Abstract
Cerebral cavernous malformation is a common human vascular disease that arises due to loss-of-function mutations in genes encoding three intracellular adaptor proteins, cerebral cavernous malformations 1 protein (CCM1), CCM2, and CCM3. CCM1, CCM2, and CCM3 interact biochemically in a pathway required in endothelial cells during cardiovascular development in mice and zebrafish. The downstream effectors by which this signaling pathway regulates endothelial function have not yet been identified. Here we have shown in zebrafish that expression of mutant ccm3 proteins (ccm3Δ) known to cause cerebral cavernous malformation in humans confers cardiovascular phenotypes identical to those associated with loss of ccm1 and ccm2. CCM3Δ proteins interacted with CCM1 and CCM2, but not with other proteins known to bind wild-type CCM3, serine/threonine protein kinase MST4 (MST4), sterile 20–like serine/threonine kinase 24 (STK24), and STK25, all of which have poorly defined biological functions. Cardiovascular phenotypes characteristic of CCM deficiency arose due to stk deficiency and combined low-level deficiency of stks and ccm3 in zebrafish embryos. In cultured human endothelial cells, CCM3 and STK25 regulated barrier function in a manner similar to CCM2, and STKs negatively regulated Rho by directly activating moesin. These studies identify STKs as essential downstream effectors of CCM signaling in development and disease that may regulate both endothelial and epithelial cell junctions.
Introduction
Cerebral cavernous malformation (CCM) is a common human vascular disease that frequently results in stroke, seizure, and cerebral hemorrhage. Familial CCMs are inherited in an autosomal dominant pattern, constitute up to 50% of CCM cases, and have been found to be associated with loss-of-function mutations in 3 genes, KRIT1 (also known as CCM1) (1–3), CCM2 (MALCAVERNIN, OSM) (4, 5), and PDCD10 (CCM3) (6, 7). Genetic studies in zebrafish have demonstrated that loss of ccm1, ccm2, or the transmembrane receptor heart of glass (heg) results in embryonic cardiovascular phenotypes characterized by a large, thin-walled heart and defective branchial arch artery development that prevents blood circulation (8–10). In mice, loss of CCM1 or CCM2 results in a lack of blood circulation due to defective branchial arch artery formation and embryonic lethality at E9, a phenotype that is reproduced by endothelial cell–specific loss of CCM2 and in Heg–/–Ccm2+/– embryos (10–13). Biochemical studies have demonstrated interaction among HEG, CCM1, and CCM2 and between CCM2 and CCM3 (10, 14–17). Human CCMs, mouse vessels lacking HEG or CCM2, and zebrafish hearts lacking heg or ccm2 display similar defects in endothelial junctions by electron microscopy (10, 18, 19, 20), a finding consistent with in vitro studies demonstrating that CCM1 and CCM2 regulate endothelial cell junctions (13, 21). These studies have culminated in the hypothesis that HEG/CCM signaling is required in endothelial cells to regulate cell-cell association during cardiovascular development and later in life. Beyond the proteins identified by these genetic studies, however, how this pathway regulates endothelial cell function is not yet established.
Of the known CCM genes, CCM3 is the least understood. Although it is highly conserved in vertebrates and has orthologs in non-vertebrates such as Drosophila and C. elegans, the CCM3 protein contains no identifiable motifs. Other than its identification as a human CCM disease gene, no biological role for CCM3 has been clearly established. In vitro studies have identified CCM3 as a potential cell death–regulatory protein (22), and CCM3 deficiency has been shown to confer a lethal “dumpy” phenotype in C. elegans (23), suggesting that CCM3 is likely to play a broader role than that implicated in CCM pathogenesis. Recent biochemical studies have demonstrated that, in addition to CCM2, CCM3 binds the GCK-III family of sterile 20–like serine/threonine kinases (STKs) that includes MST4, STK24, and STK25 (24, 25). These kinases have not been identified as part of the CCM signaling complex using proteomic analysis (16), however, and how CCM3 functions in vascular development and disease is unknown.
In the present study, we address the role of CCM3 in vivo and in vitro and identify the GCK-III family of STKs as a novel class of downstream CCM signaling effectors required in cardiovascular development in vivo and for endothelial barrier function in vitro. Our studies suggest that STK24 and STK25 control endothelial cell-cell junctions by directly regulating the activity of moesin, a negative regulator of Rho. A similar pathway has been implicated in the control of epithelial cell junctions (26), suggesting that CCM signaling may regulate a broadly used molecular mechanism of cell-cell interaction that participates in both development and disease.
Results
Zebrafish express two ccm3 alleles that encode proteins highly homologous to human CCM3.
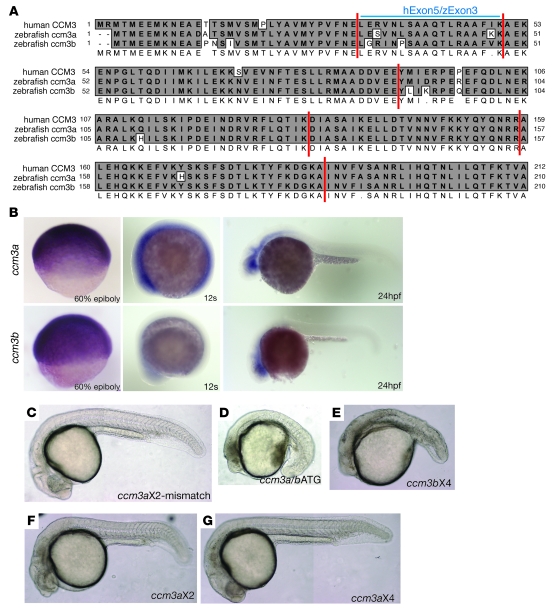
To study the role of ccm3 in cardiovascular development, and compare it with those recently established for heg, ccm1, and ccm2, we used zebrafish embryos in which loss of heg, ccm1, and ccm2 conferred identical phenotypes characterized by dilated hearts and circulatory block. Two genes encoding ccm3 orthologs exist in the fish, ccm3a and ccm3b. Alignment of the proteins encoded by the zebrafish ccm3 genes with human CCM3 revealed a very high degree of sequence conservation, with greater than 90% identity at the amino acid level (Figure 1A). In situ hybridization studies revealed that both ccm3a and ccm3b are expressed broadly, particularly in the first 24 hours of zebrafish development, and do not exhibit a cardiovascular-specific expression pattern (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI39679DS1). CCM3 is therefore a highly conserved protein that in zebrafish is encoded by 2 distinct alleles with overlapping expression patterns.
Figure 1. Zebrafish ccm3 proteins are highly conserved with the human ortholog CM3, and ccm3 is necessary during early zebrafish development.
(A) Alignment of the predicted amino acid sequences of zebrafish ccm3a and ccm3b with that of human CCM3. Identical residues are shaded in gray. Vertical lines indicate exon boundaries. The amino acids encoded by CCM3 exon 5 (hExon5) and ccm3a/b exon 3 (zExon3) are indicated. (B) Expression pattern of ccm3a and ccm3b in zebrafish embryos. ccm3a and ccm3b are ubiquitously expressed at the 60% epiboly and 12-second stages. At 24 hpf, ccm3a and ccm3b are expressed most strongly in the head. (C–G) Loss of ccm3a+ccm3b or ccm3b alone results in early embryonic lethality in zebrafish. Shown are 24-hpf zebrafish embryos following injection of ccm3aX2 mismatch control morpholino (C, 5 ng/embryo); a morpholino (ccm3a/bATG) designed to block translation of both ccm3a and ccm3b (D, 0.5 ng/embryo); or morpholinos specifically interfering the splicing of ccm3a or ccm3b: ccm3bX4 morpholino (E, 4 ng/embryo), ccm3aX2 morpholino (F, 3 ng/embryo), and ccm3aX4 morpholino (G, 4 ng/embryo). C, F, and G are composites of 2–3 images taken of the same embryos.
Mutant ccm3 proteins equivalent to those that cause human CCM confer cardiovascular phenotypes characteristic of heg, ccm1, and ccm2 deficiency in zebrafish.
To address the in vivo role of ccm3 in zebrafish, we used morpholino strategies to knock down expression of the two zebrafish ccm3 genes, ccm3a and ccm3b, both individually and together (Supplemental Figure 2). Combined loss of ccm3a and ccm3b expression with a translation blocking morpholino (ccm3a/bATG) or ccm3b expression alone with a specific splice morpholino (ccm3bX4) resulted in embryonic defects during early organogenesis and death at 24–36 hours postfertilization (hpf), precluding analysis of its role in cardiovascular development (Figure 1, C–E). In contrast, loss of ccm3a did not interfere with early embryonic development and did not confer the cardiovascular phenotypes of a dilated heart and interrupted circulation phenotypes characteristic of heg, ccm1, or ccm2 deficiency (Figure 1, F and G).
Studies of human CCM have identified a family in which the disease is caused by the expression of a CCM3 protein that lacks the 18 amino acids encoded by CCM3 exon 5 (6), suggesting that this part of the CCM3 protein may be specifically required for CCM3 participation in cardiovascular CCM signaling. To test this hypothesis, we used morpholinos to block splicing into the orthologous zebrafish exon (exon 3, Figure 1A) and express the equivalent mutant ccm3 protein in developing zebrafish embryos. Morpholinos were identified that efficiently blocked the splicing of both ccm3a and ccm3b transcripts containing exon 3 (ccm3aX3 and ccm3bX3), resulting in the expression of shorter mRNAs that encoded ccm3 proteins lacking the orthologous18 amino acids (termed ccm3Δ; Supplemental Figure 2). ccm3Δ-expressing embryos did not experience the early embryonic defects and lethality associated with complete ccm3 or ccm3b deficiency, but did exhibit the dilated, thin-walled hearts characteristic of zebrafish embryos lacking heg, ccm1, or ccm2 (Figure 2, A–C, Supplemental Figure 3, and Supplemental Videos 1 and 2). ccm3Δ-expressing embryos also exhibited circulatory block at the level of the branchial arch arteries that connect the heart to the aorta (Figure 2D) but retained normal vascular patterning (Figure 2E), phenotypes identified in both zebrafish and mouse embryos lacking heg, ccm1, or ccm2 (10, 11, 13). These cardiovascular phenotypes arose in greater than 90% of embryos injected with ccm3aX3 and ccm3bX3 morpholinos and could be rescued by coinjection of cRNAs encoding wild-type ccm3a or ccm3b (Figure 2F, Supplemental Figure 4, and Supplemental Videos 3 and 4), indicating that they arose due to specific loss of ccm3 function and that ccm3a and ccm3b are functionally redundant in this pathway. These studies demonstrate (a) that ccm3 plays an essential role in the same pathway as heg, ccm1, and ccm2 during cardiovascular development and (b) that the 18 amino acids encoded by exon 3 in zebrafish ccm3 genes and exon 5 in the human CCM3 gene are essential for the cardiovascular functions of CCM3 in this signaling pathway.
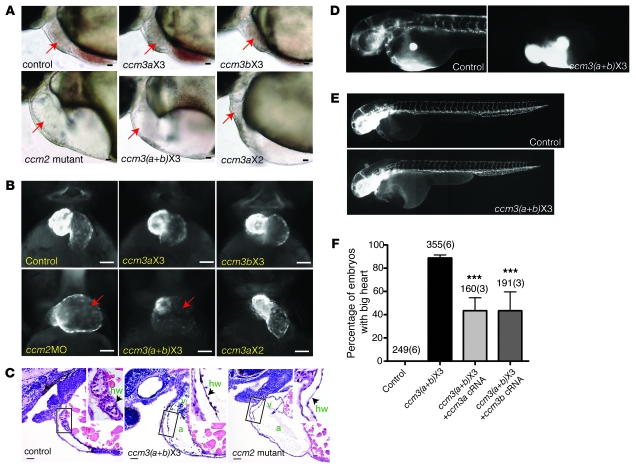
Figure 2. Expression of ccm3 proteins lacking the 18 amino acids encoded by exon 3 confers cardiovascular phenotypes characteristic of heg, ccm1, and ccm2 deficiency.
(A) Light images of the hearts of 48-hpf zebrafish control embryos, ccm2 mutant embryos, and embryos injected with morpholinos that block splicing into exon 3 of ccm3a only (ccm3aX3, 3 ng/embryo), ccm3b only (ccm3bX3, 3 ng/embryo), both ccm3a and ccm3b [ccm3(a+b)X3], or exon 2 of ccm3a (ccm3aX2, 3 ng/embryo) are shown. Arrows indicate the embryo hearts. (B) Fluorescence images of the hearts of transgenic embryos in which myocardial cells express GFP following injection of the indicated morpholinos. ccm2MO indicates a morpholino that blocks splicing of the ccm2 gene. (C) Thinned myocardium in ccm3(a+b)X3 morphants is identical to that seen in ccm2 mutants. Shown are hematoxylin/eosin-stained sagittal sections of the indicated 48-hpf embryos. a, atrium; v, ventricle; hw, heart wall. (D) Angiography of 48-hpf control and ccm3(a+b)X3 morphant embryos reveals blocked circulation at the cardiac outflow tract. (E) Vascular endothelial patterning as revealed in Tg (fli1a:EGFP)y1 embryos is undisturbed in ccm3(a+b)X3 morphant embryos. The images are composites of 2–3 images taken of the same embryos. (F) The big heart phenotype conferred by morpholinos that block splicing into exon 3 of ccm3a and ccm3b is rescued by coinjection of cRNAs (100 pg/embryo) encoding either ccm3a or ccm3b (right 2 bars). Shown are mean and SEM. The number of embryos examined is indicated above each bar, and the number of injections performed for each group shown in parentheses. ***P < 0.001 by Student’s t test. Scale bars: 20 μm.
CCM3Δ proteins interact normally with CCM1 and CCM2 but do not bind the sterile 20–like kinases MST4, STK24, and STK25.
The finding that ccm3Δ expression does not block early embryonic development but selectively confers the cardiovascular phenotypes associated with complete loss of heg, ccm1, and ccm2 suggested that the domain of ccm3 encoded by exon 3 must be required for cardiovascular CCM signaling in vivo. Recent biochemical studies have demonstrated that CCM3 binds CCM2 (14, 25), while CCM2 binds CCM1 (15, 17) and CCM1 binds the HEG receptor (10) (Figure 3A). To determine whether ccm3Δ proteins are able to complex with the known CCM signaling pathway, HA-tagged ccm2 proteins were coexpressed with myc-tagged wild-type ccm3 and ccm3Δ proteins in HEK293T cells and co-immunoprecipitation experiments performed using anti-HA and anti-myc antibodies. ccm2 but not ccm1 co-immunoprecipitated with both ccm3a and ccm3b when expressed alone, while strong immunoprecipitation of a ccm1/2/3 complex was observed when all 3 proteins were coexpressed (Figure 3B and Supplemental Figure 5A). Anti-myc immunoblots of cell lysate confirmed that ccm3Δ was expressed at levels comparable to wild-type ccm3 but was approximately 2 kDa smaller (Figure 3C and Supplemental Figure 5A). Wild-type ccm2 co-immunoprecipitated with both wild-type ccm3 and ccm3Δ proteins (Figure 3C and Supplemental Figure 5A), indicating that loss of exon 3 does not disrupt ccm2-ccm3 interaction. Both wild-type ccm3 and ccm3Δ proteins also co-immunoprecipitated with ccm2L197R (Figure 3C and Supplemental 5A), a ccm2 protein with a mutation in its phosphotyrosine binding (PTB) domain required for CCM2 interaction with CCM1 (10, 17). Thus, ccm2-ccm3 interaction is independent of ccm1 and not affected by loss of the 18 amino acids encoded by exon 3. Coexpression of ccm1 and ccm2 enhanced the formation of ccm1/2/3 complexes incorporating both wild-type and ccm3Δ proteins (Figure 3C and Supplemental Figure 5A). These studies demonstrate that ccm3Δ proteins are expressed and interact with the known CCM signaling proteins in a wild-type manner.
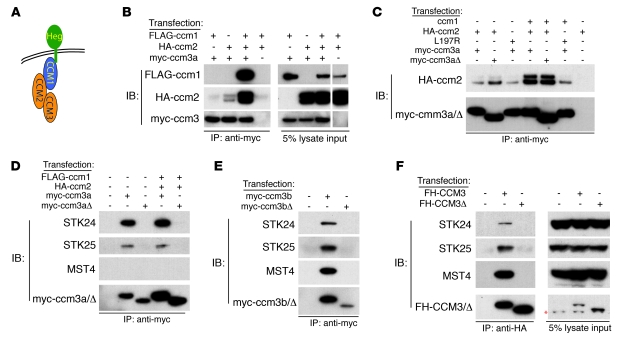
Figure 3. ccm3Δ proteins form a complex with ccm1 and ccm2 but fail to bind the GCK-III family of sterile 20–like kinases.
(A) A working model in which HEG receptors bind CCM1, CCM1 binds CCM2, and CCM2 binds CCM3 is shown. (B) Co-immunoprecipitation of a zebrafish ccm1/ccm2/ccm3 protein complex. FLAG-ccm1, HA-ccm2, and myc-ccm3 were coexpressed and ccm3 immunoprecipitations performed (left). Protein expression is shown by immunoblot analysis (right). The white line indicates the boundary of a noncontiguous lane run on the same gel. (C) Formation of the zebrafish ccm1/ccm2/ccm3a complex requires the ccm2 PTB domain but not the 18 amino acids encoded by ccm3 exon 3. ccm proteins were coexpressed and ccm3 immunoprecipitations performed. ccm2L197R contains a point mutation in the PTB domain that blocks interaction with ccm1 (10). ccm3aΔ proteins lack the 18 amino acids encoded by exon 3. (D) Zebrafish ccm3a interaction with STK24 and STK25 requires the 18 amino acids encoded by exon 3. ccm3 immunoprecipitations were performed as described above, and co-immunoprecipitated endogenous human STK24, STK25, and MST4 were detected with specific antibodies. (E) Zebrafish ccm3b interaction with STK24, STK25, and MST4 requires the 18 amino acids encoded by exon 3. (F) Human CCM3 interaction with STK24, STK25, and MST4 requires the 18 amino acids encoded by exon 5. CCM3 and CCM3Δ double-tagged with Flag and HA (FH) were expressed and CCM3-STK co-immunoprecipitation experiments performed as described for D and E. The red asterisk indicates a background band present in all lanes.
Two-hybrid screening and biochemical studies have recently identified the GCK-III family of sterile 20–like STKs MST4, STK24, and STK25 as potential CCM3 binding partners (24, 25). To test the ability of ccm3Δ to bind this family of proteins, myc-tagged wild-type ccm3 and ccm3Δ proteins were expressed in HEK293T cells and immunoprecipitated with anti-myc antibodies and co-immunoprecipitated endogenous MST4, STK24, and STK25 proteins were detected with specific antibodies. STK24 and STK25 co-immunoprecipitated with wild-type ccm3a, while MST4, STK24, and STK25 co-immunoprecipitated with wild-type ccm3b (Figure 3, D and E). In contrast to ccm3 interaction with ccm2, ccm3 interaction with MST4, STK24, and STK25 was not altered by coexpression of ccm1 and ccm2 (Figure 3D). However, MST4, STK24, and STK25 failed to co-immunoprecipitate with myc-tagged ccm3Δ proteins (Figure 3, D and E, and Supplemental Figure 5, B and C). MST4, STK24, and STK25 were also found to interact with human CCM3 proteins but not with the CCM3Δ protein identified as a cause of human CCM (Figure 3F). These findings demonstrate that CCM3Δ proteins couple normally to the known CCM signaling complex but are unable to bind the GCK-III family of STKs.
ccm3/STK signaling is required in the same pathway as heg, ccm1, and ccm2 during cardiovascular development in vivo.
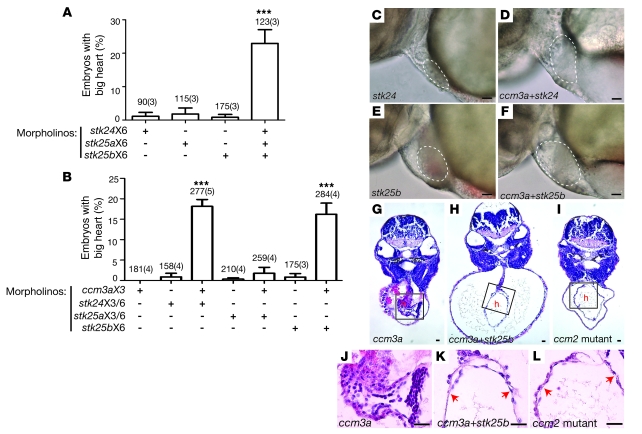
The finding that CCM3Δ proteins couple normally to the known CCM signaling components but not to STKs suggested that STKs might lie directly downstream of HEG, CCM1, CCM2, and CCM3 in a linear pathway required for cardiovascular CCM signaling. Analysis of the zebrafish genome identified two stk25 orthologs (stk25a and stk25b) and a single ortholog for both stk24 and mst4. The proteins encoded by these genes are highly conserved with their mammalian counterparts and highly homologous to each other (data not shown), suggesting that they may play redundant roles in vivo. To determine whether stks lie downstream of ccm3 in the ccm pathway in vivo, we therefore adopted a combinatorial deficiency strategy and tested the ability of morpholino knockdown of stks alone or stks plus ccm3a to confer the cardiovascular phenotype characteristic of heg, ccm1, and ccm2 deficiency (see Supplemental Figure 6). High-level deficiency of stks resulted in early embryonic lethality (prior to 12 hpf, data not shown). Low-level knockdown of any single stk gene did not confer the big heart phenotype characteristic of CCM signaling deficiency states (Figure 4, A, C, and E), but combined low-level knockdown of stk24, stk25a, and stk25b conferred the characteristic big heart phenotype in more than 20% of embryos tested (P < 0.001, Figure 4A and data not shown). Since loss of ccm3a alone does not confer the characteristic CCM deficiency phenotypes, to test functional interaction between ccm3 and the STKs in this pathway, we next combined knockdown of individual STKs with loss of ccm3a. Dilated hearts characteristic of CCM-deficient embryos were observed in a significant number (16%–18%) of zebrafish embryos injected with morpholinos to block ccm3a plus stk24 or ccm3a plus stk25b but not with ccm3a plus stk25a (P < 0.001, Figure 4, B, D, and F). Histologic analysis confirmed that dilatation resulted from severe thinning of the myocardial layer like that seen in ccm2 mutant hearts and did not merely reflect heart failure and pericardial edema (Figure 4, G–L). No phenotypes were observed in control embryos injected with identical concentrations of mismatch control morpholinos (data not shown). These findings demonstrate that STKs and ccm3/STK signaling play essential roles in the CCM cardiovascular signaling pathway in vivo.
Figure 4. ccm3 and GCK-III family STKs interact in the CCM pathway in vivo.
(A) STK deficiency confers the big heart phenotype characteristic of CCM deficiency in zebrafish embryos. Conferral of the big heart phenotype was scored following injection of low-dose morpholinos targeting stk24 (stk24X6, 3 ng/embryo), stk25a (stk25aX6, 3 ng/embryo), stk25b (stk25bX6, 3 ng/embryo) alone or a combination of all 3 morpholinos. Shown are mean and SEM. The number of embryos examined is indicated above each bar, and the number of separate injections performed with the indicated morpholinos is indicated in parentheses. ***P < 0.001 by Student’s t test. (B) ccm3 and STKs functionally interact in the CCM pathway in zebrafish embryos. Low-dose morpholinos against ccm3a (ccm3aX3, 3 ng/embryo) and stk24 (stk24X3+stk24X6, 2 ng/embryo each), stk25a (stk25aX3+stk25aX6, 2 ng/embryo each) or stk25b (stk25bX6, 2 ng/embryo) were injected separately or in combination into wild-type embryos and the presence of the big heart phenotype characteristic of CCM signaling deficiency was scored at 48 hpf. ***P < 0.001 by Student’s t test. (C–F) Representative light microscopic images of the big heart phenotypes conferred by the morpholinos described in B. The atrial margins are outlined by white dashed lines. (G–I) Representative histologic cross sections of 48-hpf morphant (ccm3aX3 and ccm3aX3+stk25bX6) and mutant (ccm2) embryos stained with hematoxylin and eosin. h, heart. (J–L) High-power images of the boxed regions of hearts shown in G–I. Scale bars: 20 μm.
CCM3 and STKs regulate endothelial cell junctions.
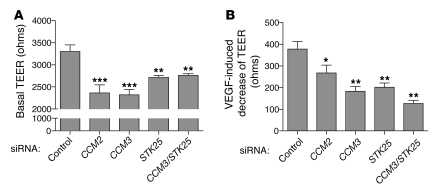
Genetic studies in mice and zebrafish support a role for CCM signaling in regulating cardiovascular development and function through effects on endothelial cell junctions (10, 13, 21). Endothelial cells lacking CCM2 have previously been shown to exhibit a loss of barrier function (13), providing functional evidence for regulation of endothelial junctions by this pathway. To determine whether CCM3 and STKs are also required in endothelial cells to regulate barrier function, we measured the electrical resistance across monolayers of primary human microvascular endothelial cells (HMVECs) transfected with control siRNA and siRNAs targeting CCM2, CCM3, and STK25 (the dominant GCK-III STK in these cells). Transfection of siRNA resulted in a 60%–80% reduction in the expression of CCM2, CCM3, and STK25 (Supplemental Figure 7). As previously reported, CCM2 deficiency resulted in a significant drop in basal resistance (Figure 5A). Significantly, knockdown of CCM3 and/or STK25 resulted in an equivalent drop in basal endothelial cell resistance (Figure 5A). Consistent with reduced basal barrier function, HMVECs lacking CCM2, CCM3, STK25, or CCM3 and STK25 also exhibited significantly smaller responses to VEGF, an agent known to loosen endothelial junctions and reduce basal endothelial barrier function (Figure 5B). These studies demonstrate that CCM3 and STK25, like CCM2, are required in endothelial cells for normal barrier function.
Figure 5. CCM3 and STKs regulate endothelial junctions.
(A) Basal TEER was measured in HMVEC monolayers following treatment with siRNAs targeting the indicated genes. (B) The change in TEER in response to VEGF was measured in HMVECs treated with the indicated siRNAs. n = 12. ***P < 0.001, **P < 0.01, *P < 0.05 by 1-way ANOVA and Bonferroni’s correction. Data shown are representative of 4 independent experiments.
STKs regulate endothelial cell actin and Rho activity like CCM proteins.
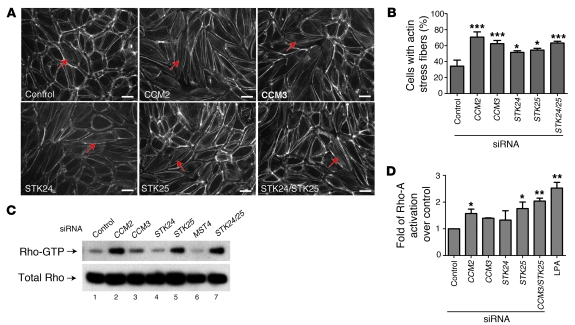
Recent studies of endothelial cells lacking CCM1 or CCM2 (10, 13, 21) have demonstrated an increased formation of actin stress fibers and an increase in the activity of Rho, a small GTPase known to negatively regulate both endothelial and epithelial cell junctions (26–29). We therefore next measured the effect of loss of CCM3 and the STKs on stress fiber formation and Rho activity. Deficiency of CCM2, CCM3, STK24, and STK25 conferred similar increases in cellular stress fiber formation, measured using Alexa Fluor 568–conjugated phalloidin to identify F-actin in the central part of the cell (Figure 6, A and B). Consistent with previous studies of CCM1 and CCM2 deficiency in endothelial cells (13, 29), we observed a significant upregulation of Rho-GTP levels in endothelial cells lacking CCM2, CCM3, or STK25, with no change in total cellular Rho levels (Figure 6C). The level of Rho-A activation observed with knockdown of CCM3 and/or STK25 was similar to that of endothelial cells treated with lysophosphatidic acid (LPA), a potent negative regulator of endothelial cell junctions (30) (Figure 6D). These findings indicate that STKs regulate endothelial cell function in vitro in a manner similar to the known CCM proteins and suggest that negative regulation of Rho activity is an important downstream effect of CCM/STK signaling in endothelial cells.
Figure 6. STK deficiency, like CCM deficiency, results in actin stress fiber formation and elevated Rho-A activity in human endothelial cells.
(A) Phalloidin staining of actin stress fibers in HUVECs treated with control siRNA and siRNAs targeting CCM2, CCM3, STK24, STK25, or STK24+STK25 is shown. (B) The percentage of cells with central actin stress fibers following the siRNA treatments described in A is shown. Shown are mean and SEM. n = 6; *P < 0.05, ***P < 0.0001 by Student’s t test. (C) Upregulation of Rho-A activity is associated with deficiency of CCM proteins or STKs. Rho-A–GTP levels were measured in endothelial cells treated with the indicated siRNAs (top bands) and compared with total cellular Rho-A (bottom bands). (D) Fold change in Rho-A activation following treatment of HUVECs with siRNAs targeting the indicated genes or with LPA (10 μM, 30 minutes). Shown are mean and SEM. n = 3. *P < 0.05, **P < 0.01 by Student’s t test. Scale bars: 20 μm.
STK25 and STK24 directly phosphorylate and activate moesin, a negative regulator of Rho.
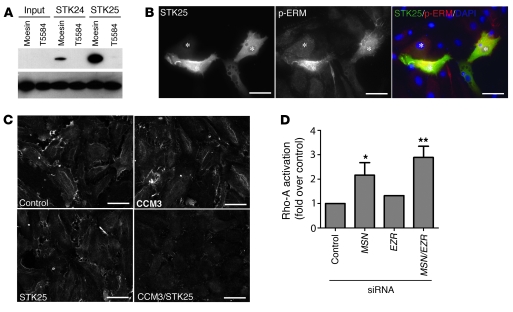
The finding that loss of STK signaling results in elevated levels of activated Rho suggested that regulation of Rho might be a primary mechanism by which this pathway controls endothelial cell junctions. Genetic studies in Drosophila have demonstrated that Slik, an STK homologous to STK24 and STK25, regulates epithelial junctions by activating moesin and downregulating Rho activity (26, 31). More recently, MST4 has been shown to directly activate ezrin, a member of the ezrin-radixin-moesin (ERM) protein family known to negatively regulate Rho (32). These studies suggested that STKs might regulate endothelial Rho and cell junctions through phosphorylation of moesin at T558, its known activation site (33, 34). To determine whether moesin is a direct substrate of STK24 and STK25, we performed in vitro kinase assays using purified STKs and HA-moesin and tested for phosphorylation at T558 using a phospho-specific monoclonal antibody. STK25 and, to a lesser extent, STK24 directly phosphorylated moesin at T558 (Figure 7A). To test the ability of STK25 to activate moesin in endothelial cells, bovine aortic endothelial cells (BAECs) were transfected with FLAG-tagged STK25, and phospho-T558 moesin was examined. Endothelial cells that overexpressed STK25 exhibited a marked increase in phospho-T558 moesin staining (Figure 7B). Conversely, HMVECs treated with siRNA to reduce the levels of CCM3 and/or STK25 exhibited reduced levels of phospho-T558 moesin at cell borders (Figure 7C), although total cellular phospho-T558 moesin was not significantly changed (data not shown). Genetic studies have demonstrated that reduced moesin activity is associated with increased Rho activation in epithelial cells, but cell studies have also demonstrated that Rho can phosphorylate ERM proteins at their activation site threonine (35). Thus, elevated phospho-T558 moesin could be upstream or downstream of activated Rho in endothelial cells. To determine whether elevated Rho activation is likely to result from loss of moesin activity in endothelial cells, we measured activated Rho in HMVECs treated with control siRNA or siRNA directed against moesin and/or ezrin. Knockdown of these ERM proteins raised the levels of activated Rho to approximately the same extent as did loss of CCM/STK signaling (Figure 7D). These findings demonstrate that STKs directly activate moesin and that loss of moesin increases Rho activity in endothelial cells.
Figure 7. STK24 and STK25 directly activate moesin, and loss of moesin increases Rho-A activity in endothelial cells.
(A) In vitro kinase assays were performed using purified STK24 or STK25 and HA-moesin or HA-moesin T558A and phosphorylation at the moesin T558 detected using an antibody against phospho-moesin (top panel). The total moesin protein used in the reactions was detected using anti-moesin antibody (bottom panel). (B) Expression of STK25 raises endothelial phospho-T558 moesin levels. BAECs transfected with plasmids to drive expression of FLAG-STK25 were stained with anti-FLAG (Alexa Fluor 488) and anti–phospho-T558 moesin (Alexa Fluor 594) antibodies as well as DAPI to detect cell nuclei. (C) Inhibition of CCM3 and STK25 reduces phospho-T558 moesin levels. siRNA was used to knock down CCM3 and/or STK25 in HMVECs and phospho-T558 moesin detected using immunofluorescence. (D) Loss of moesin increases activated Rho-A levels. siRNA was used to reduce expression of moesin (MSN) and/or ezrin (EZR) in HMVECs and activated Rho-A measured as in Figure 5D. Shown are mean and SEM. n = 2. *P < 0.05, **P < 0.01 by Student’s t test. Scale bars: 20 μm.
Discussion
Human genetic studies have identified CCM3 as a CCM disease gene, and biochemical studies have placed CCM3 downstream of CCM2 in the CCM signaling pathway, but how CCM3 signaling regulates endothelial function during cardiovascular development and disease is unknown. In the present study, we have used zebrafish to demonstrate that ccm3 plays an essential role in the pathway by which heg, ccm1, and ccm2 regulate heart and vessel formation. Investigation of the mechanism by which a CCM3 deletion mutant confers both human CCM and zebrafish cardiovascular phenotypes has identified the GCK-III family of sterile 20–like kinases including STK24 and STK25 as essential downstream effectors of CCM signaling. We find that CCM3 and STK25 control endothelial cell barrier function and that STK24 and STK25 directly activate moesin, a known regulator of Rho and epithelial cell junctions. These studies identify a molecular pathway from the cell membrane to regulation of Rho and endothelial cell junctions that begins to explain how HEG/CCM signaling participates in cardiovascular development and human vascular disease.
CCM3 has been identified as a cause of familial CCM by positional cloning studies (6, 7), but human genetic studies have revealed that CCM3 mutations are a relatively rare cause of the disease (36, 37), and forward genetic screens for cardiovascular mutants in zebrafish have failed to identify CCM3 as a regulator of cardiovascular development. Thus, whether CCM3 plays a central role in this vascular signaling pathway similar to those of CCM1 and CCM2 has been unclear. Our studies demonstrate that ccm3 plays an essential role in zebrafish cardiovascular development in the pathway previously defined for heg, ccm1, and ccm2.
The most significant role for CCM3 identified by these studies is the finding that CCM3 coupling to the GCK-III family of STKs is an essential component of cardiovascular CCM signaling. This finding is supported by conferral of the big heart and interrupted circulation phenotypes characteristic of heg, ccm1, and ccm2 deficiency following expression of ccm3Δ, a mutant ccm3 protein that binds normally to heg, ccm1, and ccm2 but not to STKs, as well as by STK and combined STK/ccm3 deficiency states. The ccm3Δ protein is equivalent to a mutant human CCM3 protein that is expressed by 3 members of a family with CCM (pedigree C043 in ref. 6), indicating that the molecular requirement for CCM3-STK interaction is conserved across species. These findings and other recent studies (38) suggest that the functional role of ccm3 during cardiovascular development in zebrafish parallels its role during human CCM pathogenesis and that CCM3/STK signaling is an essential downstream effector of this pathway in the cardiovascular system. The observation that loss of CCM3 or STK25 confers an endothelial cell barrier defect like that observed with loss of CCM2 is consistent with a mechanism in which CCM1, CCM2, CCM3, and STKs function in a linear, endothelial cell–autonomous pathway. However, future genetic studies directly testing the role of CCM3 and the STKs in endothelial cells in vivo are required to definitively test such a mechanism.
How does loss of CCM3/STK signaling confer cardiovascular defects during development and give rise to human CCMs? Recent studies have revealed that CCM1 is localized to endothelial junctions and that fish, mice, and humans deficient in the known CCM genes exhibit remarkably similar defects in endothelial junctions (10, 21). Loss of CCM1 or CCM2 in cultured endothelial cells results in destabilized junctions in association with an increase in cellular actin stress fiber formation and elevated Rho activity (13, 21), characteristics that are also conferred by loss of STK function. The biological functions of STK24 and STK25 are unknown, but genetic studies in Drosophila have shown that the kinase activity of slik, a homologous STK, is required to activate moesin and negatively regulate Rho in epithelial cells of the developing fly (26, 31, 39). Epithelial cells in flies lacking slik or moesin exhibit defective junctions and increased actin stress fibers, phenotypes similar to those of endothelial cells lacking CCM signaling that can be compensated for by reduced Rho (26, 31). MST4, the third member of the GCK-III STK family that includes STK24 and STK25, has recently been shown to directly activate the ERM protein ezrin (32), and we find that STK24 and STK25 directly activate moesin in a similar manner. Thus, CCM3/STK signaling may feed into a common pathway of STK/ERM/Rho signaling by which both epithelial and endothelial cell junctions are regulated. Future genetic and cellular studies of the GCK-III family of STKs will be required to determine whether STK regulation of Rho is a general mechanism of controlling cell junctions in these tube-forming cell types. Such studies will also be required to determine whether any of the phenotypes observed with loss of CCM3 or STK24/25 in fish and cells are attributable to signaling pathways independent of CCM1 and CCM2.
Precisely how loss of CCM signaling gives rise to dilated, fragile vascular malformations is not yet understood, and the need for an animal model to address this question and develop new approaches to disease therapy is pressing. This study and others suggest that investigation of the CCM pathway in zebrafish can provide essential clues. CCM1 and CCM2 deficiency states led to common phenotypes in zebrafish and mice, including defective endothelial junctions and a failure of branchial arch artery formation (10–13), consistent with a highly conserved role for this pathway in vertebrate cardiovascular development and function. CCM1 binds RAP1 (21, 40, 41), and studies of CCM1 function in endothelial cells suggest that it may be regulated by RAP1 at cell junctions (21). In the fish, this functional relationship is revealed by synergistic deficiency phenotypes, including the dilated heart characteristic of CCM deficiency and cerebral hemorrhage (42), that arise with low-level deficiency of both ccm1 and rap1b. Similar sensitive combinatorial phenotypes are observed with low-level knockdown of heg, ccm1, and ccm2 (8) as well as ccm3 and the STK genes. In addition, specific disease-causing CCM mutations such as CCM2L197R and CCM3Δ confer full deficiency phenotypes in fish (10). These observations suggest that future studies in zebrafish will provide a means of better understanding the role of this pathway in human CCM and perhaps even a means of testing the efficacy of new drugs designed to treat the disease.
Methods
Zebrafish studies.
Tg (fli1a:EGFP)y1 and ccm2hi296a mutant zebrafish were obtained from the Zebrafish International Resource Center (ZIRC). The Tg (i-fabp:GFP) cardiac reporter zebrafish were created by a transposon-based gene trap approach using the 192-bp zebrafish I-FABP promoter (43). Morpholino oligonucleotides were obtained from Gene Tools and were injected into the yolk of one-cell-stage embryos at the indicated dosages and combinations. To rescue the big heart phenotype conferred by ccm3 morpholinos, 100 pg of cRNA encoding ccm3a or ccm3b was coinjected with the indicated morpholino oligonucleotides. Zebrafish microangiography was performed by injecting red fluorescent (580/605 nm) 0.02 μm carboxylate-modified FluoSphere beads (Invitrogen) or 10 mg ml–1 70-kDa FITC-dextran (Sigma-Aldrich) into the sinus venosus of 48-hpf zebrafish embryos. Embryos were mounted laterally in 2% methylcellulose, and the images were acquired using an Olympus MVX10 microscope. Animal studies were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Morpholino and RT-PCR sequences are available in Supplemental Methods.
Microscopy.
Live embryos were mounted in 2% methylcellulose and the bright-field images acquired using a Nikon Eclipse 80i microscope equipped with a Nikon DXM1200F camera. Flourescence images of the heart were acquired using an Olympus MVX10 microscope. Images of hematoxylin/eosin staining of paraffin-embedded zebrafish sections were acquired using a Nikon Eclipse 80i microscope.
Whole mount in situ hybridization.
ccm3a and ccm3b antisense digoxigenin-labeled in situ probes were synthesized using the DIG RNA Labeling Kit (Roche), and digoxigenin whole mount in situ hybridization was performed at 70°C overnight following 3–5 hours prehybridization using embryos fixed with 4% paraformaldehyde. Photographs of stained embryos were captured with an Olympus MVX10 microscope. To ensure ccm3a and ccm3b specific hybridization, nonhomologous 3′ UTR regions were chosen as templates for probe synthesis.
Immunoprecipitation and immunoblotting.
Myc-tagged zebrafish ccm3a, ccm3b, ccm3aΔ, and ccm3bΔ were subcloned into the pcDNA3 vector. HA-tagged zebrafish ccm2 and ccm2 L197R were subcloned in the pCS2+ vector. FLAG-tagged zebrafish ccm1 was subcloned into the pME18S-FL3 vector. Epitope-tagged human CCM3 and CCM3Δ were subcloned into the pOZ-N vector. For co-immunoprecipitation studies, epitope-tagged proteins were transfected into HEK293T cells. After 48 hours, cells were incubated for 10 minutes in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 25 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, pH 8, containing PhosStop [Roche], and Complete protease inhibitor [Roche]), drawn through a 25-gauge needle, and cleared by centrifugation. FLAG immunoprecipitations were performed with 2 μg ml–1 M2 antibody to FLAG (Sigma-Aldrich). Proteins were detected using the following antibodies: M2 mouse monoclonal antibody to FLAG, 1:500 (Sigma-Aldrich); mouse monoclonal antibody to HA, 1:2,000 (Abcam); HRP-conjugated goat antibody to mouse IgG, 1:5,000 (Jackson ImmunoResearch Laboratories Inc.). STK proteins were detected using rabbit monoclonal anti-STK24 and anti-MST4 antibodies (1:2,000 dilution, Epitomics) and goat polyclonal anti-STK25 antibodies (1:500 dilution, Santa Cruz Biotechnology Inc.). For affinity matrix pulldowns, HEK293T and CHO cells transfected with the indicated protein-expressing plasmids were incubated for 10 minutes in lysis buffer, and 350 mg lysate was incubated with 10 mg immobilized HEG1 tail overnight. All steps were performed at 4°C.
Measurement of trans-endothelial electrical resistance (TEER).
Human dermal microvascular endothelial cell (HDMEC) barrier function was assayed by measuring the resistance of a cell-covered electrode by using an ECIS instrument (Applied BioPhysic). 8W10E plates were incubated for 15 minutes with l-cysteine (10 mM) solution, followed by gelatin 0.1% for 30 minutes at room temperature. Cells were plated on the electrode at 2 × 104 cells/well. The day after being plated, the cells were transfected with siRNA for CCM2, CCM3, STK25, and CCM3/STK25 or control siRNA at a final concentration of 20 nM. HMVECs were cultured for 62 hours before quiescing in 0.15% FBS in EBM-2 for 10 hours and then stimulated with 100 ng/ml VEGF (Genentech).
Measurement of activated Rho-A.
Measurement of activated Rho-A was performed using a Rho Activation Assay Biochem Kit or G-Lisa Rho Activation Assay Biochem Kit (Cytoskeleton). HMVECs were grown on gelatin-coated plates and transfected with 8 nM control siRNA and siRNA directed against CCM2, CCM3, STK24, STK25, or MST4 using siPORT Amine Transfection Agent (Ambion). Forty-eight hours after transfection, cells were lysed and the lysates were cleared by centrifugation. For the assay using the Rho Activation Assay Biochem Kit, lysates were incubated with Rhotekin-RBD protein–coated beads. The bound Rho-GTP was eluted and detected with anti–Rho-A antibody using immunoblot analysis. For the assay using G-Lisa Rho Activation Assay Biochem Kit, the lysates were incubated in the G-Lisa Plate, and the activated Rho-A levels were measured according to the manufacturer’s instructions.
Endothelial stress fiber formation.
HMVECs were plated on fibronectin-coated 4-well chamber slides and transfected with 8 nM siRNA directed against CCM2, CCM3, STK24, or STK25. Fifty hours after transfection the cells were fixed in 4% PFA, permeabilized with 1% NP-40, stained with Alexa Fluor 568–conjugated phalloidin, and mounted with DAPI-containing mounting buffer (Invitrogen). Images of 5–6 selected fields were analyzed, and the percentage of cells with central actin stress fibers in each image was calculated.
In vitro kinase assays.
HEK293 cells were transfected with plasmids encoding HA-tagged moesin or mutant moesin (T558A). HA-moesin and HA-T558A were precipitated with anti-HA antibodies and protein G beads (Pierce). The anti-HA antibody–protein G beads complex was suspended in 40 μl kinase assay buffer (12.5 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM EGTA, 0.5 mM Na3VO4, 2.5 mM DTT, 0.01% Triton X-100, 200 μM ATP) and incubated with 200 ng purified GST-STK24 or GST-STK25 (Invitrogen) for 10 minutes at 30°C with occasional gentle tapping. SDS sample buffer was added to stop the reaction, followed by boiling for 5 minutes, and the samples were subjected to Western blot analysis.
Immunocytochemistry.
HMVECs were grown in fibronectin-coated chamber slides and transfected with siRNA against CCM3, STK25, or CCM3 and STK25. BAECs were transfected with FLAG-tagged STK25. Forty-eight hours after transfection cells were fixed with 4% PFA, permeabilized with 1% NP-40, and incubated with primary antibodies against p-ERM or FLAG followed Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibodies. The fluorescence images were captured using a Nikon Eclipse 80i microscope.
Statistics.
P values were calculated using an unpaired 2-tailed Student’s t test unless otherwise indicated. P values for TEER data were determined by 2-way ANOVA followed by Bonferroni’s t test for multiple comparisons or 1-way ANOVA when relevant using the GraphPad Prism software. The mean and SEM are shown in the bar graphs. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Patricia Mericko for help in acquiring reagents and the members of the Kahn laboratory for thoughtful discussions of this work. These studies were supported by NIH grants T32 HL07971 (to X. Zheng) and by R01 HL094326 (to M.L. Kahn).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(8):2795–2804. doi:10.1172/JCI39679.
References
- 1.Rigamonti D, et al. Cerebral cavernous malformations. Incidence and familial occurrence. N Engl J Med. 1988;319(6):343–347. doi: 10.1056/NEJM198808113190605. [DOI] [PubMed] [Google Scholar]
- 2.Sahoo T, et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1). Hum Mol Genet. 1999;8(12):2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 3.Eerola I, Plate KH, Spiegel R, Boon LM, Mulliken JB, Vikkula M. KRIT1 is mutated in hyperkeratotic cutaneous capillary-venous malformation associated with cerebral capillary malformation. Hum Mol Genet. 2000;9(9):1351–1355. doi: 10.1093/hmg/9.9.1351. [DOI] [PubMed] [Google Scholar]
- 4.Liquori CL, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73(6):1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denier C, et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74(2):326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergametti F, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76(1):42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guclu B, et al. Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery. 2005;57(5):1008–1013. doi: 10.1227/01.NEU.0000180811.56157.E1. [DOI] [PubMed] [Google Scholar]
- 8.Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13(24):2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 9.Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133(16):3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 10.Kleaveland B, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15(2):169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131(6):1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 12.Boulday G, et al. Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech. 2009;2(3–4):168–177. doi: 10.1242/dmm.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead KJ, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl S, et al. Novel CCM1, CCM2, and CCM3 mutations in patients with cerebral cavernous malformations: in-frame deletion in CCM2 prevents formation of a CCM1/CCM2/CCM3 protein complex. Hum Mutat. 2008;29(5):709–717. doi: 10.1002/humu.20712. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Rigamonti D, Dietz HC, Clatterbuck RE. Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;60(2):353–359. doi: 10.1227/01.NEU.0000249268.11074.83. [DOI] [PubMed] [Google Scholar]
- 16.Hilder TL, et al. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6(11):4343–4355. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 17.Zawistowski JS, et al. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14(17):2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 18.Wong JH, Awad IA, Kim JH. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46(6):1454–1459. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
- 19.Clatterbuck RE, Eberhart CG, Crain BJ, Rigamonti D. Ultrastructural and immunocytochemical evidence that an incompetent blood-brain barrier is related to the pathophysiology of cavernous malformations. J Neurol Neurosurg Psychiatry. 2001;71(2):188–192. doi: 10.1136/jnnp.71.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu J, Stoodley MA, Morgan MK, Storer KP. Ultrastructural characteristics of hemorrhagic, nonhemorrhagic, and recurrent cavernous malformations. . J Neurosurg. 2005;103(5):903–909. doi: 10.3171/jns.2005.103.5.0903. [DOI] [PubMed] [Google Scholar]
- 21.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179(2):247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busch CR, Heath DD, Hubberstey A. Sensitive genetic biomarkers for determining apoptosis in the brown bullhead (Ameiurus nebulosus). Gene. 2004;329:1–10. doi: 10.1016/j.gene.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 24.Ma X, et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18(6):1965–1978. doi: 10.1091/mbc.E06-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voss K, et al. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8(4):249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 26.Speck O, Hughes SC, Noren NK, Kulikauskas RM, Fehon RG. Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature. 2003;421(6918):83–87. doi: 10.1038/nature01295. [DOI] [PubMed] [Google Scholar]
- 27.Fujita Y, Braga V. Epithelial cell shape and Rho small GTPases. Novartis Found Symp. 2005;269:144–155. [PubMed] [Google Scholar]
- 28.Brady DC, Alan JK, Madigan JP, Fanning AS, Cox AD. The transforming Rho family GTPase Wrch-1 disrupts epithelial cell tight junctions and epithelial morphogenesis. Mol Cell Biol. 2009;29(4):1035–1049. doi: 10.1128/MCB.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207(4):881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulze C, Smales C, Rubin LL, Staddon JM. Lysophosphatidic acid increases tight junction permeability in cultured brain endothelial cells. J Neurochem. 1997;68(3):991–1000. doi: 10.1046/j.1471-4159.1997.68030991.x. [DOI] [PubMed] [Google Scholar]
- 31.Hipfner DR, Keller N, Cohen SM. Slik Sterile-20 kinase regulates Moesin activity to promote epithelial integrity during tissue growth. Genes Dev. 2004;18(18):2243–2248. doi: 10.1101/gad.303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ten Klooster JP, et al. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16(4):551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Huang L, Wong TY, Lin RC, Furthmayr H. Replacement of threonine 558, a critical site of phosphorylation of moesin in vivo, with aspartate activates F-actin binding of moesin. Regulation by conformational change. J Biol Chem. 1999;274(18):12803–12810. doi: 10.1074/jbc.274.18.12803. [DOI] [PubMed] [Google Scholar]
- 34.Simons PC, Pietromonaco SF, Reczek D, Bretscher A, Elias L. C-terminal threonine phosphorylation activates ERM proteins to link the cell’s cortical lipid bilayer to the cytoskeleton. Biochem Biophys Res Commun. 1998;253(3):561–565. doi: 10.1006/bbrc.1998.9823. [DOI] [PubMed] [Google Scholar]
- 35.Matsui T, Yonemura S, Tsukita S, Tsukita S. Activation of ERM proteins in vivo by Rho involves phosphatidyl-inositol 4-phosphate 5-kinase and not ROCK kinases. Curr Biol. 1999;9(21):1259–1262. doi: 10.1016/S0960-9822(99)80508-9. [DOI] [PubMed] [Google Scholar]
- 36.Verlaan DJ, Roussel J, Laurent SB, Elger CE, Siegel AM, Rouleau GA. CCM3 mutations are uncommon in cerebral cavernous malformations. Neurology. 2005;65(12):1982–1983. doi: 10.1212/01.wnl.0000188903.75144.49. [DOI] [PubMed] [Google Scholar]
- 37.Liquori CL, et al. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat. 2006;27(1):118. doi: 10.1002/humu.9389. [DOI] [PubMed] [Google Scholar]
- 38.Voss K, et al. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum Mutat. 2009;30(6):1003–1011. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- 39.Hughes SC, Fehon RG. Phosphorylation and activity of the tumor suppressor Merlin and the ERM protein Moesin are coordinately regulated by the Slik kinase. J Cell Biol. 2006;175(2):305–313. doi: 10.1083/jcb.200608009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene. 1997;15(9):1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 41.Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J Mol Biol. 2005;348(3):741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 42.Gore AV, Lampugnani MG, Dye L, Dejana E, Weinstein BM. Combinatorial interaction between CCM pathway genes precipitates hemorrhagic stroke. Dis Model Mech. 2008;1(4–5):275–281. doi: 10.1242/dmm.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Her GM, Chiang CC, Wu JL. Zebrafish intestinal fatty acid binding protein (I-FABP) gene promoter drives gut-specific expression in stable transgenic fish. Genesis. 2004;38(1):26–31. doi: 10.1002/gene.10248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.