Abstract
The involvement of tRNA structural elements beyond the anticodon in aminoacyl-tRNA (aa-tRNA) selection by the ribosome has revealed that substrate recognition is considerably more complex than originally envisioned in the adaptor hypothesis. By combining recent breakthroughs in aa-tRNA synthesis and mechanistic and structural studies of protein synthesis, we ask if aa-tRNA recognition further extends to the amino acid, thereby explaining various translation disorders exhibited by misacylated tRNAs. Contrary to expectation, we find that natural amino acids misacylated onto natural, but non-native tRNAs are selected with efficiencies very similar to those of their correctly-acylated counterparts. Despite this, small, but reproducible differences in selection indeed demonstrate that the translational machinery is sensitive to the amino acid/tRNA pairing. These results suggest that either the ribosome is an exquisite sensor of natural versus unnatural amino acid/tRNA pairings and/or that aa-tRNA selection is not the primary step governing the amino acid specificity of the ribosome.
INTRODUCTION
The paradox of how the translational machinery processes a pool of chemically diverse aminoacyl-tRNA (aa-tRNA) substrates each with nearly equivalent catalytic efficiency1 and fidelity2,3 has long been rationalized by the adaptor hypothesis4, a central paradigm in our mechanistic understanding of protein synthesis. In this framework, it is the decoupling of tRNA anticodon recognition and amino acid participation in the chemistry of peptide bond formation that allows the ribosome to work efficiently with its diverse amino acid building blocks3,5. The seminal proof of the adaptor hypothesis is a 1962 publication in which a Cys-coding poly-UG template was translated in an unpurified cell extract using misacylated Ala-tRNACys produced by Raney Nickel reduction of wild-type Cys-tRNACys to give an Ala-rich polypeptide product6. Despite rigorous analysis of the Ala-tRNACys purity, the reaction time course, and the product yield and composition, without the ability to synthesize pure, misacylated tRNAs or make quantitative kinetic measurements within the contemporary kinetic scheme for aa-tRNA selection7 at a time resolution matched to the rate of protein synthesis (20 peptide bonds per second8), this study simply could not ask if misacylation indeed impairs the incorporation of Ala-tRNACys.
Breakthroughs in our mechanistic and structural understanding of protein synthesis now provide a molecular-resolution mechanism for aa-tRNA substrate selection in which recognition of the aa-tRNA substrate by the ribosome occurs via a series of selection steps that control access of the aminoacyl end of the aa-tRNA to the peptidyl transferase center prior to peptide bond formation2,3,9. Within this series of aa-tRNA selection steps, kinetic proofreading and induced fit mechanisms act to discriminate aa-tRNAs that are cognate to the mRNA codon from aa-tRNAs that are near- and non-cognate to the mRNA codon10. This contemporary framework for aa-tRNA selection is increasingly being used to investigate the molecular determinants of aa-tRNA selection during translation. Wild-type aa-tRNAs have been shown to traverse the aa-tRNA selection pathway with virtually indistinguishable rates in response to their cognate codons1. In contrast, near-cognate aa-tRNAs (with a single mis-match in the anticodon) are discriminated against by a factor of ~450 when competed against a cognate aa-tRNA11. Unambiguously expanding the adaptor hypothesis beyond the codon-anticodon interaction, however, the long-standing conundrum of how a single mutation in the D-arm of the Hirsh suppressor Trp-tRNA(G24A)Trp renders it able to misincorporate Trp in response to a UGA stop codon12 has been recently resolved by the demonstration that this mutation allows Trp-tRNA(G24A)Trp to activate induced fit mechanisms during aa-tRNA selection13. A similar effect has been more recently reported for a tRNAAla variant with a basepair flip mutation at the top of the anticodon loop14,15. These results are consistent with growing structural evidence that correct codon-anticodon pairing at the decoding center of the small ribosomal subunit is communicated to the GTPase-associated center of the large ribosomal subunit through extensive interactions between the tRNA and the ribosome16–18 and account for the observation that an intact aa-tRNA is required for ribosome-stimulated GTP (1) hydrolysis by EF-Tu19.
The participation of tRNA elements beyond the anticodon in aa-tRNA selection raises the intriguing possibility that aa-tRNA recognition extends all the way to the amino acid. Indeed, indirect evidence from several fields is consistent with this possibility. While the incorporation of unnatural amino acids into proteins has validated that the ribosome can accept a variety of unnatural amino acids misacylated onto a variety of tRNAs as substrates20,21, many unnatural amino acids show reduced polypeptide synthesis yields22–26 and several unnatural amino acids are simply not accepted as substrates21,22. Of particular note, even natural amino acids such as Phe (2) and Ala (3) that have been misacylated onto engineered, in vitro transcribed tRNAs exhibit lower polypeptide synthesis yields25 — with absolute yields that vary depending on the tRNA adaptor22,25,27. Providing a possible explanation, tRNAs misacylated with natural amino acids exhibit altered binding affinities to EF-Tu(GTP)28 as well as to the ribosomal A site29. Taken together with increasing evidence that the cell may have to contend with tRNAs misacylated with both natural and unnatural amino acids30, these results suggest that while a tRNA misacylated with a natural amino acid, such as Ala-tRNACys, may have been functional to the extent quantifiable in early biochemical assays, misacylation may nonetheless impair aa-tRNA selection, adversely affecting protein synthesis and, ultimately, cell viability30.
Recently, a general in vitro selected tRNA aminoacylation ribozyme has been developed which accepts a wide range of activated natural and unnatural amino acids and, critical for mechanistic studies, natural, post-transcriptionally-modified tRNAs as substrates31. Here, we exploit this state-of-the-art synthetic method and a series of biochemical assays with increasing resolution to begin a reexamination of the adaptor hypothesis within the contemporary molecular mechanism of aa-tRNA selection. Specifically, we ask whether a set of chemically-diverse, natural amino acids that have been misacylated onto natural, but non-native tRNAs are incorporated by the translational machinery as efficiently as when they are paired with their native tRNAs. We develop an experimental framework for exploring the limits of the adaptor hypothesis—progressing from measurements of peptide yield, to biochemical competition assays for measurements of relative rates of aa-tRNA selection, and finally single-molecule Förster resonance energy transfer (smFRET) assays to directly observe the dynamics of aa-tRNAs as they traverse the aa-tRNA selection pathway. Using this framework, we evaluate the specificity of the translational machinery for correctly-acylated versus misacylated tRNAs (Fig 1). Remarkably, we find that the ribosome selects both correctly-acylated and misacylated tRNAs with nearly comparable efficiencies, providing quantitative evidence that misacylation does not inherently impede aa-tRNA selection and suggesting that tRNAs misacylated with natural amino acids may be potent competitors of correctly-acylated tRNAs in vivo. Despite the nearly comparable efficiencies of correctly-acylated and misacylated tRNAs, however, we observe small, but reproducible differences in selection which provide direct evidence that the translational machinery is sensitive to the amino acid/tRNA pairing and may exploit this sensitivity in order to discriminate against unnatural amino acid/tRNA pairings.
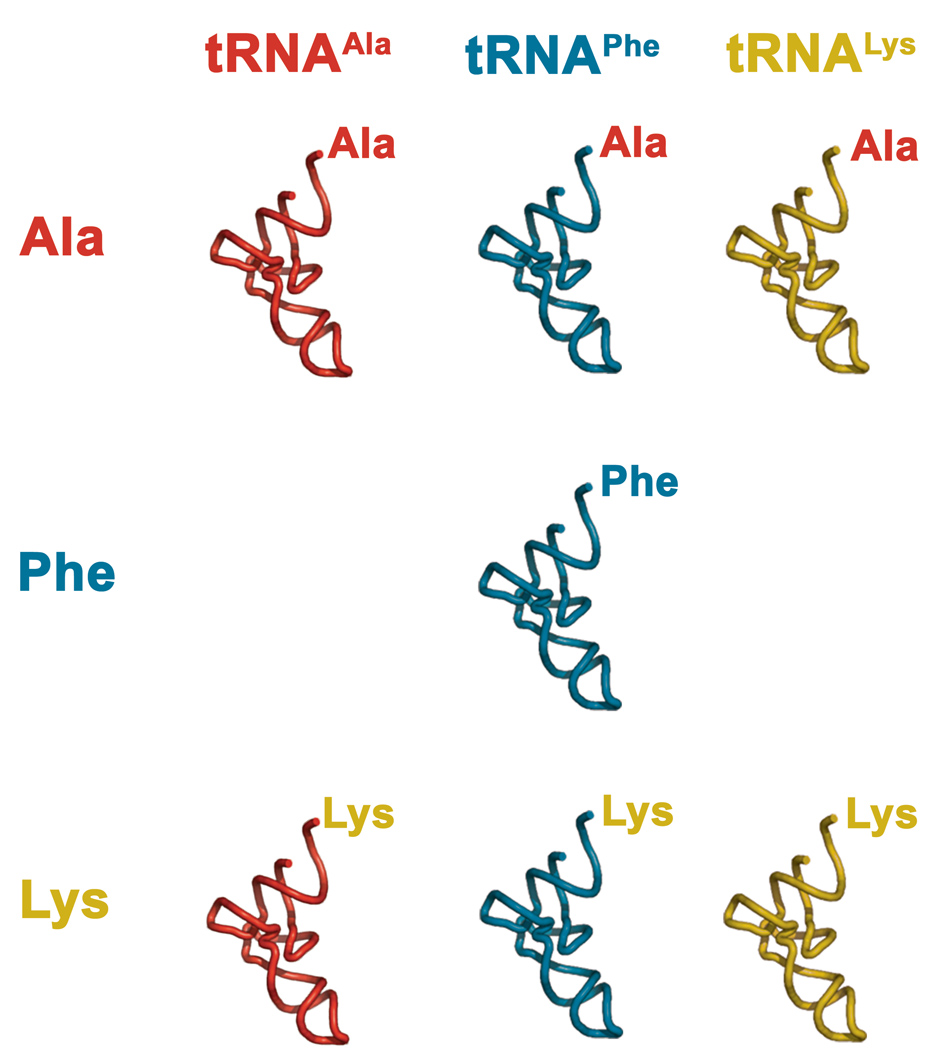
Figure 1. Design and synthesis of aminoacyl-tRNAs.
Matrix of correctly acylated and misacylated natural tRNAs used in this study to test the hypothesis that the natural amino acids require pairing with their native tRNAs for efficient selection by the translational machinery. The amino acids were chosen to cover variations in side chain size and electrostatics. Likewise, the corresponding natural tRNAs vary in their anticodon GC content and in the number and type of post-transcriptional modifications they carry. The resulting correctly-acylated and misacylated aa-tRNAs span a significant range of EF-Tu(GTP) binding affinities (Lys-tRNAAla, 5 nM; Lys-tRNAPhe, 40 nM; and Ala-tRNALys, 240 nM) 33.
RESULTS
Design and synthesis of correctly and misacylated natural aa-tRNAs
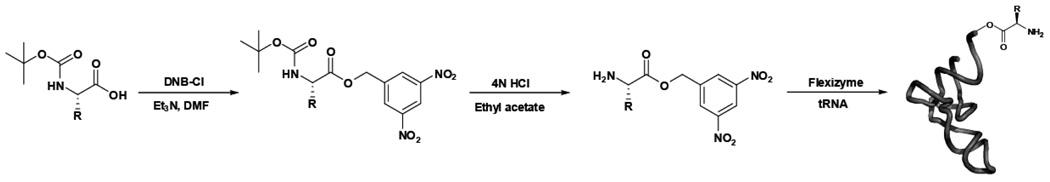
The three amino acids chosen for the misacylation matrix (Phe, Ala and Lys (4)) were chosen to span chemical space as well as to bear on the observation that misacylated aromatic amino acids generally incorporate into polypeptides with higher yields than small hydrophobic or charged amino acids22. Likewise, the natural tRNAPhe, tRNAAla, and tRNALys chosen for the matrix vary in their anticodon GC content and in their number and type of post-transcriptional modifications32. Using combinations of these amino acids and tRNAs, we have prepared a representative matrix of correctly-acylated and misacylated natural aa-tRNAs (Fig 1). The resulting aa-tRNAs span a significant range of EF-Tu(GTP) binding affinities (Lys-tRNAAla, 5 nM; Lys-tRNAPhe, 40 nM; and Ala-tRNALys, 240 nM) 33 and, consequently, can be used to test the hypothesis that the natural amino acids require pairing with their native tRNAs for efficient selection by the ribosome. The misacylated aa-tRNAs were synthesized using the dFX “Flexizyme," essentially as reported by Suga31 (Scheme 1; see Supplementary Methods and Supplementary Figures 1–4 for details).
Scheme 1. The aminoacyl-dinitrobenzyl ester substrates for the ribozyme were prepared by chemical synthesis.
Commercial N-tBOC amino acids (or otherwise appropriately protected amino acids) (5, 6) were converted to the dinitrobenzyl esters using dinitrobenzyl chloride. The tBOC protecting group was removed using concentrated hydrochloric acid. The final aminoacyl-dinitrobenzyl esters were recrystallized. tRNAs were then charged with the desired amino acid using the Flexizyme ribozyme. The tRNA and aminoacyl-active ester were incubated with the Flexizyme at high Mg2+ concentration. Charged aa-tRNA was used directly for translation reactions with no further purification. Thus, the Flexizyme ribozyme provided a general, straightforward method for the rapid synthesis of misacylated tRNAs.
Framework for evaluating ribosomal selection of aa-tRNA substrates
Three assays were chosen to study the selection of correctly and misacylated tRNAs by the ribosome. Each assay enabled us to monitor different aspects of selection. The translational endpoints of each correctly acylated and misacylated tRNA were initially evaluated with a dipeptide yield assay, while any subtle relative differences in the efficiencies of selection were assessed using a competition assay. Both assays provide a composite measure of selection since the measured output, quantity of dipeptide formed, is made after both aa-tRNA selection steps—initial selection and proofreading. Neither of the aforementioned assays provide direct information on how the ribosome contends with misacylated tRNAs at individual selection steps. Thus, in order to provide an independent measurement with which to verify the competition results, as well as to monitor individual aa-tRNA selection steps and rule out a potential change in the rate-determining step or other mechanistic subtleties, a smFRET assay was used to monitor the real-time dynamics of correctly acylated and misacylated tRNAs as they bind and are incorporated into the ribosome during aa-tRNA selection.
Misacylation does not impair dipeptide yield
The yield assay serves as a good initial measurement with which to detect significant impairment in translation of misacylated tRNAs. Most unnatural amino acids tested in this way are reported to be incorporated with reduced yields relative to their natural counterparts22. We used a dipeptide yield assay in which ternary complexes of EF-Tu(GTP)aa-tRNA were presented to preformed 70S ribosomal initiation complexes programmed with an mRNA encoding an fMet-X-Glu tripeptide (where X codes for the appropriate correctly-acylated or misacylated tRNA) and bearing f-[3H]Met-tRNAfMet at the ribosomal P site (Fig 2A). Dipeptide yield was scored as the percentage of limiting f-[3H]Met incorporated into dipeptide, f-[3H]Met-X. (For further details regarding 70S initiation complex and ternary complex preparations as well as translation reaction conditions and peptide product analysis, please see Supplemental Methods).
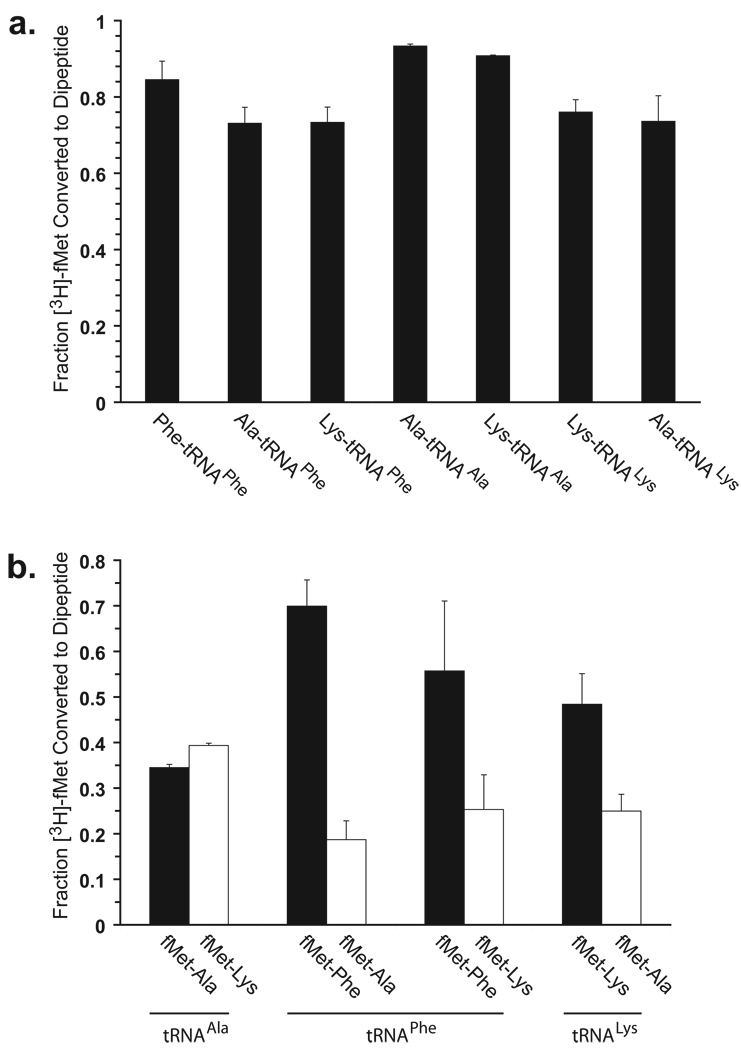
Figure 2. Dipeptide yield and competition translations.
(A) Dipeptide synthesis yields from translations using correctly-acylated and misacylated tRNAs. Yields were determined in triplicate on the basis of limiting f-[3H]Met conversion to dipeptide. Ala-tRNAAla, Lys-tRNAAla, Lys-tRNALys, Ala-tRNALys, Phe-tRNAPhe, Ala-tRNAPhe and Lys-tRNAPhe were assayed. Translation reactions were carried out by mixing preformed ternary complexes (EF-Tu(GTP)aa-tRNA) with preformed initiation complexes (70S ribosomes, mRNA and f-[3H]Met-tRNAfMet) - all preequilibrated to 37 °C. All translations were quenched after 10 sec by base hydrolysis. The reaction products were subsequently separated from unreacted f-[3H]Met by reversed-phase HPLC and quantified by liquid scintillation counting. The error bars correspond to the standard deviation from the mean. These results show that the dipeptide translation reactions all have similar apparent endpoints after 10 sec whether the tRNA is correctly-acylated or misacylated. (B) Ribosomal selection of correctly-acylated and misacylated tRNA. Translation reactions in which pairwise combinations of correctly-acylated and misacylated tRNAs compete for initiation complexes were performed. These competition reactions provide a measure of the relative overall efficiencies of aa-tRNA selection. Competition translations were performed between Phe-tRNAPhe and tRNAPhe misacylated with Ala and Lys; between Ala-tRNAAla and Lys-tRNAAla; and between Lys-tRNALys and Ala-tRNALys. The competition translations were carried out in a similar way to the dipeptide yield translations, except each ternary complex (containing correctly and misacylated tRNAs) was in 3-fold excess over the initiation complexes. These data show that misacylated tRNAs exhibit only a slight decrease in efficiency of selection relative to correctly-acylated tRNAs.
The dipeptide synthesis yield was first determined for the three correctly acylated tRNAs, Ala-tsRNAAla, Phe-tRNAPhe, and Lys-tRNALys; initial time course experiments (10 sec to 30 min) demonstrated that, as expected, the dipeptide yield reactions reached stable endpoints at or faster than the 10 sec timepoint (Supplementary Fig 5). Thus, subsequent dipeptide yield assays were only carried out as 10 sec timepoints. The following four misacylated tRNAs were then tested for dipeptide formation at 10 sec: Lys-tRNAAla, Ala-tRNAPhe, Lys-tRNAPhe, and Ala-tRNALys. In all cases, the dipeptide yields for the misacylated tRNAs were within error of those of the correctly acylated aa-tRNAs (Fig. 2A). The net dipeptide yields were all >75%, demonstrating the quality of our ribosome preparation and translation system. Thus, all dipeptide synthesis reactions reach the same apparent endpoint within 10 sec, suggesting that natural tRNAs misacylated with natural amino acids do not have the impairments in incorporation yield typical for tRNAs misacylated with unnatural amino acids22. The slightly higher yields obtained with tRNAAla may reflect higher purity and/or post-transcriptional modification of this tRNA relative to commercially overexpressed and purified tRNAs.
Misacylated tRNAs compete effectively with correctly acylated tRNAs
To probe more subtly for ribosomal discrimination between correctly acylated and misacylated tRNAs, the relative efficiency of aa-tRNA selection was assessed by monitoring dipeptide formation in experiments in which pairwise combinations of correctly acylated and misacylated tRNAs are presented in equal concentration to initiation complexes programmed with an mRNA encoding fMet-X-Glu and containing f-[3H]Met-tRNAfMet in the P site (Fig 2B). These so-called competition experiments report on all steps in the mechanism from binding of the ternary complex up to and including the point at which the aa-tRNA can no longer dissociate from the ribosome on the experimental timescale (i.e. the system becomes fully committed). Having reached this stage, the committed ribosome can no longer bind EF-Tu(GTP)aa-tRNA from solution, and the selection phase of the competition experiment is complete. Conditions were chosen to ensure that aa-tRNA was virtually entirely bound in EF-Tu(GTP)aa-tRNA ternary complexes and that ternary complexes were saturating over initiation complexes. Analogous studies in which two ternary complexes containing correctly acylated aa-tRNAs with anticodons cognate and near-cognate to the A-site codon in the initiation complex exhibited a ~450-fold selectivity against incorporation of the near-cognate aa-tRNA11. (For further details regarding reaction conditions please see Methods and Supplemental Methods).
Competition experiments were performed between Phe-tRNAPhe and tRNAPhe misacylated with Ala and Lys. Likewise, Ala-tRNAAla was competed with tRNAAla misacylated with Lys; and Lys-tRNALys was competed against tRNALys misacylated with Ala. Control experiments ensured that the competition reactions reached completion prior to the earliest timepoint taken in the experiments, 10 sec. The results demonstrate clearly that misaclyated tRNAs exhibit, at most, a four-fold decrease in the efficiency of aa-tRNA selection relative to their correctly acylated counterparts (Fig 2B). These effects are very slight compared to the ~450-fold effect seen when competing cognate against near-cognate tRNAs11. The near identical incorporation of correctly acylated and misacylated amino acids into dipeptides in these competition experiments indicates that, remarkably, the ribosome shows essentially no discrimination among these species from the initial binding through the accommodation steps of aa-tRNA selection.
Dynamics of aa-tRNA selection are not significantly perturbed by misacylation
Competition experiments report the overall selection of correctly acylated versus misacylated tRNAs, but reveal little regarding differences in the behavior of these species as they encounter the individual steps of aa-tRNA selection (Fig 3A). We therefore decided to further explore aa-tRNA selection using Ala-tRNAPhe and Lys-tRNAPhe, which exhibited detectable discrepancies in selection efficiencies relative to their correctly acylated counterpart, Phe-tRNAPhe, in the competition experiments described above. Moreover, the earliest timepoint taken in the competition experiments, 10 sec, is far longer than the expected duration of peptide bond formation, the upper limit of which is defined by accommodation, the rate-determining step in the mechanism (~8 sec−1 ref.7). To circumvent these limitations, EF-Tu(GTP)-catalyzed incorporation of Phe-tRNAPhe, Ala-tRNAPhe and Lys-tRNAPhe were monitored using a pre-steady state smFRET assay16,34,35. The smFRET assay uses fluorescently-labeled P- and A-site tRNAs to monitor the real-time movement of aa-tRNA through the ribosome as it binds to the initiation complex and is accommodated into the A site16,34,35 (Fig 3A). Previous detailed analysis of this smFRET trajectory has allowed assignment of smFRET values to individual steps in the aa-tRNA selection scheme.16,34,35
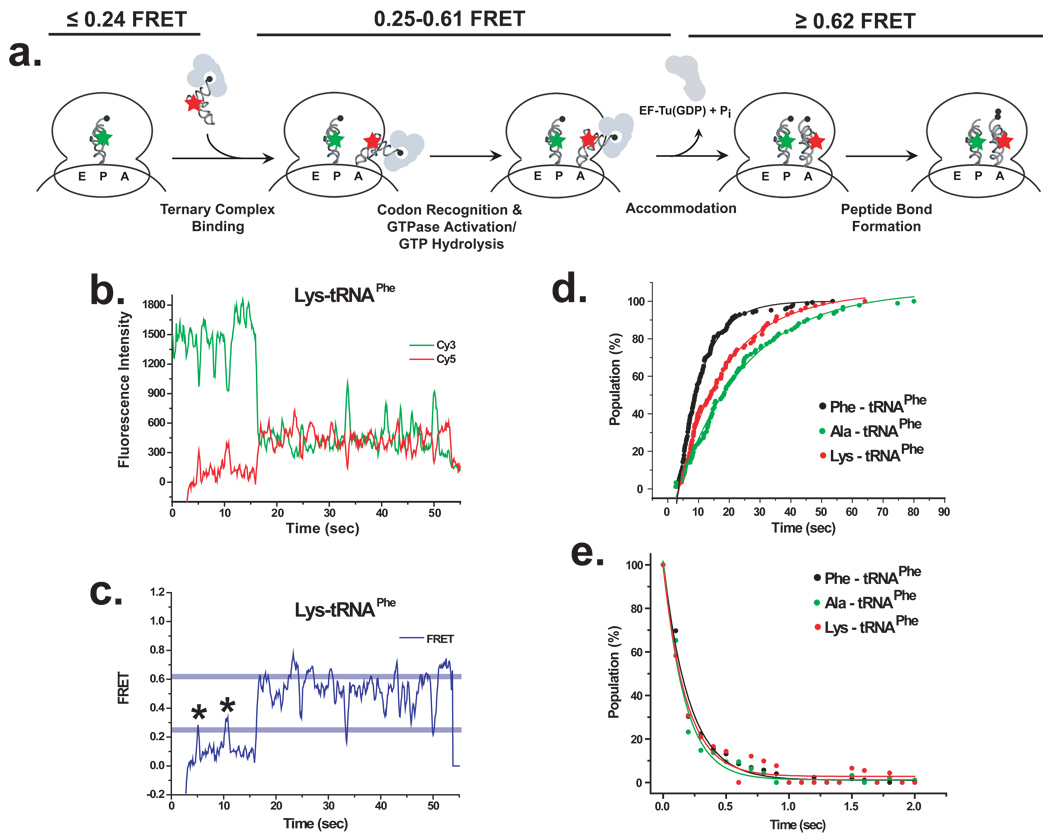
Figure 3. aa-tRNA dynamics during selection of correctly-acylated and misacylated tRNAs.
(A) Ternary complexes prepared using Phe-(Cy5)tRNAPhe, Ala-(Cy5)tRNAPhe or Lys-(Cy5)tRNAPhe as an smFRET acceptor (red star) were stopped-flow delivered to surface-immobilized initiation complexes prepared with fMet-(Cy3)tRNAfMet as an smFRET donor (green star) and an mRNA encoding fMet-Phe-Lys. (B) Representative Cy3 and Cy5 fluorescence intensities as a function of time for the binding and incorporation of Lys-(Cy5)tRNAPhe to an initiation complex are shown in green and red, respectively. (C) The corresponding smFRET signal, ICy5/(ICy3+ICy5), as a function of time, is shown in blue. Kinetic analyses were performed by defining two boundary FRET levels at 0.25 and 0.62 FRET (light blue lines). In this representative trace two A-site sampling events, in which the smFRET signal transiently samples the 0.25–0.61 FRET window before returning to the ≤0.24 FRET window (marked by asterisks) precede a productive binding event, in which the smFRET signal crosses through the 0.25–0.61 FRET window into the ≥0.62 FRET window. (D) Using only the productive events, the rates of initial ternary complex binding were estimated by plotting the increase in the population of molecules with FRET ≥0.25 as a function of time. The increase in the number of molecules with FRET ≥0.25 for Phe-(Cy5)tRNAPhe (black), Ala-(Cy5)tRNAPhe (green) and Lys-(Cy5)tRNAPhe (red) was best described by exponential growth curves of the form A1(1−exp(−t/τ1)) (Supplementary Table 1). (E) Plot of the dwell time spent between the first data point ≥0.25 FRET and the data point ≥ 0.62 FRET (i.e. within the 0.25–0.61 FRET window) for Phe-(Cy5)tRNAPhe (black) Ala-(Cy5)tRNAPhe (green) and Lys-(Cy5)tRNAPhe (red) were well described by single exponential decays of the form A1(exp(−t/τ1)) (Supplementary Table 1).
smFRET experiments were performed as previously reported 16,34,35, (Supplementary Methods). Briefly, ribosomes were enzymatically initiated such that a Cy3 donor fluorophore-labeled fMet-(Cy3)tRNAfMet was placed at the P site and a UUC codon cognate for tRNAPhe was placed into the empty A site16,34,35. smFRET versus time trajectories of EF-Tu(GTP)aa-tRNA ternary complexes prepared using (Cy5)tRNAPhe correctly acylated with Phe or misacylated with Ala or Lys were then recorded (Fig 3B and C).
Subpopulation and kinetic analyses of the smFRET trajectories requires the setting of boundary FRET levels to separate the various FRET states associated with the individual steps of aa-tRNA selection16,34,35. Here we define two such boundary FRET levels, 0.25 and 0.62 FRET, as described in Methods. The 0.25 and 0.62 boundary FRET levels define three FRET windows, ≤0.24 FRET, 0.25–0.61 FRET, and ≥0.62 FRET. Based on previous work16,34,35, the ≤0.24 FRET window corresponds to steps in the aa-tRNA selection pathway that precede codon recognition, the 0.25–0.61 FRET window encompasses a combination of codon recognition and GTPase activation/GTP hydrolysis, and the ≥0.62 FRET window corresponds to full accommodation/peptide bond formation.
Subpopulation analysis of 200 EF-Tu(GTP)Phe-(Cy5)tRNAPhe smFRET trajectories revealed that 84% of the smFRET trajectories ultimately exhibit productive binding events (Supplementary Fig 6A and 7A) where the EF-Tu(GTP)Phe-(Cy5)tRNAPhe ternary complex binds to the initiation complex, enters the 0.25–0.61 FRET window, and quickly crosses into the ≥0.62 FRET window, thus rapidly progressing to full accommodation/peptide bond formation during the observation period. For these ultimately productive trajectories, an average of 0.08 non-productive A-site sampling events, defined as events that enter the 0.25–0.61 FRET window but rapidly return to the ≤0.24 FRET window (i.e. dissociate) without crossing into the ≥0.62 FRET window, are observed per trajectory. In the remaining 16% of trajectories only A-site sampling events are detected (Supplementary Fig 6B and 7A), with an average of 1.3 A-site sampling binding events observed per trajectory (Supplementary Fig 7B). These trajectories do not productively progress into the ≥0.62 FRET window associated with full accommodation/peptide bond formation during the observation period. This subpopulation analysis is in excellent agreement with previous work in which EF-Tu(GTP)Phe-(Cy5)tRNAPhe was delivered to initiation complexes containing a cognate A-site codon under similar conditions: ~84% of the trajectories exhibited productive binding events with an average of 0.24 A-site sampling events per trajectory, whereas ~16% of trajectories exhibited only A-site sampling events during the observation period with an average of 2.5 A-site sampling events per trajectory16.
Applying the 0.25 and 0.62 boundary FRET levels only to the productive binding events, apparent rate constants for initial binding of the ternary complex, kbind, and accommodation of the aa-tRNA, kacc, can be determined by plotting the increase in the population of molecules with FRET ≥0.25 as a function of time (Fig 3D and Supplementary Table 1) and the dwell time spent between the first data point ≥0.25 FRET and the first data point ≥0.62 FRET (i.e. within the 0.25–0.61 FRET window) (Fig 3E and Supplementary Table 1), respectively. At a concentration of 2.25 nM EF-Tu(GTP)Phe-(Cy5)tRNAPhe, we obtain kbind = (5.63 ± 0.14) ×107 M−1 sec−1, in close agreement with a previously determined bimolecular rate constant (~7×107 M−1 sec−1)3. Likewise, we obtain kacc = 5.0 ± 1.2 sec−1 in good agreement with previously reported smFRET (10.8 sec−1)16 and bulk biochemical (~8 sec−1)7 values.
smFRET trajectories obtained using misacylated Ala-(Cy5)tRNAPhe and Lys-(Cy5)tRNAPhe are qualitatively very similar to those obtained for Phe-(Cy5)tRNAPhe, with the notable exception that misacylated Ala-(Cy5)tRNAPhe and Lys-(Cy5)tRNAPhe exhibit slightly increased frequencies of A-site sampling events (Fig 3B and C, Supplementary Fig 6 and 7). Subpopulation analysis of 192 Ala-(Cy5)tRNAPhe smFRET trajectories revealed that 48% of the trajectories ultimately exhibited productive binding events with an average of 0.01 A-site sampling events per trajectory, whereas 52% of the trajectories exhibited only A-site sampling events during the observation period, with an average of 1.2 A-site sampling events per trajectory. Likewise, subpopulation analysis of 153 Lys-(Cy5)tRNALys smFRET trajectories revealed that 56% of the trajectories exhibited productive binding events with an average of 0.16 A-site sampling events per trace and 44% of the trajectories exhibited only A-site sampling events during the observation period, with an average of 1.5 A-site sampling events per trajectory.
Interestingly, the 3.0- and 2.8-fold increases in the number of trajectories exhibiting only A-site sampling events for Ala-(Cy5)tRNAPhe and Lys-(Cy5)tRNAPhe relative to Phe-(Cy5)tRNAPhe, very closely matches the 3.7- and 2.2-fold decreases in the Ala-tRNAPhe and Lys-tRNAPhe versus Phe-tRNAPhe selection efficiency observed in the competition experiments. This observation strongly suggests that increased A-site sampling is the molecular basis for the decreased selection efficiency that is observed for misacylated tRNAs. Further smFRET studies, beyond the scope of the present work, are needed to explore the mechanism underlying the increased frequency of A-site sampling events observed for Lys-(Cy5)tRNAPhe and Ala-(Cy5)tRNAPhe.
Despite the increased number of A-site sampling events, measurement of kbind (Fig 3D and Supplementary Table 1) and kacc (Fig 3E and Supplementary Table 1) for Ala-(Cy5)tRNAPhe ((2.01 ± 0.07) × 107 M−1 sec−1 and 6.3 ± 2.7 sec−1, respectively) and Lys-(Cy5)tRNAPhe ((2.99 ± 0.10) × 107 M−1 sec−1 and 5.9 ± 3.8 sec−1, respectively), are in very good general agreement with the values obtained for Phe-(Cy5)tRNAPhe; nevertheless, it should be noted that the 2.8- and 1.9-fold slower kbind for Ala-(Cy5)tRNAPhe and Lys-(Cy5)tRNAPhe relative to Phe-(Cy5)tRNAPhe is consistent with the 3.0- and 2.8-fold increased frequency of A-site sampling events and 3.7- and 2.2-fold decreased aa-tRNA selection efficiencies observed in competition experiments with these misacylated tRNAs.
DISCUSSION
Based on evidence that elements of the tRNA beyond the anticodon are involved in aa-tRNA selection13–15,22–25,27, that natural amino acids misacylated onto non-native tRNAs are impaired in their binding to EF-Tu(GTP)28, and that unnatural amino acids misacylated onto natural or engineered tRNAs are poorly incorporated into full-length proteins20–22, we reasoned that natural amino acids misacylated onto non-native tRNAs might be perturbed in their ability to transit through the aa-tRNA selection steps of translation elongation. Contrary to this expectation, however, we find that natural amino acids misacylated onto non-native tRNAs pass through the aa-tRNA selection steps with rates that are nearly indistinguishable from their correctly acylated counterparts. These results suggest that differences observed in the affinities of correctly acylated versus misacylated aa-tRNAs for EF-Tu(GTP) do not significantly affect the selection of the natural amino acids. Studies using additional amino acid/tRNA pairings will be necessary to rule out the possibility that certain amino acids require pairing with their native tRNAs for efficient selection. Of particular interest is Pro, the only imino acid, which has been recently shown to be translated more efficiently on its native tRNA relative to non-native tRNAs36 and Glu and Gln, because of their biosynthetic relationship in which Gln-tRNAGln is naturally biosynthesized from a misacylated Glu-tRNAGln by a tRNA-dependent amidotransferase37.
Despite these observations, however, we do not interpret our results as supporting a widespread interpretation of the adaptor hypothesis that the ribosome is blind to the amino acid, exclusively recognizing only the tRNA anticodon38. While the effects are small (maximum 4-fold), the competition assays demonstrate reproducible differences in the rate with which misacylated tRNAs are selected relative to their correctly acylated counterparts. The results of the competition assays are corroborated by the slightly increased A-site sampling of misacylated tRNAs in the smFRET experiments. While these differences are insignificant within the aa-tRNA selection scheme (e.g. compared to the ~450-fold discrimination against aa-tRNAs in response to a near-cognate codon-anticodon interaction11), the increased A-site sampling of misacylated tRNAs observed in the smFRET experiments suggests that the translational machinery is indeed sensitive to the amino acid/tRNA pairing during the aa-tRNA selection process. While more detailed mechanistic experiments will be necessary to pinpoint exactly where along the aa-tRNA selection pathway this discrimination occurs, the ~0.30 average FRET value associated with the non-productive A-site sampling events indicates that the amino acid/tRNA pairing is recognized at a step early in the aa-tRNA selection scheme, perhaps even as early as codon recognition16,34,35. Thus, we interpret our results as supporting a model in which the translational machinery recognizes not only components of the tRNA beyond the anticodon, but that this recognition extends all the way to the amino acid and is sensitive to the amino acid/tRNA pairing; the extent to which the translational machinery exploits this recognition in order to discriminate against unnatural amino acids and specific unnatural amino acid/tRNA pairings remains to be determined.
If natural amino acids misacylated onto non-native tRNAs are selected efficiently by the translational machinery, then where do tRNAs misacylated with unnatural amino acids go wrong? There is a vast chemical space between the natural amino acid/tRNA pairings we have tested here and all potential unnatural amino acid/tRNA pairings. As articulated in recent studies of aa-tRNA binding affinities to the A site of the ribosome29, there may be a threshold response, where both the amino acid and the tRNA must be considerably unnatural-like before the composite unnatural aa-tRNA is selected against. Thus, Ala and Phe misacylated onto natural tRNAs may be selected efficiently, as observed here, but may be impaired when charged onto an engineered and/or unnatural tRNA lacking post-transcriptional modifications and/or containing a mutated anticodon, as observed in several reports from the unnatural amino acid mutagenesis field.22,25.
Alternatively, it remains formally possible that aa-tRNAs containing unnatural amino acids can also traverse unimpeded through aa-tRNA selection, but be discriminated against at other steps in the translation process that lie beyond aa-tRNA selection. Indeed, there is growing evidence for effects beyond aa-tRNA selection steps, including the side-chain dependent peptidyl transferase reactivity of P site-bound peptidyl-tRNAs39; abortive termination of translation in response misincorporated near-cogate aa-tRNAs40; and engagement of allosteric regulatory mechanisms by sequence elements in the nascent polypeptide within the ribosome exit tunnel41.
Reexamining the limits of the adaptor hypothesis in light of our contemporary mechanistic understanding of protein synthesis will broadly impact the translation field. Understanding the efficiency with which unnatural amino acid/tRNA pairs pass through each of the steps of the translation cycle will facilitate the choice and design of efficiently incorporated unnatural amino acid/tRNA pairs that can deliver ever more powerful biophysical probes42 and post-translational modification mimics43 into engineered proteins, permit the synthesis of biomedically-relevant unnatural backbone and side chain oligomers44–46, and establish a mechanistic basis for engineering a more promiscuous translational apparatus, capable of incorporating expanded sets of unnatural amino acid/tRNA pairs47,48. Furthermore, by analogy to the ribosome-targeting antibiotics49, unnatural amino acids/tRNA pairs that inhibit specific steps in the translation cycle may prove to be powerful probes for mechanistic studies of protein synthesis. Finally, these studies may provide a mechanistic rationale, and novel therapeutic targets, for potential links between tRNA misacylation and disease30. The surprisingly high efficiency with which amino acids misacylated onto non-native tRNAs pass through aa-tRNA selection, the supposed fidelity check-point of translation, begins to define the amino acid specificity of the translational machinery and raises the intriguing possibility that either the ribosome is an exquisite sensor of natural versus unnatural amino acids and/or that aa-tRNA selection is not the step governing the amino acid specificity of the ribosome.
METHODS
Dipeptide yield assays
Translation reactions were initiated by mixing 70S initiation complexes with ternary complexes to the following final concentrations: 0.5 µM 70S ribosomes, 0.55 µM f-[3H]Met-tRNAfMet, 1 µM mRNA, 5.4 µM EF-Tu and 0.9 µM aminoacyl-tRNA. Solutions were equilibrated and reactions were run at 37 °C. Reactions were quenched at ten seconds by addition of NaOH to 160 mM, and incubated for 10 minutes at 37 °C before addition of glacial acetic acid, acetonitrile, and trifluoroacetic acid to final concentrations of 10%, 1%, and 0.1%, respectively. Radiolabelled reactants were separated by reversed-phase HPLC (C18 column, 0–100% acetonitrile in 0.1 % TFA) and quantified by scintillation counting. Sample traces are presented in supplementary results (Supplementary figures 8 and 9).
Competition assays
The conditions of the competition assays were identical to those of the dipeptide yield assay with the exception that the ternary complex mixture was prepared using two aminoacyl-tRNAs, each with final reaction concentrations of 1.5 µM.
smFRET assays
Flow cells constructed from quartz microscope slides and borosilicate glass cover slips were cleaned and subsequently derivatized with a mixture of N-hydroxysuccinimidyl ester derivatives of polyethyleneglycol (PEG) and PEG-biotin in order to passivate the surface of the flow cells50. Just prior to smFRET experiments, flow cells were treated with streptavidin, and purified initiation complexes prepared with a 5'-biotinylated mRNA as described above were surface-immobilized via a biotin-streptavidin-biotin interaction50.
All smFRET experiments were conducted in Tris-polymix buffer adjusted to 3.5 mM Mg(OAc)2. The lifetimes of Cy3 and Cy5 fluorescence were extended through the addition of an oxygen scavenging system to the Tris-polymix buffer consisting of final concentrations of 1% (v/v) β-D-glucose, 25 U ml−1 glucose oxidase (Sigma, Inc.) and 250 U ml−1 catalase (Sigma, Inc.)50. In addition, 1,3,5,7-cyclooctatetraene (Aldrich) and p-nitrobenzyl alcohol (Fluka) were added to the Tris-polymix buffer to final concentrations of 1 mM each in order to quench a long-lived non-fluorescent triplet state sampled by the Cy5 fluorophore50. A laboratory-built, prism-based total internal reflection microscope, based on a Nikon TE2000-U inverted microscope, was used for all experiments. fMet-(Cy3)tRNAfMet was excited using a diode-pumped 532-nm laser (CrystaLaser) laser. Fluorescence emission was collected by a 1.2 NA 60X water-immersion objective (PlanApo, Nikon), wavelength separated using a Quad-View multichannel imaging system (MAG Biosystems), and imaged onto a back-thinned charged coupled device (Cascade II, Princeton Instruments) with 4-pixel binning at 100 msec exposure time. Stopped-flow delivery was achieved using a customized programmable syringe pump system (J-KEM Scientific); the dead time for complete mixing after delivery of substrates is estimated at ~500 ms. Ternary complex was formed as described above, diluted with Tris-polymix buffer to a final concentration of 2.25 nM, and immediately stopped-flow delivered to surface-immobilized initiation complexes.
In order to perform subpopulation and kinetic analyses of the smFRET trajectories, boundary FRET levels separating the various FRET states associated with the individual steps of aa-tRNA selection must be established. Consistent with previous reports we observe that delivery of EF-Tu(GTP)Phe-(Cy5)tRNAPhe ternary complex to initiation complexes containing a cognate codon at the A site yields a rapidly evolving smFRET signal16. As in previous work, these rapidly evolving smFRET trajectories were analyzed using two boundary FRET levels centered at 0.25 and 0.62 FRET16. The 0.25 and 0.62 boundary FRET levels define three FRET windows, ≤0.24 FRET, 0.25–0.61 FRET, and ≥0.62 FRET, which have been assigned to steps in the aa-tRNA selection pathway that precede codon recognition (≤0.24 FRET), a combination of codon recognition and GTPase activation/GTP hydrolysis (0.25–0.61 FRET), and full accommodation/peptide bond formation (≥0.62 FRET)16,34,35. We note here that unlike previous smFRET studies of aa-tRNA selection we do not perform experiments using near-cognate codons, GDPNP (a non-hydrolyzable GTP analog), or antibiotic inhibitors of aa-tRNA selection16,34,35. As a result, we do not observe the previously reported rapid and reversible rejection of ternary complexes from the codon recognition step of aa-tRNA selection nor the stable binding of biochemically-trapped ternary complexes during the GTPase activation/GTP hydrolysis step of aa-tRNA selection. Consequently, this prevents separation of the codon recognition and GTPase activation/GTP hydrolysis steps of aa-tRNA selection by a well-defined boundary FRET level and assignment of these steps to individual FRET states.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by funds to V.W.C. and R.L.G. from Columbia University as well as grants to R.L.G. from the Burroughs Wellcome Fund (CABS 1004856) and T.S.L. from the NIH (RO1 GM54469). P.R.E. was supported by Columbia University College of Physicians and Surgeons' M.D./Ph.D. Program (NIH Institutional Training Grant T32 GM07367). We would like to thank Prof. Ya-Ming Hou and Dr. Marcel Dupasquier for the kind gift of a nucleotidyl transferase overexpression strain, Prof. David Tirrell for the gift of a phenylalanyl-tRNA synthetase overexpressing strain, and Mr. Noam Prywes for cloning and purifying alanyl-tRNA synthetase as well as aiding in tRNAAla purification. We would additionally like to thank Prof. Hiroaki Suga, Prof. Rachel Green, Dr. Hani Zaher, and Dr. Lucas Gartenmann Dickson for helpful discussions and advice. Finally, we are indebted to Ms. Vanessa Mondol and Ms. Subhasree Das for managing the Cornish and Gonzalez laboratories, respectively.
Contributor Information
Philip R. Effraim, Integrated Program in Cellular, Molecular, and Biomedical Sciences, Department of Chemistry, Columbia University, New York NY 10027, pre2001@columbia.edu
Jiangning Wang, Department of Chemistry, Columbia University, New York NY 10027, jw2330@columbia.edu.
Michael T. Englander, Integrated Program in Cellular, Molecular, and Biomedical Sciences, Department of Chemistry, Columbia University, New York NY 10027, mte2002@columbia.edu
Josh Avins, Department of Chemistry, Columbia University, New York NY 10027, jla2113@columbia.edu.
Thomas S. Leyh, Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, New York 10461, leyh@aecom.yu.edu
Ruben L. Gonzalez, Jr., Department of Chemistry, Columbia University, New York NY 10027, rlg2118@columbia.edu
Virginia W. Cornish, Department of Chemistry, Columbia University, New York NY 10027, vc114@columbia.edu
REFERENCES
- 1.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 3.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 4.Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 5.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 6.Chapeville F, et al. On the role of soluble ribonucleic acid in coding for amino acids. Proc Natl Acad Sci U S A. 1962;48:1086–1092. doi: 10.1073/pnas.48.6.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. Embo J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennell D, Riezman H. Transcription and Translation Initiation Frequencies of Escherichia-Coli Lac Operon. Journal of Molecular Biology. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- 9.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 10.Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 11.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 12.Hirsh D, Gold L. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J Mol Biol. 1971;58:459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- 13.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami H, Ohta A, Suga H. Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat Struct Mol Biol. 2009;16:353–358. doi: 10.1038/nsmb.1580. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 17.Schuette JC, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. Embo J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piepenburg O, et al. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- 20.Cornish VW, et al. Site-specific incorporation of biophysical probes into proteins. Proc Natl Acad Sci U S A. 1994;91:2910–2914. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellman JA, Mendel D, Schultz PG. Site-specific incorporation of novel backbone structures into proteins. Science. 1992;255:197–200. doi: 10.1126/science.1553546. [DOI] [PubMed] [Google Scholar]
- 22.Cornish VW, Mendel D, Schultz PG. Probing protein structure and function with an expanded genetic code. Angew. Chem. Int. Ed. Engl. 1995;34:621–633. [Google Scholar]
- 23.Dougherty DA. Unnatural amino acids as probes of protein structure and function. Curr Opin Chem Biol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- 24.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Tan Z, Forster AC, Blacklow SC, Cornish VW. Amino acid backbone specificity of the Escherichia coli translation machinery. J Am Chem Soc. 2004;126:12752–12753. doi: 10.1021/ja0472174. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 27.Cload ST, Liu DR, Froland WA, Schultz PG. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem Biol. 1996;3:1033–1038. doi: 10.1016/s1074-5521(96)90169-6. [DOI] [PubMed] [Google Scholar]
- 28.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 29.Dale T, Fahlman RP, Olejniczak M, Uhlenbeck OC. Specificity of the ribosomal A site for aminoacyl-tRNAs. Nucleic Acids Res. 2009;37:1202–1210. doi: 10.1093/nar/gkn1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 31.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzl M, Hartmann T, Weber J, Blank J, Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17(Suppl):r1–r172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asahara H, Uhlenbeck OC. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry. 2005;44:11254–11261. doi: 10.1021/bi050204y. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez RL, Jr, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. Rna. 2007;13:2091–2097. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci U S A. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curnow AW, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crick FHC. The Recent Excitement in the Coding Problem. Progr Nucleic Acid Res Mol Biol. 1963;1:163–217. [Google Scholar]
- 39.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 40.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beringer M. Modulating the activity of the peptidyl transferase center of the ribosome. RNA. 2008;14:795–801. doi: 10.1261/rna.980308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J Am Chem Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 43.Rothman DM, et al. Caged phosphoproteins. J Am Chem Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- 44.Forster AC, et al. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci U S A. 2003;100:6353–6357. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohta A, Murakami H, Higashimura E, Suga H. Synthesis of polyester by means of genetic code reprogramming. Chem Biol. 2007;14:1315–1322. doi: 10.1016/j.chembiol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 46.Subtelny AO, Hartman MC, Szostak JW. Ribosomal synthesis of N-methyl peptides. J Am Chem Soc. 2008;130:6131–6136. doi: 10.1021/ja710016v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Construction of modified ribosomes for incorporation of D-amino acids into proteins. Biochemistry. 2006;45:15541–15551. doi: 10.1021/bi060986a. [DOI] [PubMed] [Google Scholar]
- 48.Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc. 2007;129:14458–14462. doi: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- 49.Spahn CM, Prescott CD. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 50.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.