Abstract
Novel immune-type receptors (NITRs) are immunoglobulin variable (V) domain-containing cell surface proteins that possess characteristic activating/inhibitory signaling motifs and are expressed in hematopoietic cells. NITRs are encoded by multi-gene families and have been identified in bony fish species. A single gene cluster, which encodes 36 NITRs that can be classified into 12 families, has been mapped to zebrafish chromosome 7. We report herein the presence of a second NITR gene cluster on zebrafish chromosome 14, which is comprised of three genes (nitr13, nitr14a and nitr14b) representing two additional NITR gene families. Phylogenetic analyses indicate that the V domains encoded by the nitr13 and nitr14 genes are more similar to each other than any other zebrafish NITR suggesting that these genes arose from a tandem gene duplication event. Similar analyses comparing zebrafish Nitr13 and Nitr14 to NITRs from other fish species indicate that the nitr13 and nitr14 genes are phylogenetically related to the catfish IpNITR13 and IpNITR15 genes. Sequence features of the chromosomal region encoding nitr13 suggest that this gene arose via retrotransposition.
Keywords: multi-gene family, cytogenetics, retrotransposition
INTRODUCTION
The novel immune-type receptors (NITRs) are a large family of cell surface receptors that have been identified in bony fish species (Strong et al. 1999;Yoder et al. 2001;Hawke et al. 2001;Yoder et al. 2002;Piyaviriyakul et al. 2006;Evenhuis et al. 2007). NITRs share structural and signaling similarities with the mammalian natural killer (NK) receptors encoded by the leukocyte receptor complex (e.g. KIRs) and possess one or two extracellular immunoglobulin (Ig) domains of the variable (V) and intermediate (I) type. Most NITRs possess cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs) and are classified as inhibitory forms. A smaller number of NITRs are considered to be activating receptors, which possess a positively charged residue within the transmembrane domain allowing partnering with and signaling via DAP12 that possesses a cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM; Yoder et al. 2007;Wei et al. 2007). The members of a third class of NITRs lack a transmembrane domain and are likely secreted proteins with unknown function (Yoder et al. 2004;Evenhuis et al. 2007). NITR transcripts are detected in multiple hematopoietic lineages including NK-like and cytotoxic T cells supporting the hypothesis that NITRs function in immunity (Hawke et al. 2001;Evenhuis et al. 2007).
Thirty-six NITR genes, which can be classified in 12 families, have been identified in a single gene cluster on zebrafish chromosome 7 (Yoder et al. 2004). We report herein the identification of an additional NITR gene cluster and confirm its presence on zebrafish chromosome 14 by cytogenetic mapping. The NITR gene cluster on chromosome 14 includes three genes (nitr13, nitr14a and nitr14b) defining two additional NITR gene families. Transcripts from nitr13 and nitr14a encode proteins that are not identifiable as inhibitory or activating forms. Phylogenetic comparisons with all zebrafish NITRs show that Nitr13 and Nitr14 proteins are most similar to each other suggesting that they share a common ancestral gene. Genomic sequence features of the nitr13 locus are consistent with this gene having arisen via retrotransposition.
MATERIALS AND METHODS
Rapid amplification of cDNA ends (RACE)
Total RNA was purified from wild-type (AB strain) adult zebrafish spleen and kidney (RNABee, Tel-test, Friendswood, TX) and RACE performed (GeneRacer™, Invitrogen, Carlsbad, CA) with Titanium™ Taq polymerase (BD Bioscience, San Jose, CA) as described (Yoder et al. 2001;Yoder et al. 2004). A single nitr13 and two alternatively spliced nitr14a cDNAs were amplified via 3′ RACE using nested, overlapping primers (and touch-down PCR as described (Panagos et al. 2006). The sequence of the primary and nested primers for nitr13 RACE were: 5′-ATGAGAATCGTGTGGATTTCTCTCATG-3′ and 5′-GGATTTCTCTCATGCTTCTATGCAGG-3′, respectively. The sequence of the primary and nested primers for nitr14a RACE were: 5′-ATGATTCTCTGGGCATTTGTTACTG-3′ and 5′-CATTTGTTACTGTTCTTTGTGTTGCGC-3′, respectively. All cDNAs were cloned into pGEM®-T (Promega, Madison, WI) or pCR®II-TOPO (Invitrogen, Carlsbad, CA) and sequenced.
Percent identity plots
Polymorphic variations between two alleles for the nitr13/nitr14/fgfrl1b gene cluster were visualized using PipMaker software (Schwartz et al. 2000). Partial DNA sequence (135,205 nucleotides) from zebrafish BAC DKEY-149K8 (GenBank AL954843) was used as the reference for comparison to sequence from zebrafish BAC RP71-24B12 (GenBank BX247870).
Fluorescence in situ hybridization (FISH)
FISH was performed as described previously (Freeman et al. 2007). Briefly, zebrafish chromosome preparations were dropped onto slides and allowed to dry overnight at 37°C. BAC DNA was labeled with either spectrum orange or spectrum green dyes (Abbott Vysis, Des Plaines, IL). Following slide pretreatment, the DNA probes were denatured for 3 min. at 70°C, added onto the slides under a coverslip, sealed with rubber cement and incubated in a dark humidified chamber at 37°C for at least 24 hours. Following incubation, slides underwent post-hybridization washes and then were counterstained with DAPI. An Olympus BX-51 fluorescence microscope, equipped with narrow band pass filters for spectrum orange and spectrum green dyes, was used to visualize the hybridization patterns. Images were captured using a Photometrics KAF1400 CCD camera and Applied Imaging Genus Software system.
Phylogenetic analyses
Predicted leader and transmembrane domains of protein sequences were identified with SMART software (http://smart.embl-heidelberg.de/; Letunic et al. 2004) and proteins aligned by ClustalW (http://www.ebi.ac.uk/clustalw/#; Chenna et al. 2003). Neighbor-joining trees (Saitou and Nei 1987) were constructed from pairwise Poissin correction distances with 2000 bootstrap replication by MEGA2.1 software (Kumar et al. 2001).
RESULTS AND DISCUSSION
Identification of zebrafish nitr13 and nitr14 genes and cDNAs
Known zebrafish NITR V regions were artificially concatenated and used as a search string to query version 5.0 of the zebrafish genome using the tBLASTn algorithm (Gertz et al. 2006). A number of hits were noted outside of the NITR locus on chromosome 7. Several gene segments mapping to BAC clone DKEY-149K8 (GenBank AL954843) and RP71-24B12 (GenBank BX247870) were identified as putative NITRs. Candidate exons for signal peptide leader sequences were identified 5’ of the V domains and a 3' rapid amplification of cDNA ends (RACE) strategy was employed to clone cDNA sequences. A single nitr13 cDNA and two alternatively spliced nitr14a cDNAs were identified in this way. A second candidate nitr14 gene, designated nitr14b, was inferred from genomic sequence encoded in BAC DKEY-149K8. As with all characterized NITRs, Nitr13, Nitr14a and Nitr14b possess authentic V domains as well as segments exhibiting significant sequence identity with Ig-type joining (J) regions (FGXGTXLXI/L) (Fig. 1a and Litman et al. 2001). A second Ig domain of the I type is encoded 3' to the V regions of most NITRs in zebrafish and other species. However, like NITRs 3b, 3d, 6, 7, 10 and 11, encoded in the NITR gene cluster on zebrafish chromosome 7, the three NITRs on chromosome 14 lack I domains C-terminal to the V region. Notably, a sequence segment encoding an I domain within the 3' untranslated region of nitr13 was identified (see below).
Figure 1. Nitr13 and Nitr14 sequences.
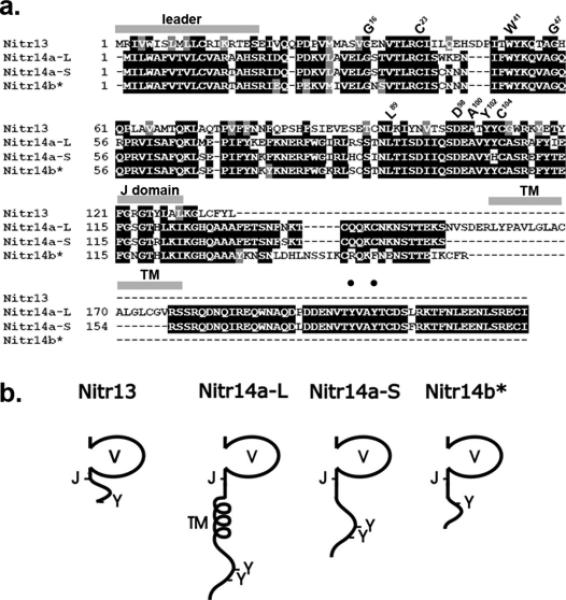
(a) The peptide sequences encoded by the nitr13 (Nitr13), nitr14a-Long (Nitr14a-L) and Nitr14a-Short (Nitr14a-S) cDNAs are aligned with the Nitr14b peptide sequence predicted from genomic sequence. Black shading indicates identical residues whereas gray shading indicates functionally similar residues. The locations of the predicted leader peptide sequences, transmembrane domains (TM), and joining (J) domains (FGXGTXLX(L/I)) are indicated with gray bars above the alignment. Residues, which are highly conserved in Ig domains (Litman et al. 2001), are indicated above the alignment using the IMGT numbering system (Giudicelli et al. 2006). Black circles indicate the locations of two tyrosines present in the cytoplasmic tail of Nitr14aL. (b) Predicted protein structures for Nitr13 and members of the Nitr14 family. Variable (V), transmembrane (TM), joining (J) and carboxy-terminal tyrosines (Y) are indicated. Only Nitr14a-L includes a transmembrane domain. The asterisk (*) next to Nitr14b indicates that this sequence is predicted from genomic sequence.
Nitr13 and Nitr14b are structurally similar to Nitr10 and Nitr11, which map to chromosome 7, in that they lack a transmembrane domain and likely are secreted proteins (Fig. 1b and Yoder et al. 2004). The two splicing variants of nitr14a include: nitr14a-long (nitr14a-L), which encodes a transmembrane domain, and nitr14a-short (nitr14a-S), which lacks a transmembrane domain as a result of mRNA splicing (Figs. 1a and 1b). Although the function of secreted NITRs remains unknown it is possible that they may dimerize with membrane bound NITRs or other membrane bound molecules that may be involved in immune recognition; alternatively, they possibly could function as decoys or bind as monomers or multimers to foreign body surfaces, potentially facilitating processes such as phagocytosis or analogous recognition mechanisms. Two tyrosines are encoded within the cytoplasmic tail of Nitr14a-L; however, neither residue is consistent with a consensus ITIM or ITAM. It presently is unknown if these tyrosines have a specific function.
Polymorphic variation of the nitr13/nitr14 gene cluster
Although nitr13 and nitr14b genes are closely linked on both genomic clones, RP71-24B12 and DKEY-149K8, only exons 1 and 2 of the nitr14a gene can readily be identified on RP71-24B12 (Fig. 2) suggesting that nitr14a has undergone less selective pressure as compared to nitr14b or nitr13. Interestingly, the FGF receptor-like 1b gene (fgfrl1b) is present on both BACs but at a distinctly different distance from the nitr13 gene due to the insertion of repetitive sequences into the DKEY-149K8 allele including a ~5kb LINE element (see 28k – 33k in Fig. 2). Such allelic/haplotypic differences are reminiscent of those described previously for the NITR gene cluster on chromosome 7 (Yoder et al. 2004). These observations suggest that the NITR genes have experienced a recent and rapid evolution as has been described for the human KIR genes (Hao and Nei 2005;Sambrook et al. 2005).
Figure 2. Allelic complexity of the NITR gene cluster on chromosome 14.
A percent identity plot (PIP) was generated using two alleles of the NITR gene cluster on chromosome 14. Sequence from BAC DKEY-149K8 was used as the reference for comparison to sequence from BAC RP71-24B12. Note that only 2 of 5 exons for nitr14a are identifiable from BAC RP71-24B12.
The nitr13/nitr14 gene cluster maps to chromosome 14
In addition to the apparent haplotypic difference seen in BACs RP71-24B12 and DKEY-149K8, their GenBank entries (BX247870 and AL954843, respectively) place these genes on chromosomes 14 and 15, respectively. In contrast, BLAST analyses of the current zebrafish genome database (Zv7: http://www.sanger.ac.uk/Projects/D_rerio/), using the BAC sequences as queries, place both genes on chromosome 14. In order to further address the chromosomal localization of the NITR gene clusters, we utilized a 2-color fluorescence in situ hybridization (FISH) strategy. The FISH data place the nitr13/nitr14 encoding BAC DKEY-149K8 onto zebrafish chromosome 14 and confirm that the previously described, main NITR gene cluster maps to chromosome 7 (Fig. 3) (Yoder et al. 2004).
Figure 3. Cytogenetic localization of NITR gene clusters to chromosomes 7 and 14.

BACs corresponding to the NITR gene cluster on chromosome 7 (DKEY-7N10) and the newly identified NITR gene cluster (DKEY-149K8) were localized to specific zebrafish chromosomes using 2-color fluorescence in situ hybridization (Freeman et al. 2007). (a) DKEY-7N10 hybridized uniquely to the subtelomeric region of chromosome 7q and is shown (orange) in relation to the near-telomeric marker for the q-arm of chromosome 7, CH211-128L16 (green). (b) DKEY-149K8 hybridized uniquely to the subtelomeric region of chromosome 14q and is shown (orange) in relation to the near-telomeric marker for the p-arm of chromosome 14, CH211-117N19 (green).
The Nitr13 and Nitr14 V domains are most similar to each other
We previously have reported phylogenetic evaluations of V domains from NITRs in zebrafish (Yoder et al. 2001;Yoder et al. 2004) and now examine the relationships of representative sequences from all 14 families. These phylogenetic analyses demonstrate that Nitr13 and both Nitr14 V domains are most similar to each other and likely share a common ancestral gene. As a group, the Nitr13 and Nitr14 V domains are more related to the V domains of Nitr3, Nitr5, and Nitr9 than to representative sequences from other zebrafish NITR gene families (Fig. 4).
Figure 4. Phylogenetic analyses of zebrafish NITR V domains.

Phylogenetic analyses of the V domains from zebrafish Nitr13, Nitr14a and Nitr14b with V domains from the zebrafish NITR families described previously (Yoder et al. 2004). Protein symbols are abbreviated (e.g. Nitr1a is represented by “1a”). Bootstrap values less than 70% are not shown. Branch lengths are measured in terms of amino acid substitutions, with the scale indicated below the tree.
In order to determine how the zebrafish nitr13 and nitr14 genes compare to NITRs in other fish species, we examined the phylogenetic relationship between all published NITRs. The V domains of zebrafish Nitr13 and Nitr14 are more similar to catfish IpNITR13 and IpNITR15 V domains than to any other known NITR (Figure 5). This relationship suggests that zebrafish nitr13/nitr14 and catfish IpNITR13/IpNITR15 likely share a common ancestry.
Figure 5. Phylogenetic analyses of NITR V domains from multiple species.

Neighbor-joining tree of V domains encoded by NITR genes in zebrafish (Danio rerio, “Dr”); channel catfish (Ictalurus punctatus, “Ip”); rainbow trout (Oncorhynchus mykiss, “Om”); Southern pufferfish (Sphoeroides nephelus, “Sn”); and Japanese flounder (Paralichthys olivaceus, “Po”). Protein symbols are abbreviated to the NITR number (e.g. zebrafish Nitr14a is shown as “Dr 14a”). In the instances of NITR gene families, only one member was included in the analysis. For example Sn 16 represents Sn 16 and 22; Sn 13 represents Sn 13 and 18; Sn 20 represents Sn 14, 15, 17, 19, 20, 21, 23-26; Ip 1 represents Ip 1 and 3; and Ip 5 represents Ip 5-11. The number assigned to each interior branch is the bootstrap value; bootstrap values less than 50 are not shown. The branch lengths are measured in terms of the number of amino acid substitutions estimated by Poisson correction, with the scale given below the tree. Note that zebrafish Nitr13 and Nitr14 V domains group with catfish IpNITR13 and 15 V domains (gray shading): these 4 V domains also group together when using maximal parsimony and UPGMA methods (not shown).
The zebrafish nitr13 gene likely arose via retrotransposition
The genomic organization of nitr13 is unique in that it encodes only two exons: exon 1, consisting of a short leader sequence, and exon 2, encoding the V domain and a 3′ untranslated region. A close inspection of exon 2 of nitr13 reveals a sequence within the 3′ untranslated region that potentially encodes an Ig I domain, which is characteristic of many NITR genes that map to chromosome 7 (Yoder et al. 2004). The putative coding sequence of the nitr13 I domain is preceded by a stop codon and a shift in the reading frame in both genomic and cDNA sequences (Fig. 6a). A second sequence within the 3′ untranslated region of nitr13, which is 3′ of the untranslated I domain, potentially encodes a transmembrane domain but is separated from the I domain by an additional shift in the reading frame. An untranslated leader sequence, which is in frame with the V domain, is located in the intronic region upstream of exon 2. The presence of an untranslated in-frame leader sequence within an intron and an untranslated I domain within the same exon as the V domain, strongly suggests that nitr13 was derived from an NITR that retrotransposed into chromosome 14 via an intron-less mRNA intermediate. The hypothetical precursor gene, nitr13p, that gave rise to this genomic change likely encoded a V and I domain (Fig. 6b).
Figure 6. The nitr13 gene originated via transposition.

(a) The nitr13 gene encodes 2 exons (top). The protein coding sequence of the nitr13 gene is comprised of 2 segments (black rectangles) in exons 1 and 2. The nitr13 mRNA possesses coding sequences for a leader (L), variable (V), intermediate (I) and transmembrane (TM) domains (middle). However a stop codon (TAA) and a reading frame shift exist between the V and I domains: a frame shift also exists between the I and the TM domains. Three non-coding sequences (gray rectangles) indicate that nitr13 arose via retrotransposition of a mature NITR mRNA: 1) a leader (L) sequence is found at the 3' end of the single intron, which is in frame with the variable (V) domain in exon 2, and 2) an Ig domain of the intermediate (I) type and 3) a transmembrane domain in the 3' untranslated region. One possible model for the nitr13 precursor mRNA is shown (bottom). (b) The protein structure of the predicted precursor to Nitr13 is compared to Nitr13.
Whether or not the second NITR cluster on chromosome 14 reflects a tetraploidization and independent diversification event in the bony fish is unclear as the nitr13/nitr14 cluster could have arisen in this manner through a translocation event or through retrotransposition. Evidence for the retrotransposition of mature mRNAs to generate new genes (or pseudogenes) has been described for more than 25 years (Lemischka and Sharp 1982) and includes a unique T cell receptor in marsupials (Parra et al. 2007). Although zebrafish nitr13/nitr14 may share a common ancestry with the catfish IpNITR13/IpNITR15 genes (see above), no genomic sequence information is available for these catfish genes barring any conclusion about a retrotransposition event in catfish NITRs. As part of a larger study, we have identified an NITR gene cluster in medaka which is physically linked to the FGFR1lB gene (Yoder unpublished obersvations); however, there is no genomic evidence for a retrotransposition event in the history of any NITRs in the medaka NITR gene cluster suggesting that the retrotransposition of nitr13 may be restricted to cypriniformes or possibly restricted to Danios or even more specifically Danio rerio. The evolutionary timing of this retrotransposition event potentially can be addressed once a more precise annotation of other bony fish genomes is achieved. Retrotransposition would join tandem gene duplication, exon swapping and alternative mRNA splicing as mechanisms for the expansion and diversification of the NITR gene family (Litman et al. 2001;Yoder et al. 2004).
The Nitr13 non-coded I domain is most similar to Nitr4, 5, 8, 9 and 12
In that numerous genes encode transmembrane and Ig domains, the presence of the untranslated I domain in the 3′ untranslated region of nitr13 is essential for classifying this gene as an NITR. Importantly, five of the six conserved cysteines that are shared features of NITR I domains are encoded by the I domain within the 3' untranslated region of nitr13 (Fig. 7a; Litman et al. 2001). Finally, a phylogenetic comparison of the nitr13 untranslated I domain with all other zebrafish NITR I domains reveals it to be most similar to the I domains of Nitr4, Nitr5, Nitr8, Nitr9 and Nitr12, albeit with a low confidence (Fig. 7b). Based on the similarities of the Nitr13 V and I domains to other NITRs (Figs. 4 and 7b) it is most likely that the nitr13 gene derived from an nitr5 or nitr9 precursor than from any other NITR thus far recognized in zebrafish.
Figure 7. Phylogenetic analyses of the untranslated I domain within the nitr13 gene.

(a) The untranslated I domain encoded in the 3' untranslated region of nitr13 (termed Nitr13P for precursor of Nitr13) is aligned with I domains from other zebrafish NITRs. Black shading indicates identical residues whereas gray shading indicates functionally similar residues. The J-related domain and residues highly conserved in Ig domains (Litman et al. 2001) are indicated above the alignment using the IMGT numbering system (Giudicelli et al. 2006). Six highly conserved cysteines, representative of NITR I domains, are indicated by asterisks below the alignment (Litman et al. 2001). (b) Phylogenetic analyses of the I domain from zebrafish Nitr13P and I domains from the zebrafish NITR families described previously (Yoder et al. 2004). Analyses and presentation are as in Fig. 4.
Attempts to predict whether nitr13 or an nitr14 gene arose first create a paradox. If nitr13 retrotransposed into this locus first and then was duplicated to form an nitr14 gene, what mechanism produced the introns in the nitr14 genes? Similarly, I domains were not detected within the nitr14 genes, suggesting that nitr13 could not have been retrotransposed from either of these neighboring genes.
Concluding remarks
Gene enumeration and physical mapping of complex multigene families can prove difficult, particularly when the individual members exhibit derived sequence features such as V family differences and variation in structural complexity, both of which are features of NITRs across species. At this point, comparisons of the genomic organization of NITR genes in other species is confounded by assembly issues as well as by the high degree of species-specific NITR variation seen in bony fish, i.e., two fish systems may have equally diversified gene families but there is only limited sequence homology across species. Nevertheless, given the current status of the zebrafish genome assembly, available search tools and a growing awareness of the diversity of NITR V regions, it is likely that the NITR gene family in this system has been defined conclusively. In the future, it will be of interest to compare the expression profiles of these two distinct NITR gene clusters as well as their functional roles in immune response. In addition, the somewhat unique nature and displacement from the primary NITR locus raises the possibility that the large multigene family encoding these transmembrane receptors may itself be diversifying, giving rise to molecules that may function in either a complementary or independent context relative to the other members of the gene family.
ACKNOWLEDGEMENTS
We are very grateful to Charles Lee for generously providing the reagents, resources and advice for FISH analyses. Sequence data from this article have been deposited with GenBank under accession numbers EU170263 (nitr13); EU170264 (nitr14a-short isoform); and EU170265 (nitr14a-long isoform). Grants from NSF (MCB-0505585 to JAY), NIH (R01 AI057559 to GWL) and the All Children's Hospital Foundation (to GWL) supported this work.
Reference List
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenhuis J, Bengten E, Snell C, Quiniou SM, Miller NW, Wilson M. Characterization of additional novel immune type receptors in channel catfish, Ictalurus punctatus. Immunogenetics. 2007 doi: 10.1007/s00251-007-0230-x. [DOI] [PubMed] [Google Scholar]
- Freeman JL, Adeniyi A, Banerjee R, Dallaire S, Maguire SF, Chi J, Ng BL, Zepeda C, Scott CE, Humphray S, Rogers J, Zhou Y, Zon LI, Carter NP, Yang F, Lee C. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. Bmc Genomics. 2007;8:195. doi: 10.1186/1471-2164-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc MP. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Nei M. Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC Class I genes. Gene. 2005;347:149–59. doi: 10.1016/j.gene.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Hawke NA, Yoder JA, Haire RN, Mueller MG, Litman RT, Miracle AL, Stuge T, Shen L, Miller N, Litman GW. Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci U S A. 2001;98:13832–7. doi: 10.1073/pnas.231418598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lemischka I, Sharp PA. The sequences of an expressed rat alpha-tubulin gene and a pseudogene with an inserted repetitive element. Nature. 1982;300:330–5. doi: 10.1038/300330a0. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Hawke NA, Yoder JA. Novel immune-type receptor genes. Immunol Rev. 2001;181:250–9. doi: 10.1034/j.1600-065x.2001.1810121.x. [DOI] [PubMed] [Google Scholar]
- Panagos PG, Dobrinski KP, Chen X, Grant AW, Traver D, Djeu JY, Wei S, Yoder JA. Immune-related, lectin-like receptors are differentially expressed in the myeloid and lymphoid lineages of zebrafish. Immunogenetics. 2006;58:31–40. doi: 10.1007/s00251-005-0064-3. [DOI] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Schwarz RS, Deakin JE, Lindblad-Toh K, Miller RD. A unique T cell receptor discovered in marsupials. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9776–81. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyaviriyakul P, Kondo H, Hirono I, Aoki T. A novel immune-type receptor of Japanese flounder (Paralichthys olivaceus) is expressed in both T and B lymphocytes. Fish Shellfish. Immunol. 2006 doi: 10.1016/j.fsi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker--a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–86. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, Rast JP, Amemiya CT, Litman GW. A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci U S A. 1999;96:15080–5. doi: 10.1073/pnas.96.26.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Zhuo J-M, Chen X, Shah R, Liu J, Orcutt TM, Traver D, Djeu JY, Litman GW, Yoder JA. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics. 2007;59:813–21. doi: 10.1007/s00251-007-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Ota T, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW. Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A. 2004;101:15706–11. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Nichols KM, Ristow SS, Thorgaard GH, Ota T, Litman GW. Cloning novel immune-type inhibitory receptors from the rainbow trout, Oncorhynchus mykiss. Immunogenetics. 2002;54:662–70. doi: 10.1007/s00251-002-0511-3. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc. Natl. Acad. Sci. USA. 2001;98:6771–6. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–64. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]



