Abstract
HLA-DM catalyzes peptide dissociation and exchange in class II MHC molecules through a mechanism that has been proposed to involve disruption of specific components of the conserved hydrogen bond network in MHC-peptide complexes. HLA-DR1 molecules with alanine substitutions at each of the six conserved H-bonding positions were expressed in cells and susceptibility to DM catalytic activity was evaluated by measuring release of CLIP. The mutants αN62A, αN69A, αR76A, and βH81A DR1 were fully susceptible to DM-mediated CLIP release, and βN82A resulted in spontaneous release of CLIP. Using recombinant soluble DR1 molecules, amino acid βN82 was observed to contribute disproportionately in stabilizing peptide complexes. Remarkably, the catalytic potency of DM with each β chain mutant was equal to or greater than that observed with wild type DR1. Our results support the conclusion that no individual component of the conserved hydrogen bond network plays an essential role in the DM catalytic mechanism.
Introduction
Class II MHC molecules initially assemble with the chaperone invariant chain (Ii) followed by transport to endosomal compartments and proteolytic cleavage of Ii, leaving a fragment, CLIP, largely buried in the peptide-binding groove. (1). HLA-DM catalyzes CLIP dissociation and peptide exchange reactions in class II molecules, accelerating the loading process for peptide antigens (2–4)and editing the repertoire of peptides presented to CD4+ T cells. DM is a nonpolymorphic MHC class II protein that is structurally similar to other class II molecules (5). However, DM does not have the capacity to bind peptide antigens and it functions as a chaperone-catalyst, stabilizing peptide-free (“empty”) class II molecules (6) and accelerating CLIP dissociation and peptide exchange through a mechanism that involves transient direct physical interaction with class II-peptide complexes. DM accelerates the rate of dissociation of all peptides (7), not just CLIP, but catalytic potency differs depending on the kinetic stability of the complex (7–10)and other less defined features of the complex (11–13). The capacity of DM to differentially “edit” peptide complexes has important biological implications through skewing the repertoire of foreign and self-peptide complexes available for activation or tolerance induction in CD4+ T cells.
The structural basis for the DM catalytic mechanism remains poorly understood. The general orientation of the physical interaction between DM and substrate MHC class II molecules (i.e. HLA-DR) has been defined by using mutational and cross-linking approaches (14–16).It is likely that DM preferentially binds to and stabilizes a relatively unpopulated conformational isomer of MHC class II-peptide complexes, characterized by a loss or weakening of noncovalent interactions that stabilize peptide binding (7, 17, 18). Two general sets of interactions are largely responsible for peptide binding; peptide sequence-dependent interactions between peptide side chains (anchors) and subsites or “pockets” in the peptide-binding groove, and a conserved hydrogen bond network formed by non-polymorphic amino acids in the MHC protein and main chain atoms in bound peptide (19). The anchor-pocket interactions are primarily responsible for determining peptide-binding specificity whereas the conserved hydrogen bond network provides a basal contribution to stability and constrains the orientation of peptide in the binding site. Destabilization of conserved hydrogen bonds has been hypothesized to be a primary component of the DM catalytic mechanism (5, 7, 20, 21). This is attractive because the hydrogen bond network is a conserved feature, consistent with the universal capacity of DM to accelerate the dissociation of peptide complexes. There is strong evidence that the network contributes greatly to stabilizing peptide complexes (22, 23). In addition, this mechanism would account for results indicating that catalytic potency is inversely proportional to kinetic stability (7). If one or more conserved hydrogen bond was the primary target for disruption in the catalytic mechanism, one might predict that the energy of stabilization would be reduced by an approximately constant factor, independent of the sequence of the bound peptide. Indeed, Narayan et al. recently proposed that DM specifically targets the hydrogen formed by the conserved histidine at position β81 in MHC class II molecules (21). HLA-DR1 molecules with an asparagine substitution at this position were reported to form highly unstable peptide complexes, and peptide dissociation was not further enhanced by DM, possibly because DM cannot further disrupt a hydrogen bond that does not exist in the mutant molecule.
In the present study, two approaches were used to systematically analyze the effect of conserved hydrogen-bond disrupting mutations on DM catalytic potency. We postulated that mutational disruption of specific hydrogen bonds targeted in the catalytic mechanism would result in reduced catalytic potency, consistent with the results reported by Narayan et al. (21). Instead, our results indicate that the conserved hydrogen bond formed by histidine β81 is not a primary target in the DM catalytic mechanism. Indeed, our findings support the conclusion that none of the conserved hydrogen bonds is a critical target necessary for DM catalyzed peptide dissociation.
Materials and Methods
Expression of mutant DR1 molecules in T2 cells
Full length DR1α and DR1β (DRA*0101/DRB1*0101) and mutant constructs were cloned into retroviral vectors pLPCX or pLXSN (Clontech). Constructs encoding full-length DM α and β chains were fused with FMDV.2A sequence(24)by PCR. The DMA-2A-DMB construct was cloned into retroviral vector MigR1, which has a GFP marker driven by an internal ribosomal entry site. T2 and Phoenix cell lines were obtained from American Type Culture Collection. High-titer retroviral supernatants were generated by transfection of Phoenix cells with pLPCX-DRA, pLXSN-DRB or MigR1-DMA-2A-DMB (25). T2-DRα-DRβ double positive cell lines were obtained by DRα retroviral infection and puromycin selection, followed by DRβ retroviral infection and G418 selection (Invitrogen). To co-express DM, cells expressing wild type or mutant DR1 were infected with DMA-2A-DMB retrovirus and sorted for GFP expression with a FACSVantage Cell Sorter (Becton Dickinson).
Antibodies and flow cytometry
Fluorophore-conjugated mAbs to HLA-DR (L243), CLIP (CerCLIP), HLA-DM (MaP.DM1) and isotype-matched negative control mAbs were purchased from BD Pharmingen. Cell surface staining was performed with a combination of mAbs according to the standard procedures. Intracellular staining was performed using BD Cytofix/Cytoperm™ kit according to the manufacturer’s instructions. Stained cells were analyzed on a FACScan flow cytometer (Becton Dickinson) and data were analyzed with FlowJo 8.4 software.
Expression and purification of soluble DR1 (sDR1) and soluble DM (sDM)
Stable S2 transfectants expressing sDR1A (residues 1–192) and sDR1B (residues 1–198) were induced for 7 days in BD Baculogold Max-XP serum free media (BD Biosciences) with 1 mM CuSO4. Cell culture supernatant was collected by centrifugation and sDR1 purified with L243 mAb affinity chromatography and aggregates were removed using a TSK-GEL G3000SW analytical gel filtration column. Soluble DM was purified from supernatants of S2 transfectants as described (4, 14).
Peptide labeling and sDR1-peptide complex formation
The peptides HA (PRFVKQNTLRLAT) and CLIP (VSKMRMATPLLMQ) were commercially synthesized with the N-terminus acetylated. Alexa Fluor 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester was used to covalently attach the Alexa Fluor 488 (AF488) fluorophore to the lysine residue in the peptide. Labeled peptides were purified by reverse phase chromatography and labeling was confirmed by MALDI mass spectrometry. sDR1-peptide complexes were formed by incubating sDR1 (5–10 μM) with 50 or 250 μM peptide in 10 mM citrate-phosphate buffer pH 5.3 and 150 mM NaCl. After an overnight incubation at 37°C, complexes were purified on a TSK-GEL G3000SW analytical gel filtration column and concentrated with a micron centrifugal device.
Fluorescence polarization peptide dissociation assays
To measure peptide dissociation rates, 50 nM sDR1 complexes containing AF488-labeled peptide was incubated with 20 μM unlabeled peptide in the absence or presence of sDM (0.25–2 μM). Assays were performed at 37°C in 10 mM citrate-phosphate buffer pH 5.0, 150 mM NaCl, and 0.05 % Tween-20 in a final volume of 75 μl. All reactions were carried out in Corning 384 well low flange black flat bottom polystyrene NBS microplates. To prevent evaporation during measurements, 10 μl of mineral oil was layered over the reactions and the plate was covered with a VIEWseal plate seal (Greiner Bio-One). Fluorescence polarization measurements were made using a Tecan Infinite F200 plate reader equipped with polarizers and two 485 nm (± 20 nm) bandwidth filters for excitation and two 535 nm (± 25 nm) bandwidth filters for emission. A total of 25 flashes were used for each reading and the integration time was set to 20 μsec. The program i-Control was used to collect data and to calculate the fluorescence polarization, p, which is defined as , where IV and IH are the intensity of the emission at polarizations both parallel and perpendicular to the excitation source, and G is a factor to correct for instrumental differences in detecting emission components. The experiments containing complexes of WT, W61A, or H81A were monitored for 1000 cycles of 432 sec. For assays containing the 488HA-N82A complex, the reactions were monitored for 1000 cycles of 8 sec in the absence of sDM and for 600 cycles of 3 sec in the presence of sDM. Rate constants were obtained by fitting the data points to the single exponential decay equation p= Ae−kt+ C using Prism 4.0 (GraphPad Software).
Results and Discussion
Susceptibility of mutant HLA-DR molecules to DM-catalyzed CLIP dissociation in cells
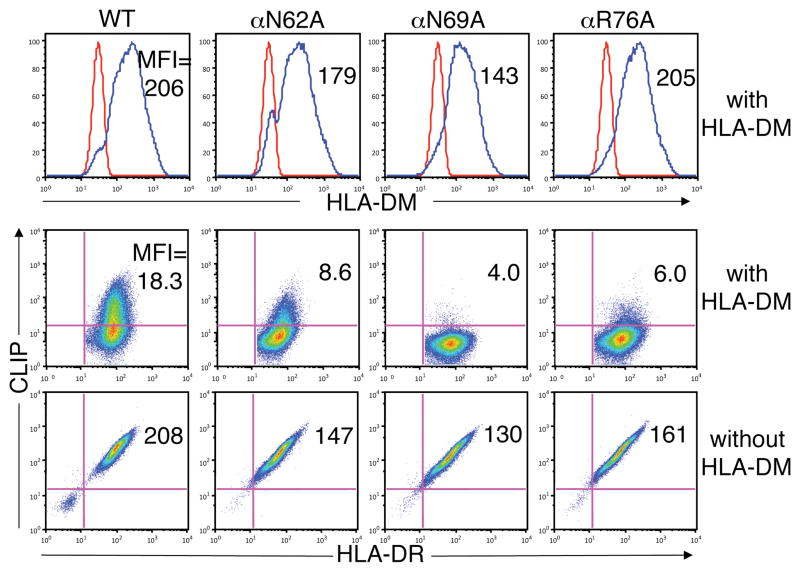
We mutated the amino acids in MHC class II molecules that participate in conserved hydrogen bonds to evaluate the effect on DM catalytic activity. As an initial approach, mutant HLA-DR1 molecules were expressed in T2 lymphoid cells in the presence or absence of DM. T2 cells express endogenous Ii but not DR or DM. Retroviral expression constructs were generated encoding DR1 molecules with alanine substitutions for each of the three conserved MHC class II α chain amino acids that form hydrogen bonds, αN62A,αN69A, and αR76A (Fig.1), thus preventing formation of specific hydrogen bonds. Cell lines were generated expressing comparable levels of total and cell surface DR1. High levels of CLIP were present on the surface of cells expressing wild type (WT) DR1 in the absence of DM (Fig.2). Spontaneous dissociation of CLIP from DR1 is inefficient and DM is required to catalyze CLIP dissociation and replacement with other peptides. CLIP expression was markedly reduced on cells co-expressing DM with WT DR1, reflecting normal DM catalytic activity. High CLIP levels were also present on cells expressing each of the α chain mutant DR1 molecules, αN62A, αN69A, or αR76A, and this phenotype was reversed in cells co-expressing DM (Fig.2). Thus, none of the five conserved hydrogen bonds formed by amino acids αN62, αN69, and αR76 is critical for stable association of CLIP with DR1, and each mutant is fully susceptible to DM-catalyzed CLIP dissociation.
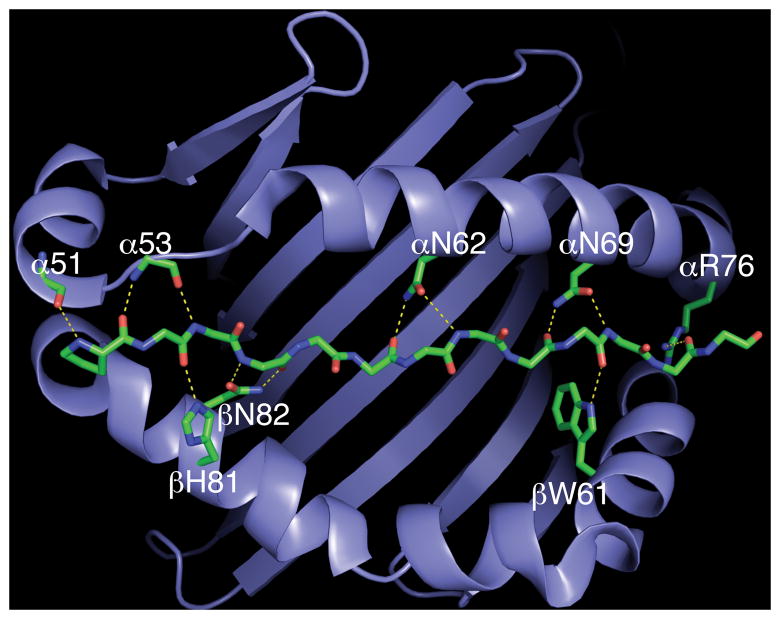
Figure 1.
The conserved hydrogen bond network in class II MHC-peptide complexes. The binding site of DR1 is represented as a ribbon diagram and the backbone of bound peptide is shown as a stick representation. Hydrogen bonds with main chain atoms of bound peptide are shown as yellow dashed lines. Main chain atoms of amino acids at DR1 positions α51 and α53 mediate hydrogen bonds, whereas conserved side chains form hydrogen bonds at other positions in the MHC molecule. The figure was generated with PyMOL software (http://www.pymol.org; PyMOL Molecular Graphics System; DeLano Scientific, San Carlos, CA) using Brookhaven Protein Data Bank coordinate file 1DLH (19).
Figure 2.
Effect of mutations in conserved positions in the DR1 alpha chain on susceptibility to DM-mediate CLIP release. Expression of DR and CLIP on the surface of T2 cells expressing WT or mutant DR1 molecules in the absence (bottom panels) or presence (middle panels) of DM was measured by flow cytometry with mAbs L243 (DR) and CerCLIP (CLIP). Total cellular expression of DM was measured in permeabilized cells by flow cytometry using mAb MaP.DM1 (top panels).
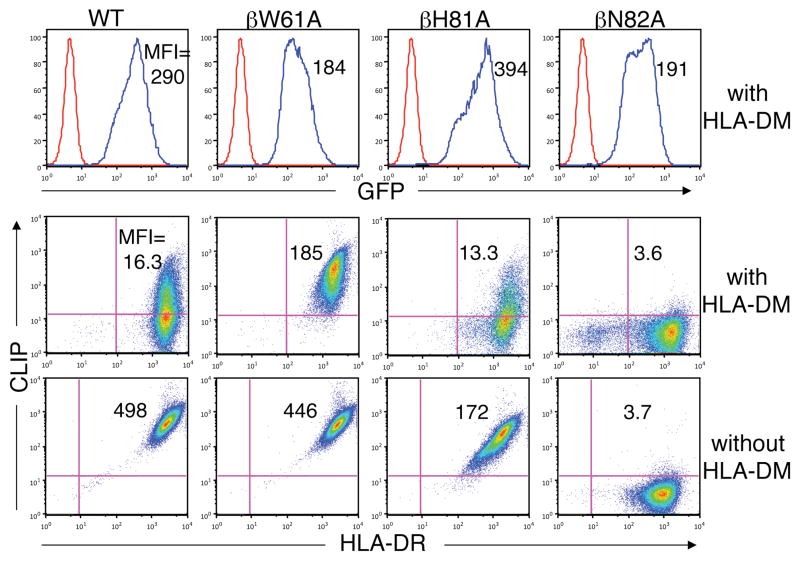
To evaluate the conserved hydrogen bonds formed by the MHC class II β chain (Fig.1), alanine substitutions were engineered at each of the three conserved positions to generateβW61A, βH81A, and βN82A DR1 molecules. The mutants were observed to assembly efficiently with Ii (data not shown). In the absence of DM, high levels of CLIP were present on cells expressing βW61A or βH81A DR1 (Fig.3). By contrast, CLIP was completely absent from the surface of cells expressing βN82A DR1, suggesting that the bidendate hydrogen bonds formed by βN82 play a disproportionate role in stabilizing CLIP-DR1 binding.
Figure 3.
Effect of mutations in conserved positions in the DR1 beta chain on susceptibility to DM-mediate CLIP release. Expression of DR and CLIP on the surface of T2 cells expressing WT or mutant DR1 molecules in the absence (bottom panels) or presence (middle panels) of DM was measured by flow cytometry. GFP fluorescence was determined by flow cytometry as a surrogate measure of DM expression (top panels).
CLIP levels were markedly reduced on cells co-expressing DM and βH81A DR1 (Fig.3), demonstrating that DM can efficiently catalyze CLIP dissociation from DR1 complexes lacking the βH81 hydrogen bond, a result that was further confirmed with the asparagine substitution mutant βH81N (Supplemental Fig.S1), directly contradicting the conclusions of Narayan et al. (21). βH81A DR1 molecules in DM expressing cells were stable in SDS detergent, evidence that CLIP is exchanged for high affinity peptides in these cells (data not shown). We were also interested in determining whether the histidine at β81 influences the pH-dependence of DM-catalyzed peptide binding, which is optimal at acidic endosomal pH (2–4). The βH81A and βH81N mutations had little or no effect on the pH-dependence of DM-catalyzed peptide binding (Supplemental Fig.S2).
By contrast to the βH81 mutations, βW61A DR1 molecules appeared to be partially resistant to DM-mediated CLIP dissociation. This was true even when subpopulations gated for identical expression of DR and DM were analyzed. Thus, the hydrogen bond formed byβW61 was a candidate target in the DM catalytic mechanism. The capacity of DM to catalyze CLIP dissociation from βN82A DR1 could not be evaluated with this approach because of the high rate of spontaneous dissociation, leaving βN82 as an additional candidate.
Effect of MHC class II beta chain mutations on DM catalytic potency
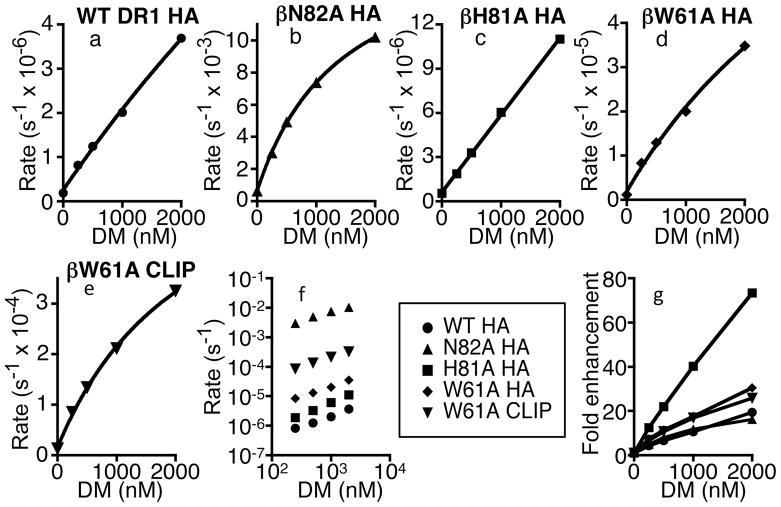
To further evaluate the role of conserved hydrogen bonds involving the MHC class II β chain in the DM catalytic mechanism, we generated soluble recombinant DR1 andβW61A, βH81A, and βN82A mutant proteins. A fluorescence polarization assay was employed to quantify the impact of DM on the kinetics of peptide dissociation. A DM concentration dependent acceleration of the rate of peptide dissociation was observed with WT DR1 complexes containing the high affinity peptide HA (Fig.4a). The βH81A mutation had essentially no effect on the intrinsic stability of the DR1-HA complex (Table 1) and the mutation did not reduce the potency of DM in catalyzing peptide dissociation (Fig.4c,f,g). Indeed, catalytic potency was somewhat greater for βH81A compared to WT DR1. Thus, the hydrogen bond formed by βH81 does not play a major role in stabilizing DR1-peptide complexes and it is not an essential target in the DM catalytic mechanism.
Figure 4.
Effect of mutational disruption of DR1 beta chain hydrogen bonds on DM catalytic potency. (a-e) Preformed complexes (50 nM) of Alexa Fluor 488 labeled HA or CLIP peptides bound to mutant sDR1 were incubated with excess unlabeled peptide (20 μM) and various concentrations of soluble DM at pH 5.0, 37oC. Peptide dissociation rates were measured using a fluorescence polarization assay as described in Materials and Methods. (f) Data for mutant sDR1 molecules are compared to wild type sDR1 (WT) in a log-log plot of peptide dissociation rates versus DM concentrations. (g) DM-catalyzed rate enhancements normalized for differences in intrinsic peptide dissociation rates.
Table I.
Peptide dissociation rates for mutant DR1 moleculesa
| DM (nM) |
||||||
|---|---|---|---|---|---|---|
| 0 | 250 | 500 | 1000 | 2000 | n | |
| DR1 (HA) | 1.9 x 10−7 | 8.2 x 10−7 | 1.2 x 10−6 | 2.0 x 10−6 | 3.7 x 10−6 | 3 |
| N82A (HA) | 6.3 x 10−4 | 3.0 x 10−3 | 4.9 x 10−3 | 7.4 x 10−3 | 1.0 x 10−2 | 3 |
| H81A (HA) | 1.5 x 10−7 | 1.9 x 10−6 | 3.3 x10−6 | 6.0 x 10−6 | 1.1 x 10−5 | 2 |
| W61A (HA) | 1.1 x 10−6 | 8.3 x 10−6 | 1.3 x 10−5 | 2.0 x 10−5 | 3.5 x 10−5 | 2 |
| W61A (CLIP) | 1.3 x 10−5 | 8.5 x 10−5 | 1.3 x 10−4 | 2.1 x 10−4 | 3.3 x 10−4 | 1 |
Rates are expressed in units of s−1.
The βW61A mutation reduced peptide complex stability, with ~5-fold increase in the intrinsic HA dissociation rate (Table 1). However, DM catalytic potency was not reduced, even after normalization for the effect on the intrinsic peptide dissociation rate (Fig.4d,f,g). These results contrasted with the finding that CLIP release from βW61A DR1 was relatively resistant to DM in cells (Fig.3). We considered the possibility that DM might selectively catalyze the dissociation of HA but not CLIP from βW61A DR1. However, DM was observed to catalyze the dissociation of a variant CLIP peptide βW61A DR1 (Fig.4e) with potency similar to that observed for HA (Fig.4f). Full-length βW61A DR1-CLIP complexes were also highly susceptible DM (data not shown). While we did not observe any gross alteration in endosomal co-localization of βW61A DR1 with DM (data not shown), it seems likely that this mutation affects the capacity of DR1-CLIP complexes to interact optimally with DM in T2 cells, either by impacting co-localization in membrane subdomains or through an effect on trafficking kinetics (26).
Strikingly, the βN82A mutation was observed to increased the intrinsic HA dissociation rate by >3000-fold (Table 1). This is consistent with the spontaneous release of CLIP observed with this mutant expressed in cells (Fig.3). Previous studies with mouse class II molecules support the idea that βN82 may in general have a dominant role in stabilizing MHC class II-peptide complexes (23, 27). Remarkably, despite the very rapid spontaneous dissociation of HA from βN82A DR1, DM further accelerated peptide dissociation (Fig.4b,f). The catalytic potency was similar to that observed with WT DR1, even with data normalized for intrinsic rates (Fig.4g). Thus, the two hydrogen bonds formed by βN82 appear to be critical for stabilizing DR1-peptide complexes, yet they are not essential targets in the DM catalytic mechanism.
Concluding Remarks
In addition to the nine hydrogen bonds that we analyzed by mutational analysis in the current study, three conserved hydrogen bonds that interact with main chain atoms in the amino terminus of bound peptide are contributed by main chain atoms of amino acids at positions α51 and α53 in DR1 (Fig.1) (19). The potential role of these hydrogen bonds in the DM catalytic mechanism cannot be analyzed by mutation. However, Stratikos, et al., used peptide truncation and amide N-methylation to prevent the formation of these hydrogenbonds in DR1-HA peptide complexes (20), demonstrating that DM catalytic potency was enhanced ~6–9-fold with peptide complexes lacking one or more of these hydrogen bonds. They observed only minor effects on catalytic potency with N-methylated peptide analogs designed to disrupt each of four additional conserved hydrogen bonds (20). It is possible that disruption of hydrogen bonds mediated by α51, α53, or β81 increases DM binding affinity by increasing DR structural flexibility, resulting in small increases in DM catalytic potency.
The central conclusion of the current study is that no individual component of the conserved hydrogen bond network plays an essential role in the DM catalytic mechanism. DM catalytic activity is preserved or even enhanced with artificial disruption of any component of the conserved network. Given the universal capacity of DM to catalyze peptide dissociation, the conserved network has been an attractive potential target in the catalytic mechanism. If this is the case, however, individual components of the network must play a redundant role. It has become increasingly clear that MHC class II-peptide binding involves cooperative interactions throughout the peptide-binding groove and the hydrogen bond network appears to be relatively isolated energetically from the anchor-pocket interactions (27–29). It is possible, for example, that DM might initially promote the destabilization of N-terminal hydrogen bonds leading to a global change in conformation that destabilizes multiple elements of the hydrogen bond network. Removing any subset or single H-bond does not prevent DM from having its catalytic effect, but instead, it might even enhance the catalytic impact. Alternatively, DM might reduce the stability of peptide complexes by impacting other global features such as the dynamics or general structural flexibility of the peptide-binding groove, possibly by altering the interaction of the groove domain with the supporting Ig domains.
Supplementary Material
Acknowledgments
We are grateful to Dr. Elizabeth Mellins for providing critical reagents.
Abbreviations
- Ii
invariant chain
- WT
wild type
Footnotes
This work was supported by National Institutes of Health Research Grants AI30554 and AI33614
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 2.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 3.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 4.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 5.Mosyak L, Zaller DM, Wiley DC. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- 6.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 8.Kropshofer H, Vogt AB, Moldenhauer G, Hammer J, Blum JS, Hammerling GJ. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- 9.van Ham SM, Gruneberg U, Malcherek G, Broker I, Melms A, Trowsdale J. Human histocompatibility leukocyte antigen (HLA)-DM edits peptides presented by HLA-DR according to their ligand binding motifs. J Exp Med. 1996;184:2019–2024. doi: 10.1084/jem.184.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallang LE, Roh S, Holm A, Bergseng E, Yoon T, Fleckenstein B, Bandyopadhyay A, Mellins ED, Sollid LM. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J Immunol. 2008;181:5451–5461. doi: 10.4049/jimmunol.181.8.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raddrizzani L, Bono E, Vogt AB, Kropshofer H, Gallazzi F, Sturniolo T, Hammerling GJ, Sinigaglia F, Hammer J. Identification of destabilizing residues in HLA class II-selected bacteriophage display libraries edited by HLA-DM. Eur J Immunol. 1999;29:660–668. doi: 10.1002/(SICI)1521-4141(199902)29:02<660::AID-IMMU660>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 13.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20:467–476. doi: 10.1016/s1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 14.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 15.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 16.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. J Exp Med. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc NatlAcad Sci U S A. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 20.Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 21.Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFarland BJ, Beeson C, Sant AJ. Cutting edge: a single, essential hydrogen bond controls the stability of peptide-MHC class II complexes. J Immunol. 1999;163:3567–3571. [PubMed] [Google Scholar]
- 23.McFarland BJ, Katz JF, Beeson C, Sant AJ. Energetic asymmetry among hydrogen bonds in MHC class II*peptide complexes. Proc Natl Acad Sci U S A. 2001;98:9231–9236. doi: 10.1073/pnas.151131498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 25.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwart W, Griekspoor A, Kuijl C, Marsman M, van Rheenen J, Janssen H, Calafat J, van Ham M, Janssen L, van Lith M, Jalink K, Neefjes J. Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity. 2005;22:221–233. doi: 10.1016/j.immuni.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 27.McFarland BJ, Katz JF, Sant AJ, Beeson C. Energetics and cooperativity of the hydrogen bonding and anchor interactions that bind peptides to MHC class II protein. J Mol Biol. 2005;350:170–183. doi: 10.1016/j.jmb.2005.04.069. [DOI] [PubMed] [Google Scholar]
- 28.Anderson MW, Gorski J. Cooperativity during the formation of peptide/MHC class II complexes. Biochemistry. 2005;44:5617–5624. doi: 10.1021/bi048675s. [DOI] [PubMed] [Google Scholar]
- 29.Ferrante A, Gorski J. Cooperativity of hydrophobic anchor interactions: evidence for epitope selection by MHC class II as a folding process. J Immunol. 2007;178:7181–7189. doi: 10.4049/jimmunol.178.11.7181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.