Summary
Repetitive antigen-stimulation by prime-boost vaccination or pathogen re-encounter increases memory CD8+ T cell numbers, however the impact on memory CD8+ T cell differentiation is unknown. Here we showed that repetitive antigen-stimulations induced accumulation of memory CD8+ T cells with uniform effector memory characteristics. However, genome-wide microarray analyses revealed that each additional antigen-challenge resulted in the differential regulation of several hundred new genes in the ensuing memory CD8+ T cell populations and therefore in stepwise diversification of CD8+ T cell transcriptomes. Thus, primary and repeatedly stimulated (secondary, tertiary, quaternary) memory CD8+ T cells differ substantially in their molecular signature while sharing expression of a small group of genes and biological pathways, which may constitute a core signature of memory differentiation. These results provide new insight into the complex regulation of memory CD8+ T cell differentiation and identify a spectrum of potential new molecular targets to dissect the function of memory cells generated by repeated antigen-stimulation.
Introduction
After the resolution of an acute infection, memory CD8+ T cells protect from re-infection with the same or related intracellular pathogens (Butz and Bevan, 1998; Harty and Badovinac, 2008; Lefrancois, 2006; Wherry et al., 2003). A plethora of pathogens including malaria sporozoites, influenza and herpes viruses infect humans repeatedly and memory CD8+ T cells bear the potential to protect hosts from all of these infections (Appay et al., 2008; McGill et al., 2008; Schmidt et al., 2008). In addition, some viruses (EBV, CMV) cause persistent, life-long infections characterized by transient episodes of antigen expression (Reeves and Sinclair, 2008). In this setting, re-activated memory CD8+ T cells play an essential role in containing the viruses and preventing systemic dissemination. These examples demonstrate that CD8+ T cells are likely to encounter their cognate antigen more than once and highlight the need to understand the function of memory CD8+ T cells that have undergone multiple rounds of antigen stimulation.
In addition to their essential role in controlling acute infections, vaccine-stimulated CD8+ T cells hold great potential for the prevention of infectious diseases (Appay et al., 2008; Miller et al., 2008; Seder et al., 2008). After vaccination, the superior protective capacity of memory CD8 T cells is closely linked to their increased abundance in both lymphoid and non-lymphoid organs and as a consequence much effort has been devoted to identifying strategies that increase the absolute numbers of memory CD8+ T cells (Harty and Badovinac, 2008; Schmidt et al., 2008). Among these strategies, prime-boost regimens are often used due to their ability to elicit large numbers of memory CD8+ T cells (Badovinac et al., 2005; Woodland, 2004). The impact of this repeated antigen exposure on memory CD8+ T cell differentiation has not been addressed in detail and it is unclear whether repeatedly stimulated memory CD8+ T cells are similar to primary memory CD8+ T cells in terms of phenotype and function.
A few recent studies have attempted to close this knowledge gap by stimulating memory CD8+ T cells multiple time with heterologous infections expressing the same antigen (Jameson and Masopust, 2009; Masopust et al., 2006; Vezys et al., 2009). These studies show that multiple antigen encounters markedly impact memory CD8+ T cell lineage, phenotype and function. In the current study, we therefore sought to analyze the impact of multiple rounds of antigen stimulation on the differentiation of memory CD8+ T cells via whole-genome microarray. Using an adoptive transfer system and homologous infection to generate highly pure splenic memory CD8+ T cell populations with a precisely defined history of antigen encounters, we show that every additional antigen stimulation (primary to quaternary) leads to an increase in the number of differentially regulated genes and thus to further differentiation of memory CD8+ T cells. As a consequence of this continuous differentiation process, additional antigen stimulation results in memory CD8+ T cell populations that possess a unique repertoire of regulated genes and biological pathways.
Results
Experimental model
The analysis of memory CD8+ T cell populations after multiple rounds of antigen stimulation requires an approach allowing for the detection and isolation of highly pure populations with a defined number of antigen encounters. Repeated infection of a host with the same infectious agent is likely to result in: a) rapid clearance of the pathogen by memory B and T cells; b) incomplete recruitment of the existing memory CD8+ T cells; and c) recruitment of new naïve CD8+ T cells and therefore in the generation of heterogeneous memory CD8+ T cell populations. Furthermore, memory T cells extracted from distinct anatomical locations may differ in phenotype and function (Jameson and Masopust, 2009). Prime-boost regimens with heterologous infections bear the potential to efficiently boost large numbers of pre-existing memory CD8+ T cells (Vezys et al., 2009) but the employment of different pathogens may complicate direct comparison of the memory CD8+ T cell populations in terms of gene expression and function.
To circumvent these complications we employed an adoptive transfer approach to compare memory cell populations from the major secondary lymphoid organ, the spleen. Physiologic numbers (5×102) of naïve Thy1.1 OT-I CD8+ T cells (Badovinac et al., 2007) were transferred into Thy1.2 hosts, which were then challenged with an OVA257-expressing strain of Listeria monocytogenes (att LM-OVA) (Figure S1A). More than 70 days after the infection, 6×104 primary (1°) memory OT-I CD8+ T cells from the spleen were again adoptively transferred whereas another group of mice received naïve OT-I CD8+ T cells prior to LM-OVA challenge. These transfers were repeated until 1°, secondary (2°), tertiary (3°) and quaternary (4°) memory CD8+ T cell populations could be studied simultaneously. The use of low numbers of adoptively transferred naive and memory CD8+ T cells ensured that all OT-I CD8+ T cells detected after infection had expanded in response to LM-OVA infection while still permitting longitudinal analysis of each responding population.
Repeated stimulation generates memory CD8+ T cell populations with uniform effector memory phenotype and function
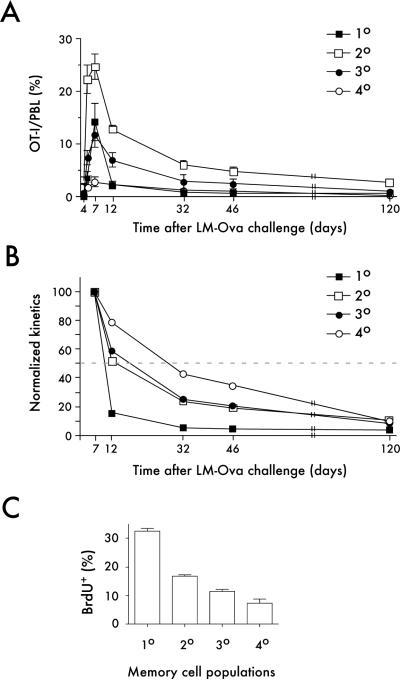
As previously described (Badovinac et al., 2007) att LM-OVA infection stimulated robust proliferative expansion of naïve OT-I T cells, which peaked at day 7, then underwent rapid contraction to memory numbers (Figure 1A,B). 1°, 2° and 3° memory cells also proliferated in response to att LM-OVA infection (Figure 1A). However, the stimulation history of these input memory cell populations had a clear impact on the magnitude of each response, which decreased as the number of antigen exposures increased (Figures 1A and S1B). Consistent with our previous report (Badovinac et al., 2003), 1° memory OT-I responding to antigen for the second time underwent delayed contraction compared to naïve OT-I undergoing a primary response and this delayed contraction was further exacerbated in 3° memory populations undergoing a quaternary response to infection (Figure 1B). However, the degree of contraction was similar for all populations when analyzed at a late memory time point (120 days, Figure 1B). Although our data did not permit analyses of individual cells, long-term survival of memory CD8+ T cell populations was apparently dependent on the number of antigen exposures (Figure S1C,D). Further analyses revealed substantial decreases in basal proliferation rates from 1° to 4° memory CD8+ T cells (Figure 1C), which may account for reduced survival of repeatedly stimulated memory populations. Thus, each additional restimulation results not only in a progressive decrease in proliferative capacity of the ensuing memory CD8+ T cell populations but also changes their rate of contraction and ability to survive.
Figure 1. Repeated antigen stimulation impacts memory CD8+ T cell kinetics and homeostatic proliferation.
Naïve (5×102) or memory (6×104) Thy1.1 OT-I T cells were adoptively transferred into naïve Thy1.2 C57Bl/6 recipient mice prior to infection with 5×106 att LM-OVA. A) OT-I T cell responses (% of OT-I T cells of all PBL) in peripheral blood. Data are mean +/− SD for 3–10 mice per group per time point. B) Percentages of OT-I T cells in PBL were normalized to the expansion peak at day 7 for each individual group. Data are mean +/− SD for 3–10 mice per group per time point. C) Proliferation of memory cells was determined by BrdU incorporation (day 70–84). Bar graphs represent the percentage of BrdU+ OT-I T cells in spleen (mean +/− SD, n=5).
Memory CD8+ T cells emerging after acute infections are currently classified into effector (Tem) or central (Tcm) lineages according to the surface expression of CD27, CD62L, CCR7 and their ability to secrete IL-2 after peptide stimulation (Hikono et al., 2007; Sallusto et al., 1999). To assess lineage commitment in repeatedly stimulated memory CD8+ T cells, we analyzed memory CD8+ T cell phenotype 90 days after infection. Surface expression of CD62L and CD27 decreased progressively from 1° to 4° memory CD8+ T cells (Figure 2A). All four memory CD8+ T cell groups efficiently produced IFNγ and TNFα in response to in vitro peptide stimulation, however, IL-2 production decreased substantially with the number of antigen exposures (Figure 2B). Consistent with their phenotype, repeatedly challenged memory CD8+ T cells were excluded from lymph nodes and accumulated in tertiary tissues (Figure 2C). These results extend previous reports that secondary and tertiary antigen challenges alter lineage commitment in favour of Tem cell populations (Jabbari and Harty, 2006; Masopust et al., 2006; Vezys et al., 2009). Importantly, this result was not a consequence of the adoptive transfer model because similar phenotypic changes (CD62Llo, KLRG-1hi) in OT-1 memory T cells were observed when repeated LM-OVA infections were performed in the same host (Figure S2A–D) and when memory CD8+ T cell phenotype was analyzed after heterologous infections in the same host (Masopust et al., 2006).
Figure 2. Repeated antigen stimulation impacts memory CD8+ T cell phenotype, function and tissue distribution.
A) Phenotype of OT-I T cells in the spleen 90 days post infection. Numbers inside dot plots represent the percentage of cells positive for the indicated molecules. B) Peptide-stimulated intracellular cytokine staining (ICS) of OT-I T cells from spleen. Numbers represent the percentage of OT-I T cells that stain positive for a given cytokine combination. C) OT-I T cell frequency in total CD8+ T cells in tissues was normalized to the frequency in PBL. Data are mean +/− SD for 3 mice per group.
Direct comparison of Tem CD8+ T cell populations revealed that CD62Llo 1° and 4° memory CD8 T cells differed substantially for surface expression of a number of markers (Figure S3A,B). This result, although limited by available antibodies, suggests the intriguing possibility that each round of Ag-stimulation further increases the complexity of memory CD8+ T cell populations in a fashion that may supersede current Tem and Tcm cell classifications.
Repetitively stimulated memory CD8+ T cells express a transcriptional `memory signature'
The increase in the frequency of Tem cell and further changes in the phenotype of Tem cell with each antigen stimulation suggested that similar gradual changes would occur in the transcriptome of the memory CD8+ T cell groups. To address this, we used flow cytometry to sort populations of 1°, 2°, 3° and 4° splenic memory OT-I T cells to high purity (>99%), performed genome-wide microarrays and normalized mRNA expression to that of naïve OT-I T cells. Using a stringent cut-off (False Discovery Rate (FDR) q-value <0.01) we detected 1528 genes with marked changes in expression in at least one of the four memory CD8+ T cell groups (Table S1). In 272 genes, changes in mRNA quantity were substantial in all four memory cell groups. Among this subset, analysis of gene expression patterns revealed two groups of genes whose transcription increased (Pattern I, n=75) or decreased (Pattern II, n=19) steadily with each additional antigen encounter (`dynamic memory signature genes' - see Figure 3A,B for representative examples and Table S2 for the complete list of genes). Molecules with increasing mRNA expression included transcription factors, apoptosis regulators and chemokine receptors (e.g. Anxa1, CCR5). In addition, these groups encompassed the effector molecules Granzyme B and IFN-γ (Pattern I) and the chemokine receptor CCR7 (Pattern II), which are known to be essential for the anti-microbial properties and trafficking patterns of Tem cells (Chtanova et al., 2005; Wherry et al., 2003). Thus, this group of markers represents a `dynamic memory signature' consisting of genes whose expression patterns parallel the observed phenotypic changes in memory CD8+ T cells generated by repeated antigen stimulation.
Figure 3. Dynamic and stable gene expression in repetitively stimulated memory CD8+ T cells.
Genome-wide mRNA expression in memory OT-I T cells was analyzed via microarray and normalized to naive OT-I T cells. An FDR q-value <0.01 defined significant changes. Genes with significant mRNA changes in all memory OT-I T cell populations (1° to 4°) were grouped according to gene expression patterns: A) transcriptional upregulation with consistent increase (pattern I), B) transcriptional downregulation with consistent decrease (pattern II), C) transcriptional upregulation with no consistent increase (pattern III), or D) transcriptional downregulation with no consistent decrease (pattern IV) from 1° to 4° memory. E) Biological pathway analysis of genes with significant mRNA changes in all memory CD8+ T cell populations was generated using KEGG pathway tool in DAVID bioinformatics resources. Arrows indicate whether genes are upregulated (↑) or downregulated (↓) in all memory CD8+ T cell populations when compared to naïve cells.
However, further analysis of gene expression patterns revealed additional groups of genes that were differentially expressed from naïve OT-I but maintained stable increased (Pattern III) or decreased (Pattern IV) mRNA levels from 1° to 4° memory (`stable memory signature genes' - Figure 3C, D and Table S2). This subset of regulated genes includes essential transcription factors (e.g. T-bet and Blimp-1, Pattern III), the trafficking molecules CD29 (Pattern III), CD103 (Pattern IV) and established memory markers (e.g. Ly6C, Pattern III).
In addition to functional annotation and expression pattern analysis of individual genes we used DAVID bioinformatics resources (Huang da et al., 2009) and KEGG pathway analysis (Kanehisa et al., 2006) of the memory signature genes ((Patterns I–IV), Table S2). These analyses reveal multiple pathways that were markedly altered in all (1° to 4°) memory CD8+ T cell populations compared to naïve cells (examples presented in Figure 3E). Despite the difference in stimulation history all memory CD8+ T cells show substantial changes in their ability to produce and respond to cytokine and chemokine-mediated signals (Figure 3E “cytokine-cytokine receptor interaction” and “JAK-STAT signaling” pathways) and general ability to traffic (“regulation of cytokine cytoskeleton” pathway) when compared to naïve cells. Finally, 14 genes with marked alterations in memory CD8+ T cells (including perforin, Granzyme B and IFN-γ transcripts) are part of a “NK-cell mediated cytotoxicity” pathway that further distinguishes all memory CD8+ T cells from their naïve counterparts (Figure 3E).
Taken together, these data suggest that some genes and biological pathways maintain the transcriptional changes observed in primary memory CD8+ T cells even under conditions of repetitive antigen stimulations. Importantly, the sustained expression pattern of genes in group III and IV suggests the possibility that these genes constitute a stimulation history independent, `stable memory signature', whose expression is also independent of the phenotypic changes associated with Tem and Tcm cell memory lineage commitment. As such, this data subset likely contains target genes that regulate shared and/or essential biological attributes of memory CD8+ T cell populations.
Repeated antigen stimulations expand the number of differentially regulated genes
The changes in mRNA expression of individual genes prompted us to test whether additional antigen stimulations also led to an increase in the absolute number of genes with significant (FDR q-value <0.01) transcriptional changes. Surprisingly, we detected multiple genes for which the changes in transcriptional regulation required two (Figure 4A,B), three (Figure 4C,D) or even four (Figure 4E,F) antigen encounters to reach statistical significance (Table S3 - complete list). It seems likely that at least some of the newly regulated genes will play important roles in determining the function of re-stimulated memory populations.
Figure 4. New genes with significant transcriptional regulation appear in memory populations after each antigen encounter.
Genes with the following expression patterns were identified: A) transcriptional upregulation or B) downregulation in 2°, 3° and 4° memory, C) transcriptional upregulation or D) downregulation in 3° and 4° memory, E) transcriptional upregulation or F) downregulation in 4° memory, G) transcriptional upregulation in 1° memory followed by decreasing mRNA expression from 1° to 4° memory. H) Protein expression of CD11b and CD11c on memory CD8+ T cells 90 days after infection (see Figure 3A+G for mRNA expression). Representative histograms are shown.
Finally, our screens identified a small group of genes whose mRNA expression increased in 1° memory CD8+ T cells but then decreased with every subsequent antigen challenge (Figure 4G and Table S3). Interestingly, this group included genes with essential roles in basal proliferation of memory CD8+ T cells (e.g. CD122) and adhesion or trafficking (ex. CD11c, CD44). These data could explain some of the functional differences between primary and repeatedly stimulated memory CD8+ T cells. These results also show that markers that are commonly used to identify “memory” CD8+ T cell populations (ex. CD44hi, CD122hi) may be compromised by repetitive antigen stimulations.
To validate that differences in gene transcription lead to similar alterations in protein quantity, surface protein expression was analyzed for a number of markers. As shown for the integrin family (Figures 4H and S5) mRNA and protein expression correlated well for most markers analyzed. Therefore, the observed changes in the transcriptome are likely to translate into differences in protein expression and thus impact memory CD8+ T cell function. These data suggest that markers identified in our screen may aid in evaluating the complexity of memory populations after multiple antigen encounters. For example, 1° memory CD8+ T cells should be CD11blo CD11chi whereas 4° memory populations will be CD11bhi CD11clo (Fig. 4H)
Repeated antigen challenges induce a stepwise diversification of memory CD8+ T cell transcriptomes
To quantify the changes in gene transcription induced by multiple antigen challenges, we next assessed the number of differentially regulated genes for each memory CD8+ T cell population. Similar to previous reports (Kaech et al., 2002; Wherry et al., 2007), we found that 1° memory CD8 T cells differentially regulated 364 genes compared to naive CD8+ T cells (Figure 5A). However, each additional antigen stimulation led to a progressive increase in the number of genes with marked mRNA changes compared to naïve CD8+ T cells, reaching 650 in 2°, 930 in 3° and 1322 in 4° memory CD8+ T cells. To compare the transcriptomes of individual memory CD8+ T cell populations we next depicted the gene expression profiles as Venn diagrams (Figure 5B). Of the 364 genes with significant changes in mRNA quantity in 1° memory CD8+ T cells, 317 were again significantly (FDR q-value <0.01) altered in 2° memory CD8+ T cells. A pool of genes with similar numbers (~300) was shared between 1° and 3° as well as 1° and 4° memory CD8+ T cells.
Figure 5. Repetitive antigen stimulation impacts global gene transcription in memory CD8+ T cell populations.
A) Genome-wide gene transcription in 1°, 2°, 3°, and 4° memory OT-I T cell (day 85–95 p.i.) populations was analyzed via microarray and compared to naïve OT-I T cells. An FDR q-value <= 0.01 defined statistically significance. Data represent the number of genes with significant regulation inside each group. B) Venn diagram presentation of genes with significant mRNA changes in indicated memory CD8+ T cell populations. Overlapping circles represent number of genes whose expression is different from naïve cells but is either shared or unique to indicated memory CD8+ T cell groups. C) Significantly altered genes were assessed for all four memory groups individually. Each memory group is represented by an individual ellipse, overlapping ellipses indicate genes shared by two or more groups.
In addition to this group of genes whose expression was preserved in all memory CD8+ T cell populations, 2° memory CD8+ T cells up- or downregulated 333 genes whose expression had not been significantly altered in 1° memory CD8+ T cells. Similarly, the number of genes with statistically significant mRNA changes increased by 412 in 3° compared to 2° memory CD8+ T cells and by 478 in 4° compared to 3° memory CD8+ T cells. Thus, each ensuing memory CD8+ T cell population differs from its predecessor by regulation of as many new genes as those distinguishing naïve from 1° memory populations. Few genes were exclusively altered in a single memory CD8+ T cell population (Figure 5C). Strikingly, > 200 genes required a second, > 300 a third and > 400 even a fourth antigen challenge to reach changes in mRNA expression that were significantly (FDR q-value <0.01) different from naïve CD8+ T cells.
These results show that the transcriptional profile of repeatedly stimulated memory CD8+ T cell populations encompasses nearly all genes that were significantly altered in the preceding memory CD8+ T cell population. However, each subsequent antigen encounter additionally leads to stepwise and substantial increases in the number of differentially regulated genes. Therefore, repeated antigen challenges broaden the spectrum of differentially expressed genes, resulting in stepwise increases in transcriptome diversity of the ensuing memory CD8+ T cell populations.
Repeated antigen stimulation differentially regulates gene expression in multiple gene families
To determine whether repeated antigen stimulation impacts gene families with known functions in T cell biology, we next used DAVID bioinformatics resource to assign biological functions to genes with significant (FDR q-value <0.01) transcriptional regulation in any of the four memory CD8+ T cell groups (1528 genes described in Table S1). More than 700 out of 1528 genes had known biological functions and were grouped into several function-related classes (Table 1). This functional annotation revealed that repetitive antigen stimulation influenced mRNA for genes involved in T cell trafficking, effector functions, regulation of transcription, lipid and protein metabolism, cell cycle regulation, intracellular signal transduction and many more.
Table 1. Repeated antigen exposure alters expression of multiple gene families in memory CD8+ T cell populations.
Biological functions were assigned to genes with significant changes in transcription in at least one of the four memory CD8+ T cell populations. To reduce the number of genes displayed, only genes with an mRNA increase >2-fold in at least two memory CD8+ T cell populations or an mRNA increase >3-fold in at least one memory CD8+ T cell population are shown. Genes were grouped according to their biological function and put in alphabetical order. Non-significant changes for a comparison are blank.
| Gene | Alias | prim | sec | tert | quart |
|---|---|---|---|---|---|
| Transcription | |||||
| Aff3 | −3.43 | −5.19 | −6.62 | ||
| Atf6 | 2.59 | 3.26 | 3.52 | ||
| Camk4 | −2.15 | −2.17 | −2.53 | ||
| Cep290 | 3.19 | 3.00 | 3.15 | ||
| Dmrta1 | Dmrt4 | 5.18 | 6.23 | 4.10 | 3.80 |
| Egr1 | −1.72 | −2.06 | −2.66 | −2.96 | |
| Egr2 | −4.35 | −4.28 | −3.77 | −4.50 | |
| Ell2 | 3.55 | 4.77 | 5.14 | 4.49 | |
| Eomes | 5.54 | 3.36 | 3.69 | 2.61 | |
| Ern1 | 2.79 | 3.29 | 4.09 | ||
| Gtf2ird1 | −2.24 | −2.07 | |||
| Hopx | 2.96 | 3.55 | 2.57 | 3.15 | |
| Id2 | 6.96 | 8.08 | 8.97 | 10.18 | |
| Id3 | −2.23 | −2.29 | −2.95 | −3.29 | |
| Ikzf2 | Helios | −3.76 | −4.20 | −3.30 | −4.18 |
| Ikzf3 | Aiolos | 2.83 | 2.02 | 2.48 | |
| Itgb3bp | 2.67 | 2.76 | 3.07 | 2.30 | |
| Lass4 | 2.79 | 3.01 | |||
| Lass6 | −2.47 | −2.98 | −3.77 | ||
| Lef1 | −2.31 | −2.72 | −3.31 | ||
| Mdfic | 1.87 | 2.26 | 3.02 | 2.88 | |
| Myb | −2.00 | −2.69 | −3.09 | ||
| Myc | −2.52 | −8.32 | −14.23 | ||
| Nr1d1 | 2.65 | 2.50 | 3.01 | ||
| Nr1d2 | 2.40 | 2.66 | 2.90 | ||
| Pcgf2 | 3.95 | 4.31 | 4.76 | ||
| Pde8a | 2.77 | 2.72 | 3.48 | ||
| Prdm1 | Blimp-1 | 4.74 | 7.19 | 6.76 | 6.18 |
| Rora | 2.22 | 3.29 | 5.65 | 6.49 | |
| Runx1 | 2.17 | 2.11 | |||
| Runx2 | 3.15 | 2.98 | 2.81 | 2.36 | |
| Satb1 | −1.53 | −2.03 | −2.76 | −4.11 | |
| Scmh1 | −2.30 | −2.27 | |||
| Smad3 | 2.25 | 2.04 | 2.07 | ||
| Smyd1 | 2.24 | 2.45 | |||
| Strm | −2.08 | −2.34 | −2.39 | ||
| Tbl1x | −1.76 | −2.31 | −2.51 | −2.38 | |
| Tbx21 | T-bet | 4.12 | 4.83 | 5.59 | 5.46 |
| Tcf7 | TCF-1 | −2.43 | −3.55 | ||
| Tef | 2.80 | 3.27 | |||
| Trip4 | 2.42 | 2.32 | |||
| Zbtb7b | 1.84 | 1.88 | 2.62 | 2.08 | |
| Zeb2 | 4.76 | 8.66 | 13.64 | 15.66 | |
| Cytokines/chemokines | |||||
| Ccl3 | MIP1-α | 3.92 | 5.84 | 6.38 | 8.04 |
| Ccl4 | MIP1-β | 2.92 | 3.61 | 4.86 | 5.17 |
| Ccl5 | RANTES | 2.57 | 2.73 | 3.25 | 3.65 |
| Ccl6 | 3.45 | ||||
| Ccl9 | 3.58 | 4.65 | 6.06 | ||
| CD163l1 | −2.75 | −2.84 | −2.17 | −2.36 | |
| Ifng | 4.21 | 5.77 | 6.21 | 6.30 | |
| Il15 | 4.25 | 3.52 | 5.23 | 6.70 | |
| Il6st | −3.69 | −5.90 | −6.45 | −6.76 | |
| Ltb | −2.78 | −2.89 | |||
| Spred1 | −3.25 | ||||
| Tnfsf14 | CD258 | 3.61 | 3.70 | 3.27 | 3.17 |
| Tnfsf8 | CD30L | −3.26 | −4.65 | ||
| Xcl1 | 3.97 | ||||
| Cytokine receptors | |||||
| Bmpr1a | 3.03 | ||||
| Ifngr2 | −4.73 | −4.26 | −4.73 | −6.35 | |
| Il10ra | 3.67 | 5.12 | 3.21 | 3.35 | |
| Il12rb2 | 2.71 | 3.61 | 4.22 | 5.65 | |
| Il18r1 | 4.07 | 4.43 | 4.88 | 5.30 | |
| Il18rap | 6.58 | 9.22 | 11.57 | 13.79 | |
| Il1rl1 | 2.29 | 2.43 | |||
| Il2ra | CD25 | 5.26 | 11.36 | 11.83 | 12.40 |
| Il2rb | CD122 | 4.05 | 3.73 | 2.16 | 1.98 |
| Il4ra | −1.84 | −2.17 | −2.18 | ||
| Il6ra | −1.76 | −2.08 | −2.35 | ||
| Tnfrsf10b | DR5 | −2.28 | −2.41 | ||
| Tnfrsf26 | −4.45 | ||||
| Cytolytic effector molecules | |||||
| Gzma | 18.02 | ||||
| Gzmb | 5.30 | 14.40 | 25.43 | 28.71 | |
| Gzmm | 2.40 | 2.79 | 1.86 | 1.82 | |
| Fasl | CD95L | 6.95 | 12.82 | 11.37 | 23.36 |
| Prf1 | Perforin | 2.19 | 3.22 | 3.82 | 3.80 |
| Selectins | |||||
| Sele | CD62E | −2.66 | −2.24 | −2.15 | |
| Sell | CD62L | −4.34 | −14.12 | −30.59 | |
| Chemokine receptors | |||||
| Ccr2 | 9.82 | 12.74 | 13.65 | 14.88 | |
| Ccr4 | −3.13 | ||||
| Ccr5 | 9.08 | 11.39 | 18.44 | 20.28 | |
| Ccr7 | −2.19 | −4.33 | −6.50 | −7.00 | |
| Ccr9 | −2.88 | −2.92 | −3.79 | ||
| Cx3cr1 | 3.14 | 6.41 | 9.79 | 10.59 | |
| Cxcr3 | 6.48 | 5.25 | |||
| Cxcr6 | 2.56 | 2.63 | 2.78 | 3.15 | |
| Integrins | |||||
| Itga1 | CD49A | 8.17 | 8.89 | 10.25 | 12.70 |
| Itga4 | CD49D | 2.01 | 2.38 | 2.84 | 2.82 |
| Itgae | CD103 | −8.00 | −6.78 | −9.33 | −7.65 |
| Itgal | CD11A | 1.78 | 2.34 | 2.45 | 2.57 |
| Itgam | CD11B | 3.24 | 3.62 | 4.36 | |
| Itgax | CD11C | 10.94 | 10.48 | 5.13 | 3.09 |
| Itgb1 | CD29 | 6.24 | 5.44 | 6.20 | 6.11 |
| Itgb2 | CD18 | 2.24 | 2.29 | 2.14 | |
| Cell adhesion | |||||
| Actn1 | −4.64 | −9.04 | −13.15 | −14.78 | |
| Alcam | 5.21 | 5.39 | 6.43 | ||
| Camk2n1 | 2.34 | 2.72 | 2.74 | 3.15 | |
| Cd22 | 2.41 | 2.48 | |||
| Cd44 | 3.28 | 2.74 | 2.20 | 1.91 | |
| L1cam | 2.25 | 2.66 | 2.93 | 2.85 | |
| Lamc1 | 2.50 | 4.04 | 5.00 | ||
| Lgals1 | 2.66 | 3.30 | 3.77 | 4.08 | |
| Mtss1 | −3.78 | −3.57 | −3.63 | −3.97 | |
| Nckap1 | 2.15 | 3.00 | 2.27 | 2.75 | |
| Nrp1 | 4.03 | 3.93 | 4.77 | 4.78 | |
| Pcdh21 | 1.92 | 3.09 | |||
| Pecam1 | −3.42 | −5.25 | −7.30 | ||
| Pros1 | 2.90 | 2.32 | 2.56 | ||
| RP23- | 2.16 | 2.40 | 2.57 | ||
| Apoptosis regulation | |||||
| Alox8 | 1.87 | 2.09 | 2.08 | 2.50 | |
| Amigo2 | −4.79 | −6.34 | −6.13 | −7.64 | |
| Bcl2a1a | 4.48 | 4.46 | 5.81 | 6.55 | |
| Bcl2a1b | 3.06 | 3.15 | 4.32 | 4.76 | |
| Bcl2a1c | 3.73 | 3.59 | 3.53 | 4.30 | |
| Bcl2a1d | 4.09 | 4.20 | 5.34 | 6.02 | |
| Bcl2l1 | 2.30 | 2.28 | 2.36 | 1.96 | |
| Capn2 | 2.51 | 3.24 | 4.04 | ||
| Casp1 | 11.90 | 11.90 | 14.16 | 14.25 | |
| Casp4 | 6.21 | 4.10 | 5.48 | 6.30 | |
| Dapk2 | 2.58 | 3.87 | 2.90 | 4.68 | |
| Egln3 | −2.25 | −2.00 | |||
| Fgl2 | −2.09 | −2.72 | |||
| Hip1 | 1.97 | 2.78 | 3.09 | 3.26 | |
| Igf1r | −2.01 | −2.11 | |||
| Lag3 | 2.15 | ||||
| Map3k5 | −2.07 | −3.32 | |||
| Naip2 | 3.30 | 3.25 | 3.70 | ||
| Nod1 | 2.03 | 1.96 | 2.00 | 1.87 | |
| Rnf216 | 2.58 | 4.43 | 5.39 | 6.37 | |
| Serpinb9 | 4.46 | 3.89 | 5.73 | ||
| Sgk3 | −4.11 | −3.92 | −5.29 | ||
| Antigen processing and presentation | |||||
| Fcgr2b | 4.07 | 9.58 | 13.80 | 16.39 | |
| H2-Oa | −2.87 | −3.87 | −3.56 | −4.20 | |
| H2-Q10 | 2.69 | 2.64 | 3.13 | 3.07 | |
| Cell division, cell cycle, mitosis | |||||
| 1190002H23Rik | −2.06 | −2.05 | |||
| Anxa1 | 5.73 | 11.46 | 14.50 | 20.09 | |
| Cd2ap | −2.24 | −2.45 | |||
| Chpt1 | 3.05 | 2.69 | |||
| Dstn | 2.18 | 2.36 | |||
| Map3k8 | 2.05 | 2.07 | 2.22 | ||
| Mapre2 | 3.56 | 3.72 | 4.07 | 4.13 | |
| Plekho1 | −3.22 | −2.55 | −2.52 | −2.65 | |
| Rassf2 | −2.01 | −2.63 | −3.84 | ||
| S100a6 | 7.22 | 7.29 | 8.51 | 10.02 | |
| Smc4 | −1.86 | −1.97 | −2.50 | −2.53 | |
| Spo11 | −2.28 | −2.37 | |||
| Killer-cell lectin-like receptors | |||||
| Klra8 | LY49H | 2.56 | 3.15 | 3.17 | |
| Klra9 | LY49I | 4.68 | 6.11 | ||
| Klra10 | LY49J | 5.23 | 7.10 | ||
| Klra14 | LY49N | 4.12 | 6.64 | ||
| Klrb1c | NK1.1 | 6.84 | 11.43 | 14.82 | 15.09 |
| Klrb1f | 4.41 | 4.10 | |||
| Klrc1 | NKG2A | 18.58 | 26.34 | 24.63 | 30.20 |
| Klrc2 | NKG2C | 6.53 | 9.32 | 11.04 | 13.90 |
| Klrc3 | NKG2E | 3.96 | 5.87 | 8.58 | 10.81 |
| Klrd1 | CD94 | 1.69 | 2.11 | ||
| Klre1 | NKG2I | 3.58 | 7.02 | 22.19 | |
| Klrg1 | KLRG-1 | 9.09 | 40.79 | 42.82 | 48.93 |
| Klri2 | 2.45 | 7.54 | |||
| Klrk1 | NKG2D | 4.31 | 4.58 | 6.98 | 8.20 |
| Lipid metabolism | |||||
| Afp | −2.45 | −2.04 | |||
| Crot | 2.18 | 2.47 | |||
| Elovl6 | −2.48 | −2.29 | |||
| Osbpl3 | 5.58 | 8.11 | 9.74 | 12.08 | |
| Prune | 2.06 | 3.06 | 1.86 | ||
| Soat2 | 3.94 | 3.71 | 4.11 | 5.02 | |
| Ig-domain or Ig like | |||||
| B430306N03Rik | −2.43 | −2.53 | −2.87 | −2.87 | |
| Cd244 | 2B4 | 5.03 | |||
| Cd80 | 2.93 | 3.55 | 4.88 | ||
| Gp49a | 7.01 | 11.54 | 7.86 | 8.59 | |
| Lair1 | 2.54 | 3.92 | 2.61 | 3.28 | |
| Lilrb4 | 4.50 | 5.95 | 5.20 | 5.74 | |
| Sema4f | 2.20 | 2.35 | |||
| Slamf1 | SLAM | 3.01 | 3.44 | ||
| Slamf6 | NTB-A | −3.12 | −5.43 | ||
| Slamf7 | CRACC | 5.83 | 6.83 | 14.61 | 17.57 |
| Intracellular signal transduction | |||||
| Als2cl | −2.37 | −2.61 | −3.24 | ||
| Dock9 | 2.87 | 3.89 | 4.80 | 4.58 | |
| Garnl4 | 2.30 | 2.59 | 2.80 | ||
| Rab3ip | −2.29 | −3.15 | |||
| Ralgps2 | −2.98 | −3.50 | |||
| Rangrf | −2.24 | −2.10 | |||
| Rap2a | 3.29 | 2.93 | 2.71 | ||
| Rapgef4 | −4.48 | −6.06 | −11.74 | −12.49 | |
| Rhobtb2 | −2.07 | −2.20 | |||
| Rras2 | −3.57 | −3.17 | −3.19 | ||
| Sos2 | 2.86 | 2.74 | |||
| Tbc1d12 | 2.62 | 2.24 | |||
| Adora2a | 2.42 | 2.93 | 2.30 | 2.34 | |
| Cmklr1 | 2.35 | 2.55 | |||
| Csprs | 3.12 | ||||
| Cysltr2 | 3.66 | ||||
| F2rl1 | −3.29 | −5.33 | |||
| F2rl2 | 2.34 | 2.92 | 3.05 | ||
| Vipr1 | −2.25 | −3.01 | −3.09 | ||
| Adcy6 | −3.19 | −3.24 | −3.68 | −4.13 | |
| Cish | 2.37 | 2.06 | 2.18 | ||
| Entpd1 | 5.59 | 9.13 | 8.62 | 12.57 | |
| Gna15 | 2.19 | 3.68 | 3.61 | ||
| Nsg2 | −3.52 | −4.25 | −4.95 | ||
| Rgs1 | 4.45 | 5.11 | 3.89 | 4.14 | |
| Rgs10 | −4.45 | −7.93 | −8.05 | ||
| Snx14 | 2.04 | 2.15 | |||
| Trat1 | −6.75 | −6.82 | −6.00 | −7.47 | |
| Proteolysis | |||||
| 4930523C11Rik | −6.84 | −7.86 | −5.29 | −4.96 | |
| Capn11 | 2.41 | −2.61 | |||
| Cd55 | −5.78 | ||||
| Ctla2a | 6.74 | 5.71 | 4.71 | 4.83 | |
| Ctla2b | 3.75 | 3.20 | |||
| Ctsd | 1.75 | 1.99 | 2.09 | 2.04 | |
| Dennd4a | 5.65 | 3.55 | 3.44 | 6.00 | |
| Dpp4 | −2.16 | −3.67 | |||
| Ephx1 | −5.87 | −7.42 | −6.06 | −4.74 | |
| Fbxl2 | 2.65 | 2.91 | |||
| Gzmk | 5.56 | 7.23 | 5.08 | 7.56 | |
| Lonrf3 | 4.20 | ||||
| Mmp25 | 3.13 | ||||
| Nedd4l | −2.06 | −2.66 | −2.87 | ||
| Prss12 | 3.60 | 3.21 | 3.74 | 3.34 | |
| Rnf144a | −2.17 | −2.66 | −3.90 | ||
| Serpina3f | 3.75 | 3.58 | 2.48 | 2.41 | |
| Serpinb1a | 2.62 | 3.40 | |||
| Tfrc | −4.30 | −3.98 | −5.83 | ||
| Usp28 | −3.91 | −5.76 | −6.07 | −5.94 | |
| Usp48 | 1.72 | 1.87 | 2.22 | 2.14 | |
| Wwp1 | 2.51 | 2.41 | 1.92 | ||
| Cst7 | 2.21 | 2.01 | 2.59 | 2.54 | |
| Protein kinases | |||||
| Cpne3 | 3.01 | ||||
| Dgka | −1.61 | −2.24 | −2.21 | ||
| Dgkh | 2.59 | 3.52 | 4.47 | ||
| Grk5 | 2.33 | 2.87 | |||
| Grk6 | −2.04 | −2.52 | |||
| Havcr2 | TIM-3 | 4.17 | 7.94 | 10.37 | |
| Ikbke | −2.08 | −2.53 | −2.56 | ||
| Insr | −5.27 | −6.42 | |||
| Lrrk1 | 2.14 | 2.52 | 3.06 | 2.89 | |
| Map4k2 | −2.03 | −2.32 | −2.37 | ||
| Map4k4 | −1.68 | −1.90 | −2.21 | −2.02 | |
| Mapkapk3 | 2.20 | 2.56 | 2.19 | 2.40 | |
| Mlkl | 3.00 | 3.42 | |||
| Myo3b | 3.39 | 2.87 | 2.29 | 2.66 | |
| Pacsin1 | −2.08 | −3.01 | −3.19 | −3.15 | |
| Pdk1 | −2.06 | −4.66 | −7.86 | −8.70 | |
| Pfkp | 1.79 | 2.13 | 2.10 | 2.40 | |
| Pik3ap | 2.56 | 4.17 | 6.01 | 6.92 | |
| Pip4k2a | −2.19 | −2.42 | |||
| Plaur | −3.28 | −3.27 | −3.13 | −3.08 | |
| Ryk | 2.05 | 2.21 | 1.96 | 2.39 | |
| Trib2 | −2.51 | −3.50 | −3.74 | −3.48 | |
| Yes1 | 5.31 | 7.01 | 6.36 | 5.42 | |
| MISCELLANEOUS | |||||
| 4631426J05Rik | −3.21 | −3.11 | −2.12 | −2.44 | |
| 5430427O19Rik | 2.92 | 3.03 | 5.24 | 4.28 | |
| Abca3 | −2.12 | −2.07 | −2.59 | −3.00 | |
| Abcb1a | 2.34 | 3.69 | 3.69 | 4.13 | |
| Ahnak | 3.80 | 4.36 | 4.11 | 4.59 | |
| Ampd1 | −5.88 | −5.33 | −6.17 | −6.21 | |
| Anxa2 | 4.75 | 6.26 | 5.24 | 5.49 | |
| Arhgap26 | 2.24 | 2.75 | 3.57 | 4.33 | |
| Arsb | 2.38 | 2.02 | 2.26 | ||
| Art2b | −3.53 | −6.30 | |||
| As3mt | 5.63 | 7.03 | 6.20 | ||
| Atp1a1 | −2.03 | −2.51 | |||
| Atp1b1 | −9.81 | −10.47 | −7.96 | −8.77 | |
| Atp2b1 | 2.54 | 2.99 | 3.46 | 3.33 | |
| Atp2b4 | 4.42 | 5.99 | 7.83 | 8.37 | |
| Atp6v0d2 | 7.08 | 12.04 | 5.17 | 1.98 | |
| Atp8b4 | 2.03 | 2.02 | 2.01 | 2.35 | |
| Atxn1 | 2.06 | 2.27 | 2.01 | 2.16 | |
| BC057170 | −2.59 | −3.03 | |||
| Bcl9 | −3.36 | −3.78 | −3.83 | −4.81 | |
| Bicd1 | 2.03 | 1.97 | 2.10 | ||
| Bspry | 2.37 | 2.72 | |||
| C230094A16Rik | 2.30 | 2.54 | |||
| Capg | 4.47 | 6.37 | |||
| Cd69 | −3.50 | −2.94 | |||
| Chsy1 | 2.05 | 2.20 | 2.24 | ||
| Cnn3 | −2.11 | −2.35 | |||
| Cyp17a1 | 3.46 | 4.67 | |||
| Ddx28 | 3.27 | 4.88 | 5.54 | ||
| Dntt | −2.16 | −2.83 | −3.25 | ||
| Dpm1 | −2.15 | −2.06 | |||
| Eea1 | 3.06 | 3.00 | 3.16 | 3.55 | |
| Emp1 | 3.40 | 4.33 | |||
| Epb4.1l2 | −1.72 | −1.90 | −3.02 | −3.87 | |
| Ext1 | −3.18 | −3.67 | −3.72 | ||
| Faah | −2.17 | −2.27 | |||
| Fads1 | −2.35 | −2.03 | |||
| Fchsd2 | −1.87 | −2.67 | −2.52 | ||
| Gabarapl2 | 2.43 | 2.41 | |||
| Galnt3 | 2.96 | 3.82 | 7.17 | 7.65 | |
| Gbp3 | 3.54 | 3.07 | |||
| Gnptab | 2.15 | 2.24 | 2.88 | 3.15 | |
| Golim4 | 2.13 | 2.15 | 2.67 | ||
| Gria3 | −7.83 | −8.90 | −5.68 | −6.33 | |
| Hook1 | −2.43 | −3.97 | −4.61 | ||
| Idh2 | −2.39 | −2.31 | −2.20 | ||
| Igfbp4 | −4.49 | −4.62 | −2.87 | −3.32 | |
| Inadl | −2.14 | −3.03 | −3.69 | ||
| Kcna2 | −2.03 | −1.92 | −1.88 | −2.04 | |
| Kcnj8 | 4.49 | 5.08 | 7.68 | 9.35 | |
| Kcnk5 | 3.25 | 3.10 | 3.52 | ||
| Krtcap2 | 1.57 | 2.23 | 2.18 | ||
| Lgals3 | 1.99 | 3.30 | 3.34 | 3.60 | |
| Lpin1 | 1.99 | 2.78 | 3.28 | 4.06 | |
| Ly6c1 | LY-6C | 3.65 | 3.96 | 5.39 | 5.90 |
| Mcoln3 | −2.08 | −1.82 | −2.40 | ||
| Mpp1 | −2.24 | −2.10 | |||
| Mtap7 | −2.27 | −2.09 | −2.61 | ||
| Myo1f | 3.16 | 3.01 | 3.52 | 3.70 | |
| Myo5a | 1.98 | 2.53 | 2.60 | 3.06 | |
| Ncald | 2.62 | 2.63 | 2.97 | ||
| Nt5e | 6.82 | 4.87 | 5.96 | 5.94 | |
| Pdlim4 | −2.44 | −2.13 | −1.74 | ||
| Plek | 9.42 | 9.11 | 8.61 | 9.29 | |
| Pole2 | −3.15 | −2.37 | |||
| Polk | 2.17 | 2.10 | 2.17 | 2.23 | |
| Prdx4 | 2.85 | 3.31 | 3.30 | ||
| Reep5 | 2.30 | 2.63 | 2.90 | ||
| Rpa2 | 2.27 | 2.40 | 1.83 | 2.13 | |
| S100a10 | 1.90 | 2.42 | 2.52 | ||
| S100a4 | 7.59 | 10.70 | 12.32 | 15.07 | |
| Samd3 | 10.99 | 12.16 | 6.66 | 5.88 | |
| Scml4 | −2.16 | −2.05 | |||
| Sdcbp2 | 2.23 | 2.66 | 2.01 | 3.13 | |
| Sec61g | 2.05 | 2.28 | |||
| Sfmbt2 | −2.51 | −2.57 | −2.33 | −2.28 | |
| Shmt2 | −2.08 | −2.07 | |||
| Slc25a24 | 3.24 | 3.42 | |||
| Slc4a7 | 2.94 | 3.46 | 3.01 | 2.99 | |
| Slc6a19 | −2.36 | −2.09 | −2.96 | −2.81 | |
| Slc9a7 | 4.84 | 7.74 | 8.46 | ||
| Slc9a9 | 1.61 | 1.82 | 2.44 | 2.69 | |
| Snx10 | 3.28 | 2.85 | 3.09 | 3.73 | |
| Srxn1 | 2.01 | 2.61 | |||
| St3gal4 | 2.00 | 2.75 | |||
| St3gal6 | 2.75 | 2.98 | 2.50 | 2.69 | |
| St6gal1 | −6.17 | −9.06 | −8.56 | −8.93 | |
| St8sia1 | −4.73 | −5.89 | −4.15 | −3.77 | |
| Sytl2 | 5.45 | 7.46 | 5.01 | 5.62 | |
| Sytl3 | 3.53 | 3.80 | |||
| Tox | −4.18 | −3.89 | −3.40 | ||
| Xdh | 1.83 | 2.36 | 2.57 | ||
| Xylt1 | 2.49 | 2.09 | 2.11 | ||
Space limitations preclude a detailed discussion of the large number of regulated genes identified in our microarray analyses. However, we identified several important classes of regulated genes that may provide targets for understanding and manipulating memory CD8+ T cell populations generated by repeated antigen stimulation. Strikingly, more than 40 transcription factors with statistically significant changes in mRNA expression were identified. Of these, genes that encode Ell2, Dmrt4, Hopx, Blimp-1, Runx2 and T-bet were upregulated in all memory populations (Pattern III), whereas others like Egr2 and Helios were downregulated (Pattern IV) and thus, reside in the “core memory signature” subset of genes. We found multiple transcription factors that displayed steady increases (Atf6, Ern1, Id2, Rorα, Zeb2, Pattern I) or steady decreases in mRNA levels (Aff3, Egr1, Lass6, Lef1, Myc, Satb1, Pattern II) after multiple antigen stimulations which make them prime candidates as potential regulators of the transcriptional diversification observed in repeatedly stimulated memory CD8+ T cells.
All four memory groups showed significant increases in the mRNA levels for proinflammatory chemokines CCL3, 4, 5 and 9 (Pattern I). Thus, the ability memory CD8+ T cells to recruit other cells to sites of infection could be enhanced by repeated antigen stimulations. Additionally, mRNA for multiple cytokine receptors was altered by repeated antigen stimulation. 4° memory CD8+ T cells had substantially lower levels of Ifngr2 mRNA but higher expression of mRNA's for Il10ra (Pattern III), Il18r1, Il18rap and the IL2Rα chain CD25 (Il2ra; Pattern I) indicating a change in the responsiveness to cytokines. The upregulation of CD25 mRNA and protein (data not shown) in repeatedly stimulated memory CD8+ T cell populations is intriguing in light of the concurrent downregulation of CD122 mRNA (Il2rb) and protein we observed in 2° to 4° compared to 1° memory CD8+ T cells. At a minimum, these data may suggest that maintenance of repeatedly stimulated CD8+ T cells is less dependent on IL-15 signalling, which requires CD122 (Jabbari and Harty, 2006; Sandau et al., 2010). Additionally, the clear evidence that regulated CD25 expression contributes to effector and memory CD8+ T cell differentiation in the primary response (Kalia et al., 2010; Pipkin et al., 2010) suggests that sustained CD25 expression may underlie some of the properties of repeatedly stimulated memory CD8+ T cell populations.
Of interest, mRNAs for numerous inflammatory chemokine receptors (CCR2, CCR5, CX3CR1, all Pattern I) and integrins (CD49d, CD11a and CD11b, Pattern I) were also highly elevated. Other markedly regulated trafficking molecules included CD18, CD29 (both Pattern III) and CD103 (Pattern IV). These data demonstrate that repeatedly challenged memory CD8+ T cells dynamically regulate multiple trafficking molecules in addition to CD62L and CCR7 and thus, acquire novel migration properties that could influence their protective capacity against infection.
Although we mention here only selected examples, our data (Table 1) demonstrate a profound impact of repeated antigen exposure on the mRNA expression of multiple gene families that are likely to play important roles in the function of memory CD8+ T cell populations. Clearly, the relative importance of these regulated genes for the biology of repeatedly stimulated memory CD8+ T cell populations will require experimental evaluation, that can now proceed from the data sets described here.
Transcriptome comparison reveals major differences between primary and quaternary memory CD8+ T cells
Much attention has focused on differences in global gene expression between naïve and primary memory T cell populations (Kaech et al., 2002; Wherry et al., 2007). The stepwise increase in the number of differentially regulated genes induced by repetitive antigen stimulation suggested that the transcriptomes of primary and repeatedly stimulated memory CD8+ T cells also differ substantially. To validate this assumption, the transcriptomes of 1° and 4° CD8+ T cells were directly compared. This analysis yielded 776 unique genes whose transcriptional regulation classified as significantly altered (FDR q-value <0.01) between the two populations (see Table S4 for a complete list). This number was surprisingly high because comparison of naïve CD8+ T cells with 1° memory CD8+ T cells only yielded 364 significant changes. Subsequently, we divided the 776 genes into two groups with either transcriptional up- or downregulation and focussed on the molecules with the most pronounced differences in expression between 1° and 4° memory (Table 2A,B).
Table 2. Comparison of gene expression between 1° and 4° memory CD8+ T cells.
Gene expression in 1° and 4° memory OT-I T cells. FDR q-value <0.01 defined statistical significance. Genes with the most pronounced A) increase or B) decrease in mRNA quantity in 4° compared to 1° memory CD8 T cells. Genes are ranked according to fold changes in mRNA quantity. Molecules in bold letters are discussed in the text.
| A) | Transcriptional upregulation quaternary vs. primary memory | ||
|---|---|---|---|
| rank | Gene Symbol | Alias | change |
| 1 | Klra10 | LY49J | 9.51 |
| 2 | Havcr2 | TIM-3 | 9.15 |
| 3 | Klre1 | NKG2I | 9.08 |
| 4 | Klra14 | LY49N | 7.79 |
| 5 | Klra9 | LY49I | 7.33 |
| 6 | Klri2 | 7.13 | |
| 7 | AA467197 | 5.96 | |
| 8 | Gzmb | 5.42 | |
| 9 | Klrg1 | KLRG-1 | 5.38 |
| 10 | Capg | 5.22 | |
| 11 | ENSMUSG00000073493 | 5.13 | |
| 12 | Gstm5 | 5.12 | |
| 13 | Car5b | 5.03 | |
| 14 | Cd244 | 2B4 | 4.63 |
| 15 | 6330403K07Rik | 4.52 | |
| 16 | Lamc1 | 4.49 | |
| 17 | EG433024 | 4.31 | |
| 18 | Ccl9 | 4.19 | |
| 19 | I830127L07Rik | 4.09 | |
| 20 | Serpinb1a | 4.05 | |
| 21 | Ms4a4a | 3.91 | |
| 22 | Fcgr2b | 3.72 | |
| 23 | Esm1 | 3.59 | |
| 24 | 4933431E20Rik | 3.58 | |
| 25 | Zeb2 | 3.42 | |
| 26 | Cx3cr1 | 3.37 | |
| 27 | Anxa1 | 3.30 | |
| 28 | Bhlhe40 | 3.29 | |
| 29 | Tspan2 | 3.22 | |
| 30 | Crybg3 | 3.22 | |
| 31 | Lonrf3 | 3.18 | |
| 32 | A930038C07Rik | 3.18 | |
| 33 | Nebl | 3.10 | |
| 34 | Itgam | CD11b | 3.09 |
| 35 | Arhgef12 | 3.09 | |
| 36 | Klra8 | LY49H | 3.08 |
| 37 | Tef | 3.06 | |
| 38 | Mmp25 | 3.06 | |
| 39 | Emp1 | 3.02 | |
| 40 | Srxn1 | 3.02 | |
| 41 | Akr1e1 | 2.98 | |
| 42 | Chn2 | 2.97 | |
| 43 | Ern1 | 2.96 | |
| 44 | Rora | 2.92 | |
| 45 | Fasl | 2.83 | |
| 46 | Tmem97 | 2.80 | |
| 47 | Pcdh21 | 2.78 | |
| 48 | Vmn2r86 | 2.78 | |
| 49 | 5730494M16Rik | 2.74 | |
| 50 | Pik3ap1 | 2.74 | |
| 51 | Klrc3 | NKG2E | 2.73 |
| 53 | Slamf7 | CRACC | 2.69 |
| 63 | Cd80 | 2.44 | |
| 70 | Slamf1 | SLAM | 2.31 |
| 76 | Ccr5 | 2.23 | |
| 79 | Il2ra | CD25 | 2.19 |
| 83 | Il18rap | 2.16 | |
| 86 | Klrc2 | NKG2C | 2.13 |
| 97 | Il12rb2 | 2.07 | |
| B) | Transcriptional downregulation quaternary vs. primary memory | ||
|---|---|---|---|
| rank | Gene Symbol | Alias | change |
| 1 | Sell | CD62L | −15.35 |
| 2 | Myc | −15.11 | |
| 3 | Tlr1 | −11.18 | |
| 4 | Wfikkn2 | −10.18 | |
| 5 | Tnfsf8 | CD30L | −9.36 |
| 6 | D17H6S56E-5 | −7.10 | |
| 7 | Insr | −6.54 | |
| 8 | Xcl1 | −6.29 | |
| 9 | F2rl1 | −6.08 | |
| 10 | Snhg1 | −5.84 | |
| 11 | Cd55 | −5.53 | |
| 12 | Mboat1 | −5.44 | |
| 13 | Dapl1 | −5.13 | |
| 14 | Sidt1 | −4.49 | |
| 15 | Slamf6 | NTB-A | −4.49 |
| 16 | Rpp38 | −4.47 | |
| 17 | Gm885 | −4.40 | |
| 18 | Treml2 | −4.16 | |
| 19 | Cxcr3 | −4.12 | |
| 20 | Rnf144a | −4.11 | |
| 21 | Pdk1 | −4.04 | |
| 22 | Aff3 | −4.03 | |
| 23 | Tnfrsf26 | Tnfrh3 | −3.94 |
| 24 | Rgs10 | −3.90 | |
| 25 | Nsg2 | −3.87 | |
| 26 | Fam169b | −3.84 | |
| 27 | Art2b | −3.62 | |
| 28 | Atp6v0d2 | −3.58 | |
| 29 | Hook1 | −3.54 | |
| 30 | Itgax | CD11c | −3.46 |
| 31 | Ms4a4c | −3.46 | |
| 32 | Insr | −3.43 | |
| 33 | Larp1 | −3.37 | |
| 34 | Ly6e | Sca-2 | −3.32 |
| 35 | Spin2 | −3.31 | |
| 36 | A530040E14Rik | −3.28 | |
| 37 | 2900016B01Rik | −3.26 | |
| 38 | Actn1 | −3.26 | |
| 39 | Ccr7 | −3.20 | |
| 40 | Qpct | −3.19 | |
| 41 | Qtrtd1 | −3.10 | |
| 42 | Capn11 | −3.10 | |
| 43 | Dpp4 | −3.08 | |
| 44 | Cdh1 | −3.05 | |
| 45 | BC059842 | −3.03 | |
| 46 | A530040E14Rik | −3.02 | |
| 47 | A530040E14Rik | −3.02 | |
| 48 | Ssbp2 | −2.98 | |
| 49 | A530040E14Rik | −2.91 | |
| 50 | A530040E14Rik | −2.91 | |
| 62 | Ltb | lymph. B | −2.66 |
| 76 | Il2 | −2.44 | |
| 82 | Tcf7 | Tcf-1 | −2.36 |
| 113 | Satb1 | −2.12 | |
In this data subset, we found that killer-cell lectin-like receptors, trafficking molecules and transcription factors were represented with multiple gene family members. Of note, 6 of the 10 genes with the most substantial transcriptional upregulation in 4° memory CD8+ T cells belonged to the killer-cell lectin-like receptor group. Of interest, expression of both inhibitory (e.g. Klra10, Klra9, Klra14, Klrg1) and activating receptors (Klri2, Klra8, Klrc1) was increased in 4° memory CD8+ T cells. The role of these molecules in memory CD8+ T cells is unknown, however, our expression data suggests that killer-cell lectin-like receptors may be important targets to modulate the function of 4° memory CD8+ T cell populations. While expression of the trafficking molecules CX3CR1 and CD11b was also increased, severe downregulation was observed in the mRNA for genes involved in steady-state trafficking into lymph nodes (e.g. CD62L, CCR7). In addition, some trafficking molecules that were initially upregulated in 1° memory CD8+ T cells (CXCR3 and CD11c) exhibited a pronounced decrease in their mRNA expression after multiple antigen exposures.
Most importantly, expression of multiple transcriptional regulators was markedly altered. Among these, Myc, a molecule involved in the downstream signalling of IL–15 (Bianchi et al., 2006) and therefore in the control of basal proliferation, exhibited the most pronounced downregulation. Furthermore, expression of Tcf7 (TCF-1), a downstream effector protein of the Wnt signalling pathway, was reduced. Activation of the Wnt pathway has recently been linked to long-lived CD4+ and CD8+ memory T cells as well as to the arrest of effector T cell differentiation and the generation of memory T cells with stem-cell like quality (Gattinoni et al., 2009; Williams et al., 2008; Zhao et al., 2010). The reduced mRNA levels of these molecules in 4° memory CD8+ T cells provides a possible explanation for the continuous decline in numbers that we have observed in repeatedly stimulated memory CD8+ T cells (Figure S1C,D). In contrast, the expression of the transcription factors Rorα (RAR related orphan receptor alpha) and Zeb2 (SIP1) increased with repetitive antigen stimulation. The ROR family possesses well-known functions in CD4+ T cell differentiation (Ivanov et al., 2006; Yang et al., 2008) but a role in CD8+ T cells has so far not been shown. Zeb2 is of particular interest since it functions as a transcriptional regulator that is able to repress cell cycle progression (Mejlvang et al., 2007) and inhibit telomerase expression (Lin and Elledge, 2003), two cellular processes that could impact the proliferative capacity of 4° memory CD8+ T cells.
To further validate and confirm the assumption that the number of antigen encounters influences the differentiation of memory CD8+ T cells we performed KEGG biological pathway analyses with all genes listed in Table S4. Of 776 input genes 675 were annotated in the DAVID database. Importantly, multiple biological pathways with significant changes between 1° and 4° memory CD8+ T cell populations were identified (examples presented in Figure 6A). Although cytokine-cytokine receptor interaction and NK-cell mediated cytotoxicity pathways were substantially altered in all memory CD8+ T cells when compared to naïve cells (Figure 3E) significant changes in these pathways (21 and 12 genes, respectively) were observed when 1° and 4° memory CD8+ T cells were compared (Figure 6A). Therefore, repeated antigen stimulations further change the ability of memory CD8+ T cells to produce and respond to cytokines and chemokines. These data also suggest that memory CD8+ T cell function (ex. cytotoxicity) might be influenced by the number of antigen encounters, a notion that may be exploited in vaccine design. 4° memory CD8+ T cells also show significant changes in cellular metabolism (polyunsaturated fatty acid biosynthesis pathway) and expression of adhesion molecules (cell adhesion molecules pathway). Finally, the most dramatic changes observed from the pathway analyses were among ribosomal subunits (Figure 6A,B). 44 ribosomal subunit genes (ribosome pathway – Figure 6A) were downregulated in 4° memory CD8+ T cells (p-value – 1.1×10−31 – Figure 6B) suggesting that the translational machinery might be impaired.
Figure 6. Similarities and differences between primary, quaternary memory and exhausted CD8+ T cell populations.
A) Genes with significant mRNA changes between 1° and 4° memory CD8 T cells (Table S4) were evaluated using KEGG pathway analysis in DAVID. Arrows indicate whether genes are upregulated (↑) or downregulated (↓) in 4° compared to 1° memory CD8 T cells. B) Fold changes in mRNA of ribosomal genes in 4° compared to 1° memory CD8+ T cells. C) GSEA was performed to compare whether exhaustion-associated genes (down (left panel) or up (right panel)) showed specific enrichment in 4° memory CD8+ T cells. Rectangles with solid lines indicate down- or up-regulated genes common to exhausted and 4° memory CD8+ T cells. Rectangles with dotted lines indicate genes showing opposite expression patterns between exhausted and 4° memory CD8+ T cells. Genes are shown in Table S5. D) Fold changes in mRNA quantity for selected inhibitory receptors (implicated in exhausted CD8+ T cells (Shin et al., 2009)) in 4° compared to 1° memory CD8+ T cells.
Recently, a similar downregulation of the ribosome pathway and poor proliferative responses were reported for exhausted CD8+ T cells when they were compared to 1° memory CD8+ T cells (Shin and Wherry, 2007; Wherry et al., 2007). To determine if 4° memory CD8+ T cells share a gene expression profile with exhausted CD8+ T cells, we performed Gene Set Enrichment Analysis (GSEA) comparing up and down regulated gene sets from exhausted memory CD8+ T cells (Wherry et al., 2007) with our 4° memory CD8+ T cell populations, both relative to 1° memory populations. GSEA uses enrichment scores to determine relative enrichment of one gene set in a ranked list of genes (Haining and Wherry, 2010; Subramanian et al., 2005). In the gene set (n=119) with downregulated mRNA expression in exhausted memory CD8+ T cells, about 1/3 were enriched in 4° memory populations. These included multiple members of the ribosomal subunit family, a Kegg pathway that is significantly downregulated in 4° memory populations as defined in Figure 6B. However, ~2/3 of the downregulated genes were not shared between these populations and in fact, ~15% of the genes downregulated in exhausted CD8+ T cells were upregulated in 4° memory (Figure 6C and Table S5). Similarly, only ~1/4 of the genes with upregulated mRNA expression in exhausted CD8+ memory (n=86) were shared by 4° memory populations and again, ~15% of the upregulated genes in exhausted memory CD8+ T cells were actually downregulated in 4° memory populations (Figure 6C and Table S5). Interestingly, although deficient in proliferation (antigen-driven expansion – Figures 1A and S1B), homeostatic proliferation (Figure 1C), and survival (Figure S1C,D) 4° memory CD8+ T cell population did not show the inhibitory receptor patterns observed in exhausted CD8+ T cells (Figure 6D). Thus, while gene expression patterns of exhausted and 4° memory populations exhibit 25–35% overlap, a large number of genes are differentially expressed between these populations, suggesting that they possess unique genetic signatures and likely, functional attributes.
Taken together, these pronounced differences in gene regulation between 1° and 4° memory CD8+ T cells suggest that the transcriptome of repeatedly stimulated memory CD8+ T cells is unique and substantially different from exhausted or 1° Tem and Tcm memory CD8+ T cells. The fact that a substantial number of the differentially regulated genes in 4° memory CD8+ T cells have no known function in CD8+ T cell biology suggests that the spectrum of genes that play a role in repeatedly stimulated memory CD8+ T cells is much broader than that of `classic' primary memory CD8+ T cells.
Discussion
The data presented here provide new insights into the process of memory differentiation in general and the influence of repeated antigen exposures on memory CD8+ T cell function and differentiation in particular. Although multiple genes are regulated during the generation of functional CD8+ T cell memory, it is unknown for most of them whether they are essential or dispensable in this process. In the current study, we have identified a core group of genes and pathways whose expression is substantially altered compared to naive CD8+ T cells regardless of the number of past antigen exposures (`stable memory signature genes'). This finding reveals a group of CD8+ T cell-related memory genes that could identify antigen-experienced memory CD8+ T cells and lead to the discovery of new molecules and pathways involved in memory differentiation. Additionally, genes from the `dynamic memory signature group', in conjunction with molecules that are differentially regulated after a specific number of antigen encounters, could be helpful in determining the number of past antigen exposures in memory CD8+ T cells.
Differences in memory CD8+ T cell function are often attributed to differential commitment to the Tem or Tcm cell lineage (Sallusto et al., 1999; Wherry et al., 2003). Our results demonstrate dynamic changes in gene expression in repeatedly stimulated memory CD8+ T cell populations that contrast with their apparent uniform Tem cell lineage commitment (CD62Llo). Therefore, changes in gene expression in repeatedly stimulated memory CD8+ T cell populations could be due to altered subset (Tem:Tcm) ratios of each memory population without major changes in gene expression within individual cells. Alternatively, repeated antigen stimulation may alter gene expression within each cell in the ensuing memory CD8+ T cell populations. Disparate studies suggest that Tem and Tcm memory CD8+ T cell subsets differ modestly (Chtanova et al., 2005) or substantially (Willinger et al., 2005) in their transcriptional profiles. GSEA analyses performed on the list of genes that are upregulated in Tem (Willinger et al., 2005) showed no progressive enrichment in `effector-memory' associated genes in 2°, 3°, and 4° memory populations (data not shown). Given the large number of genes exhibiting differential regulation between 1° and 4° memory CD8+ T cell populations, the phenotypic differences between 1° and 4° CD62Llo memory populations and the lack of progressive Tem gene enrichment in 2°, 3°, and 4° memory populations we favour the interpretation that repeated antigen-stimulations alter gene expression within each cell. Complete resolution of this issue awaits deep sequencing (mRNA-seq) on the single cell level (Tang et al., 2009). Importantly, our data also demonstrate that repeated antigen exposure evokes not only pronounced changes in the expression of individual genes but also a substantial numerical increase in differentially regulated genes. Therefore, the formation of primary memory CD8+ T cells represents but an initial step in a progressive differentiation process. In fact, each antigen stimulation increases the number of regulated genes in the ensuing memory population by as many genes as are differentially regulated between naive to 1° memory populations. Thus, our study reveals a novel and unexpected diversity in memory CD8+ T cell gene expression that is driven by repetitive antigen stimulations.
Interestingly, comparison with published data from human studies revealed that repeatedly stimulated memory cells shared many markers with senescent human T cells (Koch et al., 2007), namely CD244, CD49d, CCR7, CD27, KLRK1, KLRC3 and KLRG1. Expression of other markers like CD28, CD57 and CTLA4, however, did not correlate well with that shown for senescent human T cells. Similar to our comparison of 4° memory with exhausted CD8+ T cells, these results suggest that repeatedly challenged memory CD8+ T cells clearly represent novel populations with a unique transcriptional profile despite some similarity with senescent and exhausted CD8+ T cells.
Changes in the transcriptional profiles of multiply stimulated memory populations have important implications for the function of memory CD8+ T cells. The heterogeneity of repeatedly stimulated memory CD8+ T cells has to be considered when prime-boost regimens are employed to increase memory CD8+ T cell numbers because of the possible impact on memory CD8+ T cell function. In this regard, limited studies to date suggest that protection by memory CD8+ T cells can be enhanced by repetitive antigen stimulation (Jabbari and Harty, 2006). Therefore, it will be critical to determine how the altered transcriptomes in repetitively stimulated memory CD8+ T cells influence protection against diverse pathogens.
Additionally, our results may offer insight into the memory CD8+ T cell response to repeated infections with pathogens such as Plasmodium or reactivating latent infections with herpes viruses. Furthermore, the pronounced differences in molecular profile potentially offer unique opportunities to selectively target repeatedly stimulated memory CD8+ T cells without detrimental side effects to primary memory CD8+ T cells. The data presented here could therefore not only lead to a better understanding of the function of repeatedly stimulated memory CD8+ T cells but also to the identification of new molecular targets that help control these cells in infectious, autoimmune and oncological diseases.
Experimental Procedures
Mice and Bacteria
C57Bl/6 mice were obtained from the US National Cancer Institute. TCR-transgenic OT-I Thy1.1 mice were previously described (Hogquist et al., 1994). All mice were used at 6 –12 weeks of age. Mice were bred and maintained in the animal facilities of the University of Iowa at the appropriate biosafety levels. Attenuated actA-deficient Listeria monocytogenes expressing ovalbumin (att LM-OVA) was grown, injected i.v., and quantified as described (Haring et al., 2005). All animal experiments followed approved Institutional Animal Care and Use Committee (ACURF) protocols.
Antibodies
The following antibodies were used with the indicated specificity and the appropriate combinations of fluorochromes: Thy1.1 (OX-7 or HIS51) and CD62L (MEL-14, both BD Pharmingen), CD8 (53-6.7), CD25 (PC61.5), CD122 (5H4), CD27 (LG.7F9), CD11a (M17/4), CD11b (M1/70), CD11c (N418), CD103 (2E7), CD29 (eBioHMb1-1), CD49d (R1-2), KLRG-1 (2F1), CXCR3 (Cxcr3-173), IFN-γ (XMG1.2), TNF (MP6-XT22), IL-2 (JES6-5H4, all eBioscience) and appropriate isotype controls. For analysis of BrdU incorporation, mice were injected i.p. with 2mg BrdU at the beginning of the time period studied and given drinking water containing 0.8mg/ml BrdU. 14 days later CD8+ T cell proliferation was assessed in peripheral blood and tissues using the FITC BrdU Flow Kit (BD Pharmingen).
Adoptive transfer of OT-I
For adoptive transfer of naïve CD8+ T cells, 5×102 Thy1.1 OT-I T cells from peripheral blood were injected i.v. into naïve Thy1.2 C57Bl/6 mice (Badovinac et al., 2007). To transfer memory OT-I T cells, LM-OVA immune mice containing memory OT-I T cells were euthanized at indicated time points. Percentage of OT-I T cells in total spleen cells was determined by FACS and a spleen cell mixture containing 6×104 memory OT-I T cells was injected into naïve Thy1.2 C57Bl/6 mice. Recipient mice were immunized 24h after the adoptive transfer by i.v. injection of 5×106 CFU actA-deficient LM-OVA.
Quantification of CD8 T cell responses
CD8+ T cell responses were analyzed in peripheral blood (PBL) samples by FACS analysis for Thy1.1 transgenic OT-I T cells. For quantification of antigen-specific CD8+ T cell responses and assessment of CD8+ T cell tissue distribution, mice were euthanized at the indicated time points and perfused through the heart with PBS prior to harvesting the organs. Single-cell suspensions were prepared from organs or PBL and analyzed for Thy1.1 positive OT-I T cells by FACS analysis. Peptide-stimulated intracellular cytokine staining (ICS) was performed in peripheral blood samples as previously described (Badovinac et al., 2002).
Microarray data acquisition
1°, 2°, 3° or 4° OT-I T cells were FACS-sorted from individual mice at 85–95 days after the last infection with att LM-OVA. OT-I T cell populations were approximately 99% pure and vital. Absolute CD8+ T cell yields ranged from 6×104 to 5×105 T cells per sorted sample. Three samples from individual mice were obtained for each memory CD8+ T cell group. For 4° memory CD8+ T cells, spleens from two to three mice were pooled to obtain sufficient CD8+ T cell numbers for each of the three samples. RNA was extracted using the RNEasy Kit (Qiagen) and 5–50 ng of total RNA was used for microarray analysis. RNA quality was assessed using the Agilent Model 2100 Bioanalyzer. RNA for the microarray was processed using the NuGEN WT-Ovation Pico RNA Amplification System along with the NuGEN WT-Ovation Exon Module. Samples were hybridized and loaded onto Affymetrix GeneChip Mouse GENE 1.0 ST arrays. Arrays were scanned with the Affymetrix Model 7G upgraded scanner and data were collected using the GeneChip Operating Software (GCOS). GSEA was performed as described (Haining and Wherry, 2010; Subramanian et al., 2005).
Microarray data analysis
Data from the Affymetrix Mouse Exon 1.0 ST arrays were first quantile normalized, median polished using Roubust Multi-chip Average (RMA) background correction with log2 adjusted values. Partek batch correction was used to remove variation due to the hybridization batch. Naive cells were assayed in both hybridization batches to provide a common reference to facilitate this correction. Probe sets for exons were then summarized for a specific gene using the median value. After obtaining log2 expression values for genes, significance testing was performed using ANOVA. The ANOVA model compared the five different CD8+ T cell groups (naive, 1°, 2°, 3°, and 4° memory). Linear contrasts were used to compare specific subsets of the ANOVA model. FDR (false discovery rate) correction was applied to all of the p-values to correct for multiple testing. Significance was assessed by a FDR q-value < 0.01. Analysis and visualization was done in PartekGS software.
Functional assignment of the genes was performed using the “Functional Annotation Tools” in DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov) following recommended protocols (Huang da et al., 2009). Functional categories were listed in a priority order based on our interpretation of its relevance in CD8+ T cell functions. Genes that appeared in higher priority functional category were not included in categories with lower priority, such as in Table 1. Some additional groups (e.g. killer-cell lectin-like receptors) were added manually. The total number of genes shown was reduced by excluding genes with low absolute changes in mRNA quantity as indicated in Table 1. Enrichment of genes in known pathways were analyzed using the KEGG pathway tool and GSEA was performed as described (Haining and Wherry, 2010; Subramanian et al., 2005).
Supplementary Material
Acknowledgements
We thank Dr John Colgan (University of Iowa) for advice with cell sorting, Garry Hauser for microarray analysis (DNA Facility, University of Iowa), and Dr Stanley Perlman (University of Iowa) for critically reviewing the manuscript. The data presented herein were in part obtained at the Flow Cytometry Facility, which is a Carver College of Medicine Core Research Facilities/ Holden Comprehensive Cancer Center Core Laboratory at the University of Iowa. This work was supported by start-up funds from the Department of Pathology, University of Iowa (V.P.B.), NIH grants AI83286 (V.P.B.), AI42767, AI46653, AI50073, AI59752 (J.T.H.), AI077504 and HL095540 (H.H.X.) and the Deutsche Forschungsgemeinschaft (DFG) fellowship WI 3308/1-1 (T.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: The complete Microarray data described in this study have been deposited in the NCBI GEO (Gene Expression Omnibus) under GEO accession number GSE21360.
The authors declare no competing financial interest.
References
- Appay V, Douek DC, Price DA. CD8 T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8 T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Messingham KA, Hamilton SE, Harty JT. Regulation of CD8 T cells undergoing primary and secondary responses to infection in the same host. J Immunol. 2003;170:4933–4942. doi: 10.4049/jimmunol.170.10.4933. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8 T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8 T cells after infection. Nat Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- Bianchi T, Gasser S, Trumpp A, MacDonald HR. c-Myc acts downstream of IL-15 in the regulation of memory CD8 T-cell homeostasis. Blood. 2006;107:3992–3999. doi: 10.1182/blood-2005-09-3851. [DOI] [PubMed] [Google Scholar]
- Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8 T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T, Newton R, Liu SM, Weininger L, Young TR, Silva DG, Bertoni F, Rinaldi A, Chappaz S, Sallusto F, et al. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Haring JS, Corbin GA, Harty JT. Dynamic regulation of IFN-gamma signaling in antigen-specific CD8 T cells responding to infection. J Immunol. 2005;174:6791–6802. doi: 10.4049/jimmunol.174.11.6791. [DOI] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. Shaping and reshaping CD8 T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8 T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jabbari A, Harty JT. Secondary memory CD8 T cells are more protective but slower to acquire a central-memory phenotype. J Exp Med. 2006;203:919–932. doi: 10.1084/jem.20052237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8 T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Larbi A, Ozcelik D, Solana R, Gouttefangeas C, Attig S, Wikby A, Strindhall J, Franceschi C, Pawelec G. Cytomegalovirus infection: a driving force in human T cell immunosenescence. Ann N Y Acad Sci. 2007;1114:23–35. doi: 10.1196/annals.1396.043. [DOI] [PubMed] [Google Scholar]
- Lefrancois L. Development, trafficking, and function of memory T-cell subsets. Immunol Rev. 2006;211:93–103. doi: 10.1111/j.0105-2896.2006.00393.x. [DOI] [PubMed] [Google Scholar]
- Lin SY, Elledge SJ. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- Masopust D, Ha SJ, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol. 2006;177:831–839. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- McGill J, Van Rooijen N, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med. 2008;205:1635–1646. doi: 10.1084/jem.20080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Kriajevska M, Vandewalle C, Chernova T, Sayan AE, Berx G, Mellon JK, Tulchinsky E. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol Biol Cell. 2007;18:4615–4624. doi: 10.1091/mbc.E07-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Sandau MM, Kohlmeier JE, Woodland DL, Jameson SC. IL-15 regulates both quantitative and qualitative features of the memory CD8 T cell pool. J Immunol. 2010;184:35–44. doi: 10.4049/jimmunol.0803355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NW, Podyminogin RL, Butler NS, Badovinac VP, Tucker BJ, Bahjat KS, Lauer P, Reyes-Sandoval A, Hutchings CL, Moore AC, et al. Memory CD8 T cell responses exceeding a large but definable threshold provide long-term immunity to malaria. Proc Natl Acad Sci U S A. 2008;105:14017–14022. doi: 10.1073/pnas.0805452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8 T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Wherry EJ. CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol. 2007;19:408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, Masopust D. Memory CD8 T-cell compartment grows in size with immunological experience. Nature. 2009;457:196–199. doi: 10.1038/nature07486. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8 T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4 T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–545. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- Woodland DL. Jump-starting the immune system: prime-boosting comes of age. Trends Immunol. 2004;25:98–104. doi: 10.1016/j.it.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DM, Yu S, Zhou X, Haring JS, Held W, Badovinac VP, Harty JT, Xue HH. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J Immunol. 2010;184:1191–1199. doi: 10.4049/jimmunol.0901199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.