Abstract
Three notable members of the Harveyi clade, Vibrio harveyi, Vibrio alginolyticus and Vibrio parahaemolyticus, are best known as marine pathogens of commercial and medical import. In spite of this fact, the discrimination of Harveyi clade members remains difficult due to genetic and phenotypic similarities, and this has led to misidentifications and inaccurate estimations of a species' involvement in certain environments. To begin to understand the underlying genetics that complicate species level discrimination, we compared the genomes of Harveyi clade members isolated from different environments (seawater, shrimp, corals, oysters, finfish, humans) using microarray-based comparative genomic hybridization (CGH) and multilocus sequence analyses (MLSA). Surprisingly, we found that the only two V. harveyi strains that have had their genomes sequenced (strains BAA-1116 and HY01) have themselves been misidentified. Instead of belonging to the species harveyi, they are actually members of the species campbellii. In total, 28% of the strains tested were found to be misidentified and 42% of these appear to comprise a novel species. Taken together, our findings correct a number of species misidentifications while validating the ability of both CGH and MLSA to distinguish closely related members of the Harveyi clade.
Introduction
The eight Vibrio species currently recognized as members of the Harveyi clade (V. harveyi, V. campbellii, V. alginolyticus, V. rotiferianus, V. parahaemolyticus, V. natrigens, V. mytili and V. azureus) (Sawabe et al., 2007; Yoshizawa et al., 2009) are a subset of the Vibrio core group (Reichelt et al., 1976; Dorsch et al., 1992). Members of this clade are commonly found in marine and estuarine surface waters and sediments, as commensals on the surface or within the intestinal flora of marine animals, as opportunistic pathogens, or as primary pathogens of many commercially farmed marine invertebrate and vertebrate species (O'Brien and Sizemore, 1979; Thompson et al., 2004). In addition to thriving in similar environments, members of the Harveyi clade also share a high degree of genetic and phenotypic similarity; so much so that traditional phenotypic identification methods are often unable to confidently identify and differentiate these sister species (Sawabe et al., 2007; Cano-Gomez et al., 2009). For example, V. harveyi, V. campbellii and V. rotiferianus, which form the most recent subclade of speciation within the Harveyi clade (Pascual et al., 2009), have nearly indistinguishable phenotypes (Bryant et al., 1986; Gomez-Gil et al., 2004). These similarities have confounded typing schemes and resulted in documented misidentifications (Gauger and Gomez-Chiarri, 2002; Gomez-Gil et al., 2004). While not exceedingly problematic, these misidentifications do have the potential to overemphasize the importance of a species in a particular setting, especially since most misidentifications are initially characterized as V. harveyi.
Considering the economic importance and seemingly continually expanding host range of the Harveyi clade (Austin et al., 2005; Rosenberg et al., 2007; Cervino et al., 2008; Defoirdt et al., 2008), there remains a continued interest in the development of methods to identify and differentiate its members. In contrast with phenotypic identification methods, two genetic methods, DNA–DNA hybridization (DDH) and multilocus sequence analysis (MLSA), have successfully been applied to the study of Vibrio taxonomy and evolutionary history (Reichelt et al., 1976; Gomez-Gil et al., 2003; 2004; Thompson et al., 2005; 2007; 2008; Sawabe et al., 2007; Pascual et al., 2009). DDH in particular has been accepted for decades as the standard method for species delineation as it enables a direct assessment of overall genetic similarity and grouping by comparing the extent to which two genomes hybridize to one another (Gevers et al., 2005). Comparative genomic hybridization (CGH) using whole genome microarrays relies upon the same biophysical properties and can be considered a natural technological extension of DDH. However, CGH analyses also offer the added benefit of coding sequence (CDS) level resolution thus providing a greater number of data points that can be used to simultaneously evaluate clade or species-level genetic diversity and environment-specific genetic assemblages. Thus, CGH analyses provide high information content and discriminatory power in a format that is amenable to archiving in electronic databases for future strain comparisons (Gevers et al., 2005; Sawabe et al., 2007; Cano-Gomez et al., 2009). In this study, we employed CGH using a custom-designed Affymetrix V. harveyi BAA-1116/HY01 DNA microarray to delineate 38 geographically, environmentally and temporally distributed members of the Harveyi clade and confirmed the resulting cluster assignments using two MLSAs.
Results and discussion
CGH analysis
A total of 43 previously characterized isolates (29 V. harveyi, seven V. campbellii, two V. parahaemolyticus, three V. rotiferianus and two V. alginolyticus), from a wide temporal, geographical and environmental distribution were selected for this study (Table 1). A subset of 38 isolates were analysed via CGH using a custom-designed Affymetrix DNA microarray (Vharveyi520694F) that targets 4831 total CDS from the fully assembled and annotated V. harveyi BAA-1116 genome (Naval Research Laboratory sequencing effort GenBank CP001223-5) and 965 CDS unique to the unfinished V. harveyi HY01 genome sequence (GenBank AAWP00000000). The targeted CDS did not include insertion sequence elements, transposons or repeat sequences as they were omitted from the microarray design.
Table 1.
Geographically and temporally distributed Harveyi clade strains used in this study.
| Strain | Assigned species | Origin | Location/year | Source | Used in |
|---|---|---|---|---|---|
| CAIM 29 | harveyi | Diseased shrimp (Litopenaeus sp.) larvae isolate | Jepara, Indonesia/1990 | CAIM | CGH/MLSA |
| CAIM 148 | harveyi | Diseased shrimp (Penaeus sp.) haemolymph isolate | Valle de Matatipac, Mexico/1995 | CAIM | CGH/MLSA |
| CAIM 461 | harveyi | Shark tank water isolate | Denmark/1993 | CAIM | CGH/MLSA |
| CAIM 463 | harveyi | Sea bass (Dicentrarchus labrax) isolate | Greece/1991 | CAIM | CGH/MLSA |
| CAIM 464 | harveyi | Turbot (Scophthalmus maximus) isolate | Spain/1990 | CAIM | CGH/MLSA |
| CAIM 606a | harveyi | Japanese horse mackerel (Trachurus japonicus) isolate | Uchiura Bay, Numazu, Japan/1992 | CAIM | CGH/MLSA |
| CAIM 1075 | harveyi | Oyster (Crassostrea gigas) isolate | SCPA Burabampo SCL, Mexico/2003 | CAIM | CGH/MLSA |
| CAIM 1754 | harveyi | Puffer fish (Spheroides annulatus) heart isolate | CIAD AC, Mexico/2005 | CAIM | CGH/MLSA |
| CAIM 1766 | harveyi | Sea horse (Hipocampus ingens) liver isolate | Mazatlan Aquarium, Mexico/2005 | CAIM | CGH/MLSA |
| CAIM 1792 | harveyi | Diseased shrimp (Litopenaeus vannamei) lesion isolate | Aquastrat S.A. de C.V., Mexico/2005 | CAIM | CGH/MLSA |
| ATCC BAA-1116f | harveyi | Marine (ocean) isolate | Unknown/1993 | ATCC | CGH/MLSA |
| ATCC 33843 | harveyi | 392 [MAV] | Woods Hole, USA/1971 | ATCC | CGH/MLSA |
| ATCC 35804b | harveyi | Brown shark (Carcharhinus plumbeus) kidney isolate | Baltimore, USA/1982 | ATCC | CGH/MLSA |
| ATCC 14126c | harveyi | Dead, luminescing amphipod (Talorchestia sp.) isolate | Woods Hole, USA/1935 | ATCC | CGH/MLSA |
| D1 | harveyi | White grunt (Haemulon plumieri) gut isolate | Chub Cay, Bahamas/1992 | UWM | CGH/MLSA |
| E1 | harveyi | White grunt (H. plumieri) gut isolate | Chub Cay, Bahamas/1992 | UWM | CGH/MLSA |
| G1 | harveyi | White grunt (H. plumieri) gut isolate | Chub Cay, Bahamas/1992 | UWM | CGH/MLSA |
| H6 | harveyi | Squirrelfish (Holocentrus sp.) gut isolate | Chub Cay, Bahamas/1992 | UWM | CGH/MLSA |
| L1 | harveyi | White grunt (H. plumieri) gut isolate | Chub Cay, Bahamas/1992 | UWM | CGH/MLSA |
| 501J | harveyi | Surface water isolate | Boca Ciega Bay, Florida, USA/2001 | UWM | CGH/MLSA |
| 602L | harveyi | Surface water isolate | Boca Ciega Bay, Florida, USA/2002 | UWM | CGH/MLSA |
| 9567-98 | harveyi | Research isolate | Unknown/1998 | CDC | CGH/MLSA |
| 2415-05 | harveyi | Human blood isolate | Hawaii, USA/2005 | CDC | CGH/MLSA |
| 74F | harveyi | Diseased coral (Mussimilia braziliensis) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| PA2 | harveyi | Diseased coral (M. braziliensis) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| 1DA3 | harveyi | Diseased coral (Phyllogorgia dilatata) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| 50A | harveyi | Healthy coral (Mussimilia hispida) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH |
| 9078-83 | harveyi | Research isolate | Unknown/1983 | CDC | CGH/MLSA |
| HY01 | harveyi | Dead, luminescing shrimp isolate | Hat Yai, Thailand/2004 | PSU | CGH/MLSA |
| CAIM 115 | campbellii | Shrimp (Litopenaeus sp.) haemolymph isolate | Mexico/1999 | CAIM | CGH/MLSA |
| CAIM 198 | campbellii | Shrimp (Litopenaeus sp.) hepatopancreas isolate | Sinaloa, Mexico/1999 | CAIM | CGH/MLSA |
| CAIM 519Td | campbellii | Seawater isolate | Hawaii, USA/2005 | CAIM | CGH/MLSA |
| CAIM 1500 | campbellii | Snapper (Lutjanus guttatus) liver isolate | Sinaloa, Mexico/1972 | CAIM | CGH/MLSA |
| 2SA4 | campbellii | Healthy coral (P. dilatata) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| 42A | campbellii | Healthy coral (M. hispida) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| AND4 | campbellii | Surface water isolate | Andaman Sea, Indonesia/2000 | GenBank | MLSA |
| CAIM 577Te | rotiferianus | Rotifer (Brachionus plicatilis) water isolate | Ghent, Belgium/1999 | CAIM | MLSA |
| CAIM 994 | rotiferianus | Snapper (L. guttatus) kidney isolate | Sinaloa, Mexico/2004 | CAIM | CGH/MLSA |
| 1975 | rotiferianus | Snapper (Pagrus auratus) isolate | Unknown/1999 | UFRJ | MLSA |
| F5828 | parahaemolyticus | O3:K6 human isolate | Texas, USA/1998 | CDC | CGH/MLSA |
| RIMD2210633 | parahaemolyticus | O3:K6 human isolate | Osaka, Japan/1996 | GenBank | MLSA |
| 40B | alginolyticus | Diseased coral (M. braziliensis) isolate | Abrolhos bank, Brazil/2007 | UFRJ | CGH/MLSA |
| R-1249 | alginolyticus | Coral (Fungia sp.) yellow band lesion isolate | Sulawesi, Indonesia/2004 | PU | MLSA |
Type strain. Vibrio trachuri is a junior synonym of V. harveyi (Thompson et al., 2002).
Type strain. Vibrio carchariae is a junior synonym of V. harveyi (Pedersen et al., 1998).
Type strain. Vibrio harveyi.
Type strain. Vibrio campbellii ATCC 25920.
Type strain. Vibrio rotiferianus.
Also known as strain BB120 (Bassler et al., 1997).
CAIM, Collection of Aquatic Important Microorganisms (http://www.ciad.mx/caim); ATCC, American Type Culture Collection (http://www.atcc.org); UWM, University of Wisconsin-Milwaukee; CDC, Centers for Disease Control and Prevention, USA; UFRJ, Federal University of Rio de Janeiro; PSA, Prince of Songkla University; PU, Pace University.
Sample preparation for microarray hybridization was performed by extracting, fragmenting and biotin-labelling 3 µg of genomic DNA from each strain according to Affymetrix standard protocols. Biotinylated material was hybridized to the Vharveyi520694F microarrays for 16 h at 49°C in a GeneChip® Hybridization Oven 640 at 60 r.p.m. The microarrays were subsequently washed and stained using the GeneChip® Fluidics Station 450 and scanned using the GeneChip® Scanner 7G. All hybridization signal intensities were analysed with the GeneChip® Operating Software (GCOS) to generate raw image files (.DAT) and summary data files (.CEL). The Bioconductor/R ‘ReadAffy’ and ‘expresso’ functions were used to perform RMA background corrections and CDS summarizations using the avgdiff and MAS methods (Gentleman et al., 2004). No microarray normalization was applied. The results of both summarization methods did not differ significantly so only the avgdiff results are described. The CDS hybridization intensities of each microarray varied from 0 to 14 in log2 representations and were divided into 250 bins of width 0.0056. The number of CDS that fell into each bin was counted and plotted versus intensity. These plots were examined for the presence of two peaks for each microarray as it was expected that the majority of intensities observed for each CDS should form two clusters (present or absent). Every sample, with the exception of CAIM 29, had indications of two peaks and the data points in the immediate region of each peak were fit to a Gaussian function. The fitted centre of each peak and sigma of the Gaussian function were then used to determine cut-off values. All CDS intensities below the smaller centre +2× the sigma value of that peak (siglow) were considered ‘absent’. All CDS intensities greater than the larger centre −2× the sigma value of that peak (sighigh) were considered ‘present’. CDS intensity values between these values were considered ‘uncertain’. The ‘uncertain’ calls were further subdivided into three groups (‘intermediate low’, ‘intermediate’ and ‘intermediate high’). The ‘intermediate low’ region was defined as between +2× and +4× siglow of the low intensity peak. The ‘intermediate high’ region was defined as between −2× and −4× sighigh of the high intensity peak. The ‘intermediate’ region was defined by values that fell between the low intensity peak +4× siglow and the high intensity peak −4× sighigh.
Comparative genomic hybridization profiles were visualized with hierarchically clustered heat maps using the empirical hybridization data from V. harveyi BAA-1116 as the strain comparison outgroup (Fig. 1). The aggregate hybridization states (present, three uncertain states, or absent) of 4764 CDS from chromosomes I and II divided the 38 tested strains into four distinct subclades: the campbellii subclade which harboured the V. campbellii type strain CAIM 519T (ATCC 25920), the harveyi subclade which harboured the V. harveyi type strain ATCC 14126, the rotiferianus subclade and the parahaemolyticus/alginolyticus subclade (Fig. 1A and B). Interestingly, the hierarchical cluster analyses from both chromosomes placed six purported V. harveyi strains (BAA-1116, E1, 501J, 602L, 9078-83 and HY01) in the campbellii subclade along with the six tested V. campbellii strains. Similarly, V. harveyi strain CAIM 29 was located in the parahaemolyticus/alginolyticus subclade instead of within the harveyi subclade and V. harveyi strains D1, PA2, 1DA3 and 50A formed a distinct subclade with the sole V. rotiferianus strain (CAIM 994) tested in this analysis. Notably, these same four subclades were observed when a hierarchically clustered heat map was generated by comparing each of the strains to the 965 CDS unique to strain HY01 and absent from strain BAA-1116 (data not shown).
Fig. 1.
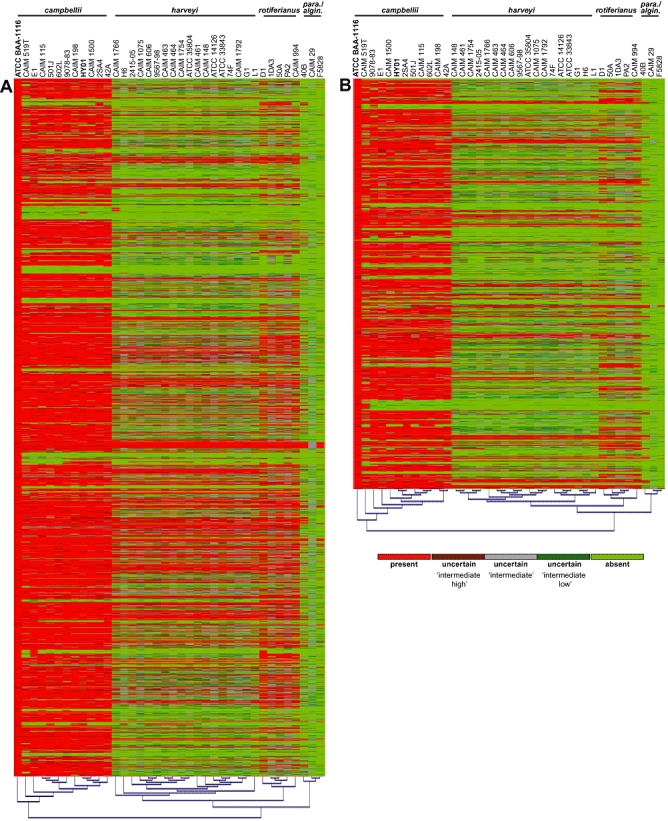
Hierarchically clustered heat maps based on CGH profiles demonstrating the presence and absence of genes within Harveyi clade members with respect to V. harveyi BAA-1116. A. Chromosome I, 2999 CDS. B. Chromosome II, 1765 CDS. The CDS in each heat map are ordered according to the genome structure of strain BAA-1116. Each CDS is depicted by one of five possible hybridization states (scale bar): (i) positive hybridization (CDS present call) = bright red bars, (ii) between positive and intermediate hybridization (uncertain ‘intermediate high’ call) = dark red bars, (iii) intermediate hybridization (uncertain ‘intermediate’ call) = grey bars, (iv) between intermediate and no hybridization (uncertain ‘intermediate low’ call) = dark green bars, and (v) no hybridization (CDS absent call) = bright green bars. Presence/absence designations generated from the hybridization profiles were calculated using the avgdiff method and clustered and visualized using MultiExperiment Viewer (MeV v4.4) software. ‘Para./algin.’ = parahaemolyticus and alginolyticus subclade.
Caution is often advised when molecular identification or phylogenetic methods result in novel or unanticipated groupings as genetic recombination may have occurred among related species thus confounding the results. This is an especially significant consideration when using too few molecular markers or when dealing with the genetically dynamic vibrios, as it is well accepted that recombination and mobile genetic elements have played a critical role in the evolution of the genus (Faruque and Mekalanos, 2003; Thompson et al., 2004; Sawabe et al., 2007; Pascual et al., 2009). However, as the hybridization state of each CDS can be considered a unique and independent data point, the scale of the CGH analysis efficiently neutralizes the effect of small-scale recombination, point mutations, horizontal gene transfer and overall genetic plasticity with respect to gene content and primary sequence identity. Thus, the CGH analyses strongly suggested that although previously characterized as V. harveyi: (i) strains E1, 501J, 602L, 9078-83 and the genome sequenced strains BAA-1116 and HY01 belong to the species campbellii, (ii) strain CAIM 29 belongs to the species parahaemolyticus, and (iii) strains D1, PA2, 1DA3 and 50A form a distinct subclade with V. rotiferianus CAIM 994.
Harveyi clade MLSAs
Multilocus sequence analysis is a sequence-based genotypic characterization method that has successfully been used to establish species-level taxonomy within the Harveyi clade (Thompson et al., 2007; Pascual et al., 2009). To validate the subclade designations generated by our CGH analyses and further solidify species assignments, we subjected the same panel of strains used in the CGH analyses to a previously validated three-gene MLSA scheme used for the classification of core Vibrio species (Thompson et al., 2007). The ftsZ (cell division protein), mreB (rod shape-determining protein) and topA (topoisomerase I) genes were PCR amplified as previously described (Thompson et al., 2007) to generate products for sequencing and the resulting sequences were concatenated (1528 nt) and subjected to phylogenetic analysis based on the Neighbour-Joining method. Sequences from four additional strains with confirmed species identities [V. rotiferianus 1975 and CAIM 577T (type strain) (http://www.taxvibrio.lncc.br/), V. parahaemolyticus RIMD 2210633 (GenBank BA000031-2) and V. alginolyticus R-1249 (Cervino et al., 2008)] were also added to strengthen the analysis. The resulting phylogeny revealed that the ftsZ-mreB-topA MLSA-derived major subclade designations were nearly identical to those seen in the CGH analyses and strains BAA-1116 and HY01 were once again found nested within the campbellii subclade (Fig. 2A). While V. harveyi and V. campbellii both formed monophyletic subclades, V. rotiferianus did not. Interestingly, the added V. rotiferianus strains (type strain CAIM 577T and 1975) did not group with strains CAIM 994, D1, PA2 and 1DA3 which were designated as the ‘rotiferianus subclade’ (Fig. 1) based on the original identification of strain CAIM 994 (Table 1). Rather, strains CAIM 994, D1, PA2 and 1DA3 formed a unique cluster that appeared to be most closely related to the harveyi subclade. A comparison of the concatenated sequence % identity found the members of this cluster to be 91.5–94.1% identical to the V. harveyi type strain (ATCC 14126) and 90.4–91.8% identical to the V. rotiferianus type strain (CAIM 577T). Strain 50A, which was also considered a member of the ‘rotiferianus subclade’ based on the CGH analyses, was omitted from the MLSA as we were unable to amplify its ftsZ gene using the previously described VftsZ75F/VftsZ800R primer pair and amplification method (Sawabe et al., 2007). Nevertheless, the use of a truncated concatenated sequence (mreB and topA only) strongly grouped strain 50A with strains CAIM 994, D1, PA2 and 1DA3 (98% bootstrap support, data not shown). Thus, the formation of this unique subclade, to the exclusion of the V. rotiferianus type strain, and its position relative to the harveyi and campbellii subclades suggests that CAIM 994 has been misidentified as V. rotiferianus and that strains CAIM 994, D1, PA2, 1DA3 and 50A likely denote a novel species within the Harveyi clade.
Fig. 2.
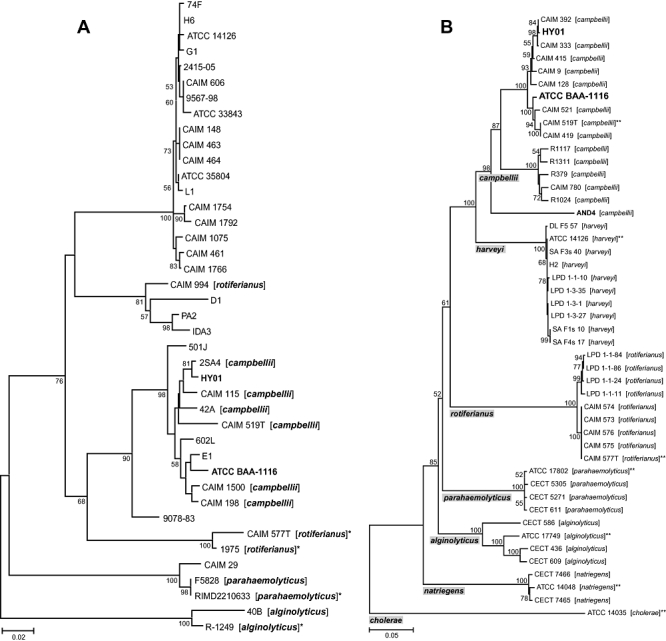
Multilocus sequence analysis (MLSA) of Harveyi clade members. A. Phylogenetic tree based on the Neighbour-Joining method using concatenated sequences from the ftsZ, mreB and topA genes (1528 nt) and MEGA software v4.0. Original species designations are in brackets. Strains lacking species designations were originally identified as V. harveyi. This analysis includes all of the strains used in the CGH analyses (with the exception of strain 50A) and four additional strains that are denoted with an asterisk ‘*’. The primary sequence information has been submitted to the GenBank database and the relevant accession numbers can be found in Table S1. B. Phylogenetic tree based on the Neighbour-Joining method using concatenated sequences from the rpoD, rctB and toxR genes (1848 nt) and MEGA software v4.0. Strain identifiers ending in ‘**’ denote type strains. With the exception of BAA-1116, HY01 and AND4 (bold type), all sequences used in this MLSA were downloaded from the ‘Taxonomy of the Vibrios’ database (http://www.taxvibrio.lncc.br/). Alignments for both analyses were generated using the clustalw program and bootstrap percentages > 50% from 1000 simulations are shown to the left of each branch point. The scale bar represents the number of substitutions per site.
As it is acknowledged that recombination may have occurred in some of the loci used in the ftsZ-mreB-topA MLSA (Thompson et al., 2007), we sought an additional confirmation of the derived classifications of the genome sequenced strains using three independent markers. The rpoD (RNA polymerase, sigma 70 factor), rctB (replication initiator protein) and toxR (virulence regulatory protein) gene sequences from strains BAA-1116 and HY01 and the genome sequenced strain V. campbellii AND4 (GenBank ABGR00000000) were concatenated (1848 nt), aligned with 44 concatenated sequences utilized in a MLSA by Pascual and colleagues (2009), and subjected to phylogenetic analysis using the Neighbour-Joining method. The resulting phylogeny, which included the type strain of each of the seven species tested, verified the V. campbellii classification of strains BAA-1116 and HY01 and parsed each species as a monophyletic subclade (Fig. 2B). Thus, the use of a second MLSA scheme with an entirely different set of Harveyi clade members with confirmed species designations (Pascual et al., 2009) corroborated the ftsZ-mreB-topA MLSA and CGH analyses findings that strains BAA-1116 and HY01 belong to the species campbellii.
CGH- and MLSA-based observations
In toto, our findings support three salient observations. First, of the 43 Harveyi clade members tested in this study, 12 (28%) appear to have been misidentified: five of which appear to represent a novel species. To some extent the misidentifications were to be expected as distinguishing members of the Harveyi clade is known to be a difficult taxonomic task (Sawabe et al., 2007) and the advent of genetic methods with high discriminatory power has previously elucidated misidentifications in a substantial percentage of strains tested [71% (Gomez-Gil et al., 2004) and 18% (Pascual et al., 2009)]. Considering the relatively recent estimated radiation time of 39 million years for V. harveyi and V. campbellii (Sawabe et al., 2007) and previous findings (Gomez-Gil et al., 2004), it was not surprising that half of misidentifications revealed in this study were V. campbellii mistaken as V. harveyi. What was clearly surprising is that the two purported V. harveyi strains that have had their genomes sequenced (strains BAA-1116 and HY01) have themselves been misidentified. The frequent misidentification of V. campbellii as V. harveyi has led to the assertion that V. campbellii is currently underestimated as an important pathogenic species of aquatic organisms (Gomez-Gil et al., 2004; Cano-Gomez et al., 2009). Our findings with strain BAA-1116, and more importantly strain HY01, which is known to be a serious shrimp pathogen (Rattanama et al., 2009), provide additional evidence to support this assertion.
Second, analysis of the CGH results indicated that 72–77% of the CDS from BAA-1116 chromosomes I and II were considered present in the campbellii subclade strains (9078-83, 501J, 602L, 2SA4, HY01, CAIM 1500, CAIM 198, CAIM 115, E1, CAIM 519T, 42A). This percent similarity is in agreement with DDH findings that have shown the intraspecies percentage similarity for V. campbellii strains to be 71–80%. This is markedly less than the 96–100% intraspecies similarity seen for V. harveyi strains (Pascual et al., 2009) suggesting that V. campbellii is more genetically diverse than V. harveyi. Fluorescent amplified fragment length polymorphism (FAFLP) analyses bolster this contention as they have previously revealed that the V. campbellii group is very diverse (FAFLP value < 10%), much more so than V. harveyi and V. rotiferianus (FAFLP value ≥ 45%) (Thompson et al., 2001; Gomez-Gil et al., 2004). In addition, both ftsZ-mreB-topA and rpoD-rctB-toxR MLSA phylogenies reveal longer branch lengths within the campbellii subclade than the harveyi subclade (Fig. 2A and B) signifying a greater genetic distance and enhanced rate of evolution within V. campbellii. Taken together, the genetic data indicate that V. campbellii is evolving at a faster rate and thus more genetically heterogeneous than V. harveyi.
Finally, although autoinduction was first described in V. harveyi using strain 392 [MAV] (ATCC 33843) (Nealson et al., 1970; Baumann et al., 1980) [previously described as MAV (Hastings et al., 1969), Photobacterium fischeri strain MAV (Nealson et al., 1970) and Beneckea harveyi strain 392 (Reichelt and Baumann, 1973)], the molecular mechanisms of V. harveyi quorum sensing have been most extensively studied in strain BAA-1116 [also known as strain BB120 (Bassler et al., 1997)] and it has consequently become a model system for quorum sensing research (Bassler et al., 1997; Henke and Bassler, 2004; Lenz et al., 2004; Waters and Bassler, 2006; Tu and Bassler, 2007; Long et al., 2009). However, as our findings identify strain BAA-1116 as V. campbellii and not V. harveyi and the quorum sensing architectures within the genus are known to be varied (Milton, 2006), it will be interesting to see how similar the V. harveyi quorum sensing system is to the well-described BAA-1116 quorum sensing system.
Concluding remarks
In this study, we have highlighted the ongoing difficulty of accurately identifying closely related Vibrio core group members. When considering the Harveyi clade, the results of this study and others suggest that a re-evaluation of the genetic or phenotypic markers commonly used to discriminate these species is needed. Comparative genomic hybridization analyses can contribute to this effort by distinguishing unique genus, species and strain-specific genetic targets for molecular identification methods development. The continued analysis of this data set to find such genetic targets, establish the V. campbellii core genome and potentially reveal the underlying genetic assemblages responsible for observed pathogenic or niche adaptation phenotypes is ongoing. Although there exists a large body of literature pertaining to the study of V. harveyi ATCC strain BAA-1116 (BB120), especially with respect to quorum sensing, these findings necessitate a change in species designation. The genome sequenced strains V. harveyi BAA-1116 and V. harveyi HY01 should hereafter be properly identified as V. campbellii BAA-1116 and V. campbellii HY01. By extension, the results also indicate that we now lack a representative genome sequence from the namesake of the Harveyi clade. Vibrio harveyi is a species that has been central to our understanding of bacterial bioluminescence and quorum sensing and continues to be a formidable pathogen in the aquaculture industry. As such, a V. harveyi genome sequencing effort is warranted.
Acknowledgments
We thank Drs Jinny Liu and Brian Barrows for critical evaluation of this manuscript. The human clinical V. harveyi isolates were kindly provided by Dr Cheryl Tarr (Centers for Disease Control and Prevention, Atlanta, GA, USA). We also thank Dr James M. Cervino (Pace University, New York, NY, USA and Woods Hole Oceanographic Institute, Woods Hole, MA, USA) for providing the V. alginolyticus R-1249 outgroup strain. This work was supported by the Office of Naval Research via US Naval Research Laboratory core funds. The opinions and assertions contained herein are those of the authors and are not to be construed as those of the US Navy, military service at large or US Government.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1. Accession numbers and source of MLSA sequences used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Austin B, Austin D, Sutherland R, Thompson F, Swings J. Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ Microbiol. 2005;7:1488–1495. doi: 10.1111/j.1462-2920.2005.00847.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Baumann L, Bang SS, Wollkalis MJ. Reevaluation of the taxonomy of Vibrio, Beneckea, and Photobacterium: abolition of the genus Beneckea. Curr Microbiol. 1980;4:127–132. [Google Scholar]
- Bryant TN, Lee JV, West PA, Colwell RR. A probability matrix for the identification of species of Vibrio and related genera. J Appl Bacteriol. 1986;61:469–480. doi: 10.1111/j.1365-2672.1986.tb04309.x. [DOI] [PubMed] [Google Scholar]
- Cano-Gomez A, Bourne DG, Hall MR, Owens L, Hoj L. Molecular identification, typing and tracking of Vibrio harveyi in aquaculture systems: current methods and future prospects. Aquaculture. 2009;287:1–10. [Google Scholar]
- Cervino JM, Thompson FL, Gomez-Gil B, Lorence EA, Goreau TJ, Hayes RL, et al. The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. J Appl Microbiol. 2008;105:1658–1671. doi: 10.1111/j.1365-2672.2008.03871.x. [DOI] [PubMed] [Google Scholar]
- Defoirdt T, Verstraete W, Bossier P. Luminescence, virulence and quorum sensing signal production by pathogenic Vibrio campbellii and Vibrio harveyi isolates. J Appl Microbiol. 2008;104:1480–1487. doi: 10.1111/j.1365-2672.2007.03672.x. [DOI] [PubMed] [Google Scholar]
- Dorsch M, Lane D, Stackebrandt E. Towards a phylogeny of the genus Vibrio based on 16S rRNA sequences. Int J Syst Bacteriol. 1992;42:58–63. doi: 10.1099/00207713-42-1-58. [DOI] [PubMed] [Google Scholar]
- Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Gauger EJ, Gomez-Chiarri M. 16S ribosomal DNA sequencing confirms the synonymy of Vibrio harveyi and V. carchariae. Dis Aquat Organ. 2002;52:39–46. doi: 10.3354/dao052039. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B, Thompson FL, Thompson CC, Swings J. Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int J Syst Evol Microbiol. 2003;53:239–243. doi: 10.1099/ijs.0.02430-0. [DOI] [PubMed] [Google Scholar]
- Gomez-Gil B, Soto-Rodriguez S, Garcia-Gasca A, Roque A, Vazquez-Juarez R, Thompson FL, Swings J. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology. 2004;150:1769–1777. doi: 10.1099/mic.0.26797-0. [DOI] [PubMed] [Google Scholar]
- Hastings JW, Weber K, Friedland J, Eberhard A, Mitchell GW, Gunsalus A. Structurally distinct bacterial luciferases. Biochemistry. 1969;8:4681–4689. doi: 10.1021/bi00840a004. [DOI] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DL. Quorum sensing in vibrios: complexity for diversification. Int J Med Microbiol. 2006;296:61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CH, Sizemore RK. Distribution of the luminous bacterium Beneckea harveyi in a semitropical estuarine environment. Appl Environ Microbiol. 1979;38:928–933. doi: 10.1128/aem.38.5.928-933.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual J, Macian MC, Arahal DR, Garay E, Pujalte MJ. Multilocus sequence analysis of the central clade of the genus Vibrio by using 16S rRNA, recA, pyrH, rpoD, gyrB, rctB, and toxR genes. Int J Syst Evol Microbiol. 2009 doi: 10.1099/ijs.0.010702-0. in press): doi:10.1099/ijs.0.010702-0. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Verdonck L, Austin B, Austin DA, Blanch AR, Grimont PAD, et al. Taxonomic evidence that Vibrio carchariae Grimes et al. 1985 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Int J Syst Bacteriol. 1998;48:749–758. [Google Scholar]
- Rattanama P, Srinitiwarawong K, Thompson JR, Pomwised R, Supamattaya K, Vuddhakul V. Shrimp pathogenicity, hemolysis, and the presence of hemolysin and TTSS genes in Vibrio harveyi isolated from Thailand. Dis Aquat Organ. 2009;86:113–122. doi: 10.3354/dao02119. [DOI] [PubMed] [Google Scholar]
- Reichelt JL, Baumann P. Taxonomy of the marine, luminous bacteria. Arch Microbiol. 1973;94:283–330. [Google Scholar]
- Reichelt JL, Baumann P, Baumann L. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch Microbiol. 1976;110:101–120. doi: 10.1007/BF00416975. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Sawabe T, Kita-Tsukamoto K, Thompson FL. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol. 2007;189:7932–7936. doi: 10.1128/JB.00693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Thompson FL, Vicente AC. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA) Int J Syst Evol Microbiol. 2008;58:617–621. doi: 10.1099/ijs.0.65461-0. [DOI] [PubMed] [Google Scholar]
- Thompson FL, Hoste B, Vandemeulebroecke K, Swings J. Genomic diversity amongst Vibrio isolates from different sources determined by fluorescent amplified fragment length polymorphism. Syst Appl Microbiol. 2001;24:520–538. doi: 10.1078/0723-2020-00067. [DOI] [PubMed] [Google Scholar]
- Thompson FL, Hoste B, Vandemeulebroecke K, Engelbeen K, Denys R, Swings J. Vibrio trachuri Iwamoto et al. 1995 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Int J Syst Evol Microbiol. 2002;52:973–976. doi: 10.1099/00207713-52-3-973. [DOI] [PubMed] [Google Scholar]
- Thompson FL, Iida T, Swings J. Biodiversity of Vibrios. Microbiol Mol Biol Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Gomez-Gil B, Vasconcelos AT, Sawabe T. Multilocus sequence analysis reveals that Vibrio harveyi and V. campbellii are distinct species. Appl Environ Microbiol. 2007;73:4279–4285. doi: 10.1128/AEM.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Wada M, Kita-Tsukamoto K, Ikemoto E, Yokota A, Kogure K. Vibrio azureus sp. nov., a luminous marine bacterium isolated from seawater. Int J Syst Evol Microbiol. 2009;59:1645–1649. doi: 10.1099/ijs.0.004283-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


