Abstract
Personalized cancer medicine is based on the concept that targeted therapies are effective on subsets of patients whose tumors carry specific molecular alterations. Several mammalian target of rapamycin (mTOR) inhibitors are in preclinical or clinical trials for cancers, but the molecular basis of sensitivity or resistance to these inhibitors among patients is largely unknown. Here we have identified oncogenic variants of phosphoinositide-3-kinase, catalytic, α polypeptide (PIK3CA) and KRAS as determinants of response to the mTOR inhibitor everolimus. Human cancer cells carrying alterations in the PI3K pathway were responsive to everolimus, both in vitro and in vivo, except when KRAS mutations occurred concomitantly or were exogenously introduced. In human cancer cells with mutations in both PIK3CA and KRAS, genetic ablation of mutant KRAS reinstated response to the drug. Consistent with these data, PIK3CA mutant cells, but not KRAS mutant cells, displayed everolimus-sensitive translation. Importantly, in a cohort of metastatic cancer patients, the presence of oncogenic KRAS mutations was associated with lack of benefit after everolimus therapy. Thus, our results demonstrate that alterations in the KRAS and PIK3CA genes may represent biomarkers to optimize treatment of patients with mTOR inhibitors.
Introduction
The PI3K/PTEN/AKT signaling pathway is frequently deregulated in human cancer (1). This pathway plays a crucial role in cell growth, survival, and proliferation; hence, its components have recently emerged as attractive therapeutic targets for cancer therapy. Accordingly, multiple drugs are being investigated either at the preclinical or clinical level to inhibit PI3K itself or its downstream effectors, such as AKT1, PDK1, and the mammalian target of rapamycin (mTOR) (2, 3). Among these novel agents, the rapamycin derivatives temsirolimus and everolimus have reached late-phase clinical development and have gained FDA approval for the treatment of metastatic renal cell carcinoma (4–6). These compounds bind to FKBP12 and inhibit mTOR complex–1 (mTORC1), resulting in suppression of both tumor growth and angiogenesis (7). Clinical trials are currently being carried out on several tumor types, such as non-Hodgkin lymphoma, glioblastoma, non-small cell lung, neuroendocrine, endometrial, colorectal, gastric, breast, and prostate carcinomas (6). mTOR inhibitors elicit predominantly disease-stabilizing, cytostatic responses, rather than tumor regression (8). Even in the best settings, such as renal cell carcinoma or neuroendocrine tumors, response rate as assessed by conventional Response Evaluation Criteria in Solid Tumors (RECIST) criteria is very low; however, these drugs significantly affect progression-free and overall survival (8). Data from early-phase studies indicate that only a subset of patients derive significant clinical benefit from treatment with mTOR inhibitors (9–12). The molecular basis of sensitivity and resistance to mTOR inhibitors such as rapamycin, temsirolimus, and everolimus is presently largely unknown. Understanding such mechanisms provides the key to select patients likely to respond, and to develop rational drug combinations to circumvent resistance.
Results
PIK3CA mutations sensitize non-transformed human cells to everolimus.
We have recently developed a panel of isogenic cell lines by employing homologous recombination to introduce (by knock-in [KI]) frequently mutated cancer alleles in human breast immortalized epithelial cells (hTERT-HME1) (13). Our previous profiling of a drug library on this panel of KI cell lines has shown that phosphoinositide-3-kinase, catalytic, α polypeptide (PIK3CA) H1047R mutated cells are selectively sensitive to a group of drugs comprising rapamycin and its derivative everolimus (13). We extended and substantiated this finding by showing that cells knocked in for the other frequently occurring PIK3CA allele, E545K, also showed an increased response to everolimus (Figure 1A). We confirmed the correlation between the KI of PIK3CA mutations and sensitization to everolimus in another cell background (MCF10A), thus suggesting that this effect could be cell type–independent (Figure 1B).
Figure 1. Genetic alterations in the PI3K and KRAS pathways are determinants of cells’ response to everolimus.
(A and B) The effect of everolimus treatment on cellular proliferation was assessed for hTERT-HME1 (A) and MCF10A (B) cells and their isogenic clones carrying the indicated PIK3CA mutations. The average cell number was measured by determining ATP content in 3 replicate wells. Results are normalized to growth of cells treated with DMSO and are represented as mean ± SD of at least 3 independent experiments. (C) Antiproliferative effects of everolimus on a panel of cancer cell lines. Cells carrying mutant or amplified PIK3CA are depicted in red or pink, respectively; cells lacking PTEN expression are represented in brown; cells carrying mutant KRAS/BRAF with or without concomitant PIK3CA mutations are depicted in blue or black, respectively. Details on specific mutations of KRAS, BRAF, and PIK3CA and the functional status of PTEN are provided in Supplemental Table 2. Results are expressed as percent viability compared with cells treated with DMSO only (control) and represent mean ± SD of at least 3 independent observations.
To better characterize the preferential effect induced by everolimus in PIK3CA KI clones, we performed FACS analysis. We found that treatment with everolimus of hTERT-HME1 cells resulted in a cytostatic effect that was more evident in PIK3CA KI clones compared with their WT counterpart (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI37539DS1). Upon treatment, all hTERT-HME1 PIK3CA KI clones accumulated in the G0/G1 phase of the cell cycle (Supplemental Figure 1B), and, accordingly, the proportions of cells in the S and G2/M phases decreased (Supplemental Table 1). Apoptosis was almost undetectable and did not vary between vehicle only– and drug-treated cells (Supplemental Figure 1B and data not shown). Of note, while vehicle only–treated cells proliferated at a comparable rate, prolonged exposure to everolimus slowed cell growth in all genotypes, with the effect being particularly evident in PIK3CA H1047R, less pronounced in PIK3CA E545K, and only minimal in WT cells (Supplemental Figure 1A). The subtle difference observed between cells carrying these two mutations might suggest a different downstream signaling of catalytic domain versus helical domain PI3K alterations, as recently suggested by Vasudevan et al. (14).
Occurrence of PIK3CA and KRAS mutations determines response of cancer cells to everolimus.
The above pharmacogenomic analysis of non-transformed cells carrying cancer alleles pointed to a relationship between the occurrence of PIK3CA mutations and response to everolimus. We next assessed whether and to what extent these findings might be applicable to human cancer cells in which mutations in the PI3K pathway occur naturally alongside additional genetic alterations. To this end, we treated with everolimus a panel of cell lines derived from glioblastoma, breast, ovarian, prostate, endometrial, and colorectal carcinomas, which are known to carry genetic alterations in PIK3CA or PTEN (phosphatase and tensin homolog) (Figure 1C and Supplemental Table 2). Interestingly, tumor cells could be classified in two main groups based on their response to everolimus. Drug-resistant cells (such as HT-29, HCT116, and DLD-1) carried mutations in both PIK3CA and KRAS/BRAF. Everolimus-sensitive cells displayed PI3K pathway alterations but no mutations in the KRAS/BRAF genes (Figure 1C).
Genetic ablation of the KRAS D13 mutation restores antiproliferative response of cancer cells to everolimus.
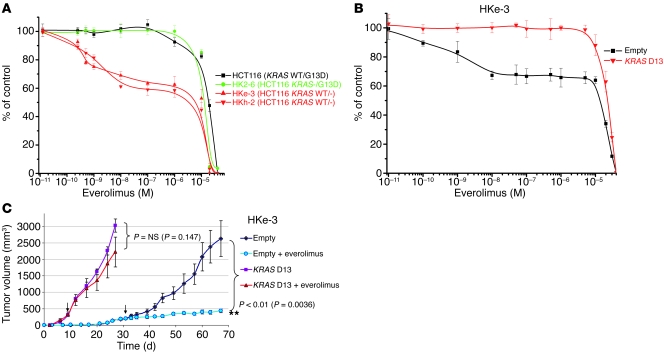
We hypothesized that genetic alterations of the KRAS pathway could represent a major genetic determinant of everolimus resistance in tumor cells carrying PIK3CA oncogenic alleles. To formally test this hypothesis, we took advantage of HCT116 cells in which the KRAS D13 mutant allele had been genetically deleted by homologous recombination (15). Strikingly, we found that HCT116-derivative cells retaining only the KRAS WT allele (named HKh-2 and HKe-3; Supplemental Table 3) were sensitive to everolimus, while the parental and the isogenic cells carrying mutated KRAS were equally resistant to this compound (Figure 2A). As a further control, we employed HCT116 cells in which the PIK3CA mutation H1047R had been deleted by targeted homologous recombination (16). As expected, since all clones retained a mutated KRAS allele, the derivative isogenic cells were nonresponsive to everolimus (Supplemental Figure 2).
Figure 2. Oncogenic KRAS D13 confers resistance to everolimus.
(A) Two independent clones of HCT116 colorectal cancer cells — in which the KRAS D13 allele was genetically deleted by homologous recombination (HKh-2 and HKe-3, depicted in red) — were more sensitive to everolimus than either their parental cells (black) or a clone in which the KRAS WT allele was knocked out but the mutated allele was retained (HK2-6, green). Data are mean ± SD of at least 3 independent observations. (B) Effect of everolimus (96 hours) on proliferation of HKe-3 (HCT116-derivative KRAS WT clone) cells infected with control or KRAS D13 lentivirus. Results are expressed as percent viability compared with cells treated with DMSO only (control) and represent mean ± SD of at least 3 independent observations. (C) NOD/SCID mice were inoculated with HKe-3 cells (5 × 106) transduced with empty or KRAS D13 lentiviral vectors; once tumors were established, animals were administered everolimus at 7.5 mg/kg every other day. The arrows indicate the time point at which drug treatment was started. Results are shown as mean ± SEM.
To further substantiate these results, we performed forward genetic rescue experiments. We found that restoration of mutated KRAS D13 expression in HCT116-derivative cells that had only the KRAS WT allele (HKe-3) prevented response to everolimus in vitro (Figure 2B).
In vivo effect of everolimus on tumors bearing PIK3CA and/or KRAS mutations.
We next evaluated the effect of everolimus in vivo by injecting subcutaneously two independent cancer cell models into immunocompromised mice. We found that HKe-3 cells (HCT116-derivative cells lacking mutated KRAS) formed tumors, albeit after a prolonged latency. Oral administration of everolimus to animals carrying HKe-3 xenografts resulted in long-term tumor growth arrest (Figure 2C). In contrast, the growth of tumors formed by HKe-3 cells transduced with oncogenic KRAS was not significantly affected by everolimus treatment (Figure 2C).
As a second cell model, we used the endometrial cancer cell line ME-180, which carries the PIK3CA E545K mutation but is WT for KRAS and is sensitive to everolimus. In vitro, expression of KRAS D13 in ME-180 cells abrogated the antiproliferative effect of everolimus, thus confirming our hypothesis (Supplemental Figure 3A). Parental and KRAS D13–expressing ME-180 cells formed tumors of comparable size when injected subcutaneously in mice. While everolimus had a prominent cytostatic effect on tumors originating from parental (PIK3CA mutated but KRAS WT) cells, it did not significantly influence the growth of tumors formed by cells expressing oncogenic KRAS D13 (Supplemental Figure 3B).
Next, we established whether the results obtained with ectopic expression of oncogenic KRAS D13 could be replicated in isogenic cells expressing physiological levels of the mutated allele. To this end, we recapitulated the genetic background of the HCT116 colorectal cancer cells in hTERT-HME1 cells by introducing via homologous recombination both KRAS G13D and PIK3CA H1047R alleles in their genome. This approach generated double-KI (hTERT-HME1 DKI) cells, in which each mutation is expressed under the corresponding gene’s promoter. When exposed to everolimus, these double mutant cells displayed a delay in cell proliferation comparable to that observed in the parental WT population (Supplemental Figure 4 and Supplemental Table 1). We found that these hTERT-HME1 isogenic cells were unable to grow as xenograft in immunocompromised mice (data not shown), which prevented us from testing the effect of everolimus on this cell model in vivo.
Effect of everolimus treatment on signal transduction.
To gain insight into the mechanisms of genotype-dependent proliferation upon everolimus treatment, we performed biochemical analysis on all hTERT-HME1 KI cells and their WT counterparts. While a wide range of concentrations was tested in proliferation experiments, a single clinically relevant dose of everolimus (50 nM) was employed for biochemical and metabolic profiling. This was chosen on the basis of plasma levels of everolimus in patients from phase I trials (11) and additional pharmacodynamic-pharmacokinetic studies showing that intratumor concentrations of everolimus can exceed by 10-fold plasma levels in animal models (17).
We analyzed the short-term effect of everolimus on the downstream effectors of RAS and PI3K (Supplemental Figure 5). Briefly, everolimus resulted in the effective inhibition of the mTORC1 effector S6 kinase (S6K1) (reviewed in ref. 18) in all genotypes (Supplemental Figure 6). Rapamycin analogs have been shown to activate indirectly mTORC2-dependent phosphorylation of AKT at Ser473 (19, 20). We found an increase in AKT phosphorylation upon short everolimus treatment in WT cells, but not in cells carrying either PIK3CA H1047R or KRAS G13D mutations (Supplemental Figure 6). In addition, all PIK3CA or KRAS G13D mutated clones showed the expected increase in basal phosphorylation of AKT (Supplemental Figure 6), when compared with parental cells. KI of oncogenic KRAS increased activated GTP-bound RAS expression (13) and resulted in augmented levels of basal ERK1/2 phosphorylation (Supplemental Figure 6). ERK1/2 is a downstream effector of active RAS, and it has been reported to be transiently phosphorylated in response to rapamycin treatment (21). However, while the levels of activated ERK1/2 increased upon treatment in parental cells, everolimus failed to induce a transient increase in ERK1/2 phosphorylation in all tested hTERT-HME1–derivative mutant clones (Supplemental Figure 6).
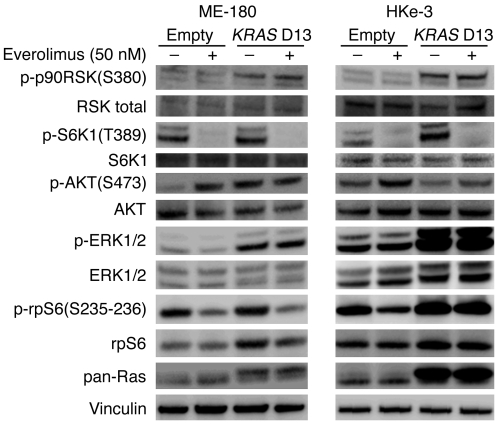
To better elucidate the molecular mechanisms responsible for resistance to everolimus by oncogenic KRAS, we extended all biochemical analyses to tumor cells employed for in vivo studies. After transduction with KRAS D13, both HKe-3 and ME-180 cells resulted in increased phosphorylation of basal ERK1/2 levels, while only ME-180 cells displayed an increase in phosphorylated AKT (Figure 3). Everolimus administration resulted in the effective inhibition of S6K1 in all treated cells. While everolimus did not change the levels of activated ERK1/2, it increased phosphorylation of AKT at Ser473 in HKe-3 and ME-180 parental but not in KRAS-transduced cells. These experiments suggested that whether short treatment with everolimus results in increased phosphorylation of AKT or MAPK depends on cell type rather than genotype. In addition to AKT and ERK1/2, we also performed immunoblotting against p90RSK to determine whether this could represent a key modulator of response. We found that all cells with oncogenic KRAS D13 displayed elevated phosphorylation levels of this protein, which were unaffected by treatment with everolimus (Figure 3). p90RSK is an AGC kinase of the RSK family that is phosphorylated and activated by ERK1/2 in response to many growth factors, hormones, and neurotransmitters (22). p90RSK phosphorylates a wide range of substrates, including members of the translational machinery such as eIF4B (23) and ribosomal protein S6 (rpS6) at Ser235/236 (24). Consistent with this, HKe-3 cells transduced with oncogenic KRAS D13 displayed not only increased levels of phosphorylated p90RSK, but also increased activation of rpS6, which was not modulated by everolimus (Figure 3). These results raised the possibility that translation could be used as a readout of everolimus inhibition and Ras activation.
Figure 3. Effect of everolimus on RAS/PI3K signaling in PIK3CA and KRAS mutant cells.
PIK3CA mutant ME-180 and HKe-3 cells were transduced with either empty or KRAS D13–expressing lentiviral vectors. All cells were treated for 30 minutes with everolimus (50 nM), and the corresponding lysates were blotted with total RSK1/RSK2/RSK3, phospho–p90RSK (Ser380), total S6K1, phospho-S6K1 (Thr389), total AKT, phospho-AKT (Ser473), total ERK1/2 and phospho-ERK1/2 antibodies, phospho-rpS6 (Ser235–236), and total rpS6. Vinculin was included as a loading control.
KRAS and PIK3CA oncogenic mutations affect everolimus-dependent translation.
RSK has been involved in activating the translational machinery by promoting recruitment of rpS6 and ribosomal subunits to the translation preinitiation complex (22). On the other hand, rapamycin and its derivatives have been shown to inhibit translation (25). Our results pinpointed p90RSK as a possible mediator of resistance to everolimus in cells with mutated KRAS. Accordingly, we tested the hypothesis that oncogenic KRAS could activate translation through an mTORC1-independent pathway and therefore could bypass everolimus-mediated mTOR inhibition.
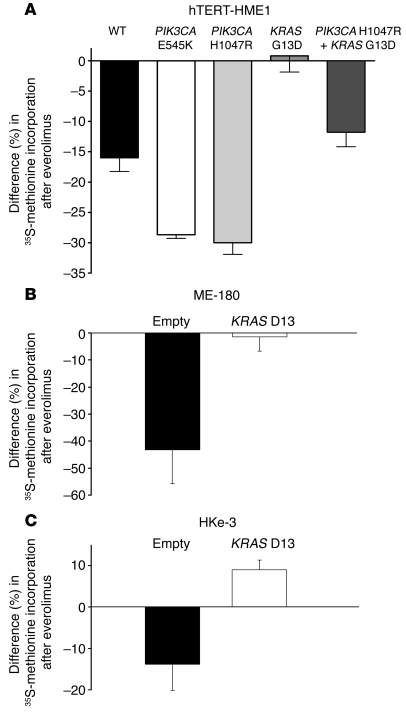
Thus, we analyzed whether the global rate of translation of hTERT-HME1 WT and KI cells was sensitive to everolimus. WT cells showed a mild sensitivity to everolimus; clones in which PI3K was constitutively active were sensitive to everolimus, and their translation rate dropped by 30%. Intriguingly, KRAS mutant cells were not affected by everolimus (Figure 4A). These data provide the first evidence to our knowledge that sensitivity of the translation rate to mTORC1 inhibition is proportional to PI3K activation and is reduced by oncogenic KRAS. Of note, the lack of sensitivity of KRAS mutants to everolimus inhibition was not due to a depression in the basal level of translation, which was comparable to that of PIK3CA mutants (Supplemental Figure 7).
Figure 4. Oncogenic KRAS and PIK3CA mutations affect translation.
(A) hTERT-HME1 cells of the indicated genotypes were treated with everolimus (500 nM) or left untreated. After 45 minutes of 35S-methionine pulse, methionine incorporation was measured in newly translated proteins. (B) ME-180 and (C) HKe-3 cells were transduced with a lentiviral vector encoding for KRAS D13 or a control vector (empty) and treated with everolimus (50 nM) or left untreated. After 45 minutes of 35S-methionine pulse, methionine incorporation was measured. All results are expressed as percent reduction in incorporation between treated and untreated cells (mean ± SEM).
We tried then to correlate the response of cancer cells to everolimus to their translation rate. Thus, we analyzed the sensitivity of translation to everolimus in the cells that we used for tumor xenografts, i.e., ME-180 with or without KRAS D13 and HKe-3 cells with or without KRAS D13. We observed that a short time pulse with everolimus (30 minutes) was sufficient to inhibit translation in ME-180 and HKe-3 cells, but not in their counterpart transduced with KRAS D13 (Figure 4, B and C). Thus, tumor cells sensitive to everolimus treatment may be predicted by a drop in the global rate of translation, after a short pulse with the drug.
Taken together, these data imply that oncogenic RAS activates translation through an mTORC1-independent pathway. Activation of the RAS-ERK1/2 cascade and of RSK1 may provide an alternative route to translational control.
Genetic alterations of the PI3K and KRAS pathways affect cancer patients’ response to everolimus.
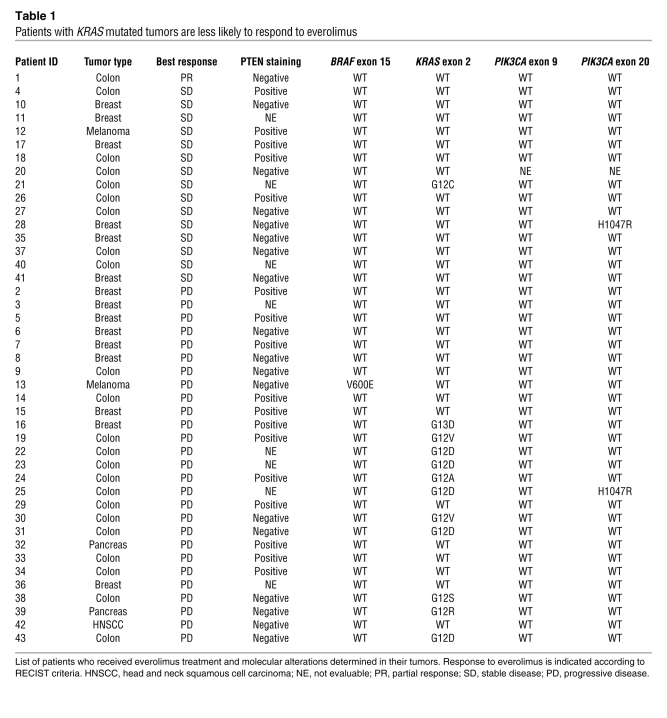
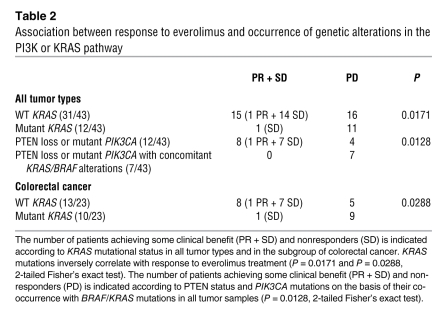
In order to determine the relevance and the applicability of the above findings to human cancer therapy, we assessed the mutational status of PIK3CA, KRAS, and BRAF in a cohort of cancer patients who had received single-agent everolimus as part of phase I and phase II studies at a single institution (11). Due to the limited amount of tumor tissue available, only KRAS exon 2, BRAF exon 15, and PIK3CA exons 9 and 20 could be amplified and subjected to direct sequencing (Table 1). These exons were selected because they include codons where the large majority of mutations occur in these genes (Catalogue of Somatic Mutations in Cancer, http://www.sanger.ac.uk/genetics/CGP/cosmic/). We also assessed the expression status of PTEN by immunostaining. In the cohort, only one partial response (PR) occurred in a patient with a heavily pretreated metastatic colorectal cancer lasting 5.3 months (disease control during 9 months). Molecular analysis of this case revealed no mutations in the sequenced exons of BRAF, KRAS, or PIK3CA, but lack of expression of PTEN protein. PIK3CA mutations could only be detected in two samples (Table 1 and Figure 5). Because of the low frequency, we were unable to perform any meaningful statistical analysis. However, it should be noted that one of the PIK3CA mutant tumors also had a KRAS mutation and did not respond to therapy (progressive disease [PD]), while the other displayed concomitant PTEN loss and resulted in disease stabilization (stable disease [SD]). PTEN staining was negative in 18 of 35 evaluable cases. When patients with SD or PR were considered, 8 individuals with tumors displaying PTEN loss had some clinical benefit from treatment with everolimus. Of the remaining 10 PTEN-negative cases that progressed, 6 were found to carry a concurrent KRAS or BRAF mutation. The combined results suggest that cancer patients whose tumors carry PIK3CA kinase domain mutations or PTEN loss of function can benefit from everolimus treatment, except when BRAF/KRAS mutations occur in concomitance with PI3K pathway alterations (Figure 5). Although this association reached statistical significance (P = 0.0128, 2-tailed Fisher’s exact test; Table 2), these results should be confirmed in additional studies including a larger number of patients. Importantly, when KRAS status was considered in univariate analysis, the occurrence of KRAS mutations negatively and significantly affected clinical benefit of everolimus. Eleven of the 12 patients with KRAS mutant tumors had disease progression, while only 16 of 31 of WT cases did not benefit from treatment (P = 0.0171, 2-tailed Fisher’s exact test; Table 2). Because the distribution of KRAS mutations is highly skewed in colorectal cancers, we restricted the analysis to this tumor subgroup and confirmed the negative predictive role of KRAS mutations (P = 0.0288, 2-tailed Fisher’s exact test; Table 2). These results strongly support our preclinical findings that oncogenic KRAS is able to bypass mTORC1 inhibition, thus representing what we believe to be a novel mechanism of resistance to rapamycin derivatives.
Table 1 .
Patients with KRAS mutated tumors are less likely to respond to everolimus
Figure 5. Oncogenic KRAS mutations are associated with clinical resistance to everolimus.
Venn diagram representation of the distribution of molecular alterations in individual cancers. Response to everolimus according to the presence of genetic abnormalities within individual tumor samples is also shown.
Table 2 .
Association between response to everolimus and occurrence of genetic alterations in the PI3K or KRAS pathway
Discussion
We present an integrated strategy to identify relationships between tumor genotypes (biomarkers) and a molecularly targeted drug (26). The approach involves as an initial step the use of homologous recombination to introduce individual or multiple cancer mutations into the genome of non-transformed epithelial cells (13). As a result, the heterozygous mutated genes are expressed under their endogenous promoters, thus closely recapitulating the lesions observed in human tumors. In the subsequent step, mutated and WT cells were exploited to identify compounds selectively targeting oncogenic PIK3CA variants. Using this approach, we found that everolimus, a rapamycin derivative, had selectivity for cells carrying PIK3CA. The strategy subsequently involves translating the findings obtained using non-transformed epithelial cells in a panel of human cancer cells in which the same mutations were found to be present. We found that tumor cells carrying oncogenic PIK3CA mutations or PTEN loss of function were sensitive to everolimus, except when KRAS or BRAF mutations were concomitantly present. Further analysis revealed that selective genetic removal of the mutated KRAS allele restored sensitivity to everolimus. These data were corroborated by forward genetic analysis in which introduction of oncogenic KRAS abrogated the everolimus response of PIK3CA mutated cells. As the isogenic cells were unable to grow as xenograft in mouse (13, 15), we were unable to test the effect of everolimus in vivo. To overcome this limitation, we transduced the KRAS D13 allele in tumorigenic and everolimus-sensitive cells carrying PIK3CA mutations. After engraftment in immunocompromised mice, oral administration of everolimus induced growth arrest in PIK3CA mutant tumors, but not in their corresponding tumors concomitantly expressing a KRAS mutation.
In the final step, we translated the findings obtained in isogenic cell models into the clinical setting by performing genetic analysis of tumors from patients treated with everolimus. Cancer patients whose tumors carried PIK3CA mutations in the kinase domain or PTEN loss of function displayed increased clinical benefit from everolimus treatment, except when KRAS mutations were present. In particular, in univariate analysis, the occurrence of oncogenic KRAS was statistically correlated with lack of response in the treated cohort. To our knowledge, this is the first time that this association has ever been reported. Data from clinical studies have mainly focused on immunohistochemical surrogate markers of early response in biopsies from treated patients (11, 27), rather than on the genetic status of tumors. It is interesting to note that occurrence of KRAS mutations is also a validated biomarker of clinical resistance to EGFR-targeted monoclonal antibodies in colorectal cancer (28).
Overall, the proposed strategy led us to conclude that the oncogenic status of KRAS plays a central role in conferring resistance to the antiproliferative effects of everolimus in human tumors harboring genetic alterations of the PIK3CA gene. Notably, previous studies also suggested that rapamycin was inefficacious in engineered mouse models for ovarian or colon cancer in which the KRAS G12D allele was expressed in a tissue-specific manner (29, 30). Two potential mechanisms of resistance to everolimus and other mTOR inhibitors have been previously proposed. The first consists of a negative feedback loop between mTOR and AKT: mTORC1 inhibition leads to AKT activation, possibly through upregulation of either receptor tyrosine kinases such as IGF-1R or substrates such as IRS-1 (19, 20, 31). In accordance with these findings, we detected increased levels of phosphorylated AKT after everolimus treatment in a number of cell lines. However, this upregulation was present in a cell-specific context and was absent in insensitive cells carrying oncogenic KRAS. It has been proposed that AKT activation might negatively interfere with the efficacy of mTOR inhibitors (32), but the mechanism of this effect needs further investigation. The second mechanism of resistance to mTOR inhibitors consists of a different feedback loop between mTOR and ERK1/2: mTORC1 inhibition leads to ERK1/2 activation through an S6K1/PI3K/RAS-dependent pathway (21). In fact, Carracedo et al. (21) showed that a dominant negative form of RAS (RAS N17) abrogates rapamycin-induced ERK1/2 activation. However, we observed a variable increase in phosphorylated ERK1/2, with this effect being present only in some cell types and not depending on genotype.
While the analysis of commonly known RAS/PI3K effectors such as ERK1/2 and AKT was not conclusive, we found that the introduction of oncogenic KRAS was accompanied by activation of the ERK1/2 effector p90RSK (22). In turn, p90RSK phosphorylates a wide range of substrates including members of the translational machinery such as eIF4B (23) and rpS6 at Ser235/236 (24). Our results pinpoint p90RSK as a key modulator of global translation in cells with oncogenic KRAS. Importantly, the refractoriness of KRAS mutant cells to everolimus was mirrored by sustained levels of global translation observed in the same cells. In contrast, in the corresponding cells with PIK3CA mutations, everolimus clearly affected translation. Recent work has showed that RSK also directly targets the mTORC1 complex by phosphorylating Raptor and thereby promoting mTORC1 kinase activity (33). We found that the expression of a KRAS D13 allele does not elicit robust mTORC1 activation (as demonstrated by the absence of increased S6K1 phosphorylation compared with control cells), thereby suggesting that RSK contributes to translation through an mTORC1-independent mechanism.
It has recently been proposed that in mutant KRAS-dependent cells, the serine/threonine kinase STK33 modulates S6K1 activity through an mTORC1-independent mechanism (34). This raises the possibility that suppression of STK33 expression could be exploited to sensitize KRAS mutated cells to mTOR inhibitors.
Overall, our results have a number of general implications. We have defined an experimental strategy (based on mutated human cells) capable of identifying biomarkers associated with response to anticancer drugs. We show that the pharmacological relationships established in non-transformed cells carrying defined cancer mutations can be translated to cancer cells in which the corresponding mutations naturally occur alongside additional genetic alterations. This indicates that the mutated cells together with their WT counterparts can be successfully used to de-convolute the complex genetic background of cancer cells. The approach defined here can be broadly applied to study how genetic relationships affect sensitivity and resistance to molecularly targeted anticancer drugs.
Methods
Plasmids and viral vectors.
All experimental procedures for targeting vector construction, AAV production, cell infection, and screening for recombinants have been described elsewhere (13). The list of primers employed to amplify the AAV vectors’ homology arms for introducing the PIK3CA E545K and H1047R alleles is detailed in Supplemental Table 4. The procedure to obtain the lentivirus expressing the KRAS G13D mutation has been described elsewhere (13).
Cells and cell culture reagents.
hTERT-HME1 and MCF10A cell lines were purchased from ATCC and were cultured in growth medium containing DMEM/F-12 (Invitrogen) supplemented with 20 ng/ml EGF, 10 μg/ml insulin, and 100 μg/ml hydrocortisone. All other cancer cell lines (U-87 MG, Ca Ski, ME-180, MCF7, BT-474, PC-3, PANC-1, HT-29, NIH:OVCAR-3, SK-OV-3, HCT116, and DLD-1) were also obtained from ATCC and cultured according to their recommendations. All cell culture media were supplemented with 5% fetal bovine serum (Sigma-Aldrich), 50 U/ml penicillin, and 50 mg/ml streptomycin. Geneticin (G418) was purchased from Gibco (Invitrogen). Isogenic HCT116 PIK3CA WT and mutant cells were provided by the B. Vogelstein/V.E. Velculescu laboratories (Johns Hopkins University, Baltimore, Maryland, USA) (16). Isogenic HCT116 KRAS WT (HKe-3, HKh-2) and mutated (HK2-6) clones have been previously described (15).
Drug proliferation assays.
Everolimus, purchased from Sigma-Aldrich (catalog 7741), was dissolved in DMSO and stored in aliquots at –80°C. The effect of the drug on cell proliferation was determined using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega), which is based on quantification of the cellular ATP level. Cells were plated in 96-well plates at a density of 2,000–5,000 cells in triplicate. The following day, cells were treated with a range of drug concentrations prepared by serial dilution. After 72–96 hours of treatment, 100 μl of luciferin luciferase–containing reagent was added to each well, and luminescence measurements were recorded by the DTX 880-Multimode plate reader (Beckman Coulter). To account for clonal variability, at least two independent clones carrying each of the mutations were generated and analyzed.
Flow cytometric analysis.
For time-course experiments, on the initial day hTERT-HME1 cells were labeled with 3 μM CFSE (Invitrogen, C1157) in PBS in the dark for 30 minutes. After washing and recording of baseline fluorescence, cells were plated in media containing 1% FBS and 2 ng/ml EGF and treatment with everolimus was initiated, with the drug replenished on a daily basis. For cell cycle analysis, trypsinized cells were washed with PBS, and cell nuclei DNA were stained with propidium iodide (PI) for at least 120 minutes using a commercial kit (DNA con 3, Consul T.S.).
All fluorescence levels were detected by flow cytometry on a FACSCalibur (BD) and analyzed using CellQuest software. The number of events collected for each sample varied between 15,000 and 50,000. After doublet exclusion, an extended analysis of the DNA content and calculations of the percentage of cells in each phase of the cell cycle were performed on ModFit LT software (Verity Software House).
Protein analysis.
Prior to biochemical analysis, all cells were grown in their specific media supplemented with 5% FBS, unless otherwise indicated. SDS-PAGE Western blotting was performed as previously described (13). The following antibodies were used for Western blotting (from Cell Signaling Technology, except where indicated): anti-AKT (BK9272); anti–phospho-AKT S473 (BK9271S); anti–phospho-S6K1 (Thr389; catalog 9205S), and anti–total S6K1 (catalog 9202); anti–phospho-p44/42 ERK (Thr202/Tyr204) (BK9101S); anti-p44/42 ERK (BK9102); phospho-p90RSK(Ser380) (catalog 9341); anti-RSK1/RSK2/RSK3 (catalog 9347); phospho-rpS6 (Ser 235–236) (catalog 2211S); anti-rpS6 (5G10) (2217); anti-actin and anti-vinculin (Sigma-Aldrich).
Mouse xenografts.
All animal procedures were approved by the Ethical Commission of the University of Turin and by the Italian Ministry of Health. Lentiviral vector–transduced HKe-3 and ME-180 cells (5 × 106 cells/mouse) were injected subcutaneously into the right posterior flanks of 7-week-old immunodeficient NOD/SCID male mice (6 mice/group; Charles River). Tumor formation was monitored twice a week, and tumor volume based on caliper measurements was calculated by the modified ellipsoid formula (tumor volume = 1/2[length × width2]). When tumors reached a volume of approximately 0.3 cm3, mice were randomized to receive either vehicle (1% methylcellulose in sterile water) or everolimus. Everolimus was administered by oral gavage every other day at a dosage of 7.5 mg/kg/treatment.
Metabolic assay.
Subconfluent hTERT-HME1 cell clones were plated in 6-well plates. Cells were incubated at 37°C with 80% methionine-free medium (Sigma-Aldrich), 20% DMEM for 45 minutes. Preincubation with everolimus was performed 30 minutes before methionine pulse. Cells were pulsed with 33 μCi per well of 35S-labeled methionine (PerkinElmer) for 45 minutes. Cells were lysed by scraping in 50 μl RIPA buffer without SDS (10 mM Tris-HCl pH 7.5, 1% Na-deoxycholate, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, proteases inhibitor cocktail). Ten-microliter extracts were TCA precipitated and counted as previously described (35). Obtained values were normalized to sample protein content, quantified by bicinchoninic acid (BCA) protein assay (Pierce). Each sample was assayed at least in triplicate.
Patient population.
We retrospectively analyzed primary or metastatic tissue samples from 43 patients with advanced solid tumors treated with single-agent everolimus at the Medical Oncology Department, Vall d’Hebron University Hospital. These patients were treated in the context of a phase I and a phase II study, and the population included patients with different tumor types refractory to standard therapies, the most frequent tumor type being colorectal cancer (11). Tumor types are summarized in Table 2. Everolimus was administered either as a single weekly oral dose (20, 50, and 70 mg) or as a continuous daily oral dose (5 and 10 mg). Treatment was continued until PD or toxicity occurred, according to the standard criteria. The Ethics Committee of Vall d’Hebron University Hospital approved the studies, which followed the Declaration of Helsinki. The molecular analysis of these samples was done at Istituto Cantonale di Patologia, Locarno, or at the Laboratory of Molecular Genetics, Institute for Cancer Research and Treatment (Turin, Italy). All patients provided informed consent for their tissues to be used for the molecular analyses that were performed in this study.
Clinical evaluation and tumor response criteria.
Objective tumor measurements were evaluated in accordance with RECIST criteria (36) after 8 weeks of treatment, and thereafter every 8 weeks. Objective tumor responses were classified as PR, SD, and PD. Patients with either SD or PR at the first workup evaluation were considered to have clinical benefit for the purpose of this molecular study.
Molecular analyses.
Formalin-fixed, paraffin-embedded tumor blocks were reviewed for quality and tumor content. Macrodissection was performed on a single representative block from the primary tumor or from metastatic lesions, in order to have at least 70% malignant cells. Genomic DNA was extracted using the QIAamp Mini kit (QIAGEN) according to the manufacturer’s instructions.
Mutational analysis of KRAS, BRAF, and PIK3CA.
We searched for KRAS (exon 2), for BRAF (exon 15) and for PIK3CA (exons 9 and 20) mutations by direct sequencing. KRAS exon 2 includes codons 12 and 13; BRAF exon 15 includes codon 600; PIK3CA exon 9 includes codons 542 and 545; and PIK3CA exon 20 includes codon 1,047, where the large majority of mutations occur. The list of primers used for mutational analyses has been previously reported (37). All samples were subjected to automated sequencing by ABI PRISM 3730 (Applied Biosystems). All mutated cases were confirmed at least twice starting from independent PCR reactions. Mutational analysis was concomitantly performed by the laboratories of Molecular Genetics, Institute for Cancer Research and Treatment (Turin, Italy) and Istituto Cantonale di Patologia, Locarno, without knowledge of clinical data or results of molecular experiments.
PTEN expression.
PTEN protein expression status by IHC on 3-μm tissue sections was performed and evaluated according to the literature (38). Anti-PTEN Ab-2 (Neomarkers) was applied at a 1:50 dilution. PTEN protein expression was detected mainly at the cytoplasmic level, although occasional nuclear positivity was present. Tumors were considered negative, i.e., with loss of PTEN expression, when absence or reduction of immunostaining was seen in more than 50% of neoplastic cells as compared with internal controls (i.e., vascular endothelial cells and nerves). Normal endometrium tissue was used as external positive control. The evaluations were performed by two independent pathologists without knowledge of clinical data or results of molecular analyses.
Statistics.
Statistical analyses were performed by the 2-tailed t test with Bonferroni’s multiple comparisons correction and the Fisher’s exact test using the Instat program (GraphPad Software). For all tests, the level of statistical significance was set at P < 0.05. Error bars throughout the figures indicate SD or SEM, as detailed in each legend.
Supplementary Material
Acknowledgments
We are grateful to Chris Torrance, Miriam Martini, Emily Crowley, and Luca Cardone for critically reading the manuscript. We thank Elena Casanova for technical assistance with FACS acquisition data; Sebastjian Hobor, Alessia Bottos, and Laura Tarditi for help with mouse handling; and Irene Marimon and José Jimenez for technical help in the collection of tumor samples. Work in the laboratories of the authors is supported by the Italian Association for Cancer Research (AIRC; A. Bardelli and S. Biffo); Italian Ministry of Health (A. Bardelli and S. Biffo); Regione Piemonte (F. Di Nicolantonio, S. Arena, S. Grosso, and A. Bardelli); Italian Ministry of University and Research (A. Bardelli); CRT Progetto Alfieri (A. Bardelli); European Union FP7 Marie Curie Programme (A. Bardelli); Association for International Cancer Research UK (A. Bardelli and S. Biffo); European Union FP6, MCSCs contract 037297 (A. Bardelli); Oncosuisse grants OCS-01921-08-2006 and OCS-02301-08-2008 (M. Frattini); and Fondazione Ticinese per la Ricerca sul Cancro (Tessin Foundation for Cancer Research; M. Frattini).
Footnotes
Conflict of interest: José Baselga and Josep Tabernero have been members of advisory boards for Novartis, the manufacturer of everolimus.
Citation for this article: J Clin Invest. 2010;120(8):2858–2866. doi:10.1172/JCI37539.
Mariangela Russo’s present address is: Horizon Discovery Ltd. Gene Engineering Laboratory, Molecular Biotechnology Center, Turin, Italy.
See the related Commentary beginning on page 2655.
References
- 1.Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436(7052):792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 2.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 3.Ma WW, Adjei AA. Novel agents on the horizon for cancer therapy. CA Cancer J Clin. 2009;59(2):111–137. doi: 10.3322/caac.20003. [DOI] [PubMed] [Google Scholar]
- 4.Hudes G, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356(22):2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 6.Atkins MB, Yasothan U, Kirkpatrick P. Everolimus. Nat Rev Drug Discov. 2009;8(7):535–536. doi: 10.1038/nrd2924. [DOI] [PubMed] [Google Scholar]
- 7.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5(8):671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 8.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27(13):2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donnell A, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral mammalian target of rapamycin inhibitor everolimus in patients with advanced solid tumors. J Clin Oncol. 2008;26(10):1588–1595. doi: 10.1200/JCO.2007.14.0988. [DOI] [PubMed] [Google Scholar]
- 10.Slomovitz BM, et al. A phase II study of oral mammalian target of rapamycin (mTOR) inhibitor, RAD001 (everolimus), in patients with recurrent endometrial carcinoma (EC). J Clin Oncol. 2008;26(34):abstr 5502. [Google Scholar]
- 11.Tabernero J, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26(10):1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 12.Yee KW, et al. Phase I/II study of the mammalian target of rapamycin inhibitor everolimus (RAD001) in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2006;12(17):5165–5173. doi: 10.1158/1078-0432.CCR-06-0764. [DOI] [PubMed] [Google Scholar]
- 13.Di Nicolantonio F, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A. 2008;105(52):20864–20869. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260(5104):85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 16.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka C, et al. Identifying optimal biologic doses of everolimus (RAD001) in patients with cancer based on the modeling of preclinical and clinical pharmacokinetic and pharmacodynamic data. J Clin Oncol. 2008;26(10):1596–1602. doi: 10.1200/JCO.2007.14.1127. [DOI] [PubMed] [Google Scholar]
- 18.Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13(6):252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.O’Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun SY, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65(16):7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 21.Carracedo A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9(10):747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 23.van Gorp AG, et al. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene. 2009;28(1):95–106. doi: 10.1038/onc.2008.367. [DOI] [PubMed] [Google Scholar]
- 24.Roux PP, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282(19):14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 26.Di Nicolantonio F, Arena S, Gallicchio M, Bardelli A. Isogenic mutant human cells: a new tool for personalized cancer medicine. Cell Cycle. 2010;9(1):20–21. doi: 10.4161/cc.9.1.10474. [DOI] [PubMed] [Google Scholar]
- 27.Reardon DA, et al. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neurooncol. 2010;96(2):219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101(19):1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung KE, et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc Natl Acad Sci U S A. 2010;107(4):1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing D, Orsulic S. A genetically defined mouse ovarian carcinoma model for the molecular characterization of pathway-targeted therapy and tumor resistance. Proc Natl Acad Sci U S A. 2005;102(19):6936–6941. doi: 10.1073/pnas.0502256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26(13):1932–1940. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Yue P, Kim YA, Fu H, Khuri FR, Sun S. Enhancing mammalian target of rapamycin (mTOR)–targeted cancer therapy by preventing mTOR/Raptor inhibition-initiated, mTOR/Rictor-independent Akt activation. Cancer Res. 2008;68(18):7409–7418. doi: 10.1158/0008-5472.CAN-08-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carriere A, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18(17):1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 34.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Grosso S, et al. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem J. 2008;415(1):77–85. doi: 10.1042/BJ20080463. [DOI] [PubMed] [Google Scholar]
- 36.Therasse P, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 37.Moroni M, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 38.Frattini M, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97(8):1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.