Abstract
The cytokine TGF-β acts as a tumor suppressor in normal epithelial cells and during the early stages of tumorigenesis. During malignant progression, cancer cells can switch their response to TGF-β and use this cytokine as a potent oncogenic factor; however, the mechanistic basis for this is poorly understood. Here we demonstrate that downregulation of disabled homolog 2 (DAB2) gene expression via promoter methylation frequently occurs in human squamous cell carcinomas (SCCs) and acts as an independent predictor of metastasis and poor prognosis. Retrospective microarray analysis in an independent data set indicated that low levels of DAB2 and high levels of TGFB2 expression correlate with poor prognosis. Immunohistochemistry, reexpression, genetic knockout, and RNAi silencing studies demonstrated that downregulation of DAB2 expression modulated the TGF-β/Smad pathway. Simultaneously, DAB2 downregulation abrogated TGF-β tumor suppressor function, while enabling TGF-β tumor-promoting activities. Downregulation of DAB2 blocked TGF-β–mediated inhibition of cell proliferation and migration and enabled TGF-β to promote cell motility, anchorage-independent growth, and tumor growth in vivo. Our data indicate that DAB2 acts as a tumor suppressor by dictating tumor cell TGF-β responses, identify a biomarker for SCC progression, and suggest a means to stratify patients with advanced SCC who may benefit clinically from anti–TGF-β therapies.
Introduction
TGF-β can act as both a potent tumor suppressor and tumor promoter in a context-dependent manner. TGF-β is considered the most potent and widespread inhibitor of cell growth known in mammals (1, 2), and resistance to TGF-β–mediated cytostasis may represent one of the fundamental hallmarks of cancer (3, 4). In contrast, there is strong evidence that TGF-β–induced pro-oncogenic effects are a common feature of advanced malignancies. TGF-β can operate to promote tumorigenesis via a combination of tumor-cell-autonomous and non–tumor-cell-autonomous effects. These include promotion of tumor cell proliferation, survival, motility, invasion, intravasation and extravasation at distant metastatic sites, promotion of angiogenesis, and inhibition of the antitumor immune response (1, 2, 4, 5). In the later stages of carcinogenesis, specific genetic and/or epigenetic changes must occur in the cancer cell that “switch” its response to autocrine- or paracrine-secreted TGF-β, yet little is known of the identity of the genes affected by these changes (6, 7).
TGF-β elicits its biological effects by activation of the canonical Smad and non-Smad pathways (8, 9). TGF-β stimulation results in the activin-like kinase 5–mediated (ALK5-mediated) c-terminal phosphorylation of the receptor-regulated Smads, Smad2 and Smad3. Following phosphorylation, Smad2 and Smad3 form heterooligomeric complexes with Smad4, accumulate in the nucleus, and regulate target gene expression (10). Efficient TGF-β–mediated Smad activation involves the action of intracellular adapter proteins, which facilitate the interaction of Smads with the activated receptor complex (11). These include Smad anchor for receptor activation (SARA) (12), cytoplasmic PML (13), and the p96 form of disabled homolog 2 (DAB2) (14). DAB2 is a multifunctional adapter protein, which acts as a regulator of clathrin-mediated endocytosis (15–18), and a negative regulator of multiple signaling pathways, including the ERK/MAPK (19), Src (20), and Wnt pathways (21). DAB2 was originally identified as DOC2 a gene downregulated in ovarian carcinomas (22, 23). Subsequent studies have shown that DAB2 downregulation occurs in prostate (24), breast (25), esophageal (26), endometrioid (27), urothelial (28), and hepatocellular carcinomas (29), suggesting that DAB2 has tumor suppressor activity.
Here we identify epigenetic transcriptional downregulation of DAB2 as a major determinant of metastatic progression and an independent predictor of clinical outcome in squamous cell carcinoma (SCC). Furthermore, we provide a mechanistic explanation of the tumor suppressor function of DAB2 and demonstrate that downregulation of DAB2 switches TGF-β from a tumor suppressor to a tumor promoter in vitro and in vivo. These studies identify what we believe to be a novel biomarker for SCC progression and patient stratification for the use of anti–TGF-β–targeted therapies.
Results
DAB2 is epigenetically downregulated in squamous carcinoma cell lines.
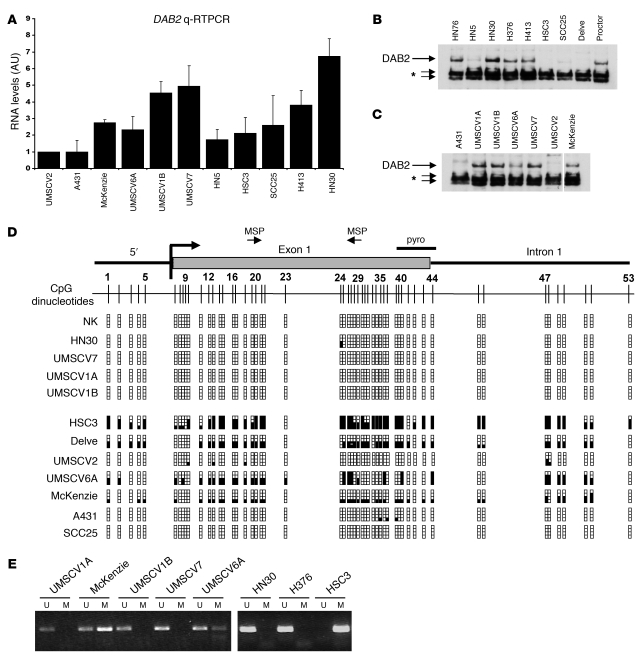
Using subtraction PCR techniques, we found that the DAB2 gene is frequently underexpressed in cDNA isolated from SCC cell lines compared with cDNA isolated from normal squamous epithelium (data not shown). We therefore analyzed DAB2 expression by quantitative RT-PCR (qRT-PCR) (Figure 1A) and Western blotting (Figure 1, B and C) in a panel of head and neck SCC (HNSCC) (Figure 1B) and vulval SCC (VSCC) (Figure 1C) cell lines. We observed a low level of DAB2 expression in the VSCC cell lines UMSCV2, A431, McKenzie, and UMSCV6A and in the HNSCC cell lines HN5, HSC3, SCC25, and Delve, compared with the VSCC cell lines UMSCV1A, UMSCV1B, and UMSCV7 and the HNSCC cell lines H413, HN30, Proctor, H376, and HN76. Where tested, DAB2 protein levels mirrored DAB2 mRNA expression (Figure 1, B and C).
Figure 1. DAB2 is epigenetically downregulated in squamous carcinoma cell lines.
(A) qRT-PCR analysis of DAB2 mRNA expression in HNSCC and VSCC cell lines. RNA levels in UMSCV2 cells were assigned the arbitrary value of 1. Determinations were performed in triplicate. Mean ± SD (n = 2) are shown. (B and C) Western blot analysis of DAB2 expression in HNSCC (B) and VSCC cell lines (C). Samples in C were run on the same gel but were noncontiguous as indicated by the white line. DAB2 is indicated on the figure. The asterisk indicates cross-reactive nonspecific bands. (D) Analysis of CpG island methylation in SCC cell lines and normal oral keratinocytes (NK). A schematic representation of the CpG island located at the 5′ end of the human DAB2 gene and each of the 53 CpG dinucleotides is shown. DAB2 promoter methylation determined by bisulphite sequencing is represented as a solid square. pyro, pyrosequencing. (E) MSP analysis of SCC cell lines using MSP primers depicted in D. U and M refer to unmethylated and methylated PCR reactions, respectively.
A CpG island is located at the 5′ end of the DAB2 gene (UCSC Genome Bioinformatics Site; http://genome.ucsc.edu/cgi-bin/hgTracks?hgsid=71021636&hgt.right2=%3E%3E+Position=chr5%3A39408502-39460703), suggesting that transcriptional silencing of DAB2 might occur via aberrant promoter methylation in squamous carcinomas. We performed bisulphite sequencing analysis of the entire CpG island in the panel of SCC cell lines and of genomic DNA isolated from normal squamous keratinocytes (NKs) (Figure 1D). There was dense methylation in the HSC3, McKenzie, UMSCV6A, and Delve cell lines, consistent with downregulation of DAB2 expression, but the CpG island was entirely unmethylated in NKs and UMSCV1A, UMSCV1B, and UMSCV7 cells, in which DAB2 was abundantly expressed. We could detect minimal to no methylation in the poorly expressing UMSCV2, A431, and SCC25 cell lines. Based on these analyses, we designed methylation-specific PCR (MSP) primers (illustrated in Figure 1D; see Supplemental Methods; supplemental material available online with this article; doi: 10.1172/JCI36125DS1), and MSP analysis was entirely consistent with bisulphite sequencing (Figure 1E). To further investigate the relationship between promoter methylation and DAB2 expression, we treated the low-level DAB2-expressing cell lines with 5-azacytidine, the histone deacetylase (HDAC) inhibitor trichostatin A (TSA), or both of these reagents (Supplemental Figure 1). qRT-PCR analysis of DAB2 mRNA expression after these treatments indicated that 5-azacytidine treatment was capable of restoring DAB2 expression in the HSC3, HN5, and A431 cell lines. TSA treatment, either alone (HSC3, UMSCV2, and HN5) or in combination with 5-azacytidine (Delve and SCC25), was also able to restore DAB2 expression, indicating that HDAC-mediated chromatin modulation may also play a role in downregulation of DAB2 expression. Compilation of these analyses revealed that epigenetic mechanisms control DAB2 expression in these cell lines (Supplemental Table 1), with direct promoter methylation occurring in 5 out of 8 of the low-level DAB2 expressors.
We next investigated whether different histone modifications at the DAB2 promoter could account for the low level of DAB2 expression (Supplemental Figure 2A) in the 3 cell lines that displayed minimal promoter methylation (A431, SCC25, and UMSCV2). Using quantitative ChIP assays, we determined the levels of histone H3 and histone H4 acetylation in 2 regions of the DAB2 promoter (illustrated in Supplemental Figure 2B). Strikingly, we found that the level of DAB2 mRNA expression correlated with the amount of H3 and H4 acetylation (modifications associated with transcriptional activation; refs. 30 and 31) at both regions. The DAB2-expressing HN30 cell line exhibited markedly greater histone acetylation than the low-level DAB2-expressing cell lines. Minimal H3 and H4 acetylation was detected in the UMSCV2 cell line that expressed the lowest amount of DAB2 (Supplemental Figure 2, C and D). These findings are consistent with the hypothesis that transcriptional silencing may play a role in downregulation of DAB2 expression in these cell lines. Polycomb complexes are instrumental in transcriptional silencing in higher eukaryotes and operate in part via methylation and recognition of histone H3 lysine 27 (32). We determined the level of H3K27 trimethylation (H3K27Me3) at the DAB2 promoter using ChIP analysis. Levels of H3K27Me3 were highest in the UMSCV2 cell line, enriched in the SCC25 cell line, and lowest in HN30 cells (Supplemental Figure 2, E and F). In contrast, all cell lines displayed comparable enrichment for the H3K27Me3 mark at the developmentally silenced γ-globin promoter (Supplemental Figure 2G). To extend these observations, we used of the compound 3-deazaneplanocin A (DZNep), which reduces protein levels of components of the cellular polycomb repressor 2 complex, including the methyltransferase EZH2 subunit, consequently acting as an inhibitor of H3K27Me3 deposition (33). A 24-hour treatment with DZNep was sufficient to reduce EZH2 protein levels in all cell lines (Supplemental Figure 2H) but could only induce DAB2 expression in the silenced cell lines, with the level of induction reflecting the initial level of H3K27Me3 (Supplemental Figure 2, H and I). Taken together, our observations indicate that DAB2 expression is transcriptionally downregulated in SCC cell lines via DNA promoter methylation and/or polycomb complex repression.
DAB2 CpG island methylation predicts metastasis and poor clinical outcome in squamous carcinomas.
We next asked whether DAB2 promoter methylation also occurred in primary squamous carcinomas. Using MSP analysis, we were able to detect DAB2 promoter methylation in 5 out of 9 archival genomic DNA samples isolated from advanced HNSCC primary tumors (Figure 2A). We then analyzed DAB2 expression in a small series of advanced HNSCC with 2 samples of patient-matched normal tissue. We assessed expression using semiquantitative RT-PCR and methylation in the CpG island using MSP and bisulphite sequencing. DAB2 mRNA was expressed in both samples of normal squamous epithelium (Figure 2B), and the CpG island was unmethylated (data not shown). DAB2 mRNA was downregulated in 2 out of 5 cases, and there was methylation, detected by both MSP and bisulphite sequencing, in the same 2 cases (Figure 2B and data not shown). These studies indicate that the methylation-dependent epigenetic downregulation of DAB2 seen in cell lines also operates in primary HNSCCs.
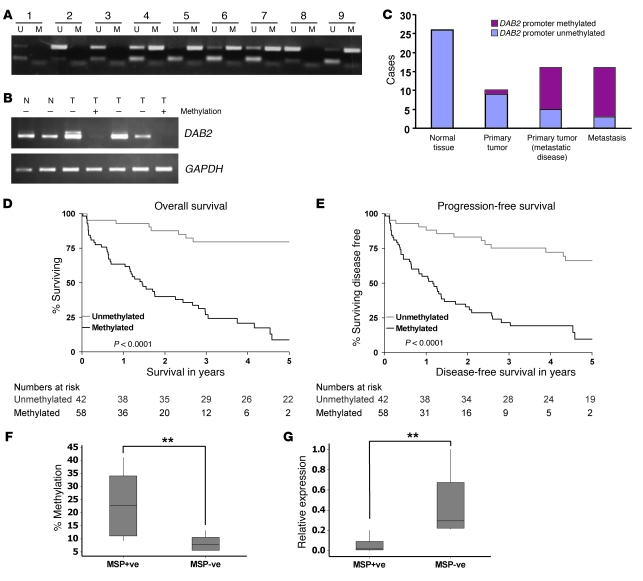
Figure 2. DAB2 promoter methylation correlates with low-level expression and predicts metastasis and poor clinical outcome in SCC.
(A) Analysis of DAB2 promoter methylation by MSP analysis in 9 HNSCC primary tumor isolates. (B) DAB2 promoter methylation correlates with low-level DAB2 expression. Analysis of DAB2 expression by RT-PCR in 2 normal (N) and 5 primary HNSCC tumor (T) samples. GAPDH RT-PCR is shown as a loading control. (C) DAB2 promoter methylation correlates with metastasis in VSCC. Graphical representation of DAB2 MSP analysis of 26 matched normal tissue and primary tumors isolated from VSCC patients. Primary tumor data is separated into patients with (16 patients) and without (10 patients) inguinal nodal involvement, and analysis of matched metastasis material is also shown. (D and E) DAB2 promoter methylation predicts poor clinical outcome in HNSCC. Log-rank survival analysis in HNSCC patients. The Kaplan-Meier curves show analyses of overall survival (D) and progression-free survival (E) in patients with HNSCC, as a function of DAB2 promoter methylation (methylated) or lacking methylation (unmethylated). The numbers under the graphs indicate the numbers of cases analyzed. (D) Overall survival is significantly worse in patients with tumors with methylation in the DAB2 promoter (P < 0.0001). (E) Progression-free survival is significantly worse in patients with tumors with methylation in the DAB2 promoter (P < 0.0001). (F) MSP analysis correlates with pyrosequencing analysis. HNSCC tumor samples were analyzed by MSP analysis as in A and by quantitative pyrosequencing of CpG dinucleotides 39–44 (depicted in Figure 1D). Data represent the average percentage of methylation at each CpG, and box plots are shown for MSP+ve and MSP–ve samples. (G) MSP analysis correlates with DAB2 expression levels. qRT-PCR analysis of DAB2 expression of samples is analyzed as in F. Expression levels are represented in relation to the highest expressing sample, which is assigned the arbitrary value of 1. (F and G) Data are analyzed by nonparametric Mann-Whitney statistics. In box plots, the 75th and 25th percentiles are represented by the top and bottom of the box, respectively. The horizontal lines refer to the mean. **P < 0.01.
Given our findings that DAB2 expression is lost in both HNSCC and VSCC cell lines, we next investigated whether DAB2 promoter methylation is also detectable in primary VSCC. We therefore performed MSP analysis on 26 VSCC primary tumor and matched normal vulval tissue samples. DAB2 promoter methylation was detected in 1 out of 10 primary tumor samples, of which the patients had no inguinal lymph node involvement, and in 11 out of 16 patients with metastatic disease but not in normal tissue samples (Figure 2C). Importantly, MSP analysis from the 16 nodal samples detected DAB2 promoter methylation in 13 out of 16 cases (Figure 2C). These data indicate that DAB2 promoter methylation in VSCC is strongly associated with the development of inguinal nodal disease (P = 0.0053, Fisher’s exact test).
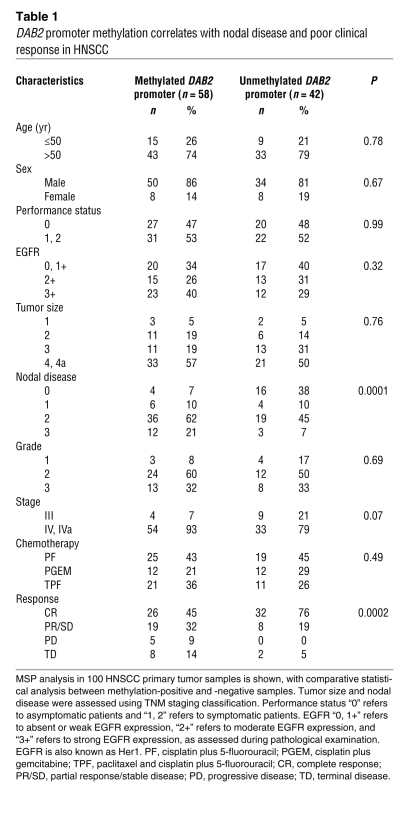
We were interested to determine whether DAB2 expression and its epigenetic regulation might also affect the clinicopathological properties and outcome in HNSCC. We therefore performed a retrospective analysis of 100 archival samples of locally advanced, stage 3 and 4 inoperable HNSCCs. Methylation in the DAB2 CpG island was detected in 58 out of 100 cases (Table 1). The frequency of DAB2 promoter methylation was significantly higher in patients with locoregional nodal metastases, compared with cases lacking nodal disease (Mantel-Haenszel χ2 test = 14.67, P = 0.0001), and DAB2 promoter methylation was also associated with poor response to radical chemoradiotherapy with cisplatin-containing chemotherapy regimens (Kendall’s t-b test = –0.31, P = 0.0002; Table 1). Next, we asked whether clinical outcome in HNSCC was influenced by DAB2 promoter methylation status. Log-rank analysis indicated that overall survival (Figure 2D) was significantly worse in patients with tumors with methylation in the DAB2 promoter (5-year survival rate, 9% [95% CI, 2%–19%] versus 80% [95% CI, 67%–92%], log-rank χ2 test = 38.54, P < 0.0001). While excluding grade due to missing data on 36% of patients, the significant predictive ability of DAB2 promoter methylation on overall survival was found to remain (hazard ratio [HR] 0.07 [95% CI, 0.03–0.15], log-rank χ2 test = 39.11; P < 0.0001) in a Cox multivariate analysis, including gender, age, performance status, EGFR, tumor size, presence of nodal disease, and tumor stage (Table 1). Similarly, progression-free survival (Figure 2E) was significantly worse in patients with tumors with methylation in the DAB2 promoter (5-year progression-free survival rate, 10% [95% CI, 1%–20%] versus 66% [95% CI, 51%–81%], log-rank χ2 test = 32.63, P < 0.0001). The significant predictive ability of DAB2 promoter methylation on progression-free survival was found to remain in a Cox multivariate analysis, including gender, age, performance status, EGFR, tumor size, presence of nodal disease, and tumor stage (HR 0.1 [95% CI, 0.05–0.21] P < 0.0001).
Table 1 .
DAB2 promoter methylation correlates with nodal disease and poor clinical response in HNSCC
Having established that detection of DAB2 CpG island methylation by MSP predicts poor survival in this retrospective study, we have initiated a prospective study of similar stage 3 and 4 inoperable HNSCC patient samples. We found that 8 out of 15 samples displayed DAB2 CpG island methylation as detected by MSP. We next interrogated these samples using pyrosequencing analysis of CpGs 39–44 (see Figure 1D) to provide a quantitative determination of methylation in this primary patient material. Samples that were scored CpG methylation positive by MSP analysis (MSP+ve) displayed a much higher mean percentage CpG methylation (Figure 2F). Samples that were MSP+ve had at least 10% and MSP–ve samples had less than 10% average methylation of CpGs 39–44 (Mann-Whitney test [95% CI, 3.65–27.5], P = 0.0092). We next determined DAB2 mRNA expression levels by qRT-PCR in these samples and found that MSP+ve samples display very low levels of DAB2 mRNA compared with MSP–ve samples (Figure 2G); MSP+ve samples had less than 0.2 and MSP–ve samples had more than 0.2 relative Dab2 mRNA expression levels (Mann-Whitney test [95% CI, –0.6659 – –0.1860], P = 0.0015). These data indicate that tumors that score positive for DAB2 promoter methylation in the MSP assay have high levels of CpG methylation and low levels of DAB2 mRNA.
Downregulation of DAB2 mRNA and protein and upregulation of TGFB2 mRNA correlate with poor survival in HNSCC.
Taken together, our studies in SCC primary tumor material indicate that methylation of the DAB2 CpG island correlates with downregulation of DAB2 mRNA, the presence of metastatic disease, and poor patient survival. We therefore reasoned that low-level DAB2 mRNA expression should correlate with poor survival. Retrospective survival data and tumor microarray gene expressions were available for 68 patients from the UK with HNSCC (T stage 1–4, clinical stage I–III). We performed univariate Cox analysis for DAB2 expression in this data set, coupled with automated discretisation (34) to separate the data set into DAB2 “high” (n = 35) and DAB2 “low” (n = 33) expressors. Kaplan-Meier analysis indicated that patients with low-level DAB2 expression had a significantly worse overall survival (low-level DAB2 expression, below average survival rate [referred to herein as DAB2 low average survival], 24.5 months, high-level DAB2 expression, above average survival rate [referred to herein as DAB2 high average survival], 31.8 months; log-rank χ2 test = 4.42, P = 0.036; Figure 3A) and provided independent verification of our methylation studies. We next sought to determine whether DAB2 protein levels correlate with survival in HNSCC patients. We performed immunohistochemistry for DAB2 on an HNSCC tissue microarray (TMA), which contained a subset of the microarray samples (35). Using the weighted histoscore system, we observed a wide range of DAB2 expression in this TMA (Supplemental Figure 3). Importantly, and consistent with the microarray analysis, we found that patients with low-level DAB2 protein expression (histoscore, <70) had a significantly worse overall survival (DAB2 low average survival, 25.03 months, DAB2 high average survival, 36.62 months; log-rank χ2 test = 4.54, P = 0.033; Figure 3B). Overall survival decreased further still with even lower DAB2 expression (histoscore, <40; DAB2 low average survival, 20.61 months, DAB2 high average survival, 35.22 months; log-rank χ2 test = 7.04, P = 0.008; Supplemental Figure 4). All of these analyses indicate that a decrease in DAB2 expression correlates with poor survival in HNSCC.
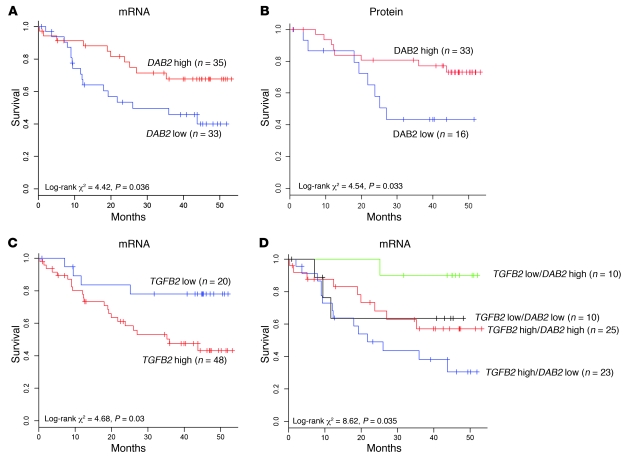
Figure 3. Low-level DAB2 and high-level TGFB2 expression correlate with poor survival in HNSCC.
(A, C, and D) Microarray analyses of 68 primary HNSCC tumor samples were assessed for expression of DAB2 (A), TGFB2 (C), and both (D). Tumor samples were separated after automated discretization into high- and low-level groups, and overall patient survival was assessed using univariate Cox and Kaplan-Meier survival analysis. (A) Low-level DAB2 mRNA expression correlates with poor survival (P = 0.036). (B) A subset of tumors analyzed in A were assessed for DAB2 protein levels by immunohistochemistry and histoscore analysis. Patients were separated into high-level groups (DAB2 histoscore, >70) and DAB2 low-level groups, and overall patient survival was assessed using univariate Cox and Kaplan-Meier survival analysis. Low-level DAB2 protein expression correlates with poor survival (P = 0.033). (C) High-level TGFB2 mRNA expression correlates with poor survival (P = 0.03). (D) Patients harboring tumors that express low-level DAB2 mRNA and high-level TGFB2 mRNA have the worst prognosis (P = 0.035).
Recent observations have indicated that DAB2 may play a role in TGF-β signaling (14, 36, 37). We therefore investigated whether changes in TGFB mRNA levels correlated with patient survival, using automated discretisation analysis on the microarray data set, with probe sets for TGFB1, TGFB2, and TGFB3. Patients expressing high levels of TGFB1 and TGFB3 appeared to fare worse, although this failed to reach statistical significance (Supplemental Figure 5). However, patients expressing high-level TGFB2 (n = 48) exhibited a statistically significantly worse overall survival than patients classified as TGFB2 low (n = 20) (TGFB2 high average survival, 25.6 months, TGFB2 low average survival, 34.9 months; log-rank χ2 test = 4.68, P = 0.03; Figure 3C). Combination of the 2 TGFB2 groups with the 2 DAB2 groups created 4 groups (TGFB2 low, DAB2 high, n = 10; TGFB2 low, DAB2 low, n = 10; TGFB2 high, DAB2 high, n = 25; TGFB2 high, DAB2 low, n = 23) that have significantly different survival prognosis (log-rank χ2 test = 8.62, P = 0.035; Figure 3D). Importantly, this analysis indicated that patients who express high-level TGFB2 and low-level DAB2 had the worst prognosis (average survival, 23.9 months; Figure 3D), suggesting that loss of DAB2 may possibly modulate TGF-β signaling.
Loss of DAB2 expression does not preclude Smad2 or Smad3 activation.
In HT1080 fibrosarcoma cells, DAB2 acts as an essential adapter, linking Smad2, Smad3, and the TGF-β receptor complex (14). We determined the ability of TGF-β to stimulate phosphorylation of Smad2 and Smad3 in the SCC cell lines (Figure 4, A and B). Unexpectedly, TGF-β clearly stimulated Smad2 phosphorylation in all cell lines tested, irrespective of DAB2 expression levels (Figure 4, A and B). For example, in HSC3, which lacks detectable endogenous DAB2 due to dense CpG methylation, there was reproducibly robust TGF-β–mediated Smad2 activation. Similarly, TGF-β stimulated Smad3 phosphorylation in all of the cell lines apart from the UMSCV2 and HN5 cell lines, which express very low levels of endogenous Smad3 (Figure 4, A and B). Consistent with these results, immunofluorescence analysis revealed that TGF-β treatment resulted in nuclear accumulation of Smad2/3 irrespective of DAB2 status (Supplemental Figure 6). These findings indicate that TGF-β–dependent activation of Smad2/Smad3 occurs in SCC cell lines, even in the absence of detectable endogenous DAB2 protein. To formally address whether DAB2 expression is absolutely required for Smad phosphorylation, we genetically deleted Dab2 expression in mouse embryonic fibroblasts (MEFs) isolated from Dab2Fl/– mice (16) by infection with a retroviral expression vector for Cre recombinase. Western blotting analysis revealed that, despite complete loss of Dab2 expression, these MEFs were capable of activating both Smad2 and Smad3 following TGF-β stimulation and, if anything, exhibited a slightly longer phospho-Smad2 response when compared with control vector–infected cells (Supplemental Figure 7).
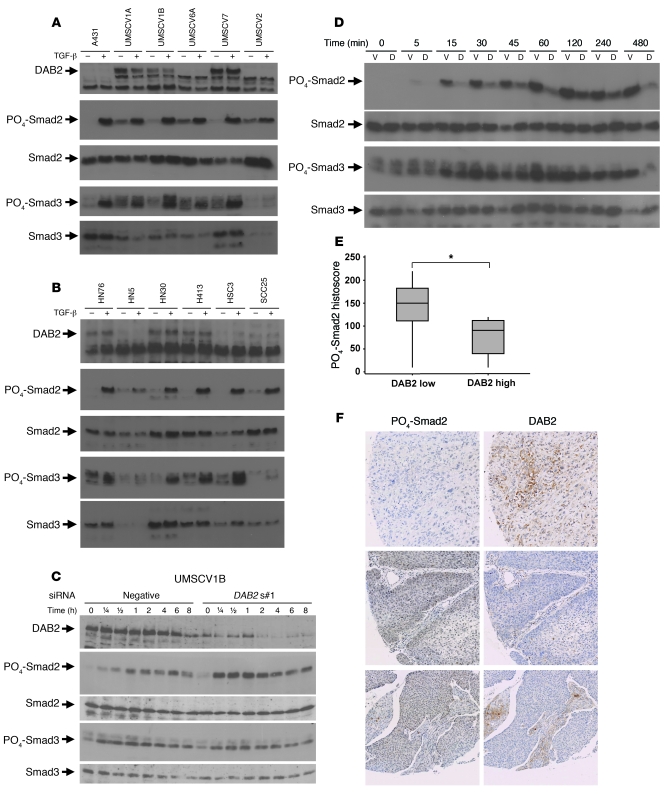
Figure 4. DAB2 represses TGF-β–mediated Smad2 activation.
(A and B) Low-level DAB2 expression does not preclude Smad2 and Smad3 phosphorylation. VSCC cell lines (A) and HNSCC cell lines (B) were treated for 1 hour with or without 1 ng/ml TGF-β prior to Western blotting for activation of Smad2 and Smad3, using PO4-Smad2 and PO4-Smad3 antibodies. DAB2, Smad2, and Smad3 Western blotting analysis is also shown. (C) siRNA-mediated knockdown of DAB2 enhances TGF-β–mediated Smad2 activation. Western blotting analysis of TGF-β–mediated activation of Smad2 and Smad3 phosphorylation in UMSCV1B cells, following transient siRNA transfection with nonsilencing control (negative) and DAB2-specific siRNA (DAB2 s#1) and TGF-β treatment (1 ng/ml) for the indicated time points. Knockdown of DAB2 was assessed by DAB2 Western blotting, and analysis of PO4-Smad2, PO4-Smad3, total Smad2, and Smad3 levels is shown. (D) DAB2 stable reexpression inhibits TGF-β–mediated Smad2 activation. Western blotting analysis of A431 vector control (V) and Dab2 stable cell lines (A431D2#1 [D] analyzed as in C). (E and F) DAB2 protein levels inversely correlate with PO4-Smad2 levels. Serial sections of a HNSCC TMA were stained with DAB2 and PO4-Smad2 antibodies, respectively, and protein levels were determined by histoscore. DAB2 low samples (histoscore, <70) have a higher level of PO4-Smad2 (*P < 0.05) than DAB2 high samples. In box plots, the 75th and 25th percentiles are represented by the top and bottom of the box, respectively. The horizontal lines refer to the mean. (F) Regions of serial sections of 3 example tumor TMA cores stained with the indicated antibodies, revealing mutually exclusive staining. Original magnification, ×20.
DAB2 suppresses TGF-β–mediated Smad2 activation.
Next, we assessed the effect of inhibiting or restoring DAB2 expression on Smad activation in the SCC cell lines. Time course analysis following siRNA knockdown of DAB2 expression in UMSCV1B cells (Figure 4C) and HN30 cells (Supplemental Figure 8A) revealed that TGF-β stimulation of Smad2 phosphorylation was markedly enhanced, whereas Smad3 activation was unaffected in knockdown cells, compared with negative control nonsilencing siRNA-transfected cells (Figure 4C and Supplemental Figure 8A). We next examined the effect of restoring DAB2 expression on Smad activation. We generated stable cell lines expressing Flag-tagged DAB2 in the A431 VSCC cell line and in the SKOV3 ovarian carcinoma cell line, previously identified as expressing low levels of DAB2 (22). We generated 2 A431 (A431D2#1 and A431D2#2) and 2 SKOV3 (SKOV3D2#1 and SKOV3D2#2) cell lines, in which DAB2 expression was higher than parental and corresponding vector control (A431V and SKOV3V) cell lines, as assessed by Western blotting (Supplemental Figure 8B). Time course analysis of Smad activation following TGF-β treatment revealed the opposite effects observed in the siRNA experiments. DAB2 reexpression markedly inhibited TGF-β–dependent Smad2 phosphorylation in both the A431D2#1 and SKOV3D#1 cell lines, compared with the corresponding vector control cell lines A431V and SKOV3V, while having little effect on relative Smad3 phosphorylation (Figure 4D and Supplemental Figure 8C). Similar effects were observed in the A431D2#2 and SKOV3D2#2 cell lines (data not shown). We next assessed the ability of TGF-β to regulate target gene expression in the A431D2#1 and A431V cell lines. TGF-β induced expression of the Smad3/Smad4 target genes junB and Smad7 (38, 39) equally in both cell lines. Recently, it has been shown that TGF-β induces expression of SnoN in a Smad2-dependent fashion (40). Consistent with this observation, we found that TGF-β stimulated SnoN expression in the A431V cell line but failed to do so in the A431D2#1 cell line (Supplemental Figure 9). Interestingly, we also observed similar regulation of the CXCR4 gene (Supplemental Figure 9). These studies indicate that in SCC cell lines DAB2 acts to repress Smad2 activation. We next sought to determine whether this also occurs in primary patient samples in vivo. We first optimized phospho-Smad2 staining using Western blotting and formalin-fixed, paraffin-embedded cell pellets of cells treated with and without the ALK5 inhibitor SB-431542 (41) and with and without TGF-β (Supplemental Figure 10). We next stained serial sections of a commercially available TMA containing samples from 18 HNSCC patients with both the DAB2 and phospho-Smad2 antibodies and analyzed expression levels using weighted histoscore analysis. Twelve of the eighteen tumors on this array exhibited low-level DAB2 staining (histoscore, <70). These tumors exhibited a higher level of phospho-Smad2 staining when compared with the tumors expressing a high level of DAB2 protein (Mann-Whitney test [95% CI, 0.03–110] P = 0.044; Figure 4E). Furthermore, we frequently observed many areas of tumors that contained inversely correlated levels of staining, showing either a high level of DAB2 staining or a high level of phospho-Smad2 staining (examples shown in Figure 4F). These findings are consistent with our cell line studies and suggest that in SCC tumors DAB2 can act as a suppressor of Smad2 activation.
DAB2 loss correlates with loss of TGF-β–dependent growth suppression.
Having established that DAB2 acts as an endogenous inhibitor of TGF-β–mediated Smad2 phosphorylation, we wished to investigate the consequences of DAB2 downregulation on TGF-β–driven biological responses. We first investigated whether DAB2 expression affects the cytostatic response to TGF-β in our SCC cell line panel. Cell lines lacking DAB2 promoter methylation and that express high levels of DAB2 universally responded to TGF-β treatment by a decrease in DNA synthesis (Figure 5A) and an inhibition of cell proliferation (Supplemental Figure 11). In contrast, cell lines expressing low or undetectable levels of DAB2 failed to exhibit a decrease in DNA synthesis (Figure 5B) and exhibited an increase (HN5), no change (HSC3, UMSCV2, and A431), or a modest decrease (UMSCV6A, SCC25, and McKenzie) in proliferation (Supplemental Figure 12).
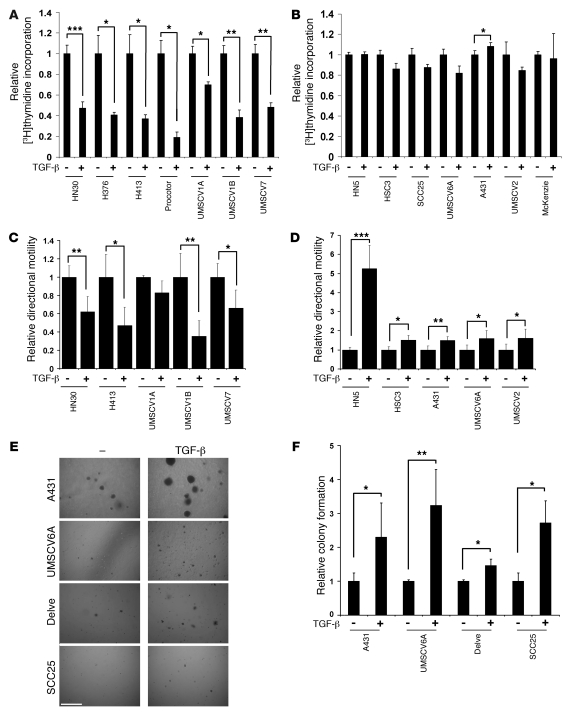
Figure 5. DAB2 levels correlate with TGF-β responses in SCC cell lines.
(A and B) Cell lines were treated with 1 ng/ml TGF-β for 48 hours, and [3H]thymidine incorporation assays were performed. Results are represented as the relative [3H]thymidine incorporation, where untreated samples are assigned an arbitrary mean value of 1. Data are represented as mean ± SD (n = 3). TGF-β inhibits proliferation in cell lines that express high levels of DAB2 (A) but does not in cell lines that express low levels of DAB2 (B). (C and D). Indicated cell lines were grown to confluency and serum starved, and then scratch assays were performed and cell motility was monitored using time lapse microscopy. Unstimulated directional motility rates were assigned an arbitrary mean value of 1. Data are represented as the mean ± SD (n = 6). TGF-β inhibits motility in cell lines that express high levels of DAB2 (C) and promotes motility in cell lines that express low levels of DAB2 (D). (E and F) TGF-β promotes anchorage-independent growth in cell lines with low-level DAB2 expression. (E) Representative images of soft agar assays from the indicated cell lines untreated (–) or treated with TGF-β. Scale bar: 0.5 mm. (F) Graphical analysis of soft agar assays. Data represent the mean ± SD (n = 3). Statistical analyses were performed with paired 2 tailed t tests throughout. *P < 0.05, **P < 0.01, ***P < 0.001.
TGF-β–mediated regulation of cell motility and anchorage-independent growth correlates with DAB2 expression levels.
We next assessed the effect of DAB2 expression on TGF-β–mediated regulation of cell motility in quantitative wound-healing scratch assays. TGF-β inhibited cell motility in the majority of DAB2-expressing lines analyzed (Figure 5C). In contrast, TGF-β induced a 5-fold stimulation of the motility rate in HN5 and a modest but statistically significant increase in motility rate in all other cell lines expressing low levels of DAB2 (Figure 5D).
TGF-β was originally identified by virtue of its ability to promote anchorage-independent growth of transformed fibroblasts (42, 43). We seeded the entire SCC cell line panel into soft agar and assessed their ability to grow in an anchorage-independent fashion. Only cell lines expressing low levels of DAB2 formed colonies in soft agar, and TGF-β treatment increased anchorage-independent growth in each case (Figure 5, E and F).
Silencing of DAB2 blocks TGF-β–mediated cytostasis, switches the TGF-β motility response, and promotes anchorage-independent growth.
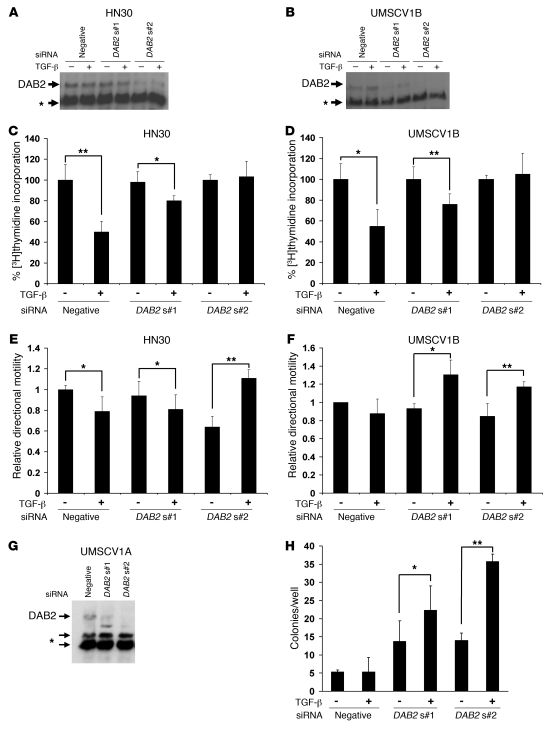
Our results imply that DAB2 expression levels dictate the TGF-β response of SCC cell lines and that DAB2 is required for TGF-β–mediated tumor suppressive effects. We used siRNA to knockdown DAB2 expression in both HNSCC and VSCC cell lines to test these hypotheses. We achieved modest knockdown with one siRNA (DAB2s#1) and more efficient knockdown with a second siRNA (DAB2s#2) in transiently transfected HN30 (Figure 6A) and UMSCV1B (Figure 6B) cells. The level of DAB2 expression correlated closely with the degree of TGF-β–mediated inhibition of DNA synthesis, with efficient knockdown completely abrogating this response (Figure 6, C and D). We next assessed the effect of DAB2 silencing on TGF-β–mediated regulation of cell motility, using the quantitative wound-healing assay. In both the HN30 and UMSCV1B cell lines, knockdown of DAB2 switched the TGF-β response from inhibition to promotion of cell motility (Figure 6, E and F). Finally, we investigated the effect of DAB2 knockdown on the ability of the UMSCV1A cell line to grow in soft agar. Knockdown of DAB2 (Figure 6G) both promoted and enabled TGF-β–mediated stimulation of anchorage-independent growth (Figure 6H).
Figure 6. Knockdown of DAB2 switches the TGF-β response.
(A and B) Western blot analysis of DAB2 expression following siRNA-mediated knockdown of DAB2 expression with 2 different siRNAs (DAB2 s#1, DAB2 s#2) or a nonsilencing control siRNA (negative) in HN30 (A) and UMSCV1B cells (B). The asterisk denotes cross-reactive bands, which serve as loading controls in A, B, and G. Cells were transfected with the indicated siRNAs, and after 24 hours were seeded into separate dishes for wound-healing scratch assay analysis and [3H]thymidine incorporation assays. Whole cell lysates were prepared after the completion of the scratch assays, and Western blot analysis is shown from a representative experiment. (C and D) [3H]thymidine analysis performed and analyzed as in Figure 5. Knockdown of DAB2 blocks TGF-β–mediated proliferative arrest in HN30 (C) and UMSCV1B cells (D). (E and F) Wound-healing scratch motility assays performed and analyzed as described in Figure 5. Knockdown of DAB2 switches the TGF-β response from inhibition to promotion of motility in HN30 (E) and UMSCV1B cells (F). (G and H) Knockdown of DAB2 permits TGF-β–promoted anchorage-independent growth in UMSCV1A cells. Cells were transiently transfected with the indicated siRNAs and then seeded for Western blot analysis (G) or into soft agar assays with or without 1 ng/ml TGF-β (H). The total number of colonies per well is shown. Mean ± SD (n = 3) is shown throughout. Statistical analyses were performed with paired 2 tailed t tests throughout. *P < 0.05, **P < 0.01.
Reexpression of DAB2 switches TGF-β from a tumor promoter to tumor suppressor.
We next performed reciprocal experiments by ectopic expression in cell lines with low endogenous levels of DAB2. We generated an A431 TetOn cell line and derivatives that expressed a high level of DAB2 (A431 TDAB2#1) and a lower level of DAB2 (A431 TDAB2#2) following doxycycline treatment (Figure 7A). Treatment of the A431 and A431 TetOn cell lines with TGF-β resulted in a modest increase in cell proliferation (Supplemental Figure 13). The leakier A431 TDAB2#1–inducible cell line failed to exhibit this increase, and cotreatment of the A431 TDAB2#1 cell line with TGF-β and doxycycline restored the ability of TGF-β to inhibit cell proliferation and abrogated this increase in the A431 TDAB2#2 cell line (Supplemental Figure 13), indicating that under these conditions a high level of DAB2 expression is required for TGF-β–mediated cytostasis. We next assessed the ability of these cell lines to respond to TGF-β and grow in an anchorage-independent manner. Consistent with our earlier findings, both the A431 and A431 TetOn cell lines readily formed colonies in soft agar, and TGF-β treatment enhanced anchorage-independent growth (Figure 7B). Both DAB2-inducible cell lines were able to form colonies in soft agar to a similar degree to that of the A431 cell line but fewer than the parental A431 TetOn cell line. Both cell lines switched their response to TGF-β, with TGF-β treatment now acting to inhibit anchorage-independent growth in a DAB2 expression level–dependent fashion (Figure 7B). TGF-β treatment inhibited colony formation in the A431 TDAB2#1 cell line, even in the absence of doxycycline, whereas TGF-β only inhibited colony formation in the A431 TDAB2#2 cell line in the presence of doxycycline. These findings indicate that a level of DAB2 expression above the baseline expression observed in the A431 TDAB2#2 cell line but less than or equal to the baseline expression observed in the A431 TDAB2#1 cell line is required to restore this activity of TGF-β.
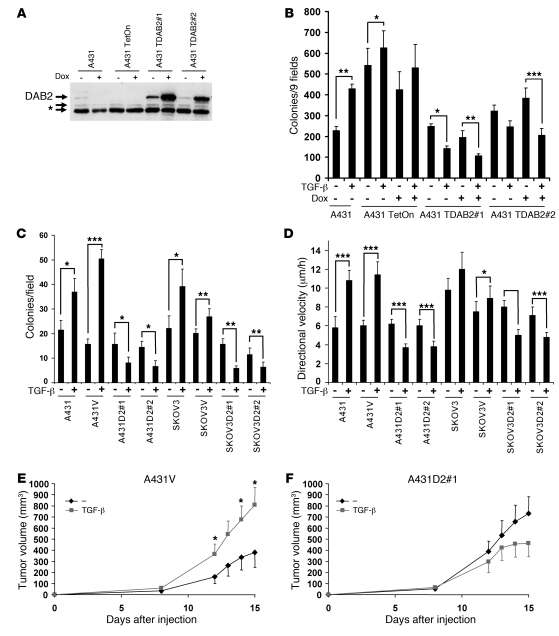
Figure 7. DAB2 reexpression switches the TGF-β response in vitro and in vivo.
(A) Western blotting analysis for DAB2 expression in A431 doxycycline-inducible cell lines. The asterisk indicates cross-reactive bands. A431 TetOn is a clonal derivative of the parental A431 cell line that stably expresses the Tet transactivator, from which the 2 DAB2-inducible cell lines were derived (A431 TDAB2#1, A431TDAB2#2). Dab2 expression was assessed after treatment with 1 μg/ml doxycycline (Dox) for 24 hours. (B and C) DAB2 expression switches the TGF-β response from promotion to inhibition of anchorage-independent growth. (B) Cells were seeded into soft agar, with or without TGF-β and/or doxycycline treatment. Data represent the mean ± SD (n = 4). (C) Stable cell lines, described in Figure 4 and Supplemental Figure 8, were seeded into soft agar and treated or not with 1 ng/ml TGF-β. Data represent the mean ± SD (n = 3). (D) Stable DAB2 expression switches the TGF-β response from promotion to inhibition of motility. Data represent the mean ± SD directional velocity of 8 cells tracked from each of 2 independent wounds. (E and F) DAB2 expression switches the TGF-β response in vivo. A431 vector control (E) and a DAB2 stable clone (F) were pretreated and mixed with 1 ng/ml TGF-β and injected into the flanks of CD1 nude mice. Tumor volumes were then calculated at the indicated time points for up to 15 days after injection. Data represent the mean volume ± SEM (n = 6). Statistical analyses were performed with paired 2-tailed t tests throughout. *P < 0.05, **P < 0.01, ***P < 0.001.
We recapitulated these findings in the A431 and SKOV3 cell lines stably expressing Flag-tagged DAB2. Soft agar analysis revealed that TGF-β promoted anchorage-independent growth in the parental and vector control cell lines, whereas enforced DAB2 expression switched this response as TGF-β inhibited colony formation in all 4 DAB2-expressing cell lines (Figure 7C). Wound-healing analysis demonstrated that TGF-β readily stimulated cell motility in the A431 and A431V cell lines and modestly stimulated cell motility in the SKOV3V cell line. In each of the cell lines ectopically expressing DAB2, TGF-β treatment now efficiently inhibited cell motility (Figure 7D).
Together, these results indicate that restoration of DAB2 expression to carcinoma cell lines of either squamous (A431) or glandular origin (SKOV3) switches the TGF-β response from tumor promoting to tumor suppressing. We next tested whether similar effects occur in vivo. A431V and A431D2#1 cell lines were pretreated for 4 days with or without TGF-β, harvested, and mixed, with or without TGF-β, and then equal numbers of cells were injected subcutaneously into the flanks of CD1 nude mice. We observed that TGF-β acted as a tumor promoter in the A431V cell line and enhanced tumor growth (Figure 7E). In contrast, restoration of DAB2 expression abrogated the protumorigenic effects of TGF-β, and, if anything, TGF-β–treated A431D2#1-derived tumors proliferated at a slower rate, although this failed to reach statistical significance (Figure 7F).
Discussion
We identified DAB2 as a candidate tumor suppressor in SCC, using subtraction hybridization techniques. These results are consistent with previous observations, demonstrating that DAB2 is downregulated in numerous other human tumor types (22–29). Analysis of the promoter region of the DAB2 gene revealed the presence of 53 CpG dinucleotides within a predicted CpG island, prompting to us to investigate aberrant promoter methylation as a potential mechanism of DAB2 silencing. Using bisulphite sequencing and MSP analysis, we found that hypermethylation of the DAB2 promoter correlated with low-level DAB2 expression in HNSCC and VSCC cell lines. In a subset of cell lines, we also found that polycomb-mediated repression may contribute to DAB2 downregulation. Importantly our MSP studies in primary tumor tissue revealed that DAB2 promoter methylation acted as a predictor of the development of metastatic disease in both VSCC and HNSCC and as a highly significant independent predictor of poor prognosis in HNSCC. To the best of our knowledge, this is the first demonstration of a distinct clinical cancer phenotype associated with loss of DAB2. We have begun to extend these studies by prospectively collecting HNSCC samples and analyzing DAB2 expression levels, using qRT-PCR, and CpG island methylation, using quantitative pyrosequencing and MSP analysis. So far our studies indicate that MSP+ve samples exhibit quantitatively higher CpG island methylation and lower DAB2 expression. Consistent with these observations, retrospective analysis of DAB2 expression levels determined by microarray analysis in a collected, independent set of patients from the UK revealed that low DAB2 levels correlate with poor survival. Furthermore, immunohistochemistry analysis on a subset of tumors derived from these patients indicated that low DAB2 protein levels in the tumor cells themselves also correlated with poor survival, with patients harboring tumors containing the lowest level of DAB2 expression performing the worst.
Despite the emerging consensus that DAB2 has tumor suppressor activity, the mechanistic basis for this is unclear. We observed both correlations between loss of DAB2 and the development of metastatic disease in SCC and between high-level TGFB2 expression and poor prognosis. Since TGF-β can act as a potent promoter of metastasis (2, 44) and DAB2 may be involved in TGF-β signaling (14), we focused our efforts on investigating the role of DAB2 in TGF-β responses. Microarray analysis indicated that HNSCC patients expressing a low level of DAB2 and a high level of TGF-β2 exhibited the worst prognosis, indicating that loss of DAB2 may modulate TGF-β responses. Using a panel of SCC cell lines and DAB2 siRNA and reexpression studies, we show that DAB2 is required for TGF-β to act as a tumor suppressor in vitro and in vivo. In the presence of high/normal levels of DAB2, TGF-β acts to inhibit cell proliferation, motility, anchorage-independent growth, and tumor growth in vivo. Fuchs and coworkers recently demonstrated that targeted deletion of TβRII in mouse skin revealed an enhanced motility rate in isolated KO fibroblasts (45), and it has been suggested that TGF-β may act as a tumor suppressor by regulating “locostasis” (46). Here we show that TGF-β can also inhibit tumor cell motility in the presence of DAB2 and our results strongly support the notion that this represents a tumor-suppressive function of TGF-β. Taken together, our data indicate that in SCC, TGF-β–mediated tumor suppressive actions require DAB2 expression, and we propose that loss of DAB2 expression by epigenetic or other means may represent an important mechanism of resistance to TGF-β–regulated tumor suppression in many other human tumor types.
Despite the well-documented role of TGF-β as a promoter of tumor progression and metastasis (2, 44, 47), the mechanistic basis for these properties remains unclear. We demonstrate here that downregulation of DAB2 switches the TGF-β response from tumor suppressing to tumor promoting and enables TGF-β to promote proliferation, motility, anchorage-independent growth, and tumor growth in vivo. To our knowledge, this is the first example of a single epigenetic event that is capable of both abrogating the tumor-suppressive function of TGF-β and facilitating the tumor promotion function of TGF-β.
Mechanistically, we demonstrate that, unlike previously published observations on the HT1080 cell line (14), DAB2 is not required for Smad2/3 phosphorylation in MEFs or SCC cell lines. Instead, DAB2 acts as a selective endogenous suppressor of TGF-β–mediated Smad2 phosphorylation in the tumor cell lines, and DAB2 levels inversely correlate with phospho-Smad2 levels in HNSCC tumor samples. It remains to be determined whether DAB2-mediated selective modulation of Smad signaling dynamics is sufficient to account for the switch of TGF-β responses. Support for this possibility comes from the demonstrations that siRNA-mediated knockdown of Smad2 attenuates TGF-β–mediated stimulation of cell motility (48) and retroviral transduction of dominant-active Smad2 promotes cell migration (49). Furthermore, elevated levels of phospho-Smad2 cooperate with mutant Ha-Ras in driving tumor progression and metastasis in a mouse model of tumor progression (49), correlate with poor prognosis in glioma (47), and are detectable in breast cancer metastases (50). Our limited gene analysis indicates that TGF-β–mediated activation of SnoN and CXCR4 expression is facilitated by loss of DAB2 expression. Intriguingly, TGF-β–mediated regulation of SnoN is Smad2 dependent and is required for TGF-β to promote anchorage-independent growth in transformed fibroblasts (40), and elevated CXCR4 is a marker of poor prognosis in several human tumor types (51). The effect of DAB2 status on the TGF-β transcriptomic response, the contribution of differentially regulated target genes to the pro-oncogenic switch in TGF-β signaling, and the potential involvement of DAB2 in TGF-β non-Smad signaling pathways clearly merit additional study.
The ability of TGF-β to promote malignant progression and metastasis implies that it is an attractive pharmacological target (52, 53). However, the clinical use of TGF-β inhibitors may be limited by disruption of the normal homeostatic and tumor suppressor functions of TGF-β. As such, biomarkers predictive of cellular response to inhibitors of TGF-β would clearly be valuable. Here we present evidence that DAB2 may act as a metastasis suppressor in SCC patients by virtue of its facilitation of the tumor suppressor function of TGF-β and that loss of DAB2 may confer a TGF-β–driven promotion of metastatic disease. This may explain why patients exhibiting both high-level TGF-β2 expression and low-level DAB2 expression exhibit the worst prognosis in our analyses. We therefore propose that patients exhibiting loss of DAB2 expression are likely to represent prime candidates for the use of TGF-β–targeted therapeutics in the management of their disease.
Methods
Plasmids, antibodies, and reagents.
pSG5Flag-DAB2 was a gift from Philip Howe (Cleveland Clinic Lerner College of Medicine, Cleveland Clinic Foundation, Cleveland, Ohio, USA) (14). pTRE2pur-DAB2 was generated as described in Supplemental Methods. Recombinant human TGF-β1 (Peprotech) was resuspended in 1 mM HCl/1 mg/ml BSA and used at appropriate concentrations. Antibodies were mouse monoclonals against DAB2 and Smad2/3 (both from BD Biosciences) and Smad4 (Santa Cruz Biotechnology Inc.); rabbit monoclonal against phospho-Smad2 (ser465/476), rabbit polyclonal antibodies against phospho-Smad2 (ser465/476), and phospho-Smad3 (ser433/435) (all from Cell Signaling Technology); and Smad2 and Smad3 (both from Zymed). Secondary antibodies were HRP-conjugated goat anti-rabbit Ig (Dako) and HRP-conjugated sheep anti-mouse Ig (Dako).
Cell lines.
We used the following squamous carcinoma cell lines: head and neck cancer cell lines HN5, HN30, H376, HN76, H413, HSC3, SCC25, Proctor, and Delve; vulval cancer cell lines A431, UMSCV1A, UMSCV1B, UMSCV2, UMSCV6A, UMSCV7, McKenzie; and ovarian cancer cell line, SKOV3. Cell lines were obtained from the CRUK cell line bank and cultured in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 100 U/ml penicillin and streptomycin. Stable cell lines were generated as described in Supplemental Methods.
Patients and tissues.
Human tissues were obtained with informed patient consent and with approval of Comitato Etico Azienda Ospedaliera S. Croce e Carle for Italian samples and the research ethics committees of Kings College London, University of Oxford, and Manchester University, Manchester, United Kingdom for the United Kingdom samples. The patient population described in Table 1 was composed of patients with stage 3 and 4, locally advanced, unresectable squamous carcinomas of the head and neck, and tissue samples were collected by the S. Croce General Hospital in Cuneo, Italy, between 1999 and 2005. The patients were randomly selected for analysis, and there is, therefore, no selection bias. All patients were treated with radical chemoradiotherapy, with cisplatin-based chemotherapy regimens. Response to treatment was assessed using standard clinical and radiological parameters. The study was retrospective and tissues, were obtained as archival, paraffin-embedded specimens. Tissues were usually diagnostic biopsies taken from either primary sites of disease or locoregional nodal disease. Vulval squamous carcinoma tissue was obtained as resection specimens, from primary sites of disease and from inguinal lymph node sites with histologically confirmed metastatic disease. The prospective study was performed on 15 patients with stage 3 or 4 HNSCC, and tissue samples were collected by S. Croce General Hospital. The patient samples analyzed by microarray analysis were collected from 71 HNSCC patients with T1-T4 disease from 2 locations in the UK and have been described in detail elsewhere (refs. 35 and 54). In all cases, the presence of adequate neoplastic tissue in samples for analysis was confirmed by histopathological examination of H&E-stained sections prior to molecular genetic analysis.
Subtraction PCR.
Total RNA was prepared from tissues using RNAzolB (Ambion), according to the manufacturer’s instructions. Poly(A)+ RNA was subsequently isolated using Poly(A)+ spin columns (New England Biolabs). Subtraction PCR was performed with 2 μg poly(A)+ RNA for tester and driver using the PCR select system (Clontech), with the modifications previously described (55).
Nucleic acid isolation and qRT-PCR.
The RNeasy kit (Qiagen) and TRIzol were used to isolate total RNA from the relevant cell lines. RNA was obtained from paraffin tissue sections using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion), according to the manufacturer’s instructions. Genomic DNA was isolated from tissue sections using extended proteinase K digestion. cDNA and qRT-PCR reactions were prepared using the SYBR Green 2-step qRT-PCR Kit (Finnzymes). Primers for each gene were from Qiagen, unless noted otherwise in the Supplemental Methods. Amplified products were analyzed by the Chromo4 Continuous Fluorescence Detector (Bio-Rad) and Opticon Monitor3 software (cell line samples) or the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) and the Assay-on-Demand software (Applera) (tumor RNA samples). RNA levels were normalized to 18S rRNA (cell line samples) or β2 microglobulin (tumor samples). Uninduced transcript levels were assigned the arbitrary value of 1.
Analysis of CpG methylation, bisulphite sequencing, MSP analysis, and pyrosequencing.
Genomic DNA was prepared from primary tumor samples, and CpG methylation in the DAB2 promoter was analyzed using bisulphite sequencing, MSP, and pyrosequencing. DNA (1 μg) was subjected to modification with sodium bisulphite, using the EZ DNA methylation kit (Zymo) according to the manufacturer’s instruction. Modified DNA was then subjected to MSP and bisulphite sequencing. Biotage Sample Prep Kit and primers designed for amplifying a 196-bp fragment across the CpG island of Dab2 gene and optimized for software dedicated to methylation analysis were used for pyrosequencing. After the pyrosequencing run, analysis was performed using Pyro Q-CpG Software (Biotage). MSP, bisulphite sequencing, pyrosequencing primers, and PCR conditions are described in Supplemental Methods.
Microarray analysis and data processing.
RNAs from the UK primary tumor cohort were processed and hybridized to Affymetrix U133Plus2 chips (54,675 probe sets/features) at the CRUK microarray facility, Paterson Institute. Raw data files are available from the Paterson Institute MIAME-VICE website ( http://bioinformatics.picr.man.ac.uk/vice/PublicProjects.vice). Three of the seventy-one patients exhibited much longer survival times than the rest of the cohort and were therefore excluded from our analysis. There were 2, 4, 2, and 6 probes available in the microarray data for TGFB1, TGFB2, TGFB3, and DAB2, respectively. First, they were treated as continuous variables. Association between probe expression and clinical parameters was examined. Spectral clustering was applied to a gene expression matrix of 14 probes to 68 samples in order to observe relationships of these probes (56, 57), followed by calculation of pair-wise correlation, both showing high correlation of between DAB2 probes (Pearson correlation 0.77–0.98) and between 3 of TGFB2 probes (Pearson correlation 0.90–0.95) and no correlation of TGFB1 probes (Pearson correlation –0.07) and TGFB3 probes (Pearson correlation –0.12). We carried out univariate Cox analysis on all 14 probes and examined Cox PH models involving 3 variables: pairs of TGFB2 and DAB2 probes and their interaction (all 3 P values < 0.01, log-rank tests < 0.01). The probes from the best model were selected for Kaplan-Meier survival curves. For each of the probes, patients were split into high-versus-low expression groups, according to the results of an automated search for the most significant discretisation (34). This analysis was performed in R and then verified in Partek.
Western blotting.
Whole cell lysates and Western blotting were performed exactly as previously described (58). Gels were loaded with 50 μg whole cell lysate per lane. Bound immunocomplexes were detected by enhanced chemiluminescence (ECL; Amersham).
Transfections and siRNA knockdown.
Cell lines were transfected with plasmids using Lipofectamine 2000 and transient siRNA knockdowns were performed using oligofectamine, as recommended by the manufacturers (Invitrogen). DAB2 and Silencer Negative control #1 siRNAs (Ambion; see Supplemental Methods) or Allstars negative control (Qiagen) were used at a final concentration of 50 nM.
[3H]thymidine assays.
[3H]thymidine assay cells were seeded into 96-well plates and then treated with or without 1 ng/ml TGF-β for 48 hours, followed by a 4-hour pulse with 1 μCi [3H]thymidine (GE Healthcare). Cells were trypsinized and harvested onto glass fiber filters (Wallac), using a Skatron 96-well plate cell harvester (Skatron Instruments). Incorporated [3H]thymidine was then measured by scintillation counting, using Optiphase Supermix scintillation fluid, 1450 LSC, Luminescence Counter Microbeta TriLux (Perkin Elmer).
Wound-healing motility studies.
Cell lines were grown to complete confluency and then serum starved overnight in DMEM supplemented with 0.1% FCS. Wounds were made to the monolayer using a p200 pipette tip, cells were then washed in starvation media, and TGF-β was added as appropriate. Wound closure was measured by time lapse microscopy. Cells were imaged with a ×10 objective and an inverted microscope (Axiovert S100, Carl Zeiss Microimaging Inc.) in an atmosphere of 5% CO2 at 37°C. Images were collected every 15 minutes for 24 hours, and directional cell motility rates were quantified by image analysis using ImageJ software ( http://rsbweb.nih.gov/ij/) (Broken Symmetry Software). For siRNA wound-healing studies, cells were treated as above 24–48 hours after transfection.
Soft agar assays.
Cells were seeded at a density of 2 × 104/well in 6-well plates, in growth media supplemented with 0.45% agarose with or without TGF-β on top of bottom layer of growth media, supplemented with 0.9% agarose in 200 μl DMEM with or without 0.1–1 ng/ml TGF-β, which was added to each well twice weekly. Colonies were counted 2–4 weeks after seeding. Colonies of more than 80 μm in diameter were scored, and the indicated number of 5,250 μM2 fields/well were analyzed using an Olympus CKX41 microscope fitted with a ×4 objective and an eyepiece graticule. Images were captured using a Qimaging Retiga EXi digital camera and QCapture Pro software (QImaging Corporation).
Nude mouse xenografts.
Nude mouse subcutaneous xenograft experiments were carried out under the UK Animal (Scientific Procedures) Act of 1986, with approval from the research ethics committee of University of Glasgow. A431V and A431D2#1 cell lines were treated with or without 1 ng/ml TGF-β for 4 days, harvested, and resuspended in 1xHBSS (Gibco), with or without 1 ng/ml TGF-β, at a concentration of 1 × 107 cells per ml. Then, 1 × 106 cells were injected subcutaneously into the flank of CD1 nude mice (Charles Rivers), and 6 injections per cell line per condition were performed. Palpable tumors were observed 8 days after injection, and tumor volumes were calculated using calliper measurement and the formula V = (W2 × L)/2, where W is width and L is length.
TMA staining.
Immunohistochemistry conditions were optimized on A431 vector control and DAB2-expressing xenografts for Dab2 and on formalin-fixed, paraffin-embedded cell pellets for phospho-Smad2. Briefly, 2 × 106 to 6 × 106 cells were treated with or without 10 μM SB431542 (Tocris) for 15 minutes, then with or without 1 ng/ml TGF-β for a further hour. Cells were washed with PBS and fixed with 10% neutral buffered formalin for 1 hour at room temperature. Fixed cells were washed with PBS and resuspended in 0.3 ml of 2% agarose. When set, the agarose cell plugs were embedded in paraffin. Then, 2.5-μm sections were cut using a microtome (RM2245, Leica) and dried for 3 hours at 60°C. Following rehydration, cell pellet sections and HNSCC TMA slides (TMA-66, ref. 35, TMA-153, protein biotechnologies) were subjected to pressure cooker–mediated antigen retrieval with EDTA buffer (1.3 mM EDTA, 4.5 mM Tris-Cl, pH 8) for the phospho-Smad2 antibody and citrate buffer pH 6 (13 mM citric acid, NaOH) for the DAB2 antibody. Slides were stained using an autostainer (Dako) and 1:50 monoclonal anti–phospho-Smad2 (catalog 3108, Cell Signalling) or 1:100 polyclonal anti-DAB2 (catalog sc-13982, Santa Cruz Biotechnology Inc.), using the ImmPRESS anti-rabbit Ig peroxidase Kit (Vector Laboratories), and counterstained with hematoxylin. Protein expression of each core was assessed using the weighted histoscore method (59). The weighted histoscore grades staining intensity as follows: 0, absent; 1, weak; 2, moderate; and 3, strong; then, the grade is multiplied by the percentage of tumor cells within each category. Examples of this are shown in Supplemental Figure 3 for DAB2 and Supplemental Figure 10 for phospho-Smad2.
Statistics.
Differences in known prognostic factors between tumors with methylated DAB2 promoter and unmethylated DAB2 promoter were assessed by use of χ2 tests, with continuity corrections where appropriate, Mantel-Haenzel χ2 tests, and the Kendall’s τ-b statistic. Progression-free survival was calculated from date of treatment start to date of progression; to date of death in patients dying without progression; and to date of censoring in patients alive and progression-free (categorized in Table 1). Overall survival was calculated from date of treatment start to date of death or date of censoring if alive (categorized in Table 1). Survival curves were constructed using Kaplan-Meier methodology. Log-rank tests were used in univariate analysis; Cox Proportional Hazards Survival Regression was used for multivariate analysis. Comparisons of MSP analysis with pyrosequencing, MSP and qRT-PCR, and PO4-Smad2 levels with DAB2 levels measured by immunohistochemistry were assessed using nonparametric Mann-Whitney U tests. Two-tailed paired t-tests were performed in cell line experiments. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Philip Howe for reagents, Tom Hamilton for invaluable help with xenograft experiments, Colin Nixon for immunohistochemistry expertise, and Kevin Ryan and Clare Isacke for helpful discussions and critically reading the manuscript. G.J. Inman is an Association for International Cancer Research (AICR) fellow. G.J. Inman and L.C. Spender are supported by AICR and CRUK. A. Hannigan, D.I. O’Brien, B. Herrera, and M.C. Frame were supported by CRUK. T. Crook is a clinical research fellow of CRUK. Work in T. Crook’s laboratory is supported by CRUK, Breakthrough Breast Cancer, and the Institute of Cancer Research. C.L. Nigro, L. Lattanzio, M. Monteverde, and M. Gasco were partially supported by the Associazione Italiana Ricerca Cancro. J.A. Cooper was supported by the US Public Health Service grant GM066257.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(8):2842–2857. doi:10.1172/JCI36125.
Gareth J. Inman’s present address is: Biomedical Research Institute, University of Dundee, Ninewells Hospital and Medical School, Dundee, United Kingdom.
References
- 1.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 2.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775(1):21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Massague J. TGF-beta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17(1–2):29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12(1):22–29. doi: 10.1016/S0959-437X(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 7.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100(15):8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 9.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 10.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 11.Mishra L, Marshall B. Adaptor proteins and ubiquinators in TGF-beta signaling. Cytokine Growth Factor Rev. 2006;17(1–2):75–87. doi: 10.1016/j.cytogfr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95(6):779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 13.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431(7005):205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 14.Hocevar BA, Smine A, Xu XX, Howe PH. The adaptor molecule Disabled-2 links the transforming growth factor beta receptors to the Smad pathway. EMBO J. 2001;20(11):2789–2801. doi: 10.1093/emboj/20.11.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra SK, Keyel PA, Hawryluk MJ, Agostinelli NR, Watkins SC, Traub LM. Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 2002;21(18):4915–4926. doi: 10.1093/emboj/cdf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris SM, Tallquist MD, Rock CO, Cooper JA. Dual roles for the Dab2 adaptor protein in embryonic development and kidney transport. EMBO J. 2002;21(7):1555–1564. doi: 10.1093/emboj/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer ME, Cooper JA. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J Cell Sci. 2006;119(pt 20):4235–4246. doi: 10.1242/jcs.03217. [DOI] [PubMed] [Google Scholar]
- 18.Teckchandani A, et al. Quantitative proteomics identifies a Dab2/integrin module regulating cell migration. J Cell Biol. 2009;186(1):99–111. doi: 10.1083/jcb.200812160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu XX, Yi T, Tang B, Lambeth JD. Disabled-2 (Dab2) is an SH3 domain-binding partner of Grb2. Oncogene. 1998;16(12):1561–1569. doi: 10.1038/sj.onc.1201678. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Scholes J, Hsieh JT. Characterization of a novel negative regulator (DOC-2/DAB2) of c-Src in normal prostatic epithelium and cancer. J Biol Chem. 2003;278(9):6936–6941. doi: 10.1074/jbc.M210628200. [DOI] [PubMed] [Google Scholar]
- 21.Hocevar BA, Mou F, Rennolds JL, Morris SM, Cooper JA, Howe PH. Regulation of the Wnt signaling pathway by disabled-2 (Dab2). EMBO J. 2003;22(12):3084–3094. doi: 10.1093/emboj/cdg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mok SC, et al. DOC-2, a candidate tumor suppressor gene in human epithelial ovarian cancer. Oncogene. 1998;16(18):2381–2387. doi: 10.1038/sj.onc.1201769. [DOI] [PubMed] [Google Scholar]
- 23.Fazili Z, Sun W, Mittelstaedt S, Cohen C, Xu XX. Disabled-2 inactivation is an early step in ovarian tumorigenicity. Oncogene. 1999;18(20):3104–3113. doi: 10.1038/sj.onc.1202649. [DOI] [PubMed] [Google Scholar]
- 24.Tseng CP, Ely BD, Li Y, Pong RC, Hsieh JT. Regulation of rat DOC-2 gene during castration-induced rat ventral prostate degeneration and its growth inhibitory function in human prostatic carcinoma cells. Endocrinology. 1998;139(8):3542–3553. doi: 10.1210/en.139.8.3542. [DOI] [PubMed] [Google Scholar]
- 25.Bagadi SA, Prasad CP, Srivastava A, Prashad R, Gupta SD, Ralhan R. Frequent loss of Dab2 protein and infrequent promoter hypermethylation in breast cancer. Breast Cancer Res Treat. 2007;104(3):277–286. doi: 10.1007/s10549-006-9422-6. [DOI] [PubMed] [Google Scholar]
- 26.Anupam K, Tusharkant C, Gupta SD, Ranju R. Loss of disabled-2 expression is an early event in esophageal squamous tumorigenesis. World J Gastroenterol. 2006;12(37):6041–6045. doi: 10.3748/wjg.v12.i37.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang DH, et al. Disabled-2 heterozygous mice are predisposed to endometrial and ovarian tumorigenesis and exhibit sex-biased embryonic lethality in a p53-null background. Am J Pathol. 2006;169(1):258–267. doi: 10.2353/ajpath.2006.060036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karam JA, et al. Decreased DOC-2/DAB2 expression in urothelial carcinoma of the bladder. Clin Cancer Res. 2007;13(15 pt 1):4400–4406. doi: 10.1158/1078-0432.CCR-07-0287. [DOI] [PubMed] [Google Scholar]
- 29.Calvisi DF, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117(9):2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 33.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenssen TK, Kuo WP, Stokke T, Hovig E. Associations between gene expressions in breast cancer and patient survival. Hum Genet. 2002;111(4–5):411–420. doi: 10.1007/s00439-002-0804-5. [DOI] [PubMed] [Google Scholar]
- 35.Winter SC, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 36.Hocevar BA, Prunier C, Howe PH. Disabled-2 (Dab2) mediates transforming growth factor beta (TGFbeta)-stimulated fibronectin synthesis through TGFbeta-activated kinase 1 and activation of the JNK pathway. J Biol Chem. 2005;280(27):25920–25927. doi: 10.1074/jbc.M501150200. [DOI] [PubMed] [Google Scholar]
- 37.Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor beta-induced epithelial to mesenchymal transition (EMT). J Biol Chem. 2005;280(17):17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 38.Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273(33):21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 39.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger EP. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor beta. J Biol Chem. 2000;275(15):11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q, Pearson-White S, Luo K. Requirement for the SnoN oncoprotein in transforming growth factor beta-induced oncogenic transformation of fibroblast cells. Mol Cell Biol. 2005;25(24):10731–10744. doi: 10.1128/MCB.25.24.10731-10744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 42.Roberts AB, Lamb LC, Newton DL, Sporn MB, De Larco JE, Todaro GJ. Transforming growth factors: isolation of polypeptides from virally and chemically transformed cells by acid/ethanol extraction. Proc Natl Acad Sci U S A. 1980;77(6):3494–3498. doi: 10.1073/pnas.77.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moses HL, Branum EL, Proper JA, Robinson RA. Transforming growth factor production by chemically transformed cells. Cancer Res. 1981;41(7):2842–2848. [PubMed] [Google Scholar]
- 44.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 45.Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12(4):313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakefield LM, Stuelten C. Keeping order in the neighborhood: new roles for TGFbeta in maintaining epithelial homeostasis. Cancer Cell. 2007;12(4):293–295. doi: 10.1016/j.ccr.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Bruna A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 48.Jazag A, et al. Single small-interfering RNA expression vector for silencing multiple transforming growth factor-beta pathway components. Nucleic Acids Res. 2005;33(15):e131. doi: 10.1093/nar/gni130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4(7):487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 50.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 51.Koizumi K, Hojo S, Akashi T, Yasumoto K, Saiki I. Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci. 2007;98(11):1652–1658. doi: 10.1111/j.1349-7006.2007.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arteaga CL. Inhibition of TGFbeta signaling in cancer therapy. Curr Opin Genet Dev. 2006;16(1):30–37. doi: 10.1016/j.gde.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Saunier EF, Akhurst RJ. TGF beta inhibition for cancer therapy. Curr Cancer Drug Targets. 2006;6(7):565–578. doi: 10.2174/156800906778742460. [DOI] [PubMed] [Google Scholar]
- 54.Thurlow JK, et al. Spectral clustering of microarray data elucidates the roles of microenvironment remodeling and immune responses in survival of head and neck squamous cell carcinoma. J Clin Oncol. 2010;28(17):2881–2888. doi: 10.1200/JCO.2009.24.8724. [DOI] [PubMed] [Google Scholar]
- 55.Atalay A, Crook T, Ozturk M, Yulug IG. Identification of genes induced by BRCA1 in breast cancer cells. Biochem Biophys Res Commun. 2002;299(5):839–846. doi: 10.1016/S0006-291X(02)02751-1. [DOI] [PubMed] [Google Scholar]
- 56.Kalna G, Vass JK, Higham DJ. Multidimensional partitioning and bi-partitioning: Analysis and application to gene expression data sets. Int J Comput Math. 2008;85(3–4):475–485. doi: 10.1080/00207160701210158. [DOI] [Google Scholar]
- 57.Hunter KD, et al. Divergent routes to oral cancer. Cancer Res. 2006;66(15):7405–7413. doi: 10.1158/0008-5472.CAN-06-0186. [DOI] [PubMed] [Google Scholar]
- 58.Inman GJ, Nicolás FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3 and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10(2):283–294. doi: 10.1016/S1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 59.van Netten JP, et al. Multiple microsample analysis of intratumor estrogen receptor distribution in breast cancers by a combined biochemical/immunohistochemical method. Eur J Cancer Clin Oncol. 1987;23(9):1337–1342. doi: 10.1016/0277-5379(87)90117-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.