Abstract
The Ewing’s sarcoma (EWS) protein is a ubiquitously expressed RNA chaperone. The EWS protein localizes predominantly to the nucleus. Previous reports have suggested that the EWS protein is capable of dimerizing. However, to date this has not been confirmed. Here, using a novel panel of recombinant proteins, we have performed an in vitro biomolecular interaction analysis of the EWS protein. We have demonstrated that all three arginine-glycine-glycine (RGG) motifs are capable of binding directly to the survival motor neuron protein, a Tudor domain containing EWS binding partner. We have also confirmed EWS is capable of self-associating, and we have mapped this binding domain to the RGG motifs. We have also found that self-association may be required for EWS nuclear import. This is the first direct evidence of RGG domains being involved in self-association and has implications on all RGG-containing proteins.
Keywords: EWS, dimerization, RGG domain, self-association
The Ewing’s sarcoma (EWS) protein is a 90 kDa protein that is believed to be involved in ribonucleoprotein assembly and RNA processing (1). The EWS protein consists of an N-terminal transcription activation domain (TAD) that interacts directly with RNA polymerase II, and a C-terminal RNA binding domain (2–4). The C-terminal domain contains six conserved motifs: an 85 amino acid RNA recognition motif (RRM) that has been identified in several other RNA-binding proteins, and is thought to interact specifically with poly-G and poly-U sequences (5); a Ran-binding protein 2 (RanBP2)-like zinc finger motif that is involved in nuclear (6); an IQ domain that interacts directly with calmodulin and is phosphorylated by protein kinase C (7); and three arginine/ glycine/glycine (RGG)-rich motifs (8).
RGG motifs are found in many nuclear RNA binding proteins, including Sm core proteins (9), fibrillarin (10), GAR1 (10) and p80-coilin (11). The RGG domains are thought to mediate transcription initiation (12), RNA binding (5) and nuclear import (12). The RGG domain within many proteins is involved in regulation of functional interactions between proteins. For example, the RGG domain of EWS is required for its interaction with the survival motor neuron (SMN) protein (8). Under certain situations, the arginine residues of the RGG-domains are symmetrically dimethylated. The protein N-arginine methyltransferase (PRMT) family of enzymes perform this methylation (13–15). The exact function of this event is not clear. Although there is some evidence that this methylation alters the binding kinetics of RGG-mediated interactions (14, 16, 17), this is not always the case (8). In addition, methylation has been shown to increase cell surface targeting of RGG motifs (18), suggesting it could play a functional role in subcellular trafficking.
A previous study suggests that the EWS protein is capable of self-associating (2). However, neither the mechanisms that mediate this association or the domains involved have been identified, nor have reasons why EWS self-associates been suggested. Here, using a novel panel of EWS proteins, we have used surface plasmon resonance (SPR) on a BiacoreX (GE healthcare) machine to map the self-association domains. Initially, we have confirmed all three RGG motifs are capable of associating with the Tudor domain of the SMN protein. Reduced expression of functional SMN protein triggers childhood spinal muscular atrophy (SMA) (19, 20). We have then shown that domains containing at least one RGG motif is capable of associating with EWS, and that removing individual RGG motifs reduces this self-association. We have further demonstrated this using recombinant protein pull down experiments. We have also implied that removing more than one RGG domain from the protein reduces nuclear import efficiency. In all, this provides the first direct evidence that EWS is capable of self-associating, and suggests that the RGG motifs mediate this association.
Materials and Methods
cDNA constructs
All EWS constructs were cloned into the pET32b (Merck Chemicals Ltd.) vector in a stepwise manner using primers. All truncated products were amplified by PCR using full-length EWS cDNA cloned into pET32b (Novagen) as a template. The ΔRGG constructs were constructed in a stepwise manner. For example, for ΔRGG1+2 the N-terminal section (5′ of RGG1) was amplified using primers For 1 and Rev 4, and cloned into the pET32a vector using a 5′ EcoRV; 3′ EcoRI site. The section between RGG1 and RGG2 was then amplified using primers For 2 and Rev 3, and cloned downstream of the N-terminal fragment using a 5′ EcoRI; 3′ SacI site. Finally, the C-terminal domain (including RGG3) was amplified with primers For 3 and Rev 1, and cloned downstream of the N-terminal and central fragments using a 5′ SacI; 3′ XhoI site. Positive clones were screened by restriction enzyme digest. pET32 constructs were transformed into Rosetta bacterial strain for recombinant protein production. For transient transfection, the constructs were then sub-cloned into pEGFP using primers. Again positive clones were screened by restriction enzyme digest. Supercoiled DNA was produced using Qiagen midi prep kit according to manufacturers’ instructions.
Recombinant protein induction and extraction
Recombinant proteins were expressed by inducing transformed Rosseta bacteria with IPTG. Cultures were incubated between 4 and 12 h at 37°C shaking. The resulting culture was pelleted and re-suspended in 5 ml of ice-cold native lysis buffer (500 mM NaCl, 20 mM Tris pH 7.8, 5 mM imidazole). The sample was vortexed and sonicated three times on ice for 5 s and incubated for 1 h on ice. Insoluble material, including inclusion bodies, was pelleted by centrifugation. Protein was solubilized from the insoluble pellet by a sequential urea extraction. Initially protein was extracted using lysis buffer containing 8 M urea (BiacoreX and protein pull down experiments) or 4 M urea (size exclusion chromatography; SEC). Extracted proteins were purified using Ni2+ NTA resin (Qiagen). Proteins extracted in 8 M urea were first bound to Ni2+ NTA resin, then the incubation buffer exchanged sequentially from 6to 2 M urea. Proteins extracted in 4 M urea were washed in IP buffer (150 mM NaCl, 25 mM Tris pH 7.4, 0.1% Triton X-100) to remove non-his-tagged protein from the sample. Protein was eluted from the resin by the addition of 2 ml elution buffer in 100 mM urea (100 mM Urea, 500 mM NaCl, 20 mM Tris pH 7.8, 500 mM Imidazole).
SEC
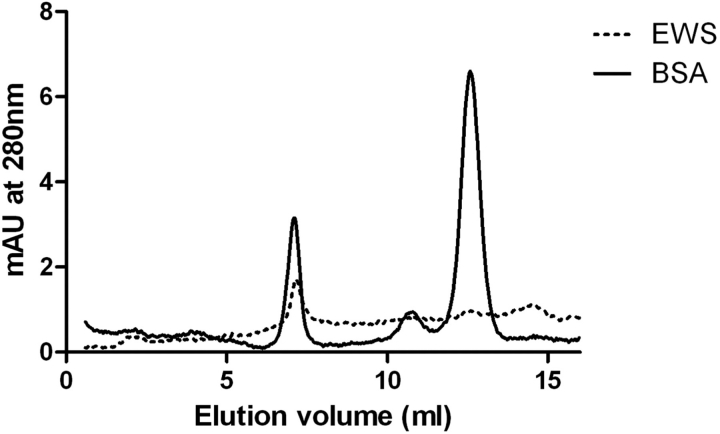
In order to establish if purified recombinant EWS is predominantly monomeric, dimeric or multimeric, histidine (His)-tagged EWS was run on a Superdex 200 precision column PC 3.2/30 (GE healthcare, Buckinghamshire, UK). Initially purified protein was concentrated down to 100 µl using an Amicon concentrator. Protein was filtered using a Millex® LH syringe driven filter. The column was then equilibrated with elution buffer at a flow rate of 40 µl/min. This was continued until the system was completely free of air bubbles (minimum of 4.8 ml). The sample (100 µl) was applied at a flow rate of 40 µl/min. BSA standard was loaded as a control (67 kDa). The volume of buffer required to elute the proteins from the column (elution volume) for EWS and BSA were noted. Note that in these experiments BSA multimers, dimers and monomers can be seen (Fig. 1).
Fig. 1.
EWS runs as a multimeric protein on size exclusion chromatography. Line graph indicating SEC of recombinant EWS protein eluted in 100 mM urea and BSA control. Full line indicates BSA control. Dotted line indicates EWS protein in 100 mM urea. The x-axis indicates the volume of buffer (ml) required to move the protein through the column (elution volume). The y-axis represents the arbitrary units at an absorbance of 280 nm. The EWS protein appears in an initial peak corresponding to an elution volume of 7 ml. When the control BSA and EWS graph are overlaid, it appears that the dominant peak produced during the EWS SEC is at the same size as mutimeric BSA indicating a minimal size of 200 kDa. The tagged EWS protein is 105 kDa in size suggesting that this peak is dimeric/multimeric EWS.
TNT in vitro transcription/translation kit
The TNT® quick coupled transcription/translation system (Promega) was used to produce protein labelled with biotinylated lysine (Transcend™ Biotinylated tRNA) as per the manufacturer’s instructions. Biotinylated lysine/non-labelled lysine were added at a ratio of 1:10. Theoretically this means 1 in 10 lysines are biotinylated and that this is a random process. Samples were then purified using Ni–NTA resin, and used in Biacore experiments (see below).
Preparation of SA-Biacore chips
Streptavadin (SA) Biacore chips were prepared by injecting 50 µl 2 M NaCl. Equilibrated chips were then pulsed with biotinylated protein until 200 RUs of protein had bound. Chips were then pulsed with 0.2% SDS for 10 s to remove any non-specifically bound protein. Chips were then screened with monoclonal antibodies specific to captured proteins to confirm protein-binding immobilization had occurred. For biotinylated SMN chips, MANSMA1 was used with anti-EWS antibodies (Santa Cruz) as a non-specific control antibody. The same antibodies were used to screen biotinylated EWS chips. Bound antibodies were used to screen chips for appropriate regeneration buffer.
In vitro Biacore binding experiments
Recombinant proteins were expressed and purified as stated above. Each protein was quantified and diluted down to stock concentrations in PBS (0.1, 0.05 and 0.01 µg/µl). Five microlitre of each sample was then pulsed over the chip, at a flow rate of 5 µl/min, resulting in exposure to 0.5 µg (0.1 µg/µl), 0.25 µg (0.05 µg/µl) and 0.05 µg (0.01 µg/µl for each corresponding dilution. Following injections, chips were regenerated by pulsing with 0.2% SDS. Binding injections were repeated in triplicate, and the mean values were taken. The standard deviation between repeats was also calculated to record the in test variability.
Standardization of binding values with regards to size
Biacore analysis measures changes in mass associated with the surface of a Biacore chip. As a result raw data generated during experiments using proteins of varying sizes can be used for yes/no binding experiments, but are not directly comparable. For example, in a study with two proteins (X and Y), where X is 10 kDa and Y is 20 kDa, if twice as much X binds as Y, the response unit (RU) increase will be the same for both experiments, which may lead to misleading interpretation. Therefore, all binding values were standardized in relation to the smallest protein used in each respective study. Due to the presence of multiple binding domains, we could not be sure of the binding ratios (1 : 1, 1 : 2, 1 : 3, etc.) and therefore binding kinetics were not measured.
Statistical evaluation
We have performed a two-way ANOVA with Bonferroni post-tests in order to obtain the P-values for the binding data. All binding data was compared to construct EWS 1–264 that acted as a negative control.
IMFs and transfections
Human cervical carcinoma (HeLa) cells were grown in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% (v/v) foetal calf serum (FCS) and 1% (w/v) each of penicillin and streptomycin. The cells were grown in 5% (v/v) CO2 at 37°C.
Subconfluent HeLa cells were grown on cover slips and transfected with 1–2 µg cDNA using GeneJuice (Novagen) according to the manufacturer’s recommendations. Briefly, for a 2 µg cDNA transfection, 100 µl of DMEM lacking FCS was incubated with 6 µl of GeneJuice at room temperature for 5 min following vortexing. Two microgram of supercoiled DNA was then added, mixed by gentle pipetting, and incubated at room temperature for 10 min. The mixture was then added drop-wise to HeLa cells in DMEM lacking FCS. Cells were incubated for 6–8 h in 5% (v/v) CO2 at 37°C, then re-fed with DMEM containing 10% FCS and incubated for a further 16–24 h. Following 16–24 h transfected cells were washed three times in pre-warmed phosphate buffered saline (PBS) pH 7.4 and then fixed using 50% acetone/50% methanol (v/v). Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI).
Microscopy
Fluorescence microscopy was performed using a Nikon TE 2000-U microscope and a Hamamatsou camera. Images were acquired by taking Z-stacks and de-convolved using Openlab version 4.0 (Improvision). The images were then processed using Adobe Photoshop 5.5.
Recombinant protein pull downs
M-280 Streptavidin Dynabeads were used for the pull-down experiment. Briefly beads were washed three times in 0.01% PBST (Tween-20). Biotinylated protein of 200 pmol (RGG or lysine-glycine-glycine (KGG)) was diluted in 100 µl PBS and added directly to the beads. This was incubated at room temperature for 30 min with continuous rotation. Following incubation, beads were washed five times in PBS containing 0.1% BSA. One hundred microlitre of 0.5 µg/µl purified recombinant protein was added to the beads and incubated with rotation at room temp for 1 h. Following washing in PBST, beads were re-suspended in 2× SDS sample buffer. Samples were run on a 10% SDS–PAGE gel then transferred to PVDF membrane by semi-dry western blot. Blots were developed using α-His antibody (Sigma) at manufacturer’s recommended dilution followed by a α-mouse-HRP (Santa Cruz) secondary antibody at manufacturer’s recommended dilution. Blots were visualized using Pierce Femto Chemiluminescence substrate on a chemidoc.
Results and discussion
This study was prompted by two previous reports: one identified a direct association between the Tudor domain of the SMN protein and the first RGG domain (RGG1) of EWS (8); the second suggested that EWS was capable of forming multimeric complexes, and therefore, may contain a self-association domain (2). Using SEC and SPR we set out to: (i) determine if EWS is a monomeric, dimeric or multimeric protein; (ii) determine if the SMN association with EWS was specific to RGG1 or whether the other two RGG motifs could also associate with the Tudor domain of SMN; (iii) identify and map the potential self-association domain(s) in the EWS protein.
EWS is a multimeric protein
In order to determine if EWS is a mutimeric protein, we performed SEC. Recombinant EWS (His-EWS) was expressed, purified and concentrated as described elsewhere (8), and the resulting protein was run on an SEC gel filtration column using BSA as a size elution control (BSA is 70 kDa and forms multimeric, dimeric and monomeric complexes) (Fig. 1). These experiments suggest that His-EWS is predominantly eluted in an initial peak corresponding to an elution volume of 7 ml (Fig. 1). This peak is well after the void volume for the column indicating that it is not aggregated protein. When the control BSA and EWS graphs are overlaid, it appears that the dominant peak produced during the EWS SEC is at the same size as mutimeric BSA (Fig. 1). This indicates that this is a minimal size of 200 kDa. The tagged EWS protein is 105 kDa in size suggesting that this peak is dimeric/multimeric EWS. Again, this confirms previous studies that suggest EWS is capable of self-associating. Again, as this complex appears to be ∼200 kDa and is not present in the void volume, it suggests it is a true dimer/multimer and is not due to non-specific aggregation of unfolded, denatured protein.
EWS is capable of associating with SMN through all three RGG motifs
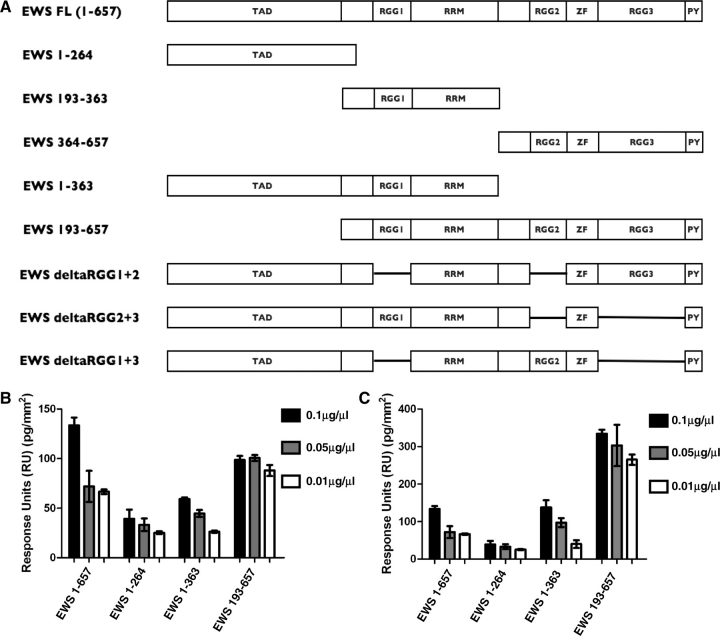
To screen for additional SMN binding sites within the EWS protein we produced a biotinylated (BA)-SMN Biacore chip. Biotinylated SMN was produced using the in vitro transcription and translation coupled kit (Promega). The SMN protein was expressed from the pET32 vector, producing a His-tagged recombinant protein of 50 kDa. This protein was purified using Ni–NTA resin, and then immobilized on a Biacore streptavidin chip to produce the BA-SMN chip (data not shown). As an experimental technique, this is one of the first examples of producing SA-protein chips using biotinylated proteins expressed in vitro using the cell free transcription/translation kit (Promega). This chip was then used to confirm whether SMN associates with EWS through all three of the RGG motifs (RGG1, 2 and 3), or whether this association is restricted to RGG1. A panel of pET32-derived recombinant EWS proteins (Fig. 2A) were expressed and purified (data not shown). Purified proteins were quantified and diluted to 0.1, 0.05 and 0.01 µg/µl. These samples were then injected over the BA-SMN chip and RU increases were measured. Initially, we screened four constructs: (i) EWS 1–657 (containing RGG1, 2 and 3); (ii) EWS 1–264 (containing no RGG domains); (iii) EWS 1–363 (contain RGG1); and (iv) EWS 193–657 (containing RGG1, 2 and 3) (Fig. 2B). When values are standardized for size (see ‘Materials and Methods’ section for an explanation of why standardization is needed), these experiments confirm that the RGG motifs mediate the SMN association, but also that the presence of RGG2 and RGG3 increases the amount of EWS protein binding to SMN (Fig. 2B). This suggests one of two things: (i) that the other RGG domains are capable of binding to SMN; and/or (ii) the C-terminus of EWS contains a self-association domain, meaning its presence allows multimeric EWS complexes to firstly form, and then associate (through RGG1) with SMN.
Fig. 2.
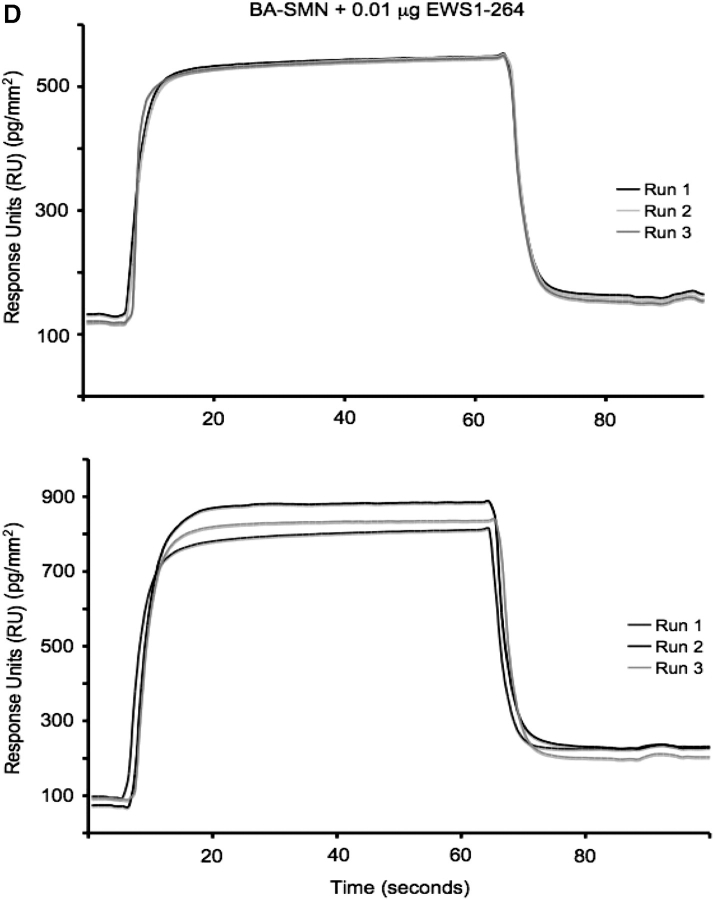
EWS-derived proteins containing one or more RGG motifs interact directly with SMN. (A) Schematic diagram of EWS panel constructs labelled by amino acid numbers. The transactivation domain (TAD), the arginine-glycine rich (RGG) motifs (RGG1, RGG2 and RGG3), the zinc-finger motif (ZF) and the C-terminal nuclear localization signal (PY) are all shown. The panel consists of the following constructs: (i) EWS full-length (EWS FL; 1–657); (ii) EWS amino acids 1–264 (EWS 1–264); (iii) EWS amino acid 193–363 (EWS 193–363); (iv) EWS amino acids 364–657 (EWS 364–657); (v) EWS amino acids 1–363 (EWS 1–363); (vi) EWS amino acids 193–657 (EWS 193–657); (vii) EWS delta RGG1 and RGG2 (EWS delta RGG1+2); (viii) EWS delta RGG2 and RGG3 (EWS delta RGG2+3); and (ix) EWS delta RGG1 and RGG3 (EWS delta RGG1+3). (B and C) BA-SMN was immobilized on a SA-Biacore chip, and pulsed with EWS 1–657 (containing RGG1, 2 and 3); EWS 1–363 (containing RGG 1); EWS 193–657 (RGG1, 2 and 3), and EWS 1–264 (no RGG-domains; negative control). (A) A bar graph comparing the binding values for 1–657, 1–363, 193–657 and 1–264 on the SMN chip. Protein binding is represented as response units (RU), with one RU representing 1 pg/mm2 of protein binding to the chip surface. The error bars show the standard deviation (SD) between the three repeats. (B) BA-SMN was immobilized on a SA-Biacore chip, and pulsed with EWS 1–657 (containing RGG1, 2 and 3); EWS 1–264 (containing no RGG domains); EWS 193–363 (containing RGG1) and EWS 364–657 (containing RGG2 and 3). Protein binding is represented as RU, with one RU representing 1 pg/mm2 of protein binding to the chip surface. The error bars show the SD between the three repeats. At a protein concentration of 0.1 µg/µl EWS 1–657, EWS 193–363 and EWS 364–657 all bind significantly greater than EWS 1–264 with a P < 0.001. (D) Sample sensorgrams for the binding of EWS 1–264 (top panel) and EWS 364–657 (bottom panel) to the BA-SMN chip. The traces show the real-time binding of both proteins to the BA-SMN chip in response units (pg/mm2). Binding experiments were repeated three times and the sensorgrams for all three runs are overlaid and presented.
To confirm RGG2 and/or RGG3 are able of binding to SMN, the BA-SMN chip was pulsed with two additional constructs: (i) EWS 193–363 (containing only RGG1) and (ii) EWS 364–657 (containing both RGG2 and RGG3) (Fig. 2C). Again, as before EWS 1–657 (containing all three RGG motifs) and EWS 1–264 (containing no RGG motifs) were used as positive and negative controls respectively (Fig. 2C). We found that, to varying degrees, all constructs containing RGG motifs were captured on the SMN chip (Fig. 2C). The ability of EWS 364–657, which lacks RGG1, to bind SMN, shows the interaction to the SMN Tudor domain is not restricted to RGG1. Sample sensorgram traces for the BA-SMN chip exposed to: (i) EWS 1–264 (Fig. 2D; top); and (ii) EWS 364–657 (Fig. 2D; bottom) are shown. These include overlays of the three individual runs used to determine the mean binding values presented (Fig. 2C).
Further analysis of these binding results highlights that after standardizing for size, SMN captured significantly more EWS 364–657 than both EWS 1–657 and EWS 193–363 (Fig. 2C and D). This is difficult to explain, mainly because all the domains present in EWS 364–657 are also present in EWS 1–657. Considering that previous reports have suggested that EWS is capable of self-associating and with numerous reports demonstrating other RGG proteins are capable of self-associating, we hypothesized that this could be due to the presence of a C-terminal self-association site. This would mean EWS 364–657 bound at significantly higher levels because it forms multimeric complexes in vitro, and although EWS 1–657 could also form these complexes, their size may be physically restricted through the presence of the N-terminus and central domains of EWS. In our previous study we used a Biacore to calculate the binding kinetics of the RGG1/SMN complex (8).
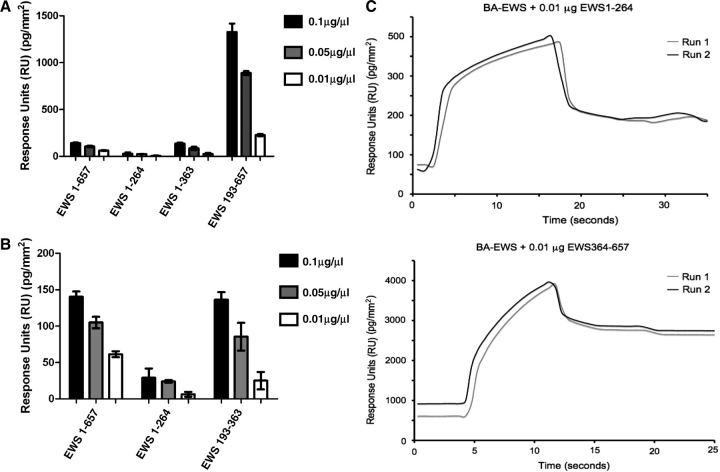
EWS contains a strong self-associates domain within its C-terminus
To test this hypothesis, we screened our panel to see if any of them are capable of binding directly to EWS 1–657 immobilized on a SA-Biacore chip. As with the BA-SMN chip, biotinylated EWS 1–657 (BA-EWS 1–657) was expressed in vitro using the TnT kit and then immobilized on a SA-chip. Initially, this was then pulsed with the EWS 1–657 (full-length), EWS 1–264 (N-terminal domain), EWS 193–363 (central domain) and EWS 364–657 (C-terminal domain) (Fig. 3A). As with the SMN-binding experiments, all constructs containing RGG motifs were captured by the BA-EWS chip (Fig. 3A). The only protein that did not bind was EWS 1–264 (Fig. 3A). EWS 193–363, EWS 364–657 and EWS 1–657 bound to varying degrees (Fig. 3A). After standardizing for size, substantially more EWS 364–657 bound than any other protein (equivalent to approximately five times as much EWS 364–657 bound as EWS 1–657; Fig. 3A). This would suggest that EWS 364–657 is forming oligomeric complexes that then bind the BA-EWS chip. The ability of EWS 193–363 to self-associate was comparable to the EWS 1–657. This is demonstrated more effectively when you remove EWS 364–657 from the graph (Fig. 3B). However, EWS 1-264 only weakly self-associates (Fig. 3B). This is shown in the sample sensorgrams (Fig. 3C), where almost 10× as much EWS 364–657 binds compared with EWS 1–264 [Fig. 3C; ∼200 pg/mm2 (EWS 1–264), compared with ∼2000 pg/mm2 (EWS 364–657)].
Fig. 3.
EWS-derived proteins containing one or more RGG motifs form heterodimers with full-length EWS. (A) BA-EWS was immobilized on a SA-Biacore chip, and pulsed with full-length EWS (EWS 1–657), EWS 1–264 (no RGG domains), EWS 193–363 (one RGG domain) and EWS 364–657 (two RGG domains). Protein stocks were diluted to 0.1, 0.05 and 0.01 µg/µl, and 5 µl of each were injected onto the chip. Injections were repeated three times and the presented figures are the mean values. Values have been standardized for size, using the smallest fragment (EWS 193–363) as a reference. The error bars represent the standard deviations (SD) between the three repeats. (B) The binding values for EWS 1–657, EWS 1–264 and EWS 193–363 in the absence of EWS 364–657 to enable analysis of their comparative binding levels. For all experiments protein binding is represented as RU, with one RU representing 1 pg/mm2 of protein binding to the chip surface. (C) Sample sensorgrams for the binding of EWS 1–264 (top panel) and EWS 364–657 (bottom panel) to the BA-EWS chip. The traces show the real-time binding of both proteins to the BA-EWS chip in response units (pg/mm2). Binding experiments were repeated three times and the sensorgrams for two of these runs are overlaid and presented.
It is possible that the EWS 363–657 protein is forming non-specific hydrophobic complexes that are binding to the Biacore chips non-specifically. In fact, this would account for the high levels of EWS 364–657 captured compared with EWS 1–657 (Fig. 3A). However, if this association were non-specific binding to the Biacore chip, a similar level of association would have been seen with the SA-SMN chip (Fig. 2). This was not the case, with approximately four times more EWS 364–657 captured on the SA-EWS chip than on the SA-SMN chip. We believe this shows the high levels of captured EWS 364–657 is specific to the SA-EWS chip.
Removing the RGG domains reduce EWS self-association
Our results suggest that there are at least two independent self-association domains, one in EWS 193–363 and another in EWS 364–657. The only common domains in these regions are the RGG motifs (Fig. 2A). This led us to hypothesis that EWS self-association is mediated through the RGG motifs. To test this hypothesis, a panel of RGG deletion constructs (EWS delta RGG1+2, EWS delta RGG2+3, and EWS RGG1+3; Fig. 4) were produced and pulsed over the SA-EWS chip. When compared with full-length EWS binding (EWS 1–657), all three constructs were captured at reduced levels (Fig. 4A), with removal of RGG2 appearing to have the most pronounced effect (Fig. 4), with significantly less EWS delta RGG1+2 and EWS delta RGG2+3 captured than EWS delta RGG1+3 (Fig. 4; P < 0.001). This would suggest: (i) removal of any of the RGG motifs reduces binding, suggesting that the RGG domains are at least partially involved in self-association; and (ii) that removal of RGG-2 has the most dramatic effect on binding, suggesting RGG2 may have the highest affinity, and therefore be the true self-association site. Interestingly, the RGG motifs are not homologous with RGG1 containing five RGG repeats, RGG2 containing only three repeats and RGG3 containing 12 repeats. This shows RGG-repeat number does not directly relate to amount of captured protein, with loss of the RGG motif with the fewer repeats having the most pronounced effect. This suggests that although the RGG motifs appear to mediate the association, the sequence in and around these repeats must enhance binding affinities. These observations show that removing the RGG domains has a deleterious effect on self-association, implying that the RGG domains mediate the self-association. However, we are currently expanding these studies to confirm that each of these domains is capable of directly associating with EWS, and whether symmetrical dimethylation of the arginines within the motifs alter the binding affinities.
Fig. 4.
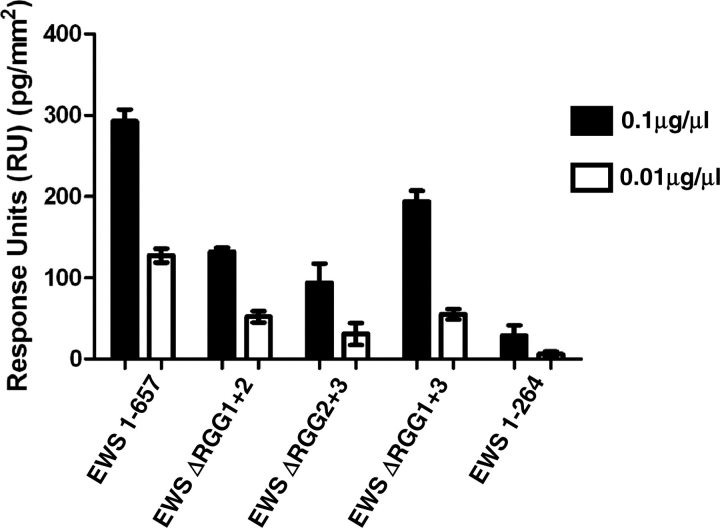
Removal of RGG domains blocks the formation of EWS heterodimers. BA-EWS was immobilized on a SA-Biacore chip, and pulsed with: (i) full-length EWS (EWS 1–657; containing RGG1, 2 and 3); (ii) EWS 1–264 (containing no RGG domains); (iii) EWS lacking the first and second RGG-domains (EWS delta RGG1+2, which contains RGG3); (iv) EWS lacking the second and third RGG domains (EWS delta RGG2+3, which contains RGG1); and (v) EWS lacking the first and third RGG domains (EWS delta RGG1+3, which contains RGG2). Protein stocks were diluted to 0.1 and 0.01 µg/µl, and 5 µl of each were injected onto the chip. Injections were repeated three times and the presented figure is the mean value. For all experiments protein binding is represented as RU, with one RU representing 1 pg/mm2 of protein binding to the chip surface, and the values have been standardized for size, using the smallest fragment (1–264), as a reference. Each presented value is the mean of three repeats. At a protein concentration of 0.1 µg/µl EWS 1–657, EWS ΔRGG1+2, and EWS ΔRGG2+3 and EWS ΔRGG1+3 all bind significantly greater than EWS 1–264 with a P < 0.001.
Removing RGG domains prevents efficient nuclear import of EWS
It has previously been shown that the RGG domains of EWS are required for nuclear import (6). Our findings here might suggest that self-association might play an important role in the nuclear import of EWS. We have previously shown that self-association plays an important role in the import of the EWS associated protein, SMN (21). If self-association were important for efficient nuclear import, removing the RGG motif, either individually, in tandem or completely, would alter EWS targeting. To study this, an additional panel of green fluorescent protein (GFP)-tagged constructs was transiently expressed in HeLa cells. The panel included: (i) ΔRGG1+2 (containing RGG3); (ii) ΔRGG2+3 (containing RGG1); (iii) ΔRGG1+3 (containing RGG2) and (iv) ΔRGG1–3 (containing no RGG domains). Compared with full-length EWS, which is exclusively nuclear, each of the deletion constructs were to a certain degree retained in the cytoplasm (Fig. 5). This shows that although all the RGG domains are needed for the efficient import of EWS, their complete removal does not completely inhibit their import. This finding also fits in with previous studies that suggest the N-terminus of EWS also contains a weak import sequence (22).
Fig. 5.
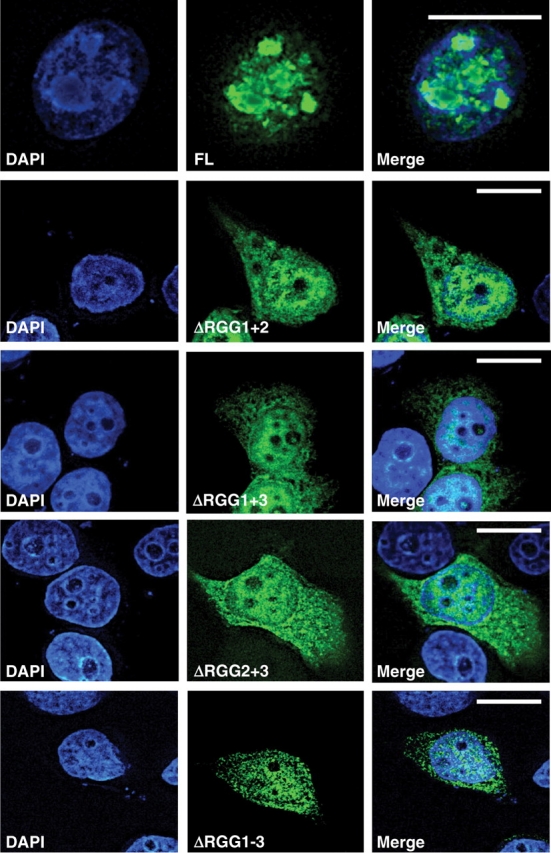
EWS requires > 1 RGG domain for efficient nuclear import. A panel of EWS constructs lacking multiple RGG domains was cloned into the pEGFP C3 vector (BD Bioscience Clontech) and transiently transfected into HeLa cells using GeneJuice (Novagen). Removal of >1 RGG domain prevents efficient nuclear entry of a GFP-tagged EWS construct.
Following our study here, we can state that the domains that mediate self-association are also important for nuclear import (Figs 2–5), and we can speculate that this shows the RGG–RGG complex forms a nuclear import signal. Previous studies have shown self-association is important for nuclear localization. For example, receptor interacting protein 3 (RIP3) has an unconventional NLS that mediates RIP3 self-association (23), as does the export region of class II transactivator (CIITA), which regulators expression of major histocompatibility complex class II genes. Point mutations within this nuclear export domain prevent both nuclear import and reduce self-association (24). These studies demonstrate that self-association and nuclear import are integrally linked. This may also be the case with regards to the EWS protein, with the RGG domains mediating self-association and nuclear entry.
However, it is possible that a more complex mechanism is involved, with the EWS–RGG domains binding to an import adaptor/intermediate instead of the RGG–RGG association forming a direct import signal. Import of the U snRNP complexes, which form the backbone of the spliceosome, display complex import mechanisms (25). This is a bipartite import signal that involves two separate complexes: (i) the 5′-trimethyl-guanosine cap (m3G)/snurportin-1/importin-β trimeric complex; and (ii) the RGG-Sm core proteins/SMN/importin-β complex (25). Because SMN also associates directly with the RGG domains of EWS, it is possible that SMN also acts as an adaptor molecule for EWS, linking EWS with importin-β and mediating import. However, we have previously shown that reducing EWS protein levels in HeLa cells does not alter the cellular distribution and nuclear targeting of SMN (8). Although not conclusive, this does suggest SMN targeting is not dependent on the presence of EWS.
EWS fragments bind RGG and KGG biotinylated peptides
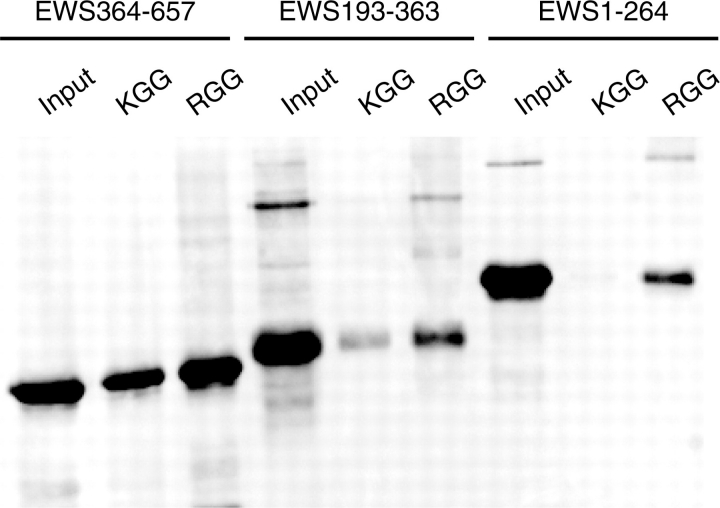
To confirm our initial BiacoreX experiments and to determine if self-association is dependent on the arginine residue of the RGG domain, we performed addition in vitro binding assays using biotinylated RGG- and KGG peptides (Fig. 6). The RGG peptide corresponded to the RGG-1 motif of the EWS protein, while the control KGG peptide was the same motif with the arginine residues substituted with lysines. We have previously used these RGG and KGG peptides to show the R-residues are essential for the SMN–RGG association (8). Interestingly, EWS 193–363 and EWS 364–657 were able to bind both the RGG and KGG peptides (Fig. 6). This suggests that the association is mediated through the glycine residues, while the arginines are dispensable. However, EWS 1–264 associated specifically (but weakly) with RGG, but failed to associate with KGG. This suggests that as well as a nuclear import sequence; the N-terminus of EWS may also contain a weak self-association domain that is RGG specific. However, compared with the strong association identified between RGG and KGG with EWS 364–657 (Fig. 6), the binding of EWS 1–264 could be non-specific.
Fig. 6.
Removal of the arginine residues from the RGG motif does not prevent EWS binding. RGG and KGG biotinylated peptides (200 pmol) were immobilized on M280-streptavidin beads. These were then incubated with 50 µg of purified recombinant EWS 1–264; EWS 193–363; or EWS 364–657. The complex was then washed with PBS-T and bound fractions were eluted in SDS sample buffer. The extracts were then run on SDS–PAGE and transferred to PVDF. Recombinant proteins were then identified by western immunoblot analysis using an anti-histidine (His) monoclonal antibody (Sigma). Five percent of the recombinant protein use for each binding reaction was loaded as an input control (input).
Conclusions
Here, we demonstrate for the first time that the C-terminus of EWS is capable of associating with SMN in vitro, and also provides the first evidence that EWS does self-associate, that this is mediated through the RGG motifs and may be important for nuclear import. This is an important observation, as it may suggest that all RGG motif proteins are capable, at least partially, of self-associating through their RGG domains.
Funding
Andrew’s Buddies and an IBCS studentship support (to D.J.S.); IBCS studentship (A.G.T.); fellowship from the Vandervell Foundation (to P.J.Y. and R.M.); ARC (EO543 and 16537 to P.E.); Devon Northcott Medical Foundation (m1263A to P.J.Y. and P.E.); FightSMA (to C.L.L.); National Institutes of Health (R01 NS41584; R01 HD054413).
Conflict of interest
None declared.
Acknowledgements
The authors thank Prof. Glenn E. Morris for comments and Biacore access.
Glossary
Abbreviations
- ARC
arthritis and rheumatism council
- BA
biotinylated
- BIA
biomolecular interaction analysis
- CIITA
class II transactivator
- EWS
Ewing's sarcoma
- GFP
green fluorescent protein
- His
histidine
- IBCS
institute of biomedical and clinical science
- KGG
lysine-glycine-glycine
- m3G
5′-trimethyl-guanosine cap
- RGG
arginine-glycine-glycine
- RIP3
receptor interacting protein 3
- SEC
size exclusion chromatography
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- SPR
surface plasmon resonance
References
- 1.Azuma M, Embree LJ, Sabaawy H, Hickstein DD. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS ONE. 2007;2:e979. doi: 10.1371/journal.pone.0000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolotti A, Bell B, Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno T, Rao VN, Reddy ES. EWS/Fli-1 chimeric protein is a transcriptional activator. Cancer Res. 1993;53:5859–5863. [PubMed] [Google Scholar]
- 6.Shaw DJ, Morse R, Todd AG, Eggleton P, Lorson CL, Young PJ. Identification of a tripartite import signal in the Ewing Sarcoma protein (EWS) Biochem. Biophys. Res. Commun. 2009;390:1197–1201. doi: 10.1016/j.bbrc.2009.10.120. [DOI] [PubMed] [Google Scholar]
- 7.Deloulme JC, Prichard L, Delattre O, Storm DR. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J. Biol. Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- 8.Young PJ, Francis JW, Lince D, Coon K, Androphy EJ, Lorson CL. The Ewing's sarcoma protein interacts with the Tudor domain of the survival motor neuron protein. Brain Res. Mol. Brain Res. 2003;119:37–49. doi: 10.1016/j.molbrainres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 11.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev. Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 12.Araya N, Hirota K, Shimamoto Y, Miyagishi M, Yoshida E, Ishida J, Kaneko S, Kaneko M, Nakajima T, Fukamizu A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 13.Miranda TB, Khusial P, Cook JR, Lee JH, Gunderson SI, Pestka S, Zieve GW, Clarke S. Spliceosome Sm proteins D1, D3, and B/B' are asymmetrically dimethylated at arginine residues in the nucleus. Biochem. Biophys. Res. Commun. 2004;323:382–387. doi: 10.1016/j.bbrc.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 14.Meister G, Eggert C, Bühler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 15.Brahms H, Meheus L, de Brabandere V, Fischer U, Lührmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimmler M, Bauer L, Nousiainen M, Körner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- 19.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 21.Morse R, Shaw DJ, Todd AG, Young PJ. Targeting of SMN to Cajal bodies is mediated by self-association. Hum. Mol. Genet. 2007;16:2349–2358. doi: 10.1093/hmg/ddm192. [DOI] [PubMed] [Google Scholar]
- 22.Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J. Mol. Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Feng S, Wu M. Multiple roles for nuclear localization signal (NLS, aa 442-472) of receptor interacting protein 3 (RIP3) Biochem. Biophys. Res. Commun. 2008;372:850–855. doi: 10.1016/j.bbrc.2008.05.144. [DOI] [PubMed] [Google Scholar]
- 24.Kretsovali A, Spilianakis C, Dimakopoulos A, Makatounakis T, Papamatheakis J. Self-association of class II transactivator correlates with its intracellular localization and transactivation. J. Biol. Chem. 2001;276:32191–32197. doi: 10.1074/jbc.M103164200. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum. Mol. Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]