, fragmentation N-terminal to proline is significantly reduced.
Discussion
We report discovery of four types of in vitro protein modifications. Addition of 108 Da to cysteine and removal of 19 Da from lysine have not been reported before. Modifications by glycerol and ethylation increase the masses of the substrate residues by 74 and 28 Da, respectively, which was reported previously for glutamate,22 but not for aspartate.
The identified in vitro modifications could be confused with other known protein modifications and, therefore, could result in misassignment during protein identification and mapping of modification sites by automatic sequence alignment. For example, ethylation of aspartate and glutamate leads to a mass shift of +28 Da, which is the same as formylation or the sum of two methylations. Therefore, when methylation is included in a search as a variable modification, ethylation of aspartate and glutamate could be misassigned as two methylation sites by protein sequence database algorithms. Such misassignment would happen more easily when the candidate methylation residues are adjacent to the ethylated aspartate and glutamate residues. Likewise, the loss of 19 Da from lysine has a similar mass shift to protein modifications associated with loss of water, and to changes from serine to dehydroalanine, from cysteine to formylglycine, from glutamate to pyroglutamate, and from aspartate to succinimide, all of which have a mass shift of −18 Da. Accordingly, these 6 types of protein modifications could be mistaken for one another due to their similar mass shifts. Two peptides that have the same sequence but undergo different modifications that have the same mass shift will have similar fragmentation patterns and will probably lead to similar statistical scores when identified by statistics-based protein alignment algorithms. Therefore, exclusive mapping of the protein modification site, complete assignment of all major product ions in the MS/MS spectrum, and exact matching of mass shifts are critical to ensure the accuracy of peptide identification and mapping of modification sites. This argument also suggests the importance of emphasizing unmatched peaks to remove false peptide identifications, as we described previously.17
Supplementary Material
Acknowledgment
This work was supported by NIH grants to Y.Z. (CA126832).
Footnotes
Supporting Information Available: Supplemental information on MS/MS spectra of in vitro modified peptides identified in BSA and Histone H4; figure of an in vitro modified peptide with a mass shift of −19 at lysine, and tables listing the number of peptides identified in BSA with either Glumethylation or Glu-ethylation induced by various destaining/ washing solutions during in-gel digestion, and the number of peptides identified with any one of four types of modifications following in-gel or in-solution tryptic digestion. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sadygov RG, Cociorva D, Yates JR., III Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat. Methods. 2004;1(3):195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 2.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods. 2007;4(10):798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Pevzner PA, Mulyukov Z, Dancik V, Tang CL. Efficiency of database search for identification of mutated and modified proteins via mass spectrometry. Genome Res. 2001;11(2):290–9. doi: 10.1101/gr.154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebler DC, Hansen BT, Davey SW, Tiscareno L, Mason DE. Peptide sequence motif analysis of tandem MS data with the SALSA algorithm. Anal. Chem. 2002;74(1):203–10. doi: 10.1021/ac0155512. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Chen W, Cobb M, Zhao Y. PTMaps—a novel sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. 2008 doi: 10.1073/pnas.0811739106. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson SD, Aebersold R. Mass spectrometric approaches for the identification of gel-separated proteins. Electrophoresis. 1995;16(10):1791–814. doi: 10.1002/elps.11501601299. [DOI] [PubMed] [Google Scholar]
- 8.Clauser KR, Hall SC, Smith DM, Webb JW, Andrews LE, Tran HM, Epstein LB, Burlingame AL. Rapid mass spectro-metric peptide sequencing and mass matching for characterization of human melanoma proteins isolated by two-dimensional PAGE. Proc. Natl. Acad. Sci. U.S.A. 1995;92(11):5072–6. doi: 10.1073/pnas.92.11.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.le Maire M, Deschamps S, Moller JV, Le Caer JP, Rossier J. Electrospray ionization mass spectrometry on hydrophobic peptides electroeluted from sodium dodecyl sulfate-polyacrylamide gel electrophoresis application to the topology of the sarcoplasmic reticulum Ca2+ ATPase. Anal. Biochem. 1993;214(1):50–7. doi: 10.1006/abio.1993.1455. [DOI] [PubMed] [Google Scholar]
- 10.Haebel S, Jensen C, Andersen SO, Roepstorff P. Isoforms of a cuticular protein from larvae of the meal beetle, Tenebrio molitor, studied by mass spectrometry in combination with Edman degradation and two-dimensional polyacrylamide gel electrophoresis. Protein Sci. 1995;4(3):394–404. doi: 10.1002/pro.5560040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klarskov K, Roecklin D, Bouchon B, Sabatie J, Van Dorsselaer A, Bischoff R. Analysis of recombinant Schistosoma mansoni antigen rSmp28 by on-line liquid chromatography-mass spectrometry combined with sodium dodecyl sulfate polyacrylamide gel electrophoresis. Anal. Biochem. 1994;216(1):127–34. doi: 10.1006/abio.1994.1016. [DOI] [PubMed] [Google Scholar]
- 12.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25(3–4):207–18. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 13.Sprung R, Chen Y, Zhang K, Cheng D, Zhang T, Peng J, Zhao Y. Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J. Proteome Res. 2008;7(3):1001–6. doi: 10.1021/pr0705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarioglu H, Lottspeich F, Walk T, Jung G, Eckerskorn C. Deamidation as a widespread phenomenon in two-dimensional polyacrylamide gel electrophoresis of human blood plasma proteins. Electrophoresis. 2000;21(11):2209–18. doi: 10.1002/1522-2683(20000601)21:11<2209::AID-ELPS2209>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 15.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2(6):1445–57. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Zhang W, Kho Y, Zhao Y. Proteomic analysis of integral plasma membrane proteins. Anal. Chem. 2004;76(7):1817–23. doi: 10.1021/ac0354037. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 2005;4(3):998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 18.Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal. Chem. 2008;80(5):1721–9. doi: 10.1021/ac7021025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creasy DM, Cottrell JS. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2(10):1426–34. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 20.Steen H, Mann M. Similarity between condensed phase and gas phase chemistry: fragmentation of peptides containing oxidized cysteine residues and its implications for proteomics. J. Am. Soc. Mass Spectrom. 2001;12(2):228–32. doi: 10.1016/S1044-0305(00)00219-1. [DOI] [PubMed] [Google Scholar]
- 21.Stephenson RC, Clarke S. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 1989;264(11):6164–70. [PubMed] [Google Scholar]
- 22.Harwig SS, Chen NP, Park AS, Lehrer RI. Purification of cysteine-rich bioactive peptides from leukocytes by continuous acid-urea-polyacrylamide gel electrophoresis. Anal. Biochem. 1993;208(2):382–6. doi: 10.1006/abio.1993.1065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.