Abstract
We reported previously that ethanol treatment regulates D1 receptor phosphorylation and signaling in a protein kinase C (PKC) δ- and PKCγ-dependent fashion by a mechanism that may involve PKC isozyme-specific interacting proteins. Using a PKC isozyme-specific coimmunoprecipitation approach coupled to mass spectrometry, we report the identification of RanBP9 and RanBP10 as novel interacting proteins for both PKCγ and PKCδ. Both RanBP9 and RanBP10 were found to specifically coimmunoprecipitate with both PKCγ and PKCδ; however, this association did not seem to mediate the ethanol regulation of the PKCs. It is noteworthy that the D1 receptor was also found to specifically coimmunoprecipitate with RanBP9/10 from human embryonic kidney (HEK) 293T cells and with endogenous RanBP9 from rat kidney. RanBP9 and RanBP10 were also found to colocalize at the cellular level with the D1 receptor in both kidney and brain tissue. Although overexpression of RanBP9 or RanBP10 in HEK293T cells did not seem to alter the kinase activities of either PKCδ or PKCγ, both RanBP proteins regulated D1 receptor phosphorylation, signaling, and, in the case of RanBP9, expression. Specifically, overexpression of either RanBP9 or RanBP10 enhanced basal D1 receptor phosphorylation, which was associated with attenuation of D1 receptor-stimulated cAMP accumulation. Moreover, treatment of cells with select PKC inhibitors blocked the RanBP9/10-dependent increase in basal receptor phosphorylation, suggesting that phosphorylation of the receptor by PKC is regulated by RanBP9/10. These data support the idea that RanBP9 and RanBP10 may function as signaling integrators and dictate the efficient regulation of D1 receptor signaling by PKCδ and PKCγ.
Dopamine (DA) is a key signaling molecule in the brain and periphery. The actions of this neurotransmitter are mediated by dopamine receptors, which are seven transmembrane-spanning proteins belonging to the large family of G-protein-coupled receptors. Dopamine receptors are divided into two major subfamilies, referred to as D1-like and D2-like, on the basis of their structure, pharmacology, and function (Missale et al., 1998). The D1-like receptors consist of the D1 and D5 subtypes, which couple to Gs/olf proteins to activate adenylyl cyclase and promote the accumulation of intracellular cAMP. In contrast, the D2-like receptors, which consist of the D2, D3, and D4 subtypes, couple to Gi/o proteins, which tend to inhibit adenylyl cyclase and decrease intracellular cAMP levels.
The D1 receptor is abundantly expressed in the forebrain and it is not surprising that aberrant D1 receptor signaling has been linked to various neuropsychiatric disorders such as substance abuse, schizophrenia, and Parkinson's disease. For example, ethanol (EtOH) consumption is reduced in genetically modified mice that lack the D1 receptor, or wild-type mice that are administered D1 selective antagonists R-(+)-7-chloro-8-hydroxy-3-methyl-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine (SCH23390) (El-Ghundi et al., 1998; Price and Middaugh, 2004). Conversely, treatment with D1-selective agonists facilitates EtOH-related behaviors (D'Souza et al., 2003). In addition, D1-selective agonists have been shown to ameliorate the cognitive deficits associated with schizophrenia and improve parkinsonian-like symptoms in animal models (Kebabian et al., 1992; Schneider et al., 1994; McLean et al., 2009). Understanding how D1 receptors are regulated may be useful for future therapeutic interventions.
Receptor phosphorylation is an important post-translational modification that regulates D1 receptor signaling. In particular, receptor phosphorylation generally contributes to desensitization, a process that renders the receptor less sensitive to subsequent agonist stimulation (Kohout and Lefkowitz, 2003). To date, three classes of protein kinases have been reported to phosphorylate the D1 receptor. These include the G protein-coupled receptor kinases (GRKs), cAMP-dependent protein kinase (PKA), and protein kinase C (PKC). GRKs generally phosphorylate the D1 receptor under agonist-activated conditions, which results in receptor desensitization (Tiberi et al., 1996; Gardner et al., 2001; Gainetdinov et al., 2004; Rankin et al., 2006). Likewise, PKA also regulates D1 receptor signaling by modulating the rate of agonist-induced receptor desensitization and intracellular trafficking (Jiang and Sibley, 1999; Mason et al., 2002). In contrast to GRKs and PKA, very little is known about the regulation of the D1 receptor by PKC phosphorylation. We determined recently that PKC constitutively phosphorylates the D1 receptor and that this negatively regulates dopaminergic signaling (Rex et al., 2008). Moreover, we found that EtOH decreases constitutive PKC phosphorylation of the D1 receptor with a concomitant potentiation of dopaminergic signaling (Rex et al., 2008). It is noteworthy that EtOH was found to directly inhibit the enzymatic activities of PKCγ and PKCδ, but only when they were isolated from the plasma membrane fraction, an effect that was not observed for other PKC isozymes that were tested (Rex et al., 2008). The molecular mechanisms underlying the EtOH-mediated inhibition of membrane-associated PKCγ and PKCδ kinase activities and how they target the D1 receptor are at present unclear. One hypothesis for the membrane-specificity of this effect is that PKC isozyme-specific interacting proteins exist in the plasma membrane and impart EtOH-sensitivity to PKCγ and PKCδ or are themselves the targets of EtOH. It is noteworthy that a growing number of interacting proteins for PKCs have been identified (Staudinger et al., 1997; Rodriguez et al., 1999; Piontek and Brandt, 2003; Zemskov et al., 2003). Likewise, several D1 receptor-interacting proteins have also been reported that include various scaffolding/trafficking proteins (Bermak et al., 2001; Heydorn et al., 2004; Free et al., 2007; Hazelwood et al., 2008).
The aim of the current study was to use a proteomic-based approach to identify and characterize PKCγ/δ isozyme-specific interacting proteins that may be regulated by EtOH and associate with the D1 dopamine receptor. We now report the identification of RanBP9 and RanBP10 as dual-interacting proteins for both PKCγ/δ and the D1 receptor. Although RanBP10 and RanBP9 do not seem to mediate EtOH-dependent inhibition of PKCδ or PKCγ, these interacting proteins were found to modulate D1 receptor activity through increased receptor phosphorylation. Moreover, we found that the RanBP9/10-dependent increase in receptor phosphorylation is blocked by PKC inhibitor treatment, consistent with the idea that these scaffolding proteins may target PKCδ or PKCγ to the D1 receptor.
Materials and Methods
Materials.
HEK293-tsa201 (HEK293T) cells were a gift from Dr. V. Ramakrishnan. HA-PKCδ and 3X-FLAG-PKCγ were gifts from Dr. J. F. Mushinski (Mischak et al., 1993) and Dr. A. Newton, respectively. RanBP10-GFP and RanBPM-GFP were gifts from Dr. Guan Wu and Dr. Takeharu Nishimoto, respectively. PKCγ antibody was purchased from GeneTex, Inc. (Irvine, CA). PKCδ antibody was purchased from BD Biosciences (San Jose, CA) and the GFP antibody from Novus Biologicals (Littleton, CO). 3-[1-[3-(dimethylamino)propyl]-5-methoxy-1H-indol-3-yl]-4-(1H-indol-3-yl)-1H-pyrrole-2,5-dione (Gö6983) and 12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3,4-c)-carbazole (Gö6976) were purchased from Calbiochem (San Diego, CA).
Cell Culture and Transfection.
HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 50 μg/ml streptomycin, 50 U/ml gentamicin at 37°C in 5% CO2. HEK293T cells were seeded in 150-mm culture dishes and transfected 24 h later using calcium phosphate precipitation (Clontech, Mountain View, CA).
Coimmunoprecipitation and Immunoblot Analysis from HEK293T Cells.
To identify candidate PKC-interacting proteins that might be regulated by EtOH, HEK293T cells expressing FLAG-PKCγ, HA-PKCδ, or empty vector were incubated in the presence or absence of EtOH (100 mM/10 min). Cells were harvested in PBS, centrifuged at 200g for 10 min and the pellets homogenized in 1 ml of homogenization buffer (250 mM sucrose, 50 mM Tris-HCl, 10 mM EGTA, 2 mM EDTA, 30 mM NaF, and 20 mM sodium pyrophosphate) supplemented with Mini Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The samples were centrifuged at 20,000g for 30 min at 4°C, and the supernatant was discarded. The pellet was resuspended and homogenized with 1 ml of solubilization buffer containing 1% Triton X-100 and supplemented with MiniComplete protease inhibitor cocktail. The homogenate was centrifuged for 5 min at 9000g, and the supernatant was used as the “solubilized” membrane fraction. HA-PKCδ or FLAG-PKCγ was immunoprecipitated from the solubilized fraction using either HA-agarose gel or FLAG-M2 gel (Sigma-Aldrich, St. Louis, MO), respectively, and rotated overnight at 4°C. To identify candidate PKC-interacting proteins, isozyme-specific immunoprecipitates were washed three times with PBS and then sent to Applied Biomics (Hayward, CA) for two-dimensional differential in-gel electrophoresis. In brief, the samples were covalently linked to CyDye Fluors and separated by isoelectric focusing in the first dimension and SDS-PAGE in the second dimension. Image analysis was performed using DeCyder software (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK), and selected spots were chosen for mass spectrometry analysis (matrix-assisted laser desorption ionization/time of flight).
To verify the association of the PKC isozymes or the D1R with RanBP10 and RanBP9, HEK293T cells coexpressing HA-PKCδ, FLAG-PKCγ, or FLAG-D1R with vector, RanBP10-GFP or RanBP9-GFP was subjected to coimmunoprecipitation experiments as described above. The resulting immunoprecipitates were washed three times with PBS, resuspended in SDS-PAGE sample buffer, and incubated for 1 h at 37°C. Proteins were separated electrophoretically using 4 to 12% Bis-Tris NuPage gels/MOPS buffer (Invitrogen, Carlsbad, CA) and transferred to polyvinylidene difluoride (PVDF) membranes (Invitrogen). Membranes were blocked in SuperBlock (Thermo Fisher Scientific, Waltham, MA) overnight followed by incubation with primary antibodies. The blots were washed three times with 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20, incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), and visualized using SuperSignal Chemiluminescence Kit (Thermo Fisher Scientific). The optical density of the immunoblots was quantified by ImageJ software (http://rsbweb.nih.gov/ij/).
In Vitro PKC Assays.
HEK293T cells coexpressing FLAG-PKCγ or HA-PKCδ with either vector, RanBP10-GFP, or RanBP9-GFP were coimmunoprecipitated from the solubilized membrane fraction as described above. Immunoprecipitates were centrifuged at 9000g for 1 min at 4°C, and the pellets were washed three times with PBS. Each pellet was resuspended with 70 μl of PBS. Kinase assays were performed using a PKC Assay Kit (Millipore, Billerica, MA) as described previously (Rex et al., 2008). In brief, the kinase activity in 10 μl of each sample was assayed by measuring the transfer of 32Pi from [γ-32P]ATP to a specific substrate peptide (Millipore). The remaining cell lysates were retained and used to determine PKC expression for each transfection. PKC activity was measured in an assay consisting of assay dilution buffer II (end concentration, 3.33 mM MOPS, pH 7.2, 4.2 mM β-glycerol phosphate, 0.17 mM sodium orthovanadate, 0.17 mM dithiothreitol, and 0.12 mM CaCl2), 83 μM PKC substrate peptide, 0.3 μM PKA inhibitor peptide, 3.3 μM Ca2+/calmodulin-dependent protein kinase inhibitor, PKC lipid activators [80 ng/μl phosphatidyl serine (PS) and 8 ng/μl diacylglycerol (DAG)]. The addition of EtOH was made from a concentrated stock solution. Maximal PKC activity was achieved in the presence of PS and DAG. Basal activity was measured in the presence of 0.5 mM EGTA instead of PS and DAG. Nonspecific activity was determined in the absence of substrate peptide. Each condition was performed in duplicate. Kinase reactions were initiated by the addition of 10 μCi [γ-32P]ATP in Mg2+/ATP cocktail and incubated for 10 min at 30°C. The reactions were terminated by transferring 25 μl of the mixture to P81 filter papers followed by washing three times with 0.75% phosphoric acid and once with acetone. The amount of phosphorylated peptide was determined by scintillation counting and expressed as the percentage of control. To compare PKC activity between the three groups (PKCγ + vector, PKCγ + RanBP10, and PKCγ + RanBP9), the remaining cell lysates were subjected to Western blot and densitometry analysis (ImageJ) to determine the relative amounts of PKC protein. To account for any changes in PKC expression between each cotransfection, the kinase activity was divided by the relative PKC expression (arbitrary units).
cAMP Accumulation Assays.
Transfected HEK293T cells were seeded into 24-well plates coated with poly(d-lysine). Duplicate wells were exposed to dopamine dilutions that were prepared in 20 mM HEPES-buffered DMEM supplemented with 200 μM sodium metabisulfite and 30 μM 4-[(3-butoxy-4-methoxyphenyl)-methyl]-2-imidazolidinone (Ro 20-1724) (a phosphodiesterase inhibitor) (Sigma-Aldrich). Basal activity was determined in the absence of dopamine. The plates were incubated at 37°C for 20 min. The reaction was terminated by removal of the medium and the addition of 3% perchloric acid to each well for 30 min on ice. Each reaction was neutralized by the addition of 15% KHCO3. cAMP accumulation was measured using the method described by Watts and Neve (1996) based on the competitive binding of cAMP to PKA.
Radioligand Binding Assays.
D1 receptor-transfected HEK293T cells were harvested in Ca2+/Mg2+-free Earle's balanced salt solution supplemented with 5 mM EDTA and centrifuged at 200g for 10 min. Cells were lysed in a Dounce homogenizer in 5 mM Tris-HCl, pH 7.4 (at 4°C) and 5 mM MgCl2. The lysate was centrifuged at 20,000g for 30 min, and the pellet was resuspended in 50 mM Tris-HCl (crude membrane fraction). A portion of the membrane suspension was quantified using the BCA protein assay kit. The membrane fraction (100 μl) was added to tubes in triplicate containing [3H]SCH23390 (PerkinElmer Life and Analytical Sciences, Waltham, MA) at a range of concentrations. Nonspecific binding was determined in the presence of (+)-butaclamol (3 μM). Assay tubes were incubated at room temperature for 1.5 h and then terminated by rapid filtration through GF/C filters pretreated with 0.6% polyethylenimine. Bound radioactivity was quantified by liquid scintillation counting.
In Situ Phosphorylation Assays.
These assays were performed as described previously (Rankin et al., 2006). In brief, transfected HEK293T cells were seeded into six-well plates coated with poly(d-lysine). A portion of the transfection was retained in a 100-mm dish for radioligand binding assay to quantify the expression of D1 receptor expression. Forty-eight hours after transfection, the medium from each well was replaced with phosphate-free medium supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 10 μg/ml gentamicin and incubated for 1 h. The medium was replaced with 1 ml of phosphate-free DMEM containing 106 μCi/ml [32P]orthophosphoric acid (PerkinElmer Life and Analytical Sciences) for 60 min. PKC inhibitors (Gö6983 and Gö6976 at 10 μM) were included during this incubation period for select samples. Subsequently, specific wells were challenged with basal medium or media containing 10 μM dopamine for 10 min. The cells were placed on ice, rinsed with ice-cold Earle's balanced salt solution, and lysed with solubilization buffer supplemented with MiniComplete protease inhibitor cocktail for 1 h at 4°C. Cell lysates were cleared by centrifugation, and protein concentration was quantified using the BCA protein assay kit. D1 receptor expression was determined by radioligand binding assays using cells seeded in the 100-mm dish. Equal amounts of D1 receptor for each condition were incubated with anti-FLAG-M2 agarose gel overnight, washed three times, and resolved using 4 to 12% Bis-Tris NuPage gels using MOPS buffer. Dried gels were subjected to autoradiography.
Immunofluorescence and Confocal Studies of Proximal Tubule Cells in Rat Kidney.
Normotensive 12-week-old Wistar-Kyoto rats were anesthetized with pentobarbital (50 mg/kg i.p.). The kidneys were perfused with normal saline until the effluent was clear. Thin sections (3 μm) of formalin-fixed and paraffin-embedded rat kidney were deparaffinized in xylene and then rehydrated with step-down concentrations of ethanol. After antigen retrieval with citrate buffer, the D1 receptor was visualized using a monoclonal mouse anti-D1 receptor antibody (1:100; Novus) followed by Alexa Fluor 568-donkey anti-mouse IgG antibody (red; Invitrogen). RanBP9 was visualized using a polyclonal goat anti-RanBP9 antibody (1:100; Santa Cruz Biotechnology) followed by Alexa Fluor 488-donkey anti-goat IgG antibody (green; Invitrogen). As a negative control, the primary antibodies were replaced with normal mouse, rabbit, or goat IgG at an appropriate dilution. The slides were mounted in Vectashield Mounting Medium with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). The immunofluorescent images were acquired using Zeiss 510 confocal laser scanning microscope (Carl Zeiss GmbH, Jena, Germany). Colocalization of D1 receptor and RanBP9 is indicated by the development of a yellow color in the merged images.
D1 Receptor and RanBP9 Coimmunoprecipitation from Kidney Cells.
Immortalized renal proximal tubule cells (passage ∼20) from adult normotensive adult humans and Wistar-Kyoto rats were used for coimmunoprecipitation (coIP). The renal proximal tubule cells were cultured at 37°C in 95% air/5% CO2 atmosphere in DMEM/F-12 with transferrin (5 μg/ml), insulin (5 μg/ml), epidermal growth factor (10 ng/ml), dexamethasone (4 μg/ml), and 5% FBS in a 100-mm Petri dish. The cells (80% confluence) were made quiescent by incubation for 2 h in medium without FBS before the treatment with a D1-like receptor agonist, fenoldopam (1 μM, 10 min). The cells were lysed with ice-cold lysis buffer for 1 h and centrifuged at 16,000g for 30 min. Equal amounts of cell lysate proteins (supernatant; 500 μg) were used for coIP. coIP was performed according to the manufacturer's immunoprecipitation protocol (Santa Cruz Biotechnology). In brief, 2 μg of rabbit D1 receptor antibody (D1R 408), the specificity of which has been established previously (Yu et al., 2006), was incubated with immunoprecipitated matrix at 4°C on a rotator for 1 h, and then the complex was incubated with 500 μg of lysate protein at 4°C overnight. The immunoprecipitate was pelleted and washed four times with phosphate-buffered saline. The pellet was resuspended in sample buffer, boiled, and immunoblotted with the rabbit RanBP9 antibody. To determine the specificity of the bands, normal rabbit IgG was used as negative control, and cell lysate (immunoblot) was used as positive control.
Identification of RanBP9 Expression in D1 Receptor-Expressing Neurons in Mouse Striatum.
Immunohistochemical localization of RanBP9 and RanBP10 was performed to determine possible coexpression in neurons in the striatum that express the D1 receptor. Coronal brain sections were processed from mice in which enhanced green fluorescent protein (EGFP) is expressed under the control of the D1 dopamine receptor gene promoter (Gong et al., 2007). Frozen cut coronal sections of brains from animals that had been perfusion-fixed with 4% formaldehyde, postfixed overnight, and saturated with 20% sucrose were incubated overnight with dilutions of either rabbit monoclonal antibodies directed against RanBP9 (1:100; Eptiomics, Burlingame CA) or RanBP10 (1:100) (Schulze et al., 2008). Visualization of RanBP9 and RanBP10 primary antibody localization was determined with affinity-purified Cy-3 conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch Laboratories, West Grove, PA), whereas EGFP may be visualized directly under epifluorescence illumination.
Data Analysis.
For immunoblots and phosphorylation assays, the relative intensities of bands on the autoradiographs were determined by scanning and analyzing the bands using ImageJ software. Figures depict representative graphs or autoradiographs for each experimental condition. Where shown, data are presented as the mean ± S.E.M. All statistical analyses were performed with a level of significance established at p < 0.05. Statistical analyses were conducted using Prism 4 software (GraphPad Software Inc., San Diego, CA).
Results
Identification of PKCγ- and PKCδ-Interacting Proteins.
We reported previously that EtOH treatment decreased the kinase activities of membrane-associated PKCγ and PKCδ under lipid-activated conditions (Rex et al., 2008). In contrast, cytosolic PKCγ and PKCδ activities were unaffected by EtOH (Rex et al., 2008). One hypothesis for the membrane-specificity of this effect is that PKC isozyme-specific interacting proteins exist in the plasma membrane and impart EtOH-sensitivity to PKCγ and PKCδ or are the targets of EtOH themselves. To address this hypothesis, we used a PKC isozyme-specific coimmunoprecipitation approach coupled to mass spectrometry sequence analysis to identify novel PKCγ- and PKCδ-interacting proteins, including those that might be regulated by EtOH.
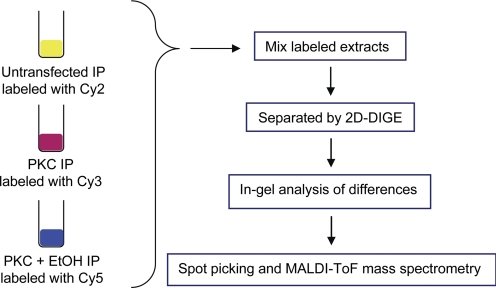
The proteomics approach that we used is illustrated in Fig. 1. FLAG-tagged PKCγ was immunoprecipitated from solubilized membrane fractions prepared from HEK293T cells that were either untreated or treated with EtOH for 15 min. As a control, untransfected HEK293T cells were used for immunoprecipitation with anti-FLAG agarose beads. Likewise, HA-tagged PKCδ was immunoprecipitated from transfected cells using HA-agarose beads under identical conditions as described above. The precipitates for each condition (control, PKCγ, and PKCδ) were separately covalently labeled with fluorochromes (Cy2, Cy3, and Cy5) and subjected to two-dimensional differential in-gel electrophoresis. Proteins were separated by isoelectric focusing in the first dimension and SDS-PAGE in the second dimension. This method allows three samples to be run on a single gel and comparison of each sample based on the wavelength of the specific fluorochrome.
Fig. 1.
Experimental scheme for identifying candidate PKC-interacting proteins. FLAG-PKCγ or HA-PKCδ constructs were expressed in HEK293T cells, whereas untransfected cells served as controls. The cells were either untreated or pretreated with ethanol (100 mM/15 min), disrupted, and the PKC constructs were immunoprecipitated (IP) from the solubilized membrane fractions using antisera to either the FLAG or HA tags, as described under Materials and Methods. The immunoprecipitates were then labeled with CyDYE fluors as follows: untransfected cells, Cy2; PKC-transfected, Cy3; PKC-transfected plus ethanol treatment, Cy5. The labeled proteins were then resolved by 2D-gel electrophoresis and identified as described under Materials and Methods.
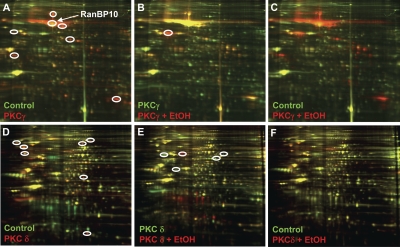
Figure 2, A and D, shows a comparison of immunoprecipitated proteins from control (untransfected) versus PKC-transfected cells. In this comparison, any unique proteins that are identified in the PKC-transfected samples are tentatively identified as specific interacting proteins for those PKCs. From the PKCγ screen, we identified seven protein spots that seemed to be specific for PKCγ (red spots) (Fig. 2A). Proteins that are common to both PKC and control immunoprecipitates were visualized as yellow spots and are presumably nonspecific in nature. From the PKCδ screen, we also identified seven protein spots that seemed to be PKCδ-specific (red spots, Fig. 2D). We were also interested in identifying any immunoprecipitated proteins in the PKC-transfected samples that increased or decreased in intensity upon treatment of the cells with EtOH. Comparing PKCγ versus PKCγ + EtOH-treated samples, a single red protein spot was present only in the EtOH-treated sample (Fig. 2B). Furthermore, this protein spot was not present in control (untransfected) cells, suggesting that the association of this protein with PKCγ might be regulated by EtOH (Fig. 2C). We identified five additional protein spots from the PKCδ + EtOH-treated samples that also seemed to be PKCδ-specific, one of which might be up-regulated by EtOH treatment (red spot, Fig. 2E). All of the circled protein spots in Fig. 2 were excised and subjected to mass spectrometry sequence analysis.
Fig. 2.
Identification of candidate PKC-interacting proteins. Each CyDYE-labeled PKC sample was simultaneously separated on a 2D-gel and scanned at specific wavelengths to reveal each of the CyDYE signals. Overlay of images reveal protein spots that are presumably specific (or nonspecific) to a given sample. Circled, protein spots selected for identification by mass spectrometry. A to C, PKCγ; D to F, PKCδ.
The candidate PKC isozyme-specific interacting proteins were identified based on comparing peptide sequences derived from the mass spectrometry analyses with protein sequences submitted to the National Center for Biotechnology Information database. In some instances a single spot on the gel corresponded to multiple parent proteins. For the PKCγ screen, eight protein spots were sequenced that corresponded to 31 different parent proteins; however, in many instances, only a single peptide sequence for that protein was identified. In general, we only considered proteins as bona fide if they were identified through multiple matching peptide fragments. Table 1 shows a list of such proteins obtained from the PKCγ and PKCδ proteomic screens. A variety of different proteins were identified ranging from scaffolding proteins to other protein kinase C isozymes. It is noteworthy that 70-kDa heat shock protein (Hsp-70) was the only candidate PKC-interacting protein that was common to both PKC screens. Heat shock proteins assist in protein folding and assembly and are frequently observed in proteomic screens, usually as nonspecific interactors.
TABLE 1.
Candidate PKCγ and PKCδ interacting proteins identified using mass spectrometry analysis
Proteins identified based on tandem mass spectrometry analysis of selected protein spots highlighted in Fig. 2.
| Candidate Proteins | Function |
|---|---|
| PKCγ-interacting | |
| RAC/CDC42 exchange factor (GEFT) | Cell signaling |
| Ran-binding protein 10 (RanBP10) | Scaffolding protein |
| Hsp-70a | Protein folding/stability |
| Glial fibrillary acidic protein | Cell structure/communication |
| PKCα | Cell signaling |
| Ephrin receptor EphB1b | Cell signaling |
| Tripartite motif proteinb | Protein localization |
| Voltage-gated sodium channel subtype III | Cell signaling |
| PKCδ-interacting | |
| Cytoplasmic dynein | Trafficking |
| Hsp-70a | Protein folding/stability |
| Hsp-90 | Protein folding/stability |
| Inositol trisphosphate receptor type 1 | Ca2+ signaling |
| Microtubule-actin crosslinking factor | Cell structure |
| Mono-ADP-ribosyltransferaseb | Protein modification |
| Valosin-containing protein | Regulation of cellular activities |
Common to both PKCγ and PKCδ proteomics screens.
Identified in EtOH-treated samples only.
Based on the sequence data for the protein spot excised from the PKCγ + EtOH-treated sample (Fig. 2B), 11 nonoverlapping peptide fragments were identified that correspond to the ephrin receptor EphB1 (Holder and Klein, 1999). Additional peptide fragments identified from this protein spot corresponded to the tripartite motif protein TRIM5 (Johnson and Sawyer, 2009). From the PKCδ screen, the single spot identified in the EtOH-treated samples (Fig. 2E) corresponds to mono-ADP-ribosyltransferase (Okada et al., 2008). Although the potential regulation of these three proteins by EtOH and their interactions with PKCγ and PKCδ should be investigated further, our initial assumption is that they are not responsible for the unique membrane-specific regulation of PKCγ and PKCδ activities observed previously (Rex et al., 2008), because none of them were identified in both PKC screens. Thus, while continuing to pursue the mechanism of EtOH-mediated regulation of PKCγ and PKCδ activities, we chose to initially focus on characterizing other candidate PKC-interacting proteins that might be especially relevant to regulating D1 receptor function. One such protein, RanBP10, and a closely related protein, RanBP9, were chosen for further characterization because RanBP9 has been shown to interact with other GPCRs (see below).
RanBP10 and RanBP9 Interact with PKCγ and PKCδ.
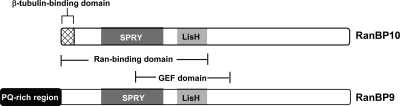
RanBP10 was among the candidate PKCγ-interacting proteins from our proteomic screen (Fig. 2A and Table 1). It is noteworthy that seven nonoverlapping peptide fragments of RanBP10 were identified by mass spectrometry sequence analysis, all of which span the majority of the protein sequence (Table 2). RanBP10 is a 67-kDa scaffolding protein that is expressed in a variety of tissues including the brain (Wang et al., 2004; Haase et al., 2008; Schulze et al., 2008) and is known to interact with several proteins that include the protein Ran and the receptor protein tyrosine kinase for hepatocyte growth factor, MET (Wang et al., 2004). It is noteworthy that there is a closely related protein, RanBP9, that has high sequence similarity to RanBP10 (Wang et al., 2004) and has been found to associate with several GPCRs, including the μ-opioid receptor and the metabotropic glutamate receptors (mGluRs) mGluR2 and mGluR8, as well as with MET (Wang et al., 2004; Seebahn et al., 2008; Talbot et al., 2009). Figure 3 shows a structural comparison of RanBP9 and RanBP10 highlighting several conserved protein-protein interactions and functional domains such as dual-specific kinase splA and ryanodine receptor, lissencephaly type-1-like homology, Ran protein binding domain, and guanine nucleotide exchange factor domain (Schulze et al., 2008). A β-tubulin binding domain has been localized to the N-terminal region of RanBP10 (Schulze et al., 2008). One major difference between RanBP10 and RanBP9 is the presence of a poly proline/glutamine tract at the N terminus of RanBP10 (Fig. 3), the role of which is unclear.
TABLE 2.
RanBP10 peptides identified in coimmunoprecipitation-mass spectrometry experiments
The protein spot indicated in Fig. 2A was picked and subjected to mass spectrometry sequencing. The seven peptides shown correspond to fragments of RanBP10. The location is indicated on the basis of multiple alignments of the peptide fragments with RanBP10. Full-length RanBP10 is 620 amino acids.
| Trypsin-Digested Peptides | Location (RanBP10) |
|---|---|
| LYPAVNQQETPLPRSWSPK | 42–N60 |
| YNYIGLSQGNLRVHYK | 63–N78 |
| ATHPIPAACGIYYFEVK | 92–N108 |
| GRDGYMGIGLSAQGVNMNRLPGWDK | 113–N137 |
| QFVEMVNGTDSEVR | 336–N349 |
| RQLCGGNQAATER | 499–N510 |
| EPVCAALNSAILESQNLPK | 566–N584 |
Fig. 3.
Structural organization of RanBP10 and RanBP9. The RanBP10 and RanBP9 proteins were aligned and compared. Conserved regions include a dual-specific kinase splA and ryanodine receptor domain, lissencephaly type-1-like homology domain, Ran binding protein domain, and guanine nucleotide exchange factor domain. RanBP10 contains a poly(proline/glutamine) tract at the amino terminus that is absent in RanBP9. [Adapted from Yudin D and Fainzilber M (2009) Ran on tracks–cytoplasmic roles for a nuclear regulator. J Cell Sci 122:587–593. Copyright © 2009 The Company of Biologists, Ltd. Used with permission.]
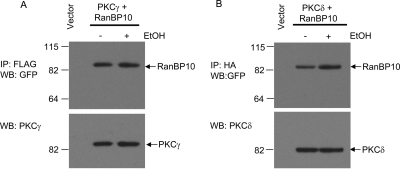
To initially verify the interaction of RanBP10 with PKCγ, we used a coimmunoprecipitation approach as shown in Fig. 4. HEK293T cells were cotransfected with FLAG-PKCγ and either empty vector or RanBP10-GFP. FLAG-PKCγ was immunoprecipitated from solubilized membrane fractions followed by one-dimensional SDS-PAGE electrophoresis and transference to PVDF membranes. The association of RanBP10 with PKCγ was visualized by Western blot analysis. In addition, the effect of EtOH treatment on the association of RanBP10 with PKCγ was examined. For these experiments, cells were pretreated with EtOH for 15 min before the preparation of the membrane fraction and subsequent immunoprecipitation. We confirmed the interaction of RanBP10 with PKCγ using this approach (Fig. 4A). Furthermore, the RanBP10-PKCγ association was not modulated by EtOH treatment. These experiments were also extended to examine the possible association of RanBP10 with PKCδ, even though RanBP10 was not identified in the initial PKCδ proteomic screen. It is noteworthy that RanBP10 was also found to coimmunoprecipitate with PKCδ, and this interaction does not seem to be modulated by EtOH either (Fig. 4B).
Fig. 4.
Association of RanBP10 with PKCγ and PKCδ. Coimmunoprecipitation and immunoblot analyses of RanBP10 with PKCγ (A) or PKCδ (B). HEK293T cells were transfected with empty vector, FLAG-PKCγ + RanBP10-GFP, or HA-PKCδ + RanBP10-GFP. PKCγ or PKCδ was immunoprecipitated from solubilized membrane fractions prepared from HEK293T cells that were either untreated or pretreated with ethanol (100 mM/15 min). As a control, the solubilized membrane fraction prepared from cells transfected with empty vector was incubated with either FLAG or HA beads. After SDS-PAGE, the gels were blotted with antisera directed to GFP (top) or to PKCγ or PKCδ. The experiment shown is representative of three independent experiments.
Based on the high amino acid sequence similarity of RanBP10 and RanBP9, we wondered whether RanBP9 might also associate with PKCγ or PKCδ. As shown in Supplemental Fig. 1, RanBP9 was also found to coimmunoprecipitate with both PCKγ and PKCδ, but EtOH treatment had no significant effect on this interaction.
Effect of RanBP10 and RanBP9 on PKCγ and PKCδ Kinase Activities.
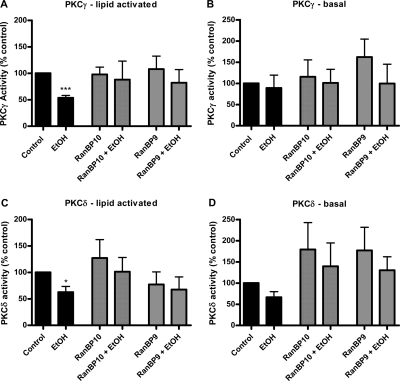
Although EtOH treatment did not seem to alter the association of either RanBP10 or RanBP9 with PKCδ or PKCγ (Figs. 4, Supplemental Fig. 1), given that both RanBP proteins interacted with both PKCs, we wondered whether RanBP10 and/or RanBP9 could still somehow be involved in the EtOH-mediated decrease in PKC activity. To directly test this possibility, we used in vitro kinase assays to evaluate PKC activities as we had performed previously (Rex et al., 2008). Our working hypothesis was that if either RanBP9 or RanBP10 were involved in imparting EtOH sensitivity to the PKCs, then overexpressing either RanBP protein might enhance the EtOH inhibition. PKCδ or PKCγ were coexpressed with RanBP10, RanBP9, or empty vector in HEK293T cells, followed by immunoprecipitation of the PKCs from a solubilized membrane preparation. The PKC immunoprecipitates were then used directly in the kinase assays. As we determined previously (Rex et al., 2008), the addition of EtOH to the kinase assays decreased the lipid-activated PKCγ and PKCδ activities without affecting basal enzyme activities (Fig. 5, A and C). In contrast, we observed that overexpression of either RanBP9 or RanBP10 blocked the EtOH-mediated inhibition of PKC activities without affecting basal or lipid-activated activities (Fig. 5). Taken together, these results suggest that neither RanBP9 nor RanBP10 imparts sensitivity of PKCγ or PKCδ to inhibition by ethanol but rather they can inhibit this response, perhaps by blocking the association of the PKCs with another distinct interacting protein.
Fig. 5.
Effect of RanBP9 overexpression on ethanol-dependent attenuation of PKC activities. Epitope-tagged PKCs were immunoprecipitated from solubilized membrane fractions prepared from HEK293T cells coexpressing either PKCγ (A and B) or PKCδ (C and D) with vector, RanBP10, or RanBP9. The kinase activities of the isozyme-specific immunoprecipitates were directly assessed using an in vitro kinase assay as described under Materials and Methods. For each condition, the kinase activities were measured under basal or lipid-activated (plus phosphatidylserine and diacylglycerol) conditions in the presence or absence of 100 mM ethanol. Data are presented as the mean ± S.E.M. of at least three independent experiments. Results that are significantly different from the control groups are indicated as ∗, p < 0.05; ∗∗∗, p < 0.001, paired Student's t test.
RanBP10 and RanBP9 Associate with the D1 Receptor.
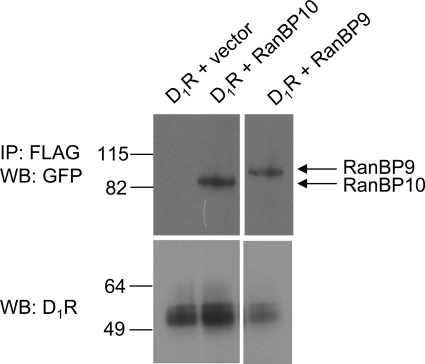
Although RanBP10 and RanBP9 do not seem to mediate the EtOH-induced attenuation of PKCγ and PKCδ kinase activities, they could function as important PKC scaffolding proteins. In fact, a role for RanBP10 and RanBP9 as scaffolding proteins has been well described previously (Wang et al., 2004; Haase et al., 2008; Talbot et al., 2009). In particular, both Ran10 and RanBP9 have been reported to associate with receptors expressed at the plasma membrane that include the MET tyrosine kinase receptor and the μ-opioid receptor, respectively (Wang et al., 2004; Talbot et al., 2009). This was of particular interest because the μ-opioid receptor is a G protein-coupled receptor, as is the D1 dopamine receptor. Furthermore, the D1 receptor is known to be phosphorylated by PKC (Gardner et al., 2001; Rex et al., 2008), which is, at least partially, mediated by PKCδ and PKCγ (unpublished observations). We hypothesized that either RanBP10 or RanBP9 might also associate with the D1 receptor and function as a scaffolding protein, perhaps to position PKCδ or PKCγ in close proximity to the receptor. To examine this possibility, we used coimmunoprecipitation analyses to determine whether either RanBP9 or RanBP10 could associate with the D1 receptor (Fig. 6). HEK293T cells were transfected with a FLAG-D1R construct and either RanBP10, RanBP9, or empty vector. A crude membrane preparation was prepared, and the D1 receptor was immunoprecipitated from the solubilized membrane preparation, separated by one-dimensional SDS-PAGE, and transferred to PVDF membranes. The presence of the D1 receptor and both RanBP proteins were detected by Western blotting. It is noteworthy that both RanBP10 and RanBP9 were found to specifically coimmunoprecipitate with the D1 receptor (Fig. 6). These results suggest that RanBP9 and RanBP10 are indeed associated with the D1 receptor when expressed in HEK293T cells.
Fig. 6.
RanBP10 and RanBP9 coimmunoprecipitate with the D1 receptor. The FLAG-D1 receptor was expressed in HEK293T cells either alone or with RanBP10-GFP or RanBP9-GFP. Membranes were prepared and solubilized followed by immunoprecipitation of the D1 receptor using anti-FLAG antisera as described under Materials and Methods. After resolution of the precipitates on SDS-PAGE, the gels were blotted with antisera directed to GFP (top) to detect RanB10 or RanBP9 or antisera directed to the D1 receptor (bottom). The experiment shown is representative of three independent experiments.
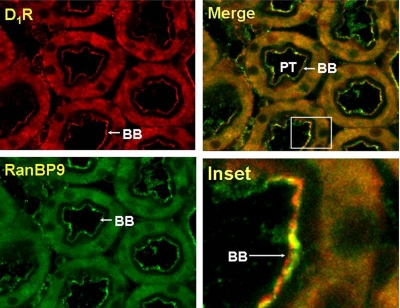
RanBP9 Associates with the D1 Receptor in Kidney Tissue.
We were interested in examining the association of RanBP9 and RanBP10 with the D1 receptor in endogenous expressing tissues. Unfortunately, only the RanBP9 antisera proved useful in kidney tissue. To determine whether RanBP9 and the D1 receptor associate endogenously, we first examined the cellular localization of these proteins in the rat kidney proximal convoluted tubule, in which the D1 receptor is expressed (Fig. 7). Both RanBP9 and the D1 receptor were observed to be colocalized in the brush border/apical membrane of the proximal convoluted tubule of the rat kidney (Fig. 7). The D1 receptor is abundantly expressed in the proximal tubule of humans and rodent kidneys, in which it negatively regulates renal sodium transport at the brush border/apical and basolateral membrane (Zeng et al., 2008). We next attempted to coimmunoprecipitate RanBP9 and the D1 receptor from renal proximal tubule cells. Figure 8 shows an experiment in which the D1 receptor was immunoprecipitated from solubilized renal proximal tubule cells, resolved by one-dimensional SDS-PAGE, and then blotted with antisera directed against RanBP9. Lane 1 shows that a protein corresponding in size to RanBP9 is present in the immunoprecipitates but absent if normal rabbit IgG is used instead of the RanBP9 antisera (Fig. 8, lane 3). This experiment was also performed subsequent to treating the cells with a D1 agonist; however, this did not alter the association of RanBP9 with the D1 receptor (Fig. 8, lane 2). Taken together, these experiments show that RanBP9 and the D1 receptor endogenously associate in the kidney.
Fig. 7.
D1 receptor and RanBP9 colocalization in rat kidney. D1R (red) and RanBP9 (green) are expressed in the brush border (BB), apical membrane, and to a lesser extent observed in the cytoplasm of the proximal tubule (PT) (original magnification, 400×). The immunofluorescence images were acquired using Zeiss 510 confocal laser scanning microscope. Colocalization of the D1 receptor and RanBP9 is indicated by the yellow color in the merged image. The experiment shown is representative of three independent experiments.
Fig. 8.
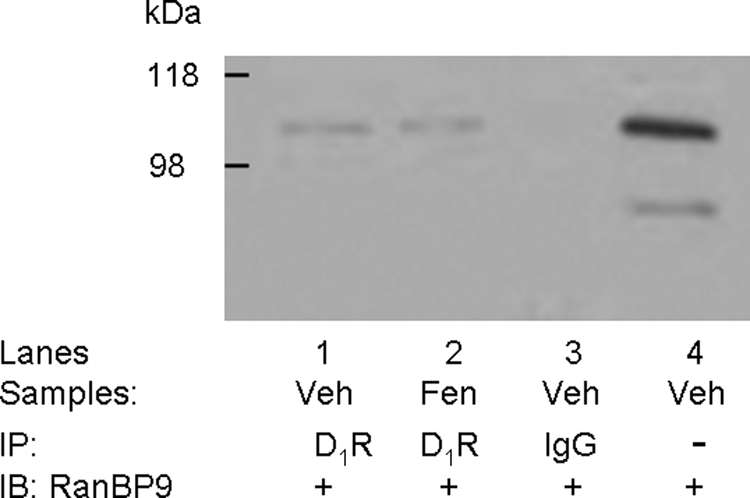
Coimmunoprecipitation of the D1 receptor and RanBP9 endogenously expressed in kidney tissue. Immortalized renal proximal tubule cells (passage, ∼20, 80% confluent), made quiescent by incubating the cells for 2 h in medium without FBS, were treated with vehicle (Veh; sterile water, 10 min, lane 1) or a D1-like agonist, fenoldopam (Fen; 1 μM, 10 min, lane 2). Lysates of treated cells or IgG were immunoprecipitated (IP) and immunoblotted (IB) as shown. To determine the specificity of the bands, normal rabbit IgG was used for immunoprecipitation and served as the negative control (lane 3), and Veh-cell lysate (immunoblot, lane 4) was used as positive control. The studies were performed three times with similar results.
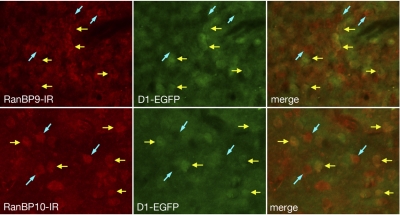
RanBP9/10 and the D1 Receptor Exhibit Cellular Colocalization in Brain Tissue.
We were also interested in examining the association of RanBP9/10 and the D1 receptor in the brain. In this study, immunohistochemical methods demonstrated widespread RanBP9 and RanBP10 immunoreactivity in the forebrain, including the majority of neurons in the cerebral cortex, olfactory bulb and striatum. This pattern of distribution of RanBP9 and RanBP10 immunoreactivity matches that reported for the mRNA for these genes as illustrated in the Allen brain atlas (available at http://www.brain-map.org/). In the striatum, half of the medium spiny neurons express the D1 receptor, which were visualized directly by their expression of EGFP in the brains of mice used for immunohistochemical localization in this study (Gong et al., 2007). As shown in Fig. 9, immunoreactivity for both RanBP9 (top) and RanBP10 (bottom), was present in striatal neurons. In addition, both antigens were present in D1-EGFP-positive neurons (yellow arrows) and D1-EGFP-negative neurons (blue arrows). These data demonstrate that both RanBP9 and RanBP10 colocalize in striatal neurons expressing the D1 receptor.
Fig. 9.
Cellular colocalization of the D1 receptor and RanBP9 in brain. Brain sections through the striatum from transgenic mice in which D1 receptors are marked with EGFP were processed for fluorescence immunohistochemical localization of RanBP9 (top) and RanBP10 (bottom). Immunoreactivity for both RanBP9 (RanBP-IR) and RanBP10 (RanBP-IR) colocalize with both D1-EGPF-positive striatal neurons (yellow arrows) and D1-EGFP-negative striatal neurons (blue arrows). Thus, both RanBP9 and RanBP10 seem to colocalize with striatal neurons expressing the D1 receptor.
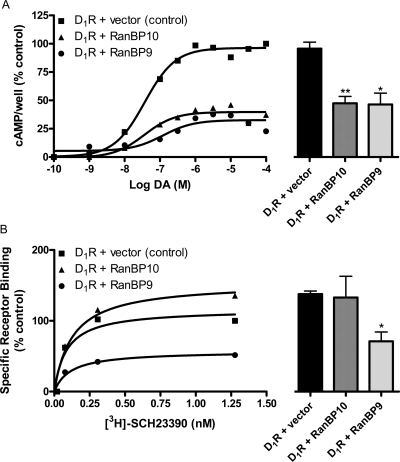
RanBP10 and RanBP9 Decrease D1 Receptor Signaling.
To determine whether RanBP9 or RanBP10 association with the D1 receptor exhibits functional consequences, we examined D1 receptor-stimulated cAMP accumulation in the absence or presence of RanBP9/RanBP10 overexpression (Fig. 10). It is noteworthy that coexpression of either RanBP10 or RanBP9 significantly decreased maximal DA-stimulated cAMP accumulation to approximately 45% of control (Fig. 10A). However, the potency of the response was not significantly altered (EC50 ± S.E.M.: control, 81 ± 58; RanBP10, 93 ± 48; RanBP9, 137 ± 12 nM). To establish whether the inhibitory effect of RanBP10 or RanBP9 was due to changes in D1 receptor binding activity, membranes prepared from cells expressing the D1 receptor ± RanBP10 or RanBP9 were assayed using the D1-selective radioligand [3H]SCH23390. It is noteworthy that coexpression of RanBP10 did not significantly alter D1 receptor expression, whereas coexpression of RanBP9 decreased D1 receptor binding by ∼50% (Fig. 10B). These results suggest that the effect of RanBP9 on D1 receptor signaling may, at least in part, be explained by decreased receptor expression. In contrast, alterations in receptor expression cannot explain the functional effect of RanBP10 expression on D1 receptor signaling.
Fig. 10.
Effect of RanBP10 and RanBP9 on D1 receptor expression and signaling. A, RanBP10 or RanBP9 was coexpressed with the D1 receptor in HEK293T cells followed by assessment of DA-stimulated cAMP accumulation. The curves on the left are representative experiments, whereas the histograms on the right represent averaged data from three experiments using 100 μM DA as the stimulus. Expression of either RanBP10 or RanBP9 decreased maximal DA-stimulated cAMP accumulation compared with control (D1 receptor + empty vector): Emax ± S.E.M. values for D1R + RanBP10 and D1R + RanBP9 were 47 ± 6 and 46 ± 10% of control, respectively. B, saturation radioligand binding experiments were performed on membranes prepared from control cells (D1 receptor + empty vector) or cells expressing the D1 receptor along with either RanBP10 or RanBP9. The curves on the left are representative experiments, whereas the histograms on the right represent averaged Bmax data from three experiments. RanBP9 coexpression reduced the D1 receptor Bmax value to 60 ± 14% of control. (∗, p < 0.05, ∗∗, p < 0.005, paired Student's t test).
RanBP10 and RanBP9 Increase Basal D1 Receptor Phosphorylation by a PKC-Dependent Mechanism.
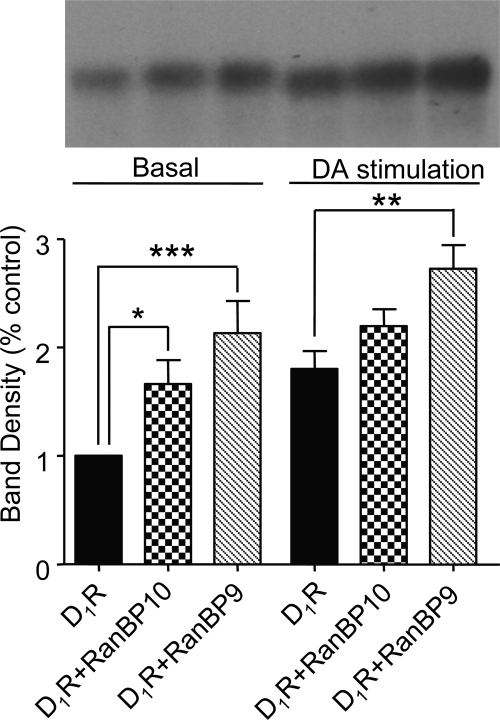
We reported previously that the D1 receptor seems to be constitutively phosphorylated by PKC in the basal state and that this attenuates agonist-induced cAMP accumulation (Rex et al., 2008). We further showed that EtOH selectively attenuated the kinase activities of PKCγ and PKCδ, decreased basal D1 receptor phosphorylation, and potentiated cAMP accumulation (Rex et al., 2008). Given that RanBP9 and RanBP10 associate with both PKCγ and PKCδ and with the D1 receptor, we wondered whether RanBP9 or RanBP10 might scaffold PKCδ and PKCγ in close proximity to the D1 receptor to facilitate receptor phosphorylation. To test this hypothesis, we examined the effect of RanBP10 and RanBP9 on D1 receptor phosphorylation under basal or agonist-stimulated conditions using an in situ phosphorylation assay. It is noteworthy that coexpression of either RanBP10 or RanBP9 with the D1 receptor significantly elevated basal D1 receptor phosphorylation (Fig. 11). As expected, DA stimulation increased D1 receptor phosphorylation compared with basal. We showed previously that the agonist-induced phosphorylation of the D1 receptor is mediated through GRK pathways (Gardner et al., 2001; Rankin et al., 2006). It is noteworthy that in the presence of RanBP9 or RanBP10, the percentage increase in phosphorylation in response to agonist was similar to or slightly less than that observed in control cells (Fig. 12), suggesting that the main effect of RanBP9 or RanBP10 was to increase the basal phosphorylation state of the D1 receptor.
Fig. 11.
RanBP10 and RanBP9 increase basal D1 receptor phosphorylation. In situ phosphorylation experiments were performed on HEK293T cells expressing the D1 receptor, D1 receptor + RanBP10, and D1 receptor + RanBP9 as described under Materials and Methods. Cells were incubated with media (control) or DA (10 μM) for 10 min. Top, autoradiogram of D1 receptor immunoprecipitates from a representative in situ phosphorylation assay. The lanes in the gel correspond to the bars at the bottom. Bottom, average values of band densities for each condition. The data are normalized as the percentage of control for each individual experiment. The histograms represent the mean ± S.E.M. from four independent experiments (∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗, p < 0.001, analysis of variance followed by Bonferroni pair-wise comparisons).
Fig. 12.
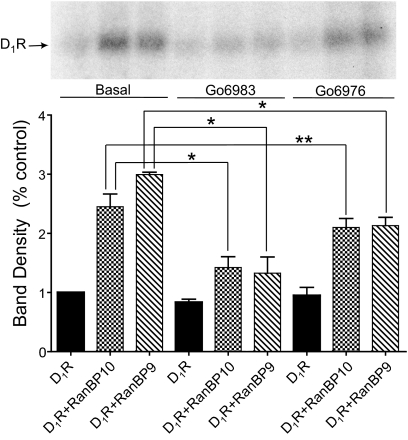
The RanBP9 and RanBP10-induced increase of D1 receptor phosphorylation is blocked by PKC inhibitors. HEK293T cells expressing the D1 receptor, D1 receptor + RanBP10, and D1 receptor + RanBP9 were treated with the PKC inhibitors Gö6983 (10 μM) or Gö6976 (10 μM) for 60 min followed by the assessment of basal phosphorylation using an in situ phosphorylation assay as described in Fig. 11. Top, autoradiogram of D1 receptor immunoprecipitates from a representative in situ phosphorylation assay. The lanes in the gel correspond to the bars at the bottom. Bottom, average values of band densities for each condition. The data are normalized as the percentage of control for each individual experiment. The histograms represent the mean ± S.E.M. from three independent experiments (analysis of variance followed by Bonferroni pair-wise comparisons; ∗, p < 0.05, ∗∗, p = 0.056).
We were particularly interested in the mechanisms responsible for the RanBP10- and RanBP9-mediated potentiation of basal D1 receptor phosphorylation and whether or not this might be mediated by PKCs as hypothesized. To test this, we used the in situ phosphorylation assay described above except that the cells were treated with the PKC inhibitors, Gö6983 and Gö6976, before incubation in basal media. Gö6983 and Gö6976 are known to inhibit several PKC isozymes including PKCδ and PKCγ (Martiny-Baron et al., 1993; Stempka et al., 1999). As described previously (Gardner et al., 2001; Rex et al., 2008), treatment with PKC inhibitors markedly decreased basal D1 receptor phosphorylation (Fig. 12). However, treatment with the PKC inhibitors significantly attenuated the RanBP10- and RanBP9-promoted increase in basal D1 receptor phosphorylation, Gö6983 exhibiting a somewhat greater effect than Gö6976 (Fig. 12). Taken together, these results suggest a role for PKCs, potentially PKCδ or PKCγ, in the RanBP10- and RanBP9-dependent modulation of D1 receptor signaling.
Discussion
The major finding of the current investigation is the identification of RanBP9 and RanBP10 as interacting proteins for protein kinase C isozymes γ and δ and for the D1 dopamine receptor. Our proteomic screen initially identified RanBP10 as an interacting protein for PKCγ, although coimmunoprecipitation analyses subsequently revealed that both PKCγ and PKCδ were also capable of interacting with the structurally related protein RanBP9. Although our initial interest was in attempting to identify interacting proteins that might impart EtOH sensitivity to PKCγ/δ, neither RanBP9 nor RanBP10 seemed to affect the kinase activities of PKCγ or PKCδ. Rather, overexpression of either RanBP protein actually blocked the EtOH-mediated inhibition of PKCγ/δ activities. Although other explanations are possible, it is tempting to speculate that this effect could be due to RanBP9 or RanBP10 preventing the interaction of the PKCs with another interacting protein that might be involved in the EtOH modulation.
Because our overall goal was the investigation of PKC-mediated regulation of D1 receptor signaling, we wondered whether either PKC-interacting protein, RanBP9 or RanBP10, could also interact with the D1 receptor and affect its function. Our coimmunoprecipitation analyses revealed that both RanBP proteins were indeed capable of interacting with D1 receptor complexes when expressed in HEK293 cells. Moreover, RanBP9 and the D1 receptor are endogenously expressed in the proximal tubule of the rat kidney, and RanBP9 and the D1 receptor were found to coimmunoprecipitate using primary cultures of human renal proximal tubule cells. We also found that both RanBP9 and RanBP10 and the D1 receptor were partially colocalized at the cellular level within brain tissue.
Functionally, the association of RanBP9 and RanBP10 with the D1 receptor seems to attenuate signaling. This was evident as a decrease in maximal cAMP accumulation elicited by receptor stimulation. It is noteworthy that overexpression of RanBP9 attenuated D1 receptor expression, whereas RanBP10 did not. This might reflect fundamental differences in the interactions of RanBP9 and RanBP10 with the receptor that will merit further investigation. Although a decrease in receptor expression by RanBP9 could contribute to the attenuated cAMP accumulation, this cannot explain the functional inhibition by RanBP10, suggesting that other mechanisms must be involved. It is noteworthy that overexpression of both RanBP9 and RanBP10 led to an increase in the basal phosphorylation state of the D1 receptor. We found previously that the D1 receptor is constitutively phosphorylated by PKC and that this attenuates receptor signaling (Rex et al., 2008). Based on our findings that RanBP9 and RanBP10 associate with PKCδ and PKCγ, we examined the possibility that the increase in basal D1 receptor phosphorylation was mediated by PKC. Indeed, HEK293T cells pretreated with select PKC inhibitors blocked the RanBP-dependent increase in basal D1 receptor phosphorylation. Taken together, we propose that RanBP9 and RanBP10 regulate D1 receptor phosphorylation and signaling though a PKC-dependent mechanism.
The identification of RanBP9 and RanBP10 as dual interacting proteins for the D1 receptor and PKCs is consistent with their function as scaffolding proteins. Both RanBP9 and RanBP10 contain multiple functional domains, and there is a growing body of evidence that they both function as scaffolding molecules in immune and neural tissues (Murrin and Talbot, 2007). Of particular interest are RanBP9's interactions with membrane receptors, especially those within the GPCR superfamily such as the μ-opioid receptor (MOR) and mGluRs. RanBP9 associates with the Gαi-coupled MOR and modulates its signaling (Murrin and Talbot, 2007). Overexpression of RanBP9 reduces MOR-stimulated ERK activation and inhibits β-arrestin-mediated receptor internalization (Talbot et al., 2009). However, in contrast to our findings with the D1 receptor, RanBP9 does not alter MOR phosphorylation or affect its regulation of cAMP production (Talbot et al., 2009). RanBP9 also associates with mGluR2 and mGluR8 in both HEK293 cells and the synaptic layers of the retina (Seebahn et al., 2008), although the role of RanBP9 in regulating glutamatergic signaling is currently unclear (Seebahn et al., 2008). Compared with RanBP9, there is little published information regarding RanBP10-dependent modulation of signaling; however, both RanBP10 and RanBP9 are described as scaffolding proteins for MET, a membrane-bound receptor tyrosine kinase for hepatocyte growth factor (Wang et al., 2004). Association of RanBP9 with MET increases recruitment of the protein SOS to MET and enhances ERK signaling. RanBP10 also associates with MET but functions as a dominant-negative to reduce SOS recruitment and decrease ERK-mediated signaling (Wang et al., 2004).
At the cellular level, we show that RanBP9 and RanBP10 can associate with the membrane-bound D1 receptor and PKCδ/γ. These findings are consistent with the site of action of the D1 receptor and regulation by PKCδ and PKCγ. It is noteworthy that Denti et al., (2004) showed that endogenous levels of RanBP9 expressed in the epithelial cells of lung, kidney, and breast are localized to the plasma membrane and cytoplasm. In the retina, RanBP9 is localized to the synaptic processes of cholinergic amacrine cells (Seebahn et al., 2008). In contrast, RanBP10 is expressed in multiple cellular locations, including the cytoplasm and nucleus of megakaryocyte cells (Schulze et al., 2008); however, there is currently no information regarding the subcellular localization of RanBP10 in other cell types, including neurons. Collectively, the subcellular locations of RanBP9 and RanBP10 seem to depend largely on the cell type.
Consistent with the notion of RanBP9 and RanBP10 functioning as scaffolding proteins, neither RanBP protein seems to modulate the kinase activities of PKCδ or PKCγ, but rather may play a role in the spatial and temporal organization of the D1 receptor-PKC signaling complex. Several PKC-interacting proteins have been described that anchor specific PKC isozymes to the appropriate signaling complex at the correct intracellular location. For example, receptor for activated C kinase 1 (RACK1) preferentially binds to PKCβii and PKCε (Ron et al., 1994; Stebbins and Mochly-Rosen, 2001; Besson et al., 2002). RACK1 is believed to stabilize the active conformation of PKCβii and shuttle the enzyme to the correct subcellular site (Ron et al., 1999). Likewise, RACK2 associates with PKCε and translocates the kinase to the Golgi membranes (Csukai et al., 1997). Protein interacting with C kinase 1 (PICK1) is a scaffolding protein that associates with presynaptic mGluR7 and PKC. It is noteworthy that PICK1 is required for the PKC-dependent phosphorylation of mGluR7 and for stable receptor expression at the cell surface (Suh et al., 2008). It is noteworthy that mGluR7-dependent neuronal plasticity is impaired in mice lacking PICK1 (Suh et al., 2008).
In summary, we report RanBP10 and RanBP9 as novel scaffolding proteins for the D1 receptors PKCδ and PKCγ. We speculate that these dual-specificity scaffolding proteins may function as signaling integrators and dictate the efficient regulation of D1 receptor signaling by PKCδ and PKCγ.
Supplementary Material
Acknowledgments
We thank the National Institute of Health National Institute of Neurological Disorders and Stroke DNA sequencing facility for generating all sequence data. We thank Dr. Harald Schulze for providing the RanBP10 antisera. We also thank Dr. Vanitha Ramakrishnan for the HEK293tsa201 cells.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This research was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grants R37-HL023081, P01-HL074940, P01-HL068686, R01-HL092196]; the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [R01-DK039308]; and Intramural Research funds from the National Institutes of Health National Institute of Neurological Disorders and Stroke and the National Institutes of Health National Institute of Mental Health.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.063727.
- PKC
- protein kinase C
- PKA
- protein kinase A
- GPCR
- G protein-coupled receptor
- HEK
- human embryonic kidney
- DA
- dopamine
- GFP
- green fluorescent protein
- PAGE
- polyacrylamide gel electrophoresis
- DMEM
- Dulbecco's modified essential medium
- FBS
- fetal bovine serum
- PVDF
- polyvinylidene difluoride
- EtOH
- ethanol
- mGluR
- metabotropic glutamate receptor
- MOR
- μ-opioid receptor
- GRK
- G protein-coupled receptor kinase
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- DAG
- diacylglycerol
- PS
- phosphatidyl serine
- coIP
- coimmunoprecipitation
- D1R
- dopamine 1 receptor
- EGFP
- enhanced green fluorescent protein
- Hsp-70
- 70-kDa heat shock protein
- ERK
- extracellular signal-regulated kinase
- PICK1
- protein interacting with C kinase 1
- RACK
- receptor for activated C kinase
- SCH23390
- R-(+)-7-chloro-8-hydroxy-3-methyl-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine
- Gö6976
- 12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3,4-c)-carbazole
- Gö6983
- 3-[1-[3-(dimethylamino)propyl]-5-methoxy-1H-indol-3-yl]-4-(1H-indol-3-yl)-1H-pyrrole-2,5-dione
- Ro 20-1724
- 4-[3-butoxy-4-methoxyphenyl)-methyl]-2-imidazolidinone.
References
- Bermak JC, Li M, Bullock C, Zhou QY. (2001) Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat Cell Biol 3:492–498 [DOI] [PubMed] [Google Scholar]
- Besson A, Wilson TL, Yong VW. (2002) The anchoring protein RACK1 links protein kinase Cepsilon to integrin beta chains. Requirements for adhesion and motility. J Biol Chem 277:22073–22084 [DOI] [PubMed] [Google Scholar]
- Csukai M, Chen CH, De Matteis MA, Mochly-Rosen D. (1997) The coatomer protein beta'-COP, a selective binding protein (RACK) for protein kinase Cepsilon. J Biol Chem 272:29200–29206 [DOI] [PubMed] [Google Scholar]
- Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. (2004) RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem 279:13027–13034 [DOI] [PubMed] [Google Scholar]
- D'Souza MS, Ikegami A, Olsen CM, Duvauchelle CL. (2003) Chronic D1 agonist and ethanol coadministration facilitate ethanol-mediated behaviors. Pharmacol Biochem Behav 76:335–342 [DOI] [PubMed] [Google Scholar]
- El-Ghundi M, George SR, Drago J, Fletcher PJ, Fan T, Nguyen T, Liu C, Sibley DR, Westphal H, O'Dowd BF. (1998) Disruption of dopamine D1 receptor gene expression attenuates alcohol-seeking behavior. Eur J Pharmacol 353:149–158 [DOI] [PubMed] [Google Scholar]
- Free RB, Hazelwood LA, Cabrera DM, Spalding HN, Namkung Y, Rankin ML, Sibley DR. (2007) D1 and D2 dopamine receptor expression is regulated by direct interaction with the chaperone protein calnexin. J Biol Chem 282:21285–21300 [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. (2004) Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci 27:107–144 [DOI] [PubMed] [Google Scholar]
- Gardner B, Liu ZF, Jiang D, Sibley DR. (2001) The role of phosphorylation/dephosphorylation in agonist-induced desensitization of D1 dopamine receptor function: evidence for a novel pathway for receptor dephosphorylation. Mol Pharmacol 59:310–321 [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. (2007) Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci 27:9817–9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Nordmann C, Sedehizade F, Borrmann C, Reiser G. (2008) RanBPM, a novel interaction partner of the brain-specific protein p42(IP4)/centaurin alpha-1. J Neurochem 105:2237–2248 [DOI] [PubMed] [Google Scholar]
- Hazelwood LA, Free RB, Cabrera DM, Skinbjerg M, Sibley DR. (2008) Reciprocal modulation of function between the D1 and D2 dopamine receptors and the Na+,K+-ATPase. J Biol Chem 283:36441–36453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Søndergaard BP, Hadrup N, Holst B, Haft CR, Schwartz TW. (2004) Distinct in vitro interaction pattern of dopamine receptor subtypes with adaptor proteins involved in post-endocytotic receptor targeting. FEBS Lett 556:276–280 [DOI] [PubMed] [Google Scholar]
- Holder N, Klein R. (1999) Eph receptors and ephrins: effectors of morphogenesis. Development 126:2033–2044 [DOI] [PubMed] [Google Scholar]
- Jiang D, Sibley DR. (1999) Regulation of D(1) dopamine receptors with mutations of protein kinase phosphorylation sites: attenuation of the rate of agonist-induced desensitization. Mol Pharmacol 56:675–683 [PubMed] [Google Scholar]
- Johnson WE, Sawyer SL. (2009) Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61:163–176 [DOI] [PubMed] [Google Scholar]
- Kebabian JW, Britton DR, DeNinno MP, Perner R, Smith L, Jenner P, Schoenleber R, Williams M. (1992) A-77636: a potent and selective dopamine D1 receptor agonist with antiparkinsonian activity in marmosets. Eur J Pharmacol 229:203–209 [DOI] [PubMed] [Google Scholar]
- Kohout TA, Lefkowitz RJ. (2003) Regulation of G protein-coupled receptor kinases and arrestins during receptor desensitization. Mol Pharmacol 63:9–18 [DOI] [PubMed] [Google Scholar]
- Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marmé D, Schächtele C. (1993) Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J Biol Chem 268:9194–9197 [PubMed] [Google Scholar]
- Mason JN, Kozell LB, Neve KA. (2002) Regulation of dopamine D(1) receptor trafficking by protein kinase A-dependent phosphorylation. Mol Pharmacol 61:806–816 [DOI] [PubMed] [Google Scholar]
- McLean SL, Idris NF, Woolley ML, Neill JC. (2009) D(1)-like receptor activation improves PCP-induced cognitive deficits in animal models: Implications for mechanisms of improved cognitive function in schizophrenia. Eur Neuropsychopharmacol 19:440–450 [DOI] [PubMed] [Google Scholar]
- Mischak H, Goodnight JA, Kolch W, Martiny-Baron G, Schaechtle C, Kazanietz MG, Blumberg PM, Pierce JH, Mushinski JF. (1993) Overexpression of protein kinase C-delta and -epsilon in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence, and tumorigenicity. J Biol Chem 268:6090–6096 [PubMed] [Google Scholar]
- Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. (1998) Dopamine receptors: from structure to function. Physiol Rev 78:189–225 [DOI] [PubMed] [Google Scholar]
- Murrin LC, Talbot JN. (2007) RanBPM, a scaffolding protein in the immune and nervous systems. J Neuroimmune Pharmacol 2:290–295 [DOI] [PubMed] [Google Scholar]
- Okada H, Tajima A, Shichiri K, Tanaka A, Tanaka K, Inoue I. (2008) Genome-wide expression of azoospermia testes demonstrates a specific profile and implicates ART3 in genetic susceptibility. PLoS Genet 4:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek J, Brandt R. (2003) Differential and regulated binding of cAMP-dependent protein kinase and protein kinase C isoenzymes to gravin in human model neurons: Evidence that gravin provides a dynamic platform for the localization for kinases during neuronal development. J Biol Chem 278:38970–38979 [DOI] [PubMed] [Google Scholar]
- Price KL, Middaugh LD. (2004) The dopamine D1 antagonist reduces ethanol reward for C57BL/6 mice. Alcohol Clin Exp Res 28:1666–1675 [DOI] [PubMed] [Google Scholar]
- Rankin ML, Marinec PS, Cabrera DM, Wang Z, Jose PA, Sibley DR. (2006) The D1 dopamine receptor is constitutively phosphorylated by G protein-coupled receptor kinase 4. Mol Pharmacol 69:759–769 [DOI] [PubMed] [Google Scholar]
- Rex EB, Rankin ML, Ariano MA, Sibley DR. (2008) Ethanol regulation of D(1) dopamine receptor signaling is mediated by protein kinase C in an isozyme-specific manner. Neuropsychopharmacology 33:2900–2911 [DOI] [PubMed] [Google Scholar]
- Rodriguez MM, Ron D, Touhara K, Chen CH, Mochly-Rosen D. (1999) RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry 38:13787–13794 [DOI] [PubMed] [Google Scholar]
- Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. (1994) Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA 91:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Jiang Z, Yao L, Vagts A, Diamond I, Gordon A. (1999) Coordinated movement of RACK1 with activated betaIIPKC. J Biol Chem 274:27039–27046 [DOI] [PubMed] [Google Scholar]
- Schneider JS, Sun ZQ, Roeltgen DP. (1994) Effects of dihydrexidine, a full dopamine D-1 receptor agonist, on delayed response performance in chronic low dose MPTP-treated monkeys. Brain Res 663:140–144 [DOI] [PubMed] [Google Scholar]
- Schulze H, Dose M, Korpal M, Meyer I, Italiano JE, Jr, Shivdasani RA. (2008) RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules. J Biol Chem 283:14109–14119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebahn A, Rose M, Enz R. (2008) RanBPM is expressed in synaptic layers of the mammalian retina and binds to metabotropic glutamate receptors. FEBS Lett 582:2453–2457 [DOI] [PubMed] [Google Scholar]
- Staudinger J, Lu J, Olson EN. (1997) Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J Biol Chem 272:32019–32024 [DOI] [PubMed] [Google Scholar]
- Stebbins EG, Mochly-Rosen D. (2001) Binding specificity for RACK1 resides in the V5 region of beta II protein kinase C. J Biol Chem 276:29644–29650 [DOI] [PubMed] [Google Scholar]
- Stempka L, Schnölzer M, Radke S, Rincke G, Marks F, Gschwendt M. (1999) Requirements of protein kinase cdelta for catalytic function. Role of glutamic acid 500 and autophosphorylation on serine 643. J Biol Chem 274:8886–8892 [DOI] [PubMed] [Google Scholar]
- Suh YH, Pelkey KA, Lavezzari G, Roche PA, Huganir RL, McBain CJ, Roche KW. (2008) Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 58:736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JN, Skifter DA, Bianchi E, Monaghan DT, Toews ML, Murrin LC. (2009) Regulation of mu opioid receptor internalization by the scaffold protein RanBPM. Neurosci Lett 466:154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberi M, Nash SR, Bertrand L, Lefkowitz RJ, Caron MG. (1996) Differential regulation of dopamine D1A receptor responsiveness by various G protein-coupled receptor kinases. J Biol Chem 271:3771–3778 [DOI] [PubMed] [Google Scholar]
- Wang D, Li Z, Schoen SR, Messing EM, Wu G. (2004) A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/Erk signaling. Biochem Biophys Res Commun 313:320–326 [DOI] [PubMed] [Google Scholar]
- Watts VJ, Neve KA. (1996) Sensitization of endogenous and recombinant adenylate cyclase by activation of D2 dopamine receptors. Mol Pharmacol 50:966–976 [PubMed] [Google Scholar]
- Yu P, Asico LD, Luo Y, Andrews P, Eisner GM, Hopfer U, Felder RA, Jose PA. (2006) D1 dopamine receptor hyperphosphorylation in renal proximal tubules in hypertension. Kidney Int 70:1072–1079 [DOI] [PubMed] [Google Scholar]
- Zemskov EA, Jana NR, Kurosawa M, Miyazaki H, Sakamoto N, Nekooki M, Nukina N. (2003) Pro-apoptotic protein kinase C delta is associated with intranuclear inclusions in a transgenic model of Huntington's disease. J Neurochem 87:395–406 [DOI] [PubMed] [Google Scholar]
- Zeng C, Armando I, Luo Y, Eisner GM, Felder RA, Jose PA. (2008) Dysregulation of dopamine-dependent mechanisms as a determinant of hypertension: studies in dopamine receptor knockout mice. Am J Physiol Heart Circ Physiol 294:H551–H569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.