Abstract
Butyrylcholinesterase (BuChE) is a stoichiometric bioscavenger against organophosphorus (OP) nerve agent poisoning, and efforts to make BuChE variants that are catalytically active against a wide spectrum of nerve agents have been ongoing for the last decade. In order to understand the structural consequences for BuChE, we carried out extensive molecular dynamics (MD) simulations on wild-type BuChE (PDB ID: 1P0I) and several known and new variants of this enzyme, but without the presence of any ligand in the active site. The MD simulations on WT-BuChE identified two labile orientations for the catalytic serine, and also showed the likelihood of a backdoor. Upon changes at the G116 position, severe alterations around the active site region were identified. Simulations on both G117H and G117N variants showed the existence of a bound water molecule that is in close proximity to S198. Modeling of the E197Q mutant suggested that Q197 can be in two distinct orientations, one similar to the E202Q-AChE crystal structure and another in proximity to G439 and E441. The double mutant, G117H/E197Q, was found to have structural characteristics of both G117H and E197Q. In light of the computational results, previous experimental observations are discussed.
Keywords: Butyrylcholinesterase, organophosphorus nerve agents, molecular dynamics simulations, G117H, E197Q
I. Introduction
Acetylcholinesterase (AChE) carries out hydrolysis of the neurotransmitter acetylcholine, and organophosphorus (OP) compounds are toxic because they inhibit this role of AChE, thereby disrupting the critical balance of acetylcholine at a neurosynaptic junction. [1,2] Such acetylcholine imbalance can lead to respiratory failures, epileptic seizures, and even death. Inactivation of AChE by OP compounds proceeds by phosphylation of the active site serine, and then (to date) irreversible aging to form the negatively charged phosphylated serine. [3] A sister protein of AChE, butyrylcholinesterase (BuChE), is available in human serum at high concentrations. As the active site of BuChE is similar to AChE, BuChE also reacts with OPs in a similar fashion. [4] The catalytic triad, S198-H438-E325, is located in a deep narrow gorge for both the enzymes; however, BuChE lacks six aromatic amino acids out of the fourteen that line the catalytic gorge of AChE. This disparity increases the available volume in the gorge of BuChE by about 200 Å3. [5,6] Thus, BuChE reacts faster with several OP compounds and can accommodate both stereoisomers of some OPs, a feature not shared with AChE. It has been suggested that high doses of BuChE in blood serum can function as a scavenger of OP nerve agents and thereby provide protection for AChE. [7,8] However, since BuChE reacts stoichiometrically, and not catalytically, with OP compounds, a high concentration of BuChE would be needed. Therefore, efforts to design a mutant of BuChE, that can catalytically hydrolyze OP compounds, have been ongoing for the last decade.
Broomfield, Millard and Lockridge tested several BuChE mutations (G115H, G117H, G227K, Q119H and G121H) at the oxyanion hole and its surroundings to introduce catalytic activity against OP exposure. [9] These efforts lead to the discovery of the G117H variant, which was found to reactivate upon inhibition by sarin (GB), VX, paraoxon, and echothiophate. [10] However, the G117H mutant was not active against soman (GD), one of the most toxic OPs. Further investigations identified a double mutant (G117H/E197Q) that was able to hydrolyze GD isomers. [11] Nevertheless, none of the above mutants had sufficiently high catalytic rate to be considered as a catalytic scavenger of OP compounds; therefore, the search for improved variants is ongoing. In order to provide rational guidance in this design strategy, it is critical to understand the structural perturbations incurred by these mutations and the effect that the local change in the mutant's active site environment has on binding of an OP. In this report, we address the first concern, and present our results from molecular dynamics (MD) simulations on wild-type (WT) BuChE, some of the known variants, and also some new mutants that were found in an initial experimental screening by Zhang and co-workers. [12]
II. Computational Details
We utilized the available crystal structure of human BuChE (PDB: 1P0I) [13] as the starting point. Missing residues, including D378, D379 and N455, were added to the structure manually, and missing hydrogens were added using AMBER's xleap [14,15] module. The protonation states of the titratable residues were determined using the pdb2pqr [16] utility for a pH of 7. The resulting structure was utilized as a template to design the other mutants. All of the molecular dynamics (MD) simulations were performed with the AMBER FF03 force field [17,18] in the presence of explicit TIP3P [19] water molecules within 8 Å of the protein in an octahedral box. The MD protocol involved a three-step minimization, followed by a pre-production step, and finally production MD simulations. In the minimization procedure, the missing residues were first minimized, followed by the water molecules, and in the third step, the entire system was allowed to relax for 2500 steps. Then the temperature of the system was raised from 0 to 300 K with a small force constant on the enzyme in order to restrict any drastic changes. Finally, this system was subjected to production MD simulations for 1 to 5 ns. The number of atoms, temperature, and pressure were kept constant for these NPT MD simulations, and periodic boundary conditions were used. Furthermore, multiple trajectories, starting from unique structures, were computed for these mutants and analyzed for consistency.
III. Results and Discussions
A. WT-BuChE
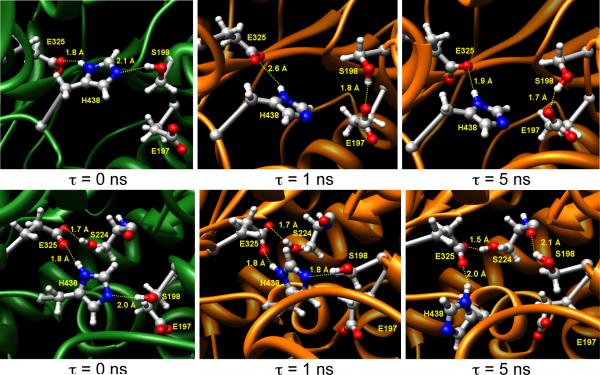
The root mean square deviation (RMSD) for three different trajectories showed no major structural changes after about 100 ps while the RMSD values for individual residues for one of the trajectories predicted that the most flexible parts of the enzyme were the ω-loop (I69-S79) that lines the primary entrance of the gorge, and the loop with the missing residues (D378-D379) from the crystal structure. The inherent flexibility of the D378-D379 loop is consistent with these residues not being resolved in the crystal structure. We found that the catalytic triad is not always in the ideal form; instead, these residues attain different orientations during the simulations, as observed earlier. [20,21] The active site serine, S198, can interact with H438 as well as with the backbone carbonyl group of S224 (Figure 1). Alternatively, the orientation of E197 is such that the carboxylate group can also stably interact with S198. The alternative H-bonding of S198 with H438 and S224 is transient, and S198 flips between these two sites very frequently.
Figure 1.
Orientations of the catalytic triad and its surroundings for different time points during the two independent MD simulation trajectories for wild-type BuChE. In these figures, τ = 0 ns is the time when the temperature of the system has been raised from 0 to 300K, and then atoms were allowed to relax. Therefore the upper and lower left panels do not have the same initial geometry at τ = 0 ns.
In the crystal structure [13] that was utilized for the simulations, a choline molecule was noted to interact with H438 via a water molecule; therefore, the authors stated that the H438 residue might not deprotonate the catalytic serine. Nevertheless, the catalytic triad attained an ideal orientation after the minimization steps for our MD protocol. Nevertheless, there were some distortions in the S198-H438 H-bonding patterns. Furthermore, besides the choline-bound crystal structure (PDB: 1P0I), several other crystal structures of BuChE [13, 21] have E197 in the exact same orientation. This supports the notion, as seen in our simulations, that the orientation of E197 remains in the proximity of the oxyanion hole and the catalytic serine, possibly affecting the orientation of S198.
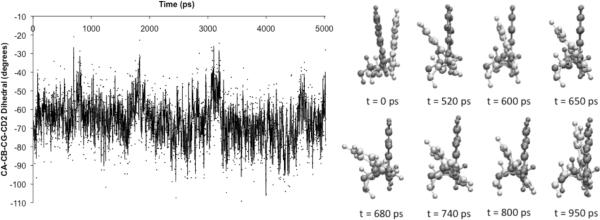
The W82 residue has been cited several times [22] as the secondary door of the active site. Possible movements of W82 that can allow the products to exit via the secondary door may also explain the very high efficiency of these cholinesterases.[10] Indeed, in our simulations, we do observe the oscillatory movements (Figure 2) of W82 in some of our trajectories, and its large flexible movements suggest the opening and closing of the secondary door.
Figure 2.
Oscillation of the backbone to aromatic unit dihedral angle of W82 with time (left) and pictorial representations (right) of the W82 conformations at different time points across the initial 1 ns of the MD simulations for one trajectory of wild-type BuChE.
B. G116H and G116S
The G116 residue is a key component of the oxyanion hole that provides a hydrogen-bond donor to the carbonyl/phosphoryl oxygen; thus, any changes to this residue may cause severe distortions around the active site. In fact, our MD simulations predict the same for both the G116H and G116S mutants. The histidine variant places the H116 residue in the gorge area such that the active site is still accessible for the substrate/inhibitor; however, the available volume does decrease. On the other hand, the G116S variant has S116 interacting with either T120 or E197, thus rendering clear access to the active site. The most noticeable feature in the G116H and G116S mutants was the distortion in H-bonding interactions in the vicinity of position 116 when compared to the WT-BuChE simulations. These changes lead to deformation of the overall structure of the oxyanion hole in both G116H and G116S. Additionally, a water molecule was also found to bridge between the oxyanion hole and H438 resulting in an unusual orientation of H438 in both mutants. Moreover, W82 was driven into the active site from its usual position, further adding to perturbation in the catalytic region. In light of these observations, we conclude that G116S and G116H will have either small or no catalytic efficiency due to the highly distorted active site and lack of the oxyanion hole. Indeed, it was found experimentally that many of the mutants at the original G116 position were not active for hydrolysis of butyrylthiocholine. [12] While expression for these mutants were variable based on ELISA analysis, the lack of activity for these mutants could not be attributed to lack of expression.
C. G117H and G117N
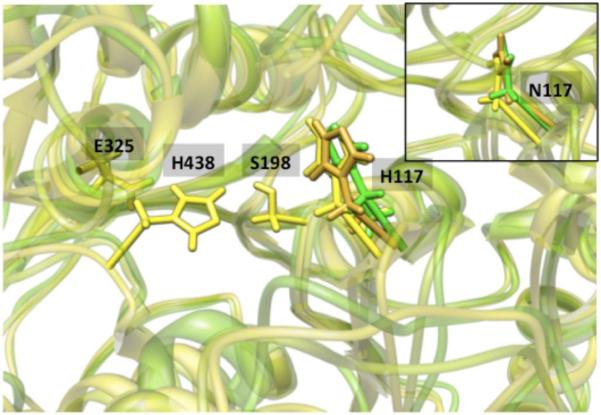
The 117 position is a critical component of the oxyanion hole, and experiments [9,10,11] showed that the G117H mutant is capable of hydrolyzing some OP compounds. We examined both the G117H and G117N mutants by MD simulations. Both H117 and N117 attained a conserved orientation in all of the independent trajectories (Figure 3), where they resided under the acyl-loop in the vicinity of the catalytic serine, but without restricting access to the catalytic triad. Furthermore, H/N117 was coordinated to a water molecule in all of the independent MD simulations. This water molecule was found to interact with another water molecule that was placed in the oxyanion hole. While it is expected that the water molecule in the oxyanion hole will be replaced by the phosphoryl oxygen during the inhibition process by an OP, the H/N117 coordinated water molecule should remain conserved, as an H-bond acceptor will still be available in the form of the phosphoryl oxygen. Thus, upon covalent binding of the OP compound to the active site, this water molecule bound to the H/N117 may be activated to perform the OP hydrolysis.
Figure 3.
Orientations of H117 with respect to the catalytic triad in G117H variant from three independent trajectories indicating that most likely, H117 lies under the acyl-loop and close to the S198 residue. (The inset shows orientations of N117 in the G117N variant from three independent trajectories, demonstrating that N117 also has a dominant conformation similar to H117 in G117H variant.
For the H/N117 mutants, other observations, such as alternate binding of S198 to H438 and S224 and perturbation of E197 from the ideal orientation of the catalytic triad, were observed. Since H/N117 resides under the acyl-loop, there will be no significant effect on the binding of a substrate unless a large side chain of the substrate has to be placed in the acyl-loop pocket. For example, the PS isomer of soman will place the large pinacolyl group towards the W82 residue while the PR isomer will place the pinacolyl group under the acyl-loop where H117 is located. Our previous modeling results [23,24] on pre-reacting complexes with BuChE can be used to visualize the orientation of stereoisomers in the active site. Accordingly, this mutant will have better selectivity for the PS isomer of soman over WT-BuChE. Indeed, this has been observed experimentally; the inhibition constant dropped more significantly for the PRCR isomer than for the PSCR isomer upon G117H mutation. [11] We predict the same effect for the G117N mutant as well. We should note that the current G117H MD structure is slightly different than presented by the partially resolved crystal structure [25], as there is no ligand present in the current scenario. We predict that the current structure will change upon binding of the ligand, as water molecule(s) occupying the oxyanion hole will be replaced.
D. E197Q and G117H/E197Q
As E197 is adjacent to the catalytic serine, substitution at E197 is likely to affect the electronic environment and conformational stability at the active site. Experimentally, the E197Q mutant has about 11 times less cholinesterase activity than WT-BuChE, while it has no increased activity against OPs. The G117H variant did not catalyze the hydrolysis of soman because of the faster aging rate relative to the dephosphylation rate. It was concluded that the G117H/E197Q variant retarded the aging rate, thus allowing for the hydrolysis of the soman-BuChE adduct to occur. It was predicted earlier that E197 participates in the aging reaction. [26,27]
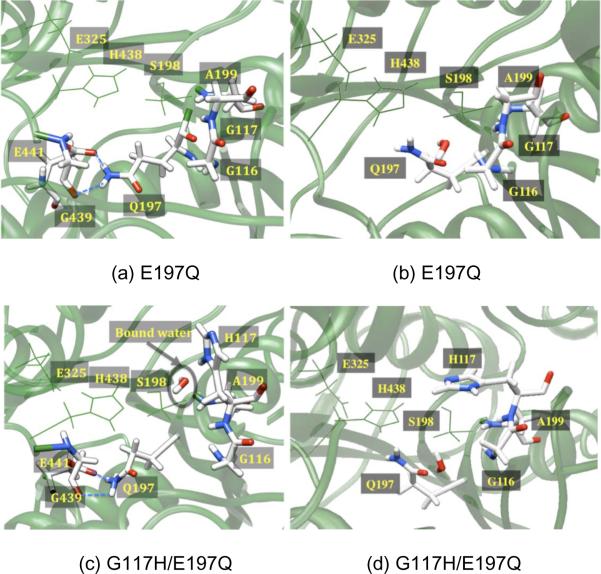
In the MD simulations of the E197Q mutant, the Q197 residue orients in a different conformation than E197 in the WT-BuChE. Q197 interacts with E441 and G439, which is in the opposite direction of the active site (Figure 4); furthermore, these results are contrary to the crystal structure of E202Q-AChE. [28] Thus, we performed MD simulations for the E197Q mutation in two random snapshots taken from the MD trajectories of WT-BuChE. Upon performing MD simulations on these snapshots, Q197 was found to interact with Y128 and E441 either directly or through a bridging water, similar to Figure 4a. In addition, a set of MD simulations was performed starting from the crystallographic structure of WT-BuChE with the E197Q mutation and explicit crystallographic waters, resulting in an orientation similar to that observed in the E202Q-AChE [28] crystal structure (Figure 4b). Observing two different orientations for Q197 from two different starting structures suggests that Q197 has some flexibility in the active site. However, at this point, it is unclear at this stage if both of these orientations can interconvert and what would be the energy barrier for this transformation.
Figure 4.
(a) For the E197Q mutant (top), a depiction of the active site showing Q197 making H-bonds with G439 and E441. (b) A second Q197 orientation was obtained for the E197Q mutant in which crystallographic waters were retained prior to the equilibration and production MD simulations. In this orientation, Q197 is H-bonded to Y128 via a number of water molecules. For the G117H/E197Q double mutant (bottom), there are similarly two orientations of Q197, depicting (c) two H-bonds of Q197 with G439 and E441 as well as H117 and the conserved water molecule, and (d) the second orientation of Q197, similar to that in the E202Q-AChE crystal structure, while H117 fluctuates under the acyl-loop.
For the G117H/E197Q double mutant, we observed similar orientations of both H117 and Q197 as seen in the respective single mutants (Figure 4). In fact, coordination of a water molecule to H117, as observed in the single mutant, was also found for the double mutant. [29] As noted above for the E197Q mutant, we observed two distinct orientations for Q197 in the double mutant (Figure 4c and 4d). Our current simulations do not provide direct evidence for the involvement of E197Q in the aging process. Nevertheless, in light of previous work [26,27] (E197 facilitates the aging process) and our computational model (Q197 can be oriented far from the active site and will not interact with the phosphorus center), these results imply that the E197Q mutation may slow down the aging process. The stereoselectivity of the G117H/E197Q double mutant is also predicted to be the same as the G117H single mutant as H117 attains the same orientation in both cases. OP compounds that must place a large side chain under the acyl-loop will be slow in binding to an H117 mutant because H117 competes for the space under the acyl-loop. Notably, it has been seen experimentally that the inhibition constant is found to decrease more for the PRCR isomer than PSCR in the double mutant when compared to WT-BuChE, perhaps because the latter cannot place the pinacolyl group under the acyl-loop. [11]
IV. Conclusions
In summary, MD simulations on WT-BuChE and many of the mutants elucidate many molecular-level details which we have attempted to correlate to the experimental observations. Across all MD simulations for the WT-enzyme as well as its mutants, the ω-loop and the D378-D379 loop were the most flexible parts of the enzyme. In addition, two transient interactions of the active site serine (S198) were observed, with H438 and S224. The possibility of a backdoor was also identified, as W82 fluctuates to open and close the door. The MD simulations identified that mutations at G116 cause significant deformation of the active site region, while the G117 position does not alter the active site. The H/N117 residue lies under the acyl-loop and is coordinated with a water molecule. Our current structural model of G117H also explains the increased stereoselectivity of the G117H variant of BuChE. The E197Q mutation placed Q197 in two different orientations, one similar to the E202Q-AChE crystal structure while another is a very different location for E197. This is possibly due to a differential orientation of water molecules around Q197. The double mutant simulations concluded that both E197Q and G117H have different effects on the protein.
V. Acknowledgments
This research was supported by grants from the NIH (U54-NS058183) and the U.S. Army (W91ZLK-06-C-0012). The authors are also grateful to the Ohio Supercomputer Center for a generous grant of computational resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
VI. References
- [1].Rosenberry TL. Acetylcholinesterase. Adv. Enzymol. 1975;43:103–218. doi: 10.1002/9780470122884.ch3. [DOI] [PubMed] [Google Scholar]
- [2].Quinn DM. Acetylcholinesterase: enzyme structure, reaction dynamics, and virtual transition states. Chem. Rev. 1987;87:955–979. [Google Scholar]
- [3].Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999;38:7032–7039. doi: 10.1021/bi982678l. [DOI] [PubMed] [Google Scholar]
- [4].Nachon F, Masson P, Nicolet Y, Lockridge O, Fontecilla-Camps JC. Comparison of the structures of Butyrylcholinesterase and acetylcholinesterase. In: Giacobini E, editor. Butyrylcholinesterase: Its functions and inhibitors. Martin Dunitz; New York NY: 2003. [Google Scholar]
- [5].Kovarik Z, Bosak A, Šinko G, Latas T. Exploring the active sites of cholinesterase by inhibition with bambuterol and haloxon. Croatica Chemica Acta. 2003;76:63–67. [Google Scholar]
- [6].Saxena A, Redman AMG, Jiang X, Lockridge O, Doctor BP. Differences in active site gorge dimensions of cholinesterases revealed by binding of inhibitors to human butyrylcholinesterase. Biochemistry. 1997;36:14642–14651. doi: 10.1021/bi971425+. [DOI] [PubMed] [Google Scholar]
- [7].Doctor BP, Saxena A. Bioscavengers for the protection of humans against organophosphate toxicity. Chem. Biol. Interact. 2005;(157-158):167–171. doi: 10.1016/j.cbi.2005.10.024. [DOI] [PubMed] [Google Scholar]
- [8].M. Cerasoli D, Griffiths EM, Doctor BP, Saxena A, Fedorko JM, Greig NH, Yu QS, Huang Y, Wilgus H, Karatzas CN, Koplovitz I, Lenz DE. In vitro and in vivo characterization of recombinant human butyrylcholinesterase (Protexia) as a potential nerve agent bioscavenger. Chem. Biol. Interact. 2005;(157-158):363–365. doi: 10.1016/j.cbi.2005.10.052. [DOI] [PubMed] [Google Scholar]
- [9].Millard CB, Lockridge O, Broomfield CA. Design and expression of organophosphorus acid anhydride anhydrolase activity in human butyrylcholinesterase. Biochemistry. 1995;34:15925–15933. doi: 10.1021/bi00049a007. [DOI] [PubMed] [Google Scholar]
- [10].Lockridge O, M. Blong R, Masson P, Froment M-T, Millard CB, A. Broomfield C. A single amino acid substitution, Gly117His, confers phosphotriesterase (organophosphorus acid anhydride anhydrolase) activity on human Butyrylcholinesterase. Biochemistry. 1997;36:786–795. doi: 10.1021/bi961412g. [DOI] [PubMed] [Google Scholar]
- [11].Millard CB, Lockridge O, Broomfield CA. Organophosphorus acid anhydride anhydrolase activity in human Butyrylcholinesterase: synergy results in a somanase. Biochemistry. 1998;37:237–247. doi: 10.1021/bi972057c. [DOI] [PubMed] [Google Scholar]
- [12].Zhang J. Human Biomolecular Research Institute, unpublished results.
- [13].Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. Crystal structure of human Butyrylcholinesterase and of its complexes with substrate and products. J. Biol. Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- [14].Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Amber, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- [15].Ponder JW, Case DA. Force fields for protein simulations. Adv. Prot. Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- [16].Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Research. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Case DA, et al. AMBER 8, University of California; San Francisco: 2004. For full citation, please refer to http://ambermd.org. [Google Scholar]
- [18].Case DA, et al. AMBER 10, University of California; San Francisco: 2008. For full citation, please refer to http://ambermd.org. [Google Scholar]
- [19].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential fluctuations for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- [20].WlodekAmber ST, Clark TW, Scott LR, McCammon JA. Molecular dynamics of Acetylcholinesterase dimer complexed with Tacrine. J. Am. Chem. Soc. 1997;119:9513–9522. [Google Scholar]
- [21].Suárez D, Field MJ. Molecular dynamics simulations of human Butyrylcholinesterase. Proteins. 2005;59:104–117. doi: 10.1002/prot.20398. [DOI] [PubMed] [Google Scholar]
- [22].Simon S, Le Goff A, Frobert Y, Grassi J, Massoulie J. The binding sites of inhibitory monoclonal antibodies on acetylcholinesterase. Identification of a novel regulatory site at the putative “back door”. J. Biol. Chem. 1999;274:27740–27746. doi: 10.1074/jbc.274.39.27740. [DOI] [PubMed] [Google Scholar]
- [23].Barakat NH, Zheng X, Gilley CB, MacDonald M, Okolotowicz K, Cashman JR, Vyas S, Beck JM, Hadad CM, Zhang J. Chemical Synthesis of Two Series of Nerve Agent Model Compounds and Their Stereoselective Interaction with Human Acetylcholinesterase and Human Butyrylcholinesterase. Chem. Res. Toxicol. 2009;22:1669–1679. doi: 10.1021/tx900096j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vyas S, Xia S, Zhang J, Hadad CM. Molecular dynamics of BuChE variants and docking simulations of OP-analogues. Xth International Meeting on Cholinesterases; Sibenik (Croatia). 20–25 September 2009; 2009. Poster P-3-35. [Google Scholar]
- [25].Masson P, Nachon F, Broomfield CA, Lenz DE, Verdier L, Schopfer LM, Lockridge O. Chem. Biol. Interact. 2008;175:273–280. doi: 10.1016/j.cbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- [26].Viragh C, Akhmetshin R, Kovach IM, Broomfield CA. Unique push-pull mechanism of dealkylation in soman-inhibited cholinesterases. Biochemistry. 1997;36:8243–8252. doi: 10.1021/bi962764q. [DOI] [PubMed] [Google Scholar]
- [27].Ordentlich A, Kronman C, Barak D, Stein D, Ariel N, Marcus D, Velan B, Shafferman A. Engineering resistance to `aging' of phosphylated human acetylcholinesterase. FEBS Lett. 1993;334:215–220. doi: 10.1016/0014-5793(93)81714-b. [DOI] [PubMed] [Google Scholar]
- [28].Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Shafferman A, Silman I, Sussman JL. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II. Acta Crystal. Sec. D. 2000;56:1385–1394. doi: 10.1107/s0907444900010659. [DOI] [PubMed] [Google Scholar]
- [29]. Interestingly, W82 slips into the active site in all of the trajectories from its original position; the origin (or ramification) of this effect is not obvious. Such movement of W82 was sometimes noted for other mutants, but was not present in all of the trajectories.