Abstract
Multiple studies hypothesize that DEAD-box proteins facilitate folding of the ai5γ group II intron. However, these conclusions are generally inferred from splicing kinetics, and not from direct monitoring of DEAD-box protein-facilitated folding of the intron. Using native gel electrophoresis and DMS structural probing, we monitored Mss 116-facilitated folding of ai5γ intron ribozymes and a catalytically active self-splicing RNA containing full length intron and short exons. We found that the protein directly stimulates folding of these RNAs by accelerating formation of the compact near-native state. This process occurs in an ATP-independent manner, although, ATP is required for the protein turnover. As Mss 116 binds RNA non-specifically, most of binding events do not result in the formation of the compact state, and ATP is required for the protein to dissociate from such non-productive complexes and rebind the unfolded RNA. Results obtained from experiments at different concentrations of magnesium ions suggest that Mss 116 stimulates folding of ai5γ ribozymes by promoting the formation of unstable folding intermediates, which is then followed by a cascade of folding events resulting in the formation of the compact near-native state. DMS probing results suggest that the compact state formed in the presence of the protein is identical to the near-native state formed more slowly in its absence. Our results also indicate that Mss 116 does not stabilize the native state of the ribozyme, but that such stabilization results from binding of attached exons.
Keywords: ribozyme, RNA folding, DEAD-box protein, tertiary structure
INTRODUCTION
In order to perform their biological functions, RNA molecules must fold into functionally active, native conformations. The folding pathway for a large RNA can be “smooth” (Azoarcus group I intron 1; 2; 3 ai5γ group II intron 4; 5 6) or “rugged” with one or more stable misfolded intermediates (“kinetic traps”)(Tetrahymena ribozyme 7; 8; 9, td group I intron 10, B. subtilis RNase P 11). Recent studies have shown that many RNAs fold slowly in vitro, with the rate-limiting step being either refolding from a kinetic trap 8; 12 or formation and stabilization of on-pathway intermediates 1; 2; 3 4; 5. These obstacles are overcome with help of protein co-factors, which often guide RNA folding in vivo. Cofactor proteins employ various strategies to facilitate RNA folding. RNA chaperone proteins (e.g. StpA) transiently bind RNA and destabilize misfolded structures in an ATP-independent manner 10; 13. Other proteins stabilize RNA structure through annealing activity (Hfq), selective binding of otherwise transient on-pathway folding intermediates (Cbp2), or binding and stabilization of tertiary structure (CYT-18) 13; 14.
Ribozymes derived from group II intron ai5γ fold slowly and directly to the near-native state under near-physiological conditions (30 °C and 3-10 mM MgCl2)15. This process occurs under kinetic control, with a very large energy barrier between the unfolded and near-native states15. Under these conditions, the native state of the ribozyme is unstable and, although the major intron scaffolding is maintained, catalytic domains dock only transiently within the core 15. The catalytically-active native state can be stabilized in vitro by employing conditions of elevated temperature (42 °C) and high magnesium (100 mM) and monovalent (500 mM) ion concentrations.
Splicing of the ai5γ intron is essential for the survival of the host organism (Saccharomyces cerevisiae), and it must occur at physiological concentrations of magnesium and monovalent ions. We have therefore hypothesized that protein cofactors assist folding of the ai5γ intron in vivo by using at least one of the following strategies15. They may accelerate formation of the near-native state by stabilizing the transition state between unfolded and near-native states and lowering the energy barrier between them. Alternatively, cofactors may stabilize the native state of the ribozyme at physiological concentrations of magnesium ions. However, direct information on these functions has been lacking.
It was recently demonstrated that DEAD-box proteins from S. cerevisiae and N. crassa (Mss 116, Ded1 and CYT-19) effectively stimulate splicing of the ai5γ intron both in vivo and in vitro under near-physiological conditions in an ATP-dependent manner 16; 17; 18. Two distinct mechanisms have been proposed to explain the function of these proteins. The first model suggests that DEAD-box proteins facilitate splicing by unwinding misfolded structures that form within the intron or the exons 18; 19; 20. The second model suggests that DEAD-box proteins may have a broader role in RNA folding and that they may sometimes stabilize folding intermediates 17. Multiple studies have generated experimental evidence in support of both models. The first model is supported by the ability of CYT-19 and Ded 1 proteins to selectively destabilize misfolded intermediates formed by the Tetrahymena ribozyme, resulting in kinetic redistribution between native and misfolded states 21 19. The second model is supported by the smooth folding pathway of the ai5γ intron ribozymes 4; 5 6, as well as the ability of a helicase - deficient Mss 116 mutant to actively facilitate the ai5γ intron splicing 17.
Although both mechanisms assume that DEAD-box proteins affect the ai5γ splicing at the level of folding, no experiments have been carried out to monitor protein-facilitated folding of the intron directly. Here we examine Mss 116-stimulated folding of various ai5γ intron constructs under different ionic conditions using a combination of native gel electrophoresis and biochemical structural probing. The results indicate that Mss 116 accelerates formation of the compact intermediate state, but it does not stabilize the native state of the ribozyme. The native state is stabilized by base-pairing interactions between the intron and adjacent exon sequences. Surprisingly, Mss 116-facilitated folding of D1 and other ai5γ constructs is not strictly ATP-dependent. In addition, Mss 116-promoted folding is severely impaired at reduced magnesium ion concentrations. Taken together, these data suggest that Mss 116 stimulates formation of a structurally unstable critical folding intermediate that is essential for productive compaction of ai5γ ribozymes.
RESULTS
Mss 116 stimulates formation of the compact intermediate in D1 RNA
In order to elucidate the effect of Mss 116 on folding of the ai5γ group II intron, we first determined whether the protein facilitates folding of intron Domain 1 (D1). D1 is the largest domain, and it folds independently of the rest of the intron 5; 22. It serves as a scaffold for docking of other domains (most importantly, catalytic domain 5 (D5)) and it recognizes 5’-exonic substrates via base-pairing interactions 23. Importantly, multiple studies have demonstrated that folding of D1 is the rate-limiting step in folding of the entire intron 5; 15; 24. Thus, if the protein helps folding of D1 RNA, it is likely to be important for folding of the entire intron.
The rate-limiting event during D1 folding is the formation of a discrete, compact species that migrates rapidly on a native polyacrylamide gel and has the same electrophoretic mobility as native species15. It has previously been shown that this species represents an on-pathway intermediate and it has been classified as the near-native state 5; 6; 15. We therefore investigated the effect of Mss 116 on D1 folding by monitoring formation of this compact state in the presence and in the absence of the protein. D1 compaction was examined using a well-characterized electrophoretic assay that has been employed extensively for studying the folding of large RNAs 15; 25; 26; 27. Experiments were carried out at 8 mM MgCl2, which is the condition generally used for studying Mss 116-facilitated splicing of the ai5γ intron 17; 18.
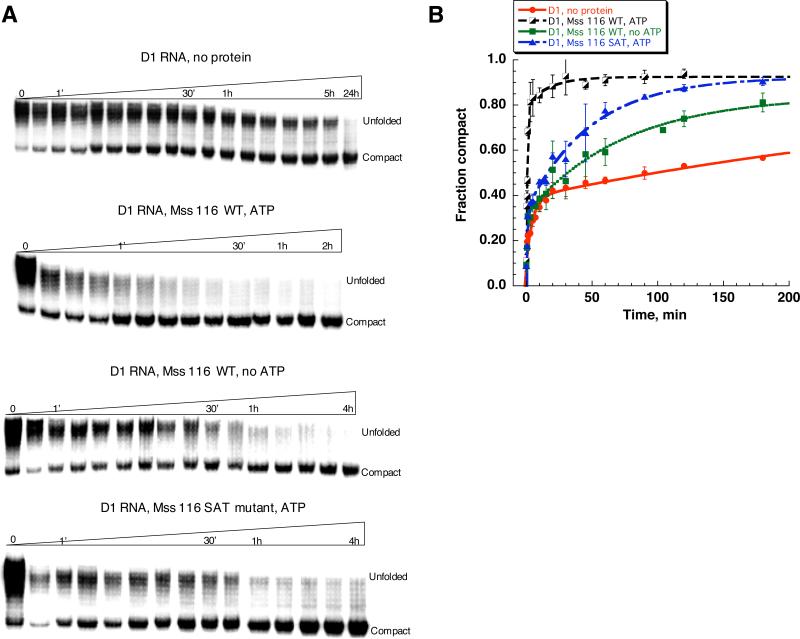
In the absence of the protein, formation of the compact intermediate is very slow (Fig.1, Table 1), as expected from previous studies.15 Notably, the unfolded species migrate on a native gel as a broad smeary band (Fig. 1A), which is consistent with multiple populations of unfolded molecules. Furthermore, when the fraction of the compact population is plotted versus time over the course of 3-5 hours, the resulting curve is biphasic, further suggesting that there are multiple populations of unfolded species, and that these form the compact state with different rate constants. The faster population (0.23±0.06 min-1) represents only ~25% of all RNA molecules (Fig. 1, Table 1), while most of the population folds with a half-time of hours. Over the course of 24 hours all RNA eventually forms the compact state (Fig. 1 A). Compaction of the remaining RNA population, which occurs between 5 and 24 hour time points, appears to be even slower than the slow-folding population, however, more detailed study is needed to fully characterize it. All time courses reveal a small population (~10%) that forms immediately after the addition of magnesium and monovalent ions (“0” time point, Fig. 1A). This initial collapse occurs much faster than compaction of the fast-folding population, and that it does not depend on the presence of Mss 116 or the concentration of magnesium ions used in our experiments (see Fig. 2).
Figure 1.
Comparison of the D1 RNA compaction time course at 8 mM MgCl2 and 30 °C in the presence and in the absence of Mss 116. Protein-facilitated compaction was studied in the presence and in the absence of ATP. D1 compaction was monitored by native gel electrophoresis (A), fraction of the compact population was plotted versus time (B) and fit to a double-exponential equation to obtain compaction rate constants and amplitudes shown in Table 1.
Table 1.
Kinetic parameters for RNA compaction under splicing conditions. D1 and D1356 RNAs (1 nM) were studied at 8 mM MgCl2 and 30 °C in the absence and in the presence of Mss 116 (WT or SAT mutant, 20 nM), as indicated.
| RNA | Conditions | kfast, min-1 | Ampl. fast, %* | kslow*102, min-1 | Ampl. slow, %* |
|---|---|---|---|---|---|
| D1 | No protein | 0.23±0.06 | 27±1 | 0.56±0.06 | 32±9 |
| Mss 116WT, ATP | 1.3±0.1 | 73±7 | 2.9±0.3 | 17±5 | |
| Mss 116WT, no ATP | 1.1±0.3 | 21±3 | 1.5±0.2 | 56±4 | |
| Mss 116SAT, ATP | 1.4±0.2 | 21±4 | 2.0±0.4 | 57±3 | |
| Mss 116SAT, no ATP | 0.71±0.04 | 26±1 | 0.85±0.07 | 60±10 | |
| D1356 | No protein | 0.16±0.03 | 46±4 | 0.6±0.2 | 20±6 |
| Mss 116WT, ATP | 1.4±0.2 | 78±3 | 3.0±0.5 | 15±5 | |
| Mss 116WT, no ATP | 0.35±0.02 | 43±4 | 1.5±0.4 | 41±1 | |
| Mss 116SAT, ATP | 1.6±0.3 | 36±7 | 4±1 | 45±7 |
These values do not include the amplitude of an initial collapse observed at a "0" time point, which constitutes ~10%.
Figure 2.
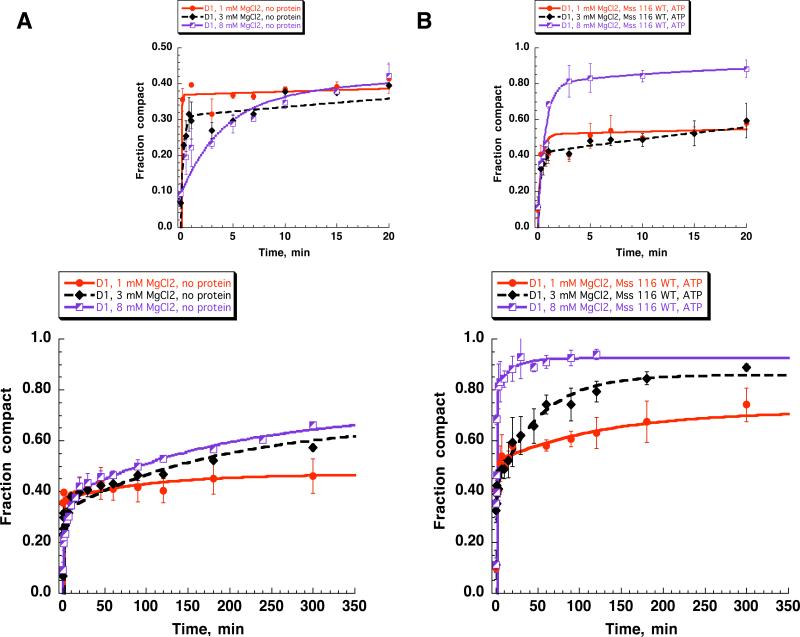
Compaction of the D1 RNA at lower magnesium concentrations (a) without Mss 116 and (b) in the presence of the WT Mss 116 and ATP. Smaller graphs on top show the first 20 minutes of respective time courses. Kinetic parameters are listed in Table 2.
Upon addition of Mss 116 and ATP, formation of the compact state drastically accelerates: after 1 minute of incubation, most of the RNA population has formed the compact state (Fig. 1, A). Under these conditions, the majority of molecules are fast-folding, suggesting that Mss116 converts the slow-folding population into the fast-folding population (Fig. 1, Table 1). In addition, the folding rate constant for this population also increases (Table 1), suggesting that Mss 116 does not merely capture a transiently-sampled state, but that it influences the folding mechanism.
Surprisingly, Mss 116 facilitates compaction of D1 even in the absence of ATP (Fig. 1, Table 1). In this case, the rate constant for folding of the fast population is almost identical to that in the presence of ATP, although the fraction of the fast population is smaller (Table 1). Treatment with hexokinase and glucose does not abolish ATP-independent compaction (Supplementary Fig. 1A), indicating that this effect is not caused by ATP contamination. These findings suggest that the mechanistic role of Mss116 in the folding pathway is independent of ATP hydrolysis, and indeed fully independent of ATP. However, the low reaction amplitude suggests that the amount of Mss116 present in the reaction mixture is insufficient to fold the entire population of D1.
Protein-dependent compaction of D1 was also monitored in the presence of an Mss116 variant that contains mutations in catalytic motif III (Mss116SAT→AAA). The (Mss116SAT→AAA) mutant retains normal levels of ATP and RNA binding, but displays reduced ATPase activity17. RNA duplex unwinding by the mutant (helicase activity) is greatly reduced, although Mss116SAT→AAA still supports robust Mss116-dependent splicing of intron ai5γ17. In the presence of Mss116SAT→AAA and ATP, D1 compaction occurs rapidly (Fig. 1A, Table 1). The rate constant for compaction of the fast population is indistinguishable from that of WT Mss116 (Fig. 1, Table 1). However, as in the case of Mss116 in the absence of ATP, the fraction of this population is small. Thus, Mss116SAT→AAA appears to provide a second metric for monitoring the ATP-independent reaction.
Although folding of D1 is central for assembly of the entire intron, it was of interest to determine whether the protein-dependence of compaction changed when additional domains of the intron were present. To this end, we monitored Mss116 and ATP-dependence of compaction for the D1356 RNA, which includes the catalytically important domains 3, 5 and 615. As expected, compaction of D1356 displays protein dependence that is similar to D1, suggesting that addition of the major catalytic domains does not change the basic mechanism of Mss 116-facilitated folding (Table 1).
Protein-dependent compaction is sensitive to varying magnesium ion concentrations
It is well established that RNA duplexes and tertiary structures are destabilized by reductions in ionic strength and reductions in magnesium ion concentration 28; 29; 30; 31. If the major barrier to folding of group II introns is the presence of a stable, misfolded structure, then reduction in magnesium ion concentration is expected to lower this barrier and make it easier for the intron to fold32; 33. Likewise, if Mss116/ATP facilitates D1 folding by acting as a helicase and unwinding a stable, misfolded structure that disrupts the folding pathway, then protein-dependent compaction is expected to be faster or more effective at reduced magnesium ion concentrations. However, two conditions must be met in order to test this hypothesis: First, it must be possible to maintain the compact state of D1 at reduced magnesium ion concentrations. Second, the ATPase activity and RNA binding capability of the protein should not be compromised at low concentrations of MgCl2.
In order to evaluate the first condition, we tested the ability of D1 RNA to exist in a compact state at lower magnesium ion concentrations by folding it to the native state at 100 mM MgCl2 and subsequently diluting the sample to lower magnesium ion concentrations (0.5 to 20 mM MgCl2). We found that D1 can exist in a compact state even at 1 mM MgCl2, however, it immediately reverts to the unfolded state at 0.5 mM MgCl2 (Supplementary Fig. 2A), which is consistent with previously published results obtained for the D1356 RNA 15.
The second condition has been partly addressed in previous work, which demonstrated that ATPase activity of Mss 116 does not decrease and even slightly increases upon lowering magnesium ion concentration from 8 to 1 mM MgCl2 18. We have observed the same trend as well (Supplementary Fig. 3). Thus, ATPase activity of the protein is not compromised at low magnesium ion concentrations.
To evaluate the magnesium ion dependence of RNA binding by Mss116, we measured dissociation constants for the Mss116-D1 complex at 1, 3 and 8 mM MgCl2. Mss 116 binds D1 RNA tightly at 8 mM MgCl2 (Kd = 1.2 ± 0.2 nM), which is in good agreement with previously published data on Mss 116 interactions with other RNAs 34. At 1 and 3 mM MgCl2, the affinity of Mss116 remains strong (Kd = 1.3 ± 0.3 and 1.9 ± 0.3 nM, respectively), indicating that RNA binding properties of the protein are unaffected under these conditions. In addition, according to previously published results,18,35 Mss 116 efficiently unwinds RNA duplexes at 1 mM and even 0.5 mM MgCl2.
Taken together, all these results indicate that mechanistic insights can be gleaned by studying Mss 116-facilitated compaction of D1 at different magnesium ion concentrations. If Mss 116 accelerates compaction of D1 RNA by unwinding kinetic traps, then lowering magnesium ion concentrations from 8 to 1 or 3 mM should stimulate Mss 116-facilitated compaction of D1 RNA. Conversely, if the protein stimulates the formation of unstable substructures that are obligate intermediates along the D1 folding pathway, decreasing the magnesium ion concentration should severely impair protein-assisted compaction of D1.
Compaction was first monitored in the absence of protein, at varying concentrations of MgCl2 (1, 3 and 8 mM, Fig. 2). The results indicate that a small population of D1 benefits from the destabilizing effect of lowering [MgCl2]: the rate constant for compaction of this population is ten-fold faster at 1 and 3 mM MgCl2 than at 8 mM MgCl2 (~ 25%, Fig. 2A, Tables 1 and 2). However, most of the population remains unfolded at 1 mM MgCl2 or it folds very slowly at 3 and 8 mM MgCl2 (Fig. 2A).
Table 2.
Kinetic parameters of the D1 RNA compaction at low magnesium ion concentrations. D1 (1 nM) was studied at 1 and 3 mM MgCl2 and 30 °C in the absence and in the presence of WT Mss 116 (20 nM) and ATP (1 mM). Values for 8 mM MgCl2 are from Table 1.
| Mss 116 | [MgCl2], mM | kfast, min-1 | Ampl fast, %* | kslow* 102,min-1 | Ampl. slow, %* |
|---|---|---|---|---|---|
| No protein | 1 | >5 | 28 ± 4 | 0.8 ± 0.2 | 10 ± 1 |
| 3 | 3 ± 1 | 26 ± 1 | 1.1 ± 0.5 | 27 ± 5 | |
| 8 | 0.23 ± 0.06 | 27 ± 1 | 0.56 ± 0.06 | 32 ± 9 | |
| Mss 116WT, ATP | 1 | >5 | 40 ± 3 | 0.7 ± 0.1 | 15 ± 7 |
| 3 | 3.1 ± 0.9 | 37 ± 7 | 1.9 ± 0.6 | 42 ± 3 | |
| 8 | 1.3 ± 0.1 | 73 ± 7 | 2.9 ± 0.3 | 17 ± 5 |
These values do not include the amplitude of an initial collapse observed at a "0" time point, which constitutes ~10%.
D1 compaction was then monitored in the presence of Mss116 and ATP, at 1, 3 and 8 mM MgCl2. At 8 mM MgCl2, Mss116 converts almost the entire slow-folding population into a very fast-folding population (Fig. 2B), thereby increasing the folding rate-constant for most of the RNA molecules by more than 60-fold. By contrast, Mss 116 has very little influence on the overall extent or rate-constant of reaction at 1 mM MgCl2 (Fig. 2B, Table 2). Similarly, Mss116 has only a small stimulatory effect at 3 mM MgCl2 (Fig. 2B, Table 2). At these low concentrations of MgCl2, the rate constant and amplitude of the fast-folding population is almost the same as that in the absence of the protein and ATP (Fig. 2 A and B, top, Table 2), indicating that Mss116 has no additional influence on the folding mechanism of that population. Taken together, these data indicate that reduced (<8 mM) magnesium ion concentrations adversely affect protein-facilitated compaction of D1 RNA.
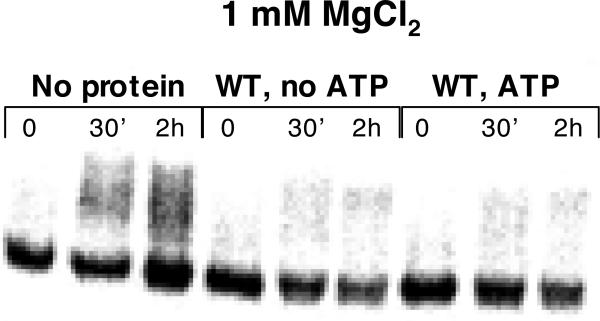
There is an alternative explanation for the adverse effect of low MgCl2 concentration on protein-induced compaction of D1: Lower MgCl2 concentrations are known to stimulate duplex unwinding (helicase) activity of Mss 116 18. Under these conditions, Mss116 may destabilize the compact state by unwinding RNA helices that are essential for the native D1 structure. To test this assumption, we preassembled the compact state of D1 RNA at 1, 3 and 8 mM MgCl2 and we monitored unfolding over time upon addition of Mss 116 and ATP (Fig. 3, Supplementary Fig. 4). We observe that the compact state remains stable at all MgCl2 concentrations tested, even after 2 hours of incubation with the protein (Fig. 3, Supplementary Fig. 4). This indicates that the adverse effect of low magnesium ion concentrations cannot be attributed to unfolding of the compact state by the protein.
Figure 3.
Mss 116 does not facilitate unfolding of the preassembled compact D1 RNA. Compact D1 RNA was obtained by folding to the native state at 100 mM MgCl2 and subsequent dilution of the samples to 1 mM MgCl2. Samples were incubated with WT Mss 116 in the presence and in the absence of ATP over the course of 2h and analyzed by native gel electrophoresis.
ATP is required for enzyme turnover (recycling)
Recent studies have suggested that ATP hydrolysis is not required for helicase activity of Mss 116 and related DEAD-box proteins 35. Rather, these proteins are thought to require ATP binding for the unwinding mechanism and ATP hydrolysis is required only for the protein dissociation from the products of unwinding and rebinding to new RNA substrates (protein recycling) 35. Recycling of the protein is essential when unwinding is carried out under conditions of substrate excess, in which Mss116 acts as a multiple-turnover enzyme.
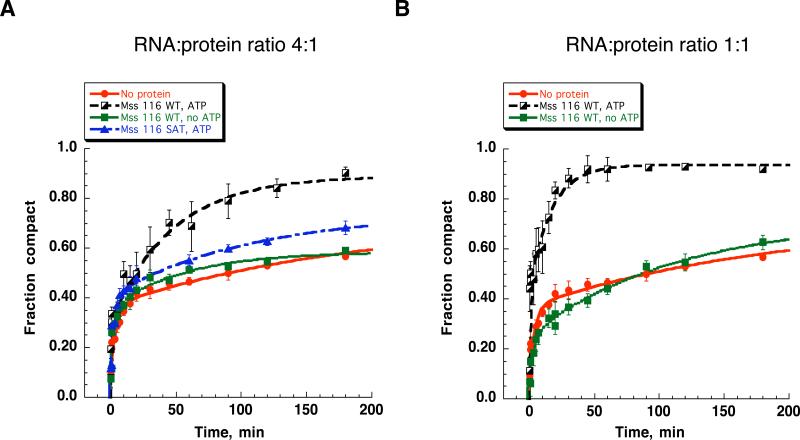
In order to determine whether Mss 116-assisted compaction of D1 requires ATP hydrolysis for protein recycling, we carried out D1 compaction experiments under multiple turnover conditions. Thus far, the experiments in this study were carried out under single turnover conditions, with a 20-fold excess of Mss116 over RNA. In the multiple-turnover experiments, D1 RNA (20 nM) was present in four-fold excess over Mss 116 (5 nM). To facilitate compaction of the entire D1 population under these conditions, Mss 116 must dissociate from the compact RNA and rebind an unfolded RNA.
In the presence of small quantities of Mss 116 and excess ATP, D1 compaction proceeds to completion. In the presence of Mss116 and the absence of ATP, D1 compaction is incomplete, proceeding only to the extent observed in the absence of the protein (Fig. 4A). Similarly, the ATPase-deficient Mss116SAT→AAA mutant17 has only a small effect on the reaction extent (Fig. 4A). These results suggest that there is an ATP requirement for recycling of the protein and for multiple-turnover activity of Mss116.
Figure 4.
Kinetic analysis of Mss 116-facilitated compaction of D1 RNA under multiple turnover conditions and at 1:1 RNA:protein ratio. Comparison of the compaction time courses at 5 nM (A) and 20 nM (B) Mss 116 and 20 nM D1 RNA for the WT Mss 116 protein in the presence and in the absence of ATP (A, B) and SAT:AAA mutant in the presence of ATP (A).
Previous studies suggest that Mss 116 binds RNA non-specifically 18; 34. Thus, many of D1-Mss 116 complexes are likely to result from nonproductive binding that plays no role in folding and does not lead to the formation of the compact state. In these cases, ATP will be required for the protein to dissociate from the nonspecific complex and rebind the RNA in a productive manner, forming an active complex. It is therefore notable that Mss 116-facilitated compaction in the absence of ATP of is inefficient even at a 1:1 RNA-protein ratio (20 nM RNA and 20 nM protein) (Fig. 4B). This suggests that initial binding of Mss 116 to D1 results in a relatively small fraction of productive RNA-protein complexes.
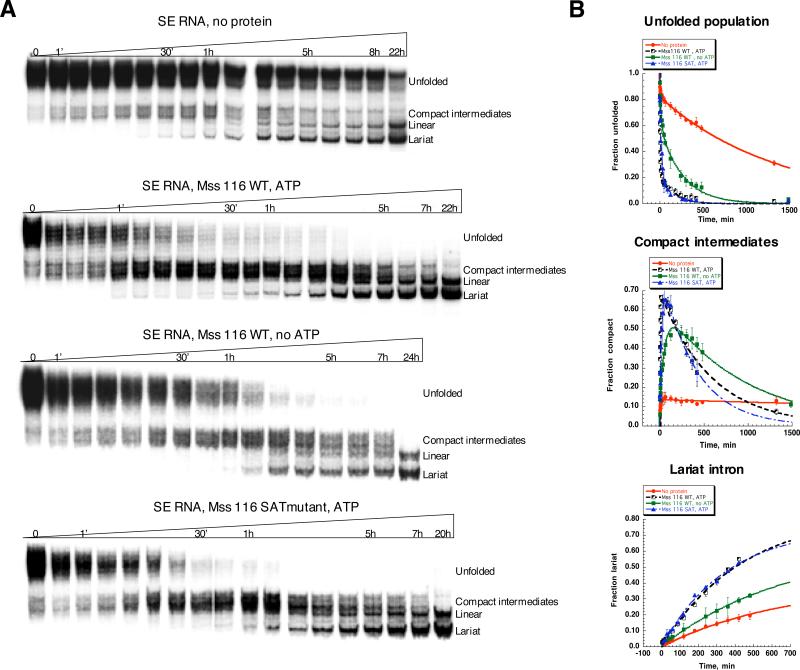
Mss 116 strongly stimulates compaction of a self-splicing intron construct
Although we have shown that Mss 116 stimulates compaction of D1 RNA, which is the rate-limiting step during folding of the entire intron, it was important to determine whether Mss116 facilitates compaction of a more complete, catalytically active intron construct. To address this issue, we monitored compaction of a self-splicing construct that contains the entire intron RNA (D123456) flanked by short (28 and 15 nt, respectively) 5’- and 3’-exonic sequences (SE RNA).
The various populations of compacted SE RNA are more complex than observed for D1. Unfolded RNA first gives rise to compact intermediates, which then evolve into spliced lariat and linear intron molecules (Supplementary Fig. 5). In the absence of Mss116 and ATP, compaction of SE RNA is remarkably slow (the rate constant for the majority of the population is 0.00075±0.00003 min-1), compact intermediates do not accumulate, and, as they form, they almost immediately react resulting in lariat and linear introns (Fig. 5). The fast-folding population in this case is very small (less than 10%, Table 3). Upon addition of Mss 116 and ATP, the formation of compact intermediates accelerates by a thousand-fold (the rate constant for the main population is 0.8±0.1 min-1) and the compact species rapidly accumulate. Mss116 substantially increases the amplitude of lariat formation: all the compact intermediate is transformed into splicing products (lariat and linear intron, Fig. 5). However, the effect of the protein on the rate constant of lariat formation is rather modest (Fig. 5, Table 3). These data clearly show that the primary role of Mss 116 is to accelerate the formation of compact, near-native intermediates and not to stimulate the subsequent rate constant for splicing or to stabilize the native state of the ribozyme.
Figure 5.
Comparison of the SE RNA compaction time course at 8 mM MgCl2 in the presence and in the absence of Mss 116. Protein-facilitated compaction was studied in the presence and in the absence of ATP. SE compaction was monitored by native gel electrophoresis (A), fraction of the unfolded precursor, compact population and lariat intron was plotted versus time (B) and fit as described in Materials and Methods to obtain compaction rate constants and amplitudes shown in Table 3.
Table 3.
Kinetic parameters for SE RNA compaction and lariat formation under splicing conditions. SE RNA (1 nM) was studied at 8 mM MgCl2 and 30 °C in the absence and in the presence of Mss 116 (WT or SAT mutant, 20 nM), as indicated.
| Conditions | kfast, min-1 | Ampl. fast, %* | kslow*102, min-1 | Ampl. slow, %* | klar*102, min-1 |
|---|---|---|---|---|---|
| No protein | 0.1±0.05 | 8±1 | 0.075±0.003 | 80±3 | 0.155±0.007 |
| Mss 116WT, ATP | 0.8±0.1 | 54±1 | 0.9±0.2 | 28±1 | 0.3±0.08 |
| Mss 116WT, no ATP | 0.05±0.01 | 26±2 | 0.5±0.1 | 62±3 | 0.25±0.06 |
| Mss 116SAT, ATP | 0.06±0.01 | 63±3 | 0.55±0.06 | 18±3 | 0.33±0.08 |
Although we observe multiple bands that correspond to compact intermediates of SE RNA, we did not quantify them separately because they form and react at the same time (Fig. 5), suggesting that they are equally active in splicing. These various compact species are likely to represent different isoforms of the same active species, which is supported by DMS structural probing (see below).
Remarkably, Mss 116 substantially facilitates compaction of SE RNA even in the absence of ATP (Fig. 5). ATP-independent compaction of the SE RNA is not abolished by hexokinase treatment, suggesting that it is not catalyzed by trace amounts of ATP in the system (Supplementary figure 1B). ATP-independent compaction of SE RNA is unlikely to involve unwinding of kinetically trapped species, because such a mechanism would require at least the binding of ATP 35. Note that Mss116 and related DEAD-box proteins require ATP, but not necessarily ATP hydrolysis, for the unwinding of RNA duplexes 35. Interestingly, the ATP-independent effect on compaction of SE RNA is more prominent than that observed for D1 RNA (compare time courses in the absence and in the presence of Mss 116 (no ATP) in Fig. 1 and Fig. 5).
Reduction in [MgCl2] inhibits Mss 116-stimulated folding of SE RNA
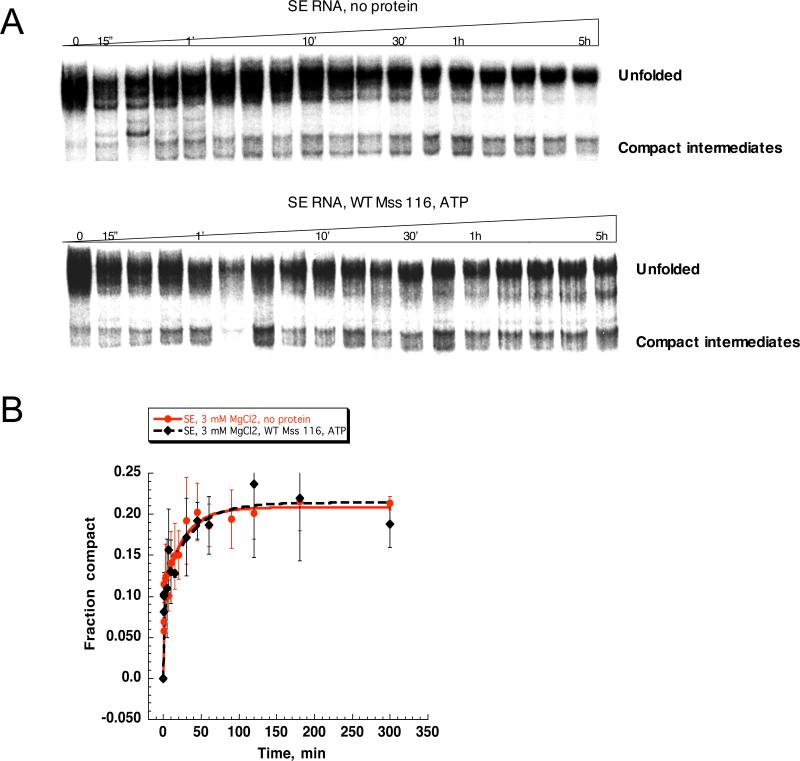
Reduction in magnesium ion concentration has an adverse effect on Mss 116-facilitated compaction of D1 RNA, which is consistent with an important stabilizing role for both Mg2+ and Mss116 in the folding of D1. To determine if these same principles apply to folding of the intact intron, we monitored compaction of SE RNA at 3 mM MgCl2 in the absence and in the presence of protein and ATP (Fig. 6). The majority of SE RNA population is unfolded at 3 mM MgCl2, and Mss 116/ATP does not display any stimulatory effect under these conditions (Fig. 6).
Figure 6.
Comparison of the SE RNA compaction time course at 3 mM MgCl2 in the presence and in the absence of Mss 116. Protein-facilitated compaction was studied in the presence of ATP. SE compaction was monitored by native gel electrophoresis (A), fraction of the compact population was plotted versus time (B) and fit as described in Materials and Methods. The rate constants for the fast-folding population (9±1%) in the absence and in the presence of Mss 116 were 3.1±0.3 and 4.1±0.4 min-1, respectively. The rate constant for the slow-folding population (11±1% in the absence of the protein and 8±3% in the presence of the protein) was 0.03± 0.01 min-1 both in the absence and in the presence of the protein.
To interpret these results, it was essential to determine whether the compact intermediate was simply inherently unstable at 3 mM MgCl2. Compact intermediate states were first generated by incubating SE RNA with Mss 116 and ATP in the presence of 8 mM MgCl2. Samples were then diluted to 3 and 1 mM MgCl2, respectively. At 3 mM MgCl2, the SE compact state is stable and does not spontaneously unfold (Supplementary Fig. 2B), although at 1 mM MgCl2, compact intermediates partly revert to the unfolded state (Supplementary Fig. 2B). These data indicate that, despite the fact that compact intermediates are ultimately stable at 3 mM MgCl2, the protein cannot facilitate the process of SE compaction under these conditions. Therefore, as in the case of D1, Mss 116 facilitates the kinetic folding pathway of SE RNA by stimulating the formation of unstable structural intermediates.
The compact state is structurally similar in the absence and presence of Mss116
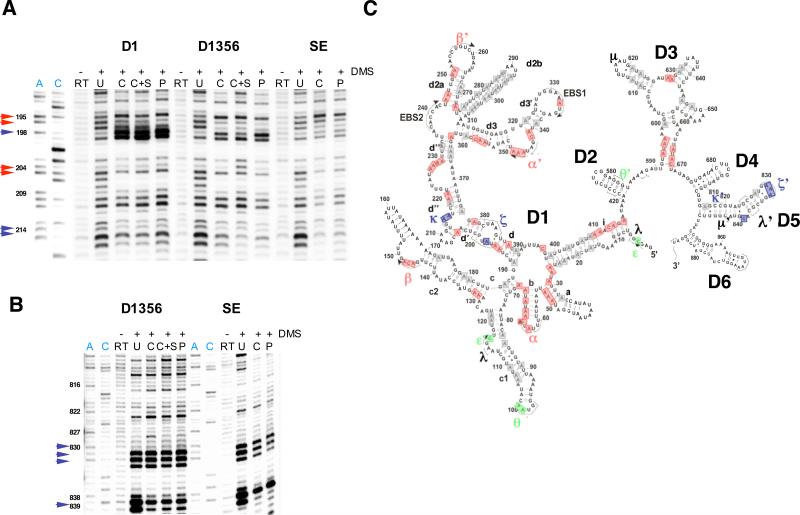
Up to this point, the study has focused on the kinetic behavior of D1, D1356 and SE RNAs as they fold in the presence and absence of Mss 116. To complement and interpret these experiments, it was necessary to examine the structural features of the compact intermediate states that arise during folding of these RNAs. We utilized chemical probing with dimethyl sulfate (DMS) to monitor specific changes in the secondary and tertiary structure of D1, D1356 and SE RNAs as they formed the compact state. DMS methylates N1 and N3 atoms of adenosine and cytidines residues, respectively, when they are not involved in hydrogen bond formation.
Compact intermediates of D1, D1356 and SE RNAs were preassembled in the presence and in the absence of the protein at 8 mM MgCl2 as described above. Because the compact state of SE RNA reacts immediately to form free intron, we probed the structure of lariat intron in the case of SE. Samples were treated with DMS, subjected to primer extension and visualized on a high-resolution polyacrylamide gel. For all the RNAs tested, compact intermediate species that form in the presence and in the absence of the protein appear to be identical (Fig. 7, Supplementary Fig. 6). Notably, the compact species formed by the SE RNA exhibit the same DMS protection pattern as the lariat intron. Analysis of the DMS protection patterns shows that the compact species contain all the known long-range tertiary interactions critical for the function of the intron (Fig. 7, Supplementary Fig. 6).
Figure 7.
DMS structural probing of compact intermediates formed at 8 mM MgCl2 in the presence and in the absence of Mss 116 by D1, D1356 and SE RNas. A, representative sequencing gels showing A and C residues in Domain 1 of the intron identically protected against DMS modifications in all three RNAs (red arrows) and A residues that become protected only in the presence of cis-exons in the SE RNA (purple arrows). B, representative sequencing gels showing A residues in Domain 5 of the intron that become protected only in the presence of cis-exons in the SE RNA (purple arrows). Sequencing lanes showing the position of A and C residues are highlighted in blue. RT, samples untreated with DMS that show natural RT stops in respective RNA molecules; U, unfolded RNA; C, compact RNA formed in the absence of the protein; C+S, compact RNA formed in the absence of the protein and in the presence of 24-mer exonic substrate added in trans; P, compact RNA formed in the presence of Mss 116. C, the secondary structure map showing A and C residues identically protected in all three RNAs (red), protected only in D1356 and SE RNAs (green) and only in SE RNA (purple). Residues that are involved in the RNA secondary structure and thus not modified by DMS in the unfolded state are shown in grey. See Materials and Methods for experimental details.
We have previously shown that, at magnesium ion concentrations lower than 100 mM, the native state of the D1356 ribozyme is not stable, and it adopts the near-native state with transiently docked catalytic domain 5 15. Interestingly, at 8 mM MgCl2 compact intermediates of the SE RNA contain a docked domain 5: the tetraloop receptor in D1 (ζ) and the tetraloop (ζ’) and bulge of D5 are well protected against DMS modification (Fig. 7). This suggests that the presence of exons helps stabilize docked D5 in the ribozyme active site. Interestingly, the addition of exonic substrate in trans does not facilitate docking of D5 in the D1356 construct (Fig. 6). Therefore the results suggest that covalently attached exons help to stabilize the native state of the ribozyme. By forming EBS-IBS duplexes, cis-exons may facilitate positioning of D5 binding sites in D1 (ζ and κ) closer to the 5’-splice site and D5, thus stabilizing the native conformation of the ribozyme.
DISCUSSION
Mss116 plays a direct role in RNA folding
Although it has long been presumed that Mss116 stimulates group II intron self-splicing by influencing the RNA folding pathway, the effect of Mss116 on folding had never been studied previously. By monitoring the formation and architectural features of ai5γ folding intermediates, we now show that Mss116 plays a direct role in facilitating proper folding of the intron. Here we show that Mss116 facilitates the very first step in the folding process: the collapse of D1 to form a compact intermediate state that serves as an obligate scaffold for subsequent rapid steps along the folding pathway (Fig. 1, Table 1) 5; 15. Importantly, the protein promotes formation of the same compact state that forms, albeit more slowly, in the absence of the protein.
Under the near-physiological conditions employed here, and in related studies of Mss116-stimulated splicing (8 mM MgCl2 and 30°C), the intron is almost incapable of folding in the absence of protein 15; 17; 18. The compact state eventually forms, but it occurs over a timescale of hours rather than minutes. Importantly, an alternative set of reaction conditions can mimic the stimulatory effect of the protein: At high concentrations of MgCl2 (100 mM), the same obligate compact intermediate state forms rapidly and completely in the absence of protein 5. This intermediate state has been well-characterized and is known to result from the formation of unstable secondary and tertiary interactions within a specific junction region within intron Domain 1 24; 36. Thus, the first step during intron folding is the collapse of Domain 1, and it can be stimulated either by high [Mg2+] or by the Mss116 protein.
The mechanism of Mss116-stimulated folding of ai5γ
Having established that Mss116 operates specifically along the folding pathway, it was important to explore the mechanistic basis for its effect. Previous studies of Mss116-stimulated splicing have suggested two different roles for the protein: One mechanism suggests that the protein acts as a helicase, which unwinds misfolded intermediates that kinetically obstruct the productive RNA folding pathway 18. The other mechanism suggests an opposite role for Mss116, whereby the protein promotes formation of weak substructures that are crucial for the formation of on-pathway folding intermediates 17.
While it is becoming increasingly clear that DEAD-box proteins can play diverse roles in the folding of even the same RNA (vida infra), a specific role for Mss116 in the folding of ai5γ is suggested by several lines of evidence
A first clue is provided by the simple fact that high [Mg2+] (100 mM) and Mss116 protein have the same effects on the folding pathway. Any RNA substructure that forms at high [Mg2+] and not at low [Mg2+] is typically unstable and it requires an optimal electrostatic environment for its formation. The literature on ribozymes and RNA binding proteins is full of examples where the electrostatic requirements of an RNA structure are met interchangeably with high [Mg2+] or a positively-charged protein 37; 38; 39.
A second clue to the mechanism of Mss116 is provided by the fact that ATP is not required for protein-stimulated compaction or folding of any group II intron RNAs that were investigated in this study. We show that ATP plays an important role in the compaction of Mss116, but only by facilitating turnover and recycling of the enzyme, as shown for other DEAD-box proteins. A comparison of single and multiple-turnover experiments shows that ATP facilitates Mss116-stimulated folding when the RNA is in excess, or when the RNA is large and contains numerous nonspecific binding sites for the protein. Mss116 binds RNA with very high affinity, and it is therefore not surprising that ATP or ATP hydrolysis may function as a conformational switch, enabling release of the protein and providing another opportunity for the formation of productive complexes. These findings have important implications for the folding mechanism since duplex unwinding by Mss116 has been shown to be strictly dependent on ATP binding (if not ATP hydrolysis) 35. It is therefore unlikely that Mss116 promotes formation of the compact intermediate state by functioning as a helicase and unwinding kinetic traps; in some instances such as described here, Mss116 behaves like an actively-stabilizing RNA-binding protein.
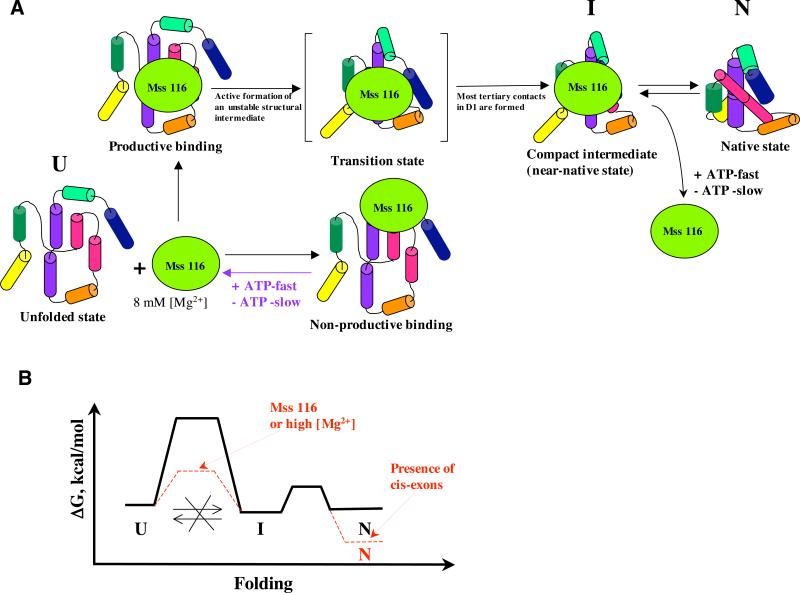
A final clue to the role of Mss116 is provided by experiments in which protein-stimulated compaction was monitored at sequentially reduced concentrations of magnesium ion. We reasoned that, if Mss116 serves to unwind stable, misfolded structures that obstruct formation of the compact state, it should be more effective at reduced [Mg2+], as observed in other kinetically-trapped systems 32; 33. For example, the related DEAD-box protein Cyt-19 is more effective at unwinding RNA tertiary structures at 1 mM MgCl2 than at 5 mM MgCl2.21 We therefore conducted our experiments at magnesium ion concentrations that are lower than the 8 mM Mg2+ that is optimal for Mss116-stimulated splicing (1 and 3 mM MgCl2). We observe that reduced MgCl2 concentrations are highly detrimental for Mss 116-assisted compaction of both D1 and SE RNAs (Fig. 2 and 6, Table 2) and that this effect is not attributable to Mss116-stimulated unwinding of the native, compact state. Together with the other lines of evidence, these findings indicate that the transition to the compact intermediate state by the main population of the intron RNA requires structural stabilization, rather than destabilization, and that this is provided by Mss116 at moderate (8 mM) concentrations of magnesium ion (Fig. 8).
Figure 8.
A, a scheme illustrating the mechanism of the Mss 116-assisted folding for the main population of the ai5γ group II intron RNA. B, a scheme of a hypothetical free energy diagram for the ribozyme folding to the near-native and native state in the presence and in the absence of Mss 116.
Although the compaction experiments are consistent with a stabilizing role for Mss116 during the ai5γ folding pathway, our results do not exclude the possibility that unwinding by Mss 116 may still play an important role during folding of constructs with longer exons, which are likely to form misfolded structures at least some of the time. In addition, our data show that ~25% of the D1 RNA population does benefit from some destabilization for efficient formation of the compact state (Fig. 2, Table 2). Therefore, the unwinding activity of Mss116 may facilitate folding by a subpopulation of intron molecules.
Mss 116 guides the D1 RNA on a one-way path to the compact state
Recent studies of CYT-19-facilitated folding of the T. thermophila-derived group I intron indicate that this DEAD box protein assists folding by promoting kinetic redistribution between misfolded and native ribozyme structures 21. The protein can unfold both misfolded and native states to the intermediate state, but it unfolds the misfolded state more efficiently. The intermediate then gets a new chance to refold to the native state. The authors have shown, that although CYT-19 is incapable of unfolding the native state at 5 mM MgCl2, it could do it at 1 mM MgCl2 as the native structure becomes less stable.
By contrast, our results show that Mss 116 cannot unfold the compact state of the D1 RNA even at 1 mM MgCl2 (Fig. 3). The compact state is not exceptionally stable under these conditions (ΔG=0.7 kcal/mol) 15, and upon further decrease of magnesium ion concentration to 0.5 mM MgCl2 it immediately reverts to the unfolded state (Supplementary figure 1A). This suggests that, rather than promoting kinetic redistribution, Mss116 guides D1 RNA along a one-way path to the compact state: once the RNA reaches the compact state, it does not unfold (Fig. 8). Although Mss 116 facilitates the formation of unstable folding intermediates, the subsequent cascade of tertiary structure formation (e.g. pairing of α and α’) appears to result in the formation of a stable compact state. Thus, every productive binding event results in formation of the compact state followed by splicing. Even if the majority of protein binding events is non-productive, this folding pathway gives Mss116 a chance to eventually convert all unfolded intron molecules into splicing products. This mechanism is potentially more biologically relevant than kinetic redistribution between native and misfolded states, because the latter involves refolding of misfolded intron RNAs, which usually degrade rather than refold in vivo 40.
The native state of the intron is stabilized by exon binding
Although this study focuses on the first folding intermediate along the pathway (the compact state), experiments on SE RNA provide additional insights into the stability of the native state. We have previously shown that the native state of D1356 is unstable under physiological ionic conditions. This is based on the observation that, in the absence of protein, docking of catalytic domain 5 within the active site is only transient 15. Docking of D5 is the final stage of native-state formation and it is critical for group II intron catalysis. Based on these findings, we hypothesized that a protein cofactor must stabilize the native state of the intron in vivo15.
However, in the present study we observe that, while Mss 116 greatly accelerates formation of compact intermediates by the SE RNA (as with all the constructs), it does not appear to influence later stages in the folding pathway or to influence the rate constant for lariat formation (Fig. 5, Table 3). Mss116 accelerates formation of SE compact intermediates by a thousand-fold (Fig. 5, Table 3) and increases the amplitude of lariat intron formation (Fig. 5), but has little effect on the rate constant of lariat formation (Fig. 5, Table 3), suggesting that the primary role of Mss 116 is to lower the energy barrier between unfolded and compact states (Fig. 8).
Furthermore, DMS probing shows that, in the compact state of SE RNA, D5 is fully docked and protected within the active site (Fig. 7). These findings suggest that, while Mss116 stimulates the early compaction phase, native-state stabilization of SE RNA is provided by RNA elements within the construct.
The major difference between SE RNA and any other construct studied thus far is that it contains flanking short exons. The presence of these exons and their covalent attachment to D1 appears to be important because the addition of exon-like oligonucleotides in trans does not stabilize the native state of D1356 RNA (Fig. 7). When the exons are present in-cis, it is possible that the resulting EBS-IBS interactions optimize the proper docking of D5 within the core. Thus, the protein does not facilitate formation of the final native state; rather, this function is performed by elements of an intact splicing construct.
Mss 116 is a multi-functional RNA-binding protein
It has been proposed that Mss 116 and other similar DEAD-box proteins (Ded1, CYT-19) assist RNA folding exclusively via their helicase activity, which results in unwinding of “kinetic traps”, thereby giving misfolded RNA a chance to refold 18; 19; 20; 21. Strong experimental evidence supports this mechanism for Ded1 and CYT-19-stimulated folding of the Tetrahymena ribozyme 19; 21. However, recent studies have demonstrated that, in addition to helicase activity, many DEAD-box proteins can have other functions, such as strand annealing activity 41, protein displacement activity 42; 43; 44 and stabilization of the exon junction complex 45; 46. Specifically, our results suggest that DEAD-box protein Mss 116 facilitates folding of the ai5γ-derived RNA constructs using a different mechanism, which involves a stabilizing rather than destabilizing function of the protein. This implies that Mss 116 and other DEAD-box proteins as well, are not just helicases, but general ATP-dependent RNA-binding proteins, which may use different mechanisms to assist folding of various RNAs.
Mss 116 is not the only example of an RNA-binding protein that exhibits different modes of interaction with different RNAs. The mitochondrial tyrosyl-tRNA synthetase CYT-18 from N. crassa is a bifunctional protein, which, in addition to aminoacylation of its cognate tRNA, has also adapted to facilitate folding of N. crassa mitochondrial group I introns via stabilization of the RNA tertiary interactions 47; 48; 49. Although biochemical studies indicated that the protein recognizes a tRNA-like structure within group I introns 47, recent crystallographic studies indicate that the protein uses completely different binding surfaces to interact with tRNA and group I intron RNAs 49. Interestingly, CYT-18 does not have the same effect on all group I introns; it may facilitate or inhibit splicing depending on the intron structure 50. Similarly, Mss 116 facilitates splicing of yeast mitochondrial group I introns but inhibits splicing of the Tetrahymena intron 18. Together, all recent findings suggest that Mss 116, CYT-18 and probably other RNA-binding proteins as well have adapted to perform dual functions on different RNAs, which facilitates their involvement in a wide variety of biological processes.
MATERIALS AND METHODS
RNA and protein preparation
The D1356, D1 and SE RNAs were transcribed as previously described from plasmids pEL30, pLJS1 and pAS10, which were linearized with BamHI (pEL30) and Hind III (pLJS1 and pAS10)51. All RNAs were purified on a denaturing 5% polyacrylamide gel. The 5’-end labeling of D1 and D1356 RNAs and internal labeling of SE RNA was carried out as described 51. Expression and purification of Mss 116 protein was carried out as previously described 17.
Analysis of global compaction
To analyze compaction of D1, D1356 and SE RNAs in the absence of protein, RNA (1 nM) was denatured in 80 mM MOPS, pH 7.0 at 95 °C for 1 minute and cooled down to 30 °C. Then KCl (50 mM) and MgCl2 (1, 3 or 8 mM) were added and the samples were incubated at 30 °C in the final volume of 60 μl. Aliquots (3 μl) were taken at indicated time points (see Figures 1-5) and analyzed on a native 6% polyacrylamide gel that contained 3 mM MgCl2 in 34 mM Tris-66 mM Hepes, 0.1 mM EDTA as described previously, and then imaged by Phosphorimager scanning.
In order to study Mss116-facilitated compaction of D1, D1356 or SE (1 nM), the RNAs were first denatured, and then KCl and MgCl2 were added as described above. ATP-MgCl2 was then added (1 mM final concentration, ATP and MgCl2 were present at equimolar ratios), together with Mss116 (20 nM final concentration). The samples were then incubated at 30 °C and 3 μl aliquots were removed at various time points. To facilitate electrophoresis, the 3 μl sample aliquots were pre-treated with 1 μl of proteinase K (30 mg/ml stock) for 45 sec at 30 °C before addition of a pre-chilled loading buffer. Electrophoresis and radioanalytic imaging were conducted as described above.
Compaction rate constants for the fast-folding and slow-folding populations of D1 and D1356 RNAs were obtained by fitting the resultant data to a double exponential equation as described previously 52. Compaction rates for the SE RNA were obtained by fitting the data to equations for a sequential reaction with one intermediate. Experimental errors were determined by calculating standard deviation from the mean values, obtained from at least three independent experiments.
To analyze global compaction under multiple turnover conditions, D1 RNA and Mss116 were present at 20 nM and 5 nM, respectively. Analysis of compaction at a 1:1 RNA:protein ratio was performed at 20 nM D1 RNA and 20 nM Mss116. Samples were prepared and analyzed as above.
Binding of Mss 116 to RNA
D1, D1356 or SE RNA (5 pM final concentration) was denatured in a buffer of 80 mM MOPS, pH 7.0 at 95 °C for 1 minute and then cooled to 30 °C. Then KCl (50 mM), MgCl2 (1, 3 or 8 mM MgCl2), and Mss116 (0.05, 0.1, 0.25. 0.5, 1, 2, 3, 5, 10, 15, 20, 30, 50, 100 or 250 nM) were added and the samples (final volume of 50 μl) were incubated at 30 °C for 30 minutes in the presence or in the absence of equimolar ATP-MgCl2 mix (1 mM final concentration). Aliquots of 40 μl were removed and analyzed by filter binding as previously described 17; 53. Data were fit to 1:1 binding isotherm and Kd values were calculated as described previously 54.
Unfolding experiments on the pre-assembled, D1 compact state
D1 RNA was first folded to the native state at 100 mM MgCl2 and 50 mM KCl as previously described 15. Then the RNA was diluted to 1, 3 or 8 mM final concentration of MgCl2 with one of the following solutions: 80 mM MOPS, pH 7.0, 50 mM KCl (“no protein” samples), 80 mM MOPS, pH 7.0, 50 mM KCl, 20 nM Mss 116 (final concentration after dilution) (“Mss 116, no ATP” samples) or 80 mM MOPS, pH 7.0, 50 mM KCl, 1 mM ATP (final concentration after dilution; ATP alone or equimolar ATP-MgCl2 mix was used in this experiment, the results were identical), 20 nM Mss 116 (final concentration after dilution) (“Mss 116 plus ATP” samples). Resulting samples were incubated at 30 °C, aliquots were taken after 0, 30 min. and 2 h and analyzed on a 6% native gel as above.
DMS footprinting
DMS footprinting of the compact state in the absence of the protein was carried out as previously described 15. In order to probe the compact state formed in the presence of protein, D1, D1356 or SE RNA (50 nM final concentration) were denatured in 80 mM potassium cacodylate (pH 7.0), and then MgCl2, equimolar ATP-MgCl2 mix and Mss 116 were added to 8 mM, 1 mM and 50 nM final concentrations, respectively. Samples were incubated at 30 °C for 60-90 min, then a DMS solution in ethanol (1:500 final dilution) was added. The resulting mixture was incubated at room temperature for 20 min, the reaction was quenched by β-mercaptoethanol, ethanol precipitated and analyzed by reverse transcription as described 55. Samples that were treated with Proteinase K (see above) prior to quenching of the reaction produced the same results as untreated samples.
Supplementary Material
AKCNOWLEDGEMENTS
We thank Gaby Drews for excellent technical assistance. O.F. is a Research Specialist and A.M.P. is an Investigator with Howard Hughes Medical Institute. This work was supported by the NIH grant GM50313.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chauhan S, Behrouzi R, Rangan P, Woodson SA. Structural rearrangements linked to global folding pathways of the Azoarcus group I ribozyme. J. Mol. Biol. 2009;386:1167–1178. doi: 10.1016/j.jmb.2008.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol. 2005;353:1199–11209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Salas U, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. J Mol Biol. 2003;329:229–238. [Google Scholar]
- 4.Su L, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J Mol Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 5.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucl. Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci U S A. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 8.Treiber D, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 9.Treiber D, Rook M, Zarrinkar P, JR W. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–6. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 10.Mayer O, Waldsich C, Grossberger R, Schroeder R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem Soc Trans. 2002;30:1175–1180. doi: 10.1042/bst0301175. [DOI] [PubMed] [Google Scholar]
- 11.Zarrinkar P, Wang J, Williamson J. Slow folding kinetics of RNase PRNA. RNA. 1996;2:564–73. [PMC free article] [PubMed] [Google Scholar]
- 12.Woodson S. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 14.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 15.Fedorova O, Waldsich C, Pyle AM. Group II Intron Folding under Near-physiological Conditions: Collapsing to the Near-native State. J. Mol. Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U S A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol. Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box Proteins in Group I and Group II Intron Splicing. Biochemical Characterization of Mss116p, ATP Hydrolysis-dependent and -independent Mechanisms, and General RNA Chaperone Activity. J. Mol. Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by Local Strand Separation Is Critical for the Function of DEAD-Box Proteins as RNA Chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol. Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin P. Studying RNA folding by fluorescence spectroscopy. Columbia University; 1999. [Google Scholar]
- 23.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 24.Waldsich C, Pyle AM. A Kinetic Intermediate that Regulates Proper Folding of a Group II Intron RNA. J. Mol. Biol. 2007;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangan P, Masquida B, Westhof E, Woodson SA. Architecture and folding mechanism of the Azoarcus group I pre-tRNA. J. Mol. Biol. 2004;339:41–51. doi: 10.1016/j.jmb.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 26.Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. II. Counterion-dependence of folding kinetics. J. Mol. Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 27.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laing LG, Gluick TC, Draper DE. Stabilization of RNA structure by Mg ions. J. Mol. Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 29.Laing LG, Draper DE. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J. Mol. Biol. 1994;237:560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- 30.Tanner MA, Cech TR. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA Ile of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- 31.Serra MJ, Baird JD, Dale T, Fey BL, Retatagos K, Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, Thirumalai D, Woodson S. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc Natl Acad Sci U S A. 1999;96:6149–54. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rook M, Treiber D, Williamson J. An optimal Mg2+ concentration for kinetic folding of the tetrahymena ribozyme. Proc Natl Acad Sci U S A. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 37.Xing Y, Draper DE. Stabilization of a ribosomal RNA tertiary structure by ribosomal protein L11. J. Mol. Biol. 1995;249:319–331. doi: 10.1006/jmbi.1995.0299. [DOI] [PubMed] [Google Scholar]
- 38.Ramaswamy P, Woodson SA. Global stabilization of rRNA structure by ribosomal proteins S4, S17 and S20. J. Mol. Biol. 2009;392:666–677. doi: 10.1016/j.jmb.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramaswamy P, Woodson SA. S16 throws a conformational switch during assembly of 30S 5' domain. Nat. Struct. Mol. Biol. 2009;16:438–445. doi: 10.1038/nsmb.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson SA, Koduvayur S, Woodson SA. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA. 2006;12:2149–2159. doi: 10.1261/rna.184206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 42.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 43.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 46.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Mohr G, Rennard R, Cherniack AD, Stryker J, Lambowitz AM. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J Mol Biol. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Mohr G, Lambowitz AM. The Neurospora crassa CYT-18 protein C-terminal RNA-binding domain helps stabilize interdomain tertiary interactions in group I introns. RNA. 2004;10:634–644. doi: 10.1261/rna.5212604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 50.Vicens Q, Paukstelis PJ, Westhof E, Lambowitz AM, Cech TR. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA. 2008;14:2013–2029. doi: 10.1261/rna.1027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pyle AM, Green JB. Building a kinetic framework for group II intron ribozyme activity: quantitation of interdomain binding and reaction rate. Biochemistry. 1994;33:2716–2725. doi: 10.1021/bi00175a047. [DOI] [PubMed] [Google Scholar]
- 52.Daniels D, Michels WJ, Pyle AM. Two competing pathways for self-splicing by group II introns; a quantitative analysis of in-vitro reaction rates and products. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 53.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fedorova O, Su LJ, Pyle AM. Group II introns: highly specific endonucleases with modular structures and diverse catalytic functions. Methods. 2002;28:323–335. doi: 10.1016/s1046-2023(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 55.Waldsich C, Masquida B, Westhof E, Schroeder R. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 2002;21:2300–2312. doi: 10.1093/emboj/cdf504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.