Abstract
Reengineering the receptor footprints of adeno-associated virus (AAV) isolates may yield variants with improved properties for clinical applications. We generated a panel of synthetic AAV2 vectors by replacing a hexapeptide sequence in a previously identified heparan sulfate receptor footprint with corresponding residues from other AAV strains. This approach yielded several chimeric capsids displaying systemic tropism after intravenous administration in mice. Of particular interest, an AAV2/AAV8 chimera designated AAV2i8 displayed an altered antigenic profile, readily traversed the blood vasculature, and selectively transduced cardiac and whole-body skeletal muscle tissues with high efficiency. Unlike other AAV serotypes, which are preferentially sequestered in the liver, AAV2i8 showed markedly reduced hepatic tropism. These features of AAV2i8 suggest that it is well suited to translational studies in gene therapy of musculoskeletal disorders.
New viral strains constantly evolve in nature through iterative mutagenesis1,2. The breadth of tissue tropism displayed by various AAV isolates, such as AAV8 and AAV9, is beneficial for gene transfer by systemic delivery3–6. In some cases, however, it would be desirable to direct homing of AAV vectors to specific organs. All naturally occurring AAV serotypes and variants tested to date have a propensity to accumulate within the liver, albeit with varying efficiency4. Consequently, strategies to redirect AAV capsids from the liver to target organs would be very useful from a clinical standpoint. Tissue-specific promoters and, more recently, microRNA-based gene regulation strategies, have been used to sharply segregate gene expression patterns among different tissue types7,8. However, such regulatory strategies do not preclude sequestration of AAV vector genomes in off-target organs such as the liver after systemic administration.
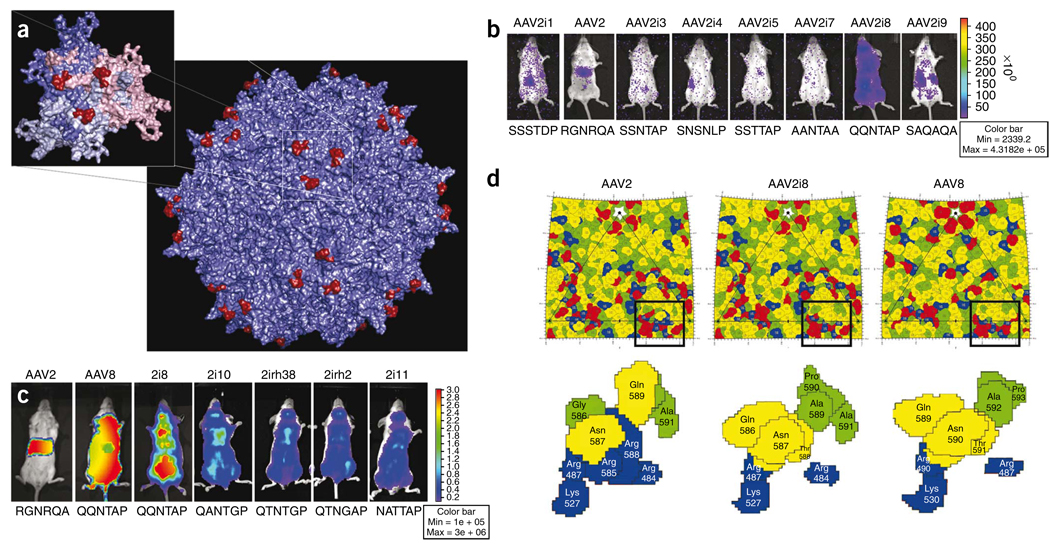
To develop AAV vectors with improved tropism for clinical applications, we reengineered the heparan sulfate receptor footprint on the AAV2 capsid surface using information available from structural studies, including crystallographic data and cryo-electron microscope analysis of AAV capsids and their cognate receptors9–12. The heparan sulfate footprint on the AAV2 capsid consists of the basic amino acid residues R484, R487, K527, K532, R585 and R588, which form a continuous basic patch10–12. R585 and R588, located within the so-called GH loop, form the inner walls of the spikes located on the icosahedral threefold axis (Fig. 1a), and other residues occupy the floor surrounding these regions12. Mutation of either R585 or R588 disrupts the basic cluster and abolishes heparan sulfate binding10,11.
Figure 1.
Structure-function correlates of AAV2i vectors with reengineered receptor footprints. (a) Three-dimensional structural model of the AAV2 capsid highlighting the 585–590 region containing basic residues implicated in heparan sulfate binding. Inset shows VP3 trimer, with residues 585-RGNRQA-590 located on the innermost surface loop highlighted in red. VP3 monomers are colored salmon, blue, and gray. Images were rendered using Pymol. (b) Representative live animal bioluminescent images of luciferase transgene expression profiles in BALB/c mice (n = 3) injected intravenously (tail vein) with AAV2i CMV-Luc vectors (dose 1 × 1010 vg in 200 µl PBS). Photographs and bioluminescent images were obtained at 1 week after injection. The overlay demonstrates decreased transduction efficiency for most AAV2i mutants with the exception of AAV2i8. Bioluminescence scale ranges from 0–4 × 105 relative light units (photons/sec/cm2). Residues within the 585–590 region in each AAV2i mutant are indicated below corresponding mouse image data. (c) Representative live animal bioluminescent images of luciferase transgene expression profiles in BALB/c mice (n = 3) injected intravenously (tail vein) with AAV2, AAV8, AAV2i8 and structurally related AAV2i mutants (dose 1 × 1011 vg in 200 µl PBS) packaging the CBA (chicken beta actin)-Luc cassette. All AAV2i mutants show a systemic transduction profile similar to that of AAV8, with AAV2i8 showing enhanced transduction efficiency. Bioluminescence scale ranges from 0–3 × 106 relative light units (photons/sec/cm2). Residues within 585–590 region in each AAV2i mutant is indicated below corresponding mouse image data. (d) Comparison of AAV2, AAV2i8 and AAV8 capsid surface residues based on schematic “Roadmap” projections. A section of the asymmetric unit surface residues on the capsid crystal structures of AAV2 and AAV8, as well as a model of AAV2i8, are shown. Close-up views of the heparan sulfate binding region and residues 585–590 reveal a chimeric footprint on the AAV2i8 capsid surface. Red, acidic residues; blue, basic residues; yellow, polar residues; green, hydrophobic residues. Each residue is shown with a black boundary and labeled with VP1 numbering based on the AAV2 capsid protein sequence.
Using site-directed mutagenesis, we substituted the hexapeptide motif 585-RGNRQA-590, which contains R585 and R588, with corresponding amino acids from different AAV serotypes and nonhuman primate isolates (Fig. 1b,c) to generate a series of AAV2 inner loop (AAV2i) mutants. Earlier studies established that mutating R585 and/or R588 on the AAV2 capsid to C, M, A or E is sufficient to attenuate heparan sulfate binding10,11. In the current study, AAV2i mutants containing Q, A, S or N in position 585 and T, N, A or G in position 588 were also unable to bind heparan sulfate under physiological conditions, as demonstrated by affinity column binding assays (Supplementary Fig. 1). In general, titers of all AAV2i mutants were similar to that of the parental AAV2, and their efficiency at transducing various cell types in vitro was reduced by several orders of magnitude (Supplementary Fig. 2).
Using live animal bioluminescence imaging, we studied vector biodistribution in normal BALB/C mice after intravenous administration at low dosage. One week after administration, most AAV2i mutants were deficient in transduction as evidenced by low bioluminescent signal (Fig. 1b). A notable exception was AAV2i8, which displayed a systemic transduction profile (Fig. 1b) regardless of the duration of gene expression or the intravenous route of administration (tail or portal vein; Supplementary Fig. 3).
Based on the above observations with AAV2i8 containing a 585-QQNTAP-590 motif, we tested several AAV2i mutants with 585-QXXTXP-590 or 585-NXXTXP-590 motifs derived from other strains of AAV. AAV2i mutants with residues Q/N585, T588 and P590 showed systemic transduction profiles similar to that of the AAV8 control (Fig. 1c). In contrast, the AAV2 control showed a greater tropism for liver, as established earlier4. The higher transduction efficiency of AAV2i8 compared with AAV2i10, AAV2i11, AAV2irh.2 and AAV2irh.38 highlights the subtle synergy between residues within the hexapeptide motif in conferring systemic tissue tropism. Notably, the 585-QQNTAP-590 motif did not result in systemic tropism when incorporated into the corresponding region on AAV1 or AAV3 capsids (Supplementary Fig. 4). Taken together, these results highlight the complexity of the structural coordinates required to attain an atypical systemic transduction profile.
To examine the surface footprint of AAV2i8, we generated model surface maps of this mutant and of parental AAV2 and AAV8 capsids using stereographic roadmap projections, which allow simultaneous projection of amino acids, charge distribution and capsid surface topology onto a two-dimensional surface13. Substitution of 585-RGNRQA-590 with QQNTAP results in disruption of the continuous basic patch (blue residues) formed by the cluster of arginine and lysine residues (Fig. 1d). In addition, our model of the AAV2i8 footprint shows an overall chimeric distribution of amino acid residues with respect to AAV2 and AAV8 (refs. 9,14). The chimeric nature of AAV2i8 is corroborated in the observation that these capsids were only modestly neutralized when exposed to anti-AAV2 serum or human serum (Supplementary Tables 1 and 2). Thus, reengineering receptor footprints on AAV capsids can simultaneously alter antigenicity.
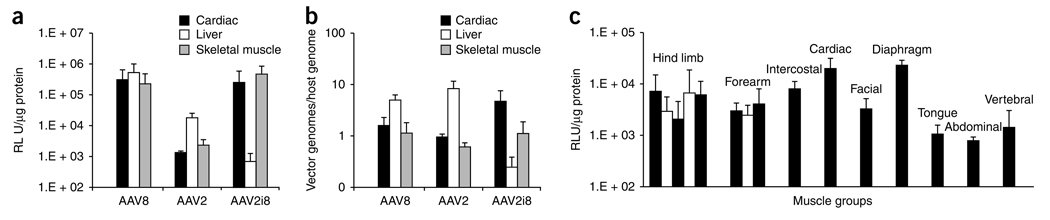
Based on its promising transduction profile, the lab-derived AAV2i8 strain was further characterized. We quantified luciferase transgene expression and genome copy numbers in cardiac, skeletal muscle and liver tissue lysates at 2 weeks after vector administration in BALB/C mice. As shown in Figure 2a, AAV8 ubiquitously transduced muscle and liver tissue with high efficiency, consistent with the systemic transduction profile in Figure 1c. AAV2 also transduced liver preferentially, although less efficiently than AAV8, and showed only modest transduction in muscle tissue. The chimeric AAV2i8 transduced cardiac and skeletal muscle tissue with a high efficiency similar to that of AAV8 and was detargeted from the liver. These findings were supported by data on biodistribution of vector genome copies in muscle and liver tissues determined by Q-PCR (Fig. 2b). For AAV2 and AAV8, high amounts of vector genome copies were recovered from liver compared with cardiac or skeletal muscle. For AAV2i8, sequestration in liver was ~40-fold lower compared with AAV2 or AAV8.
Figure 2.
Selective muscle tropism of AAV2i8. (a) Quantification of luciferase transgene expression in three major tissues: cardiac (black bars), skeletal muscle (pooled hind limb and abdominal; gray bars) and liver (white bars). Tissue lysates were obtained from BALB/c mice (n = 3) at 2 weeks after administration of AAV2, AAV2i8 and AAV8 (dose 1 × 1011 vg, tail vein) and subjected to luminometric analysis. AAV2i8 shows high transduction in cardiac and skeletal muscle and low transduction in liver. Luciferase levels are shown as relative light units normalized to protein levels determined using a Bradford assay. Error bars indicate s.d. (b) Vector genome copy numbers (luciferase transgene) in three major tissues: cardiac (black bars), skeletal muscle (pooled hind limb and abdominal; gray bars) and liver (white bars). Host genomic as well vector DNA was extracted from tissue lysates obtained from BALB/c mice (n = 3) at 2 weeks after administration of AAV2, AAV2i8 and AAV8 (dose 1 × 1011 vg, tail vein). Host and vector genome copy number were determined by Q-PCR with specific primer sets against the lamin gene and luciferase transgene, respectively. AAV2i8 shows enhanced muscle sequestration and decreased accumulation in liver tissue compared with AAV2 and AAV8. (c) Luciferase transgene expression in major muscle sub-groups obtained from BALB/c mice (n = 3) at 2 weeks after intravenous administration of AAV2i8 (dose 1 × 1011 vg, tail vein) packaging the CBA (chicken beta actin)-Luc cassette. Tissue lysates from five different muscle groups from the hind limb skeletal muscle (alternating black and white bars), three groups from the forelimb (alternating black and white bars), intercostals, cardiac, facial, diaphragm, tongue, abdominal and vertebral muscle types (black bars) were subjected to luminometric analysis. Luciferase levels are shown as relative light units normalized to protein levels determined by a Bradford assay. Error bars indicate s.d.
Further biodistribution studies confirmed the redirection of AAV2i8 from liver to muscle. AAV2i8 transduced a wide range of muscle groups in the murine forearms and hind legs as well as intercostal, facial and abdominal muscles (Fig. 2c). Cardiac and diaphragm muscle were transduced with high efficiency, whereas low levels of vector genome copies were recovered from other major organs, such as brain, lung and spleen. These results distinguish the tissue tropism of the chimeric AAV2i8 capsid from that of any naturally occurring AAV serotype or isolate characterized thus far (Supplementary Fig. 5).
Our results confirm previous findings that attenuation of heparin binding in general can result in liver detargeting and systemic dissemination of AAV2-derived vectors. Earlier studies demonstrated a strong correlation between heparin binding and liver tropism in the case of AAV2 and AAV6 (refs. 10,15). Disruption of the basic receptor footprint through mutagenesis of R585 and/or R588 residues (in AAV2) or K531 (in AAV6) attenuated heparan sulfate binding, which correlated with decreased liver tropism10,15. In addition, it was demonstrated that an R484E;R585E AAV2 mutant is detargeted from the liver and retains the ability to transduce muscle tissue with modest efficiency, similar to the parental AAV2 (ref. 16). Although the R484E;R585E AAV2 vector has not shown high transduction efficiency in larger animal models17 or been compared directly with AAV8 or AAV9, these early studies clearly show the potential to control tissue tropism by manipulating receptor-binding domains. Recently, a novel AAV mutant with cardiac-specific transduction was generated through directed evolution18. The laboratory-derived M41 clone displayed a tenfold higher transduction efficiency in cardiac tissue compared with the liver. Whereas similar trends were observed for several of our mutants, an important advantage of AAV2i8 and mutants with a 585-QXXTXP-590 motif is their ability to efficiently transduce not only cardiac muscle but the entire range of muscle groups with a transduction efficiency 2–3 orders of magnitude higher than that observed in the liver (Fig. 2c and Supplementary Fig. 5). To our knowledge, such an efficient switch in tropism from liver to muscle has not been demonstrated previously.
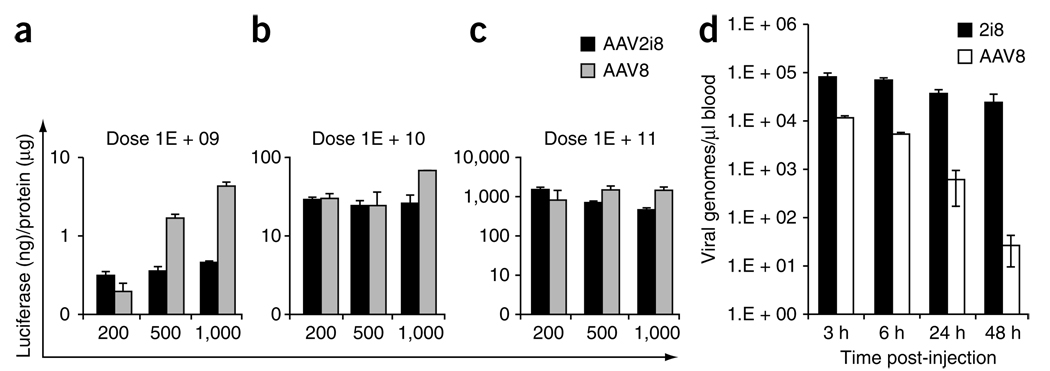
Next, we used an isolated hind limb perfusion technique19 to examine the efficiency with which AAV2i8 traverses the blood vessel barrier. AAV2i8 transduced hind limb skeletal muscle as efficiently as AAV8 at low volume of injection, at moderate and high vector dosage (Fig. 3a–c). At low vector dose, AAV8 displayed three- to tenfold increases in transduction efficiency at higher volumes of injection. However, AAV2i8 traversed blood vessels and transduced underlying skeletal muscle with high efficiency regardless of the volume of injection.
Figure 3.
Blood transport profile of AAV2i8. (a–c) Luciferase transgene expression in pooled skeletal muscle subgroups from right and left hind limb of BALB/c mice (n = 4) after isolated perfusion of AAV2i8 (black bars) or AAV8 (gray bars) into each saphenous vein. Tissue lysates prepared after administration of three different doses (1 × 109 (a), 1 × 1010 (b), 1 × 1011 (c) vg) in low (200 µl), medium (500 µl) or high (1 ml) volume of injection were subjected to luminometric analysis. Luciferase levels are shown as relative light units ormalized to protein levels determined using a Bradford assay. (d) Vector genome copy numbers recovered from blood at different time intervals after administration through the tail vein (n = 3). Whole blood DNA was extracted and analyzed by Q-PCR with primers against the luciferase transgene. AAV2i8 shows prolonged circulation compared with AAV8. Error bars indicate s.d.
The atypical tropism of AAV2i8 distinguishes it from natural AAV serotypes 8 and 9 and suggests that engineered AAV vectors can be tailored for specific clinical applications. The mechanism underlying the switch in AAV2i8 tropism from liver to muscle is currently unknown. Our results support the notion that the chimeric vector possesses a unique surface footprint that facilitates specific interactions with receptors distinct from those used by AAV2 and AAV8. Another possible explanation of our findings is that the increased circulation half-life of AAV2i8 allows sequestration in tissues other than the liver through heparan sulfate–independent uptake mechanisms. AAV2i8 showed markedly reduced blood clearance and appears to persist well over 48 h in blood (Fig. 3d). Moreover, muscle-specific luciferase transgene expression levels increased gradually over the course of several weeks (Supplementary Fig. 6). In contrast, AAV8 vector genome copy number rapidly decreased, approaching background levels within the same time period. These results and previous observations that other AAV serotypes with systemic tissue tropism have long circulation half-lives4 suggest that strategies to manipulate blood circulation time of AAV capsids might afford control over vector tropism.
From the standpoint of vector development and clinical safety, AAV2i8 is an attractive candidate for gene therapy of muscular dystrophies, which requires transduction of a wide range of muscle types after systemic administration6,20. The selective muscle tropism of AAV2i8 and its ability to evade sequestration by liver, when exploited in conjunction with transcriptional regulatory elements such as muscle- specific promoters, should allow exquisite control over vector biodistribution as well as cardiac or skeletal muscle–specific transgene expression. Preliminary isolated limb perfusion studies in nonhuman primates comparing AAV2i8 with AAV8 and AAV9 have shown promising results (data not shown) in this regard.
In summary, we have developed a strategy to engineer synthetic AAV strains with atypical transduction profiles. Extrapolation of this approach to receptor binding domains other than the heparan sulfate binding domain and to other natural AAV isolates might yield new chimeric vectors with unique tissue tropisms and antigenicity suitable for translational disease-specific applications.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the American Heart Association (A.A.; no. 0735637N); National Institute of Arthritis and Musculoskeletal and Skin Diseases (A.A. & R.J.S.; R21AR055712); National Institute of Allergy and Infectious Diseases (R.J.S. and A.A.; R01AI072176); National Heart, Lung, and Blood Institute (A.A.; R01HL089221); Senator Paul Wellstone Center for Muscular Dystrophy (R.J.S.; U54AR056953); National Institute of General Medical Sciences (M.A.-M.; R01GM082946); and Asklepios Biopharmaceutical for research support. We would also like to MirusBio Corp. for assistance with isolated limb perfusion studies.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
A.A. conceived the strategy, designed the project and analyzed data. A.A., S.M. and R.J.S. supervised the project and prepared the manuscript. J.C.C. and R.S. built capsid mutants, J.L.P. and C.L. carried out animal experiments, and J.H., S.M. and J.W. designed and carried out isolated limb perfusion studies. S.Y. carried out Q-PCR studies and N.D., H.-J.N. and M.A.-M. carried out molecular modeling studies.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Hensley SE, et al. Hemagglutinin receptor binding avidity drives Influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmenberg AC, et al. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 4.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 5.Inagaki K, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 7.Wang B, et al. Construction and analysis of compact muscle-specific promoters for AAV vectors. Gene Ther. 2008;15:1489–1499. doi: 10.1038/gt.2008.104. [DOI] [PubMed] [Google Scholar]
- 8.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 9.Xie Q, et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kern A, et al. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy HC, et al. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J. Struct. Biol. 2009;165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam HJ, et al. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, et al. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller OJ, et al. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc. Res. 2006;70:70–78. doi: 10.1016/j.cardiores.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Raake PW, et al. Cardio-specific long-term gene expression in a porcine model after selective pressure-regulated retroinfusion of adeno-associated viral (AAV) vectors. Gene Ther. 2008;15:12–17. doi: 10.1038/sj.gt.3303035. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, et al. A myocardium-tropic AAV evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. USA. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagstrom JE, et al. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Gregorevic P, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.