Abstract
Background
Escherichia coli serogroup O157:H7 has emerged as an important zoonotic bacterial pathogen, causing a range of symptoms from self-limiting bloody diarrhea to severe hemorrhagic colitis and hemolytic-uremic syndrome in humans. Beef and dairy cattle are considered the most important animal reservoirs for this pathogen. One of the important virulence characteristics of E. coli O157:H7 is the eaeA gene encoding the 97 kDa surface protein intimin. Intimin is required for attachment and effacement during the interaction of enterohemorrhagic E. coli with human and bovine neonatal enterocytes. The present study was undertaken to test the hypothesis that an adaptive mucosal immune response directed against intimin will reduce or prevent enteric colonization and fecal shedding of E. coli O157:H7 in cattle.
Results
Cattle were orally inoculated with either milk (control), milk with live attenuated Salmonella enterica serovar Dublin (vector), or milk with live attenuated recombinant S. Dublin expressing intimin (vaccinated) on days 0, 14 and 28. On day 98, all calves were challenged orally with E. coli O157:H7 to evaluate whether vaccination with the recombinant S. Dublin expressing intimin would reduce the level of E. coli O157:H7 fecal shedding.
During the first 28 days, vaccinated calves shed both the vector strain and the intimin-expressing S. Dublin strain at a similar level. The vector strain was shed for a significantly longer period as compared to the level of recombinant vaccine strain. Calves that received the intimin-expressed vaccine ceased shedding S. Dublin from day 28 to day 63. All calves were challenged with E. coli O157:H7 on day 98 to determine the effect on fecal shedding of E. coli O157:H7. The amount of E. coli O157:H7 in feces was measured for 30 days post-challenge. We observed a transient clearance of E. coli O157:H7 from the feces in the vaccinated calves. The magnitude of fecal E. coli O157:H7 shedding did not correlate with the presence of intimin-specific fecal IgA.
Conclusion
Oral vaccination with live attenuated recombinant S. Dublin expressing intimin reduced enteric colonization and fecal shedding of E. coli O157:H7. However, the transient clearance of E. coli O157:H7 was not associated with an enhanced IgA-mediated mucosal immune response.
Background
Escherichia coli serogroup O157:H7 (E. coli O157:H7) is a zoonotic bacterial pathogen that causes symptoms ranging from self-limiting bloody diarrhea to severe hemorrhagic colitis in humans [1,2]. E. coli O157:H7 infection can also cause extra-intestinal illness, most importantly hemolytic-uremic syndrome (HUS). The majority of E. coli O157:H7-associated fatalities results from renal failure, neurologic manifestations, or other complications of HUS [3-5]. E. coli O157:H7 is mainly a food borne pathogen. Beef and dairy cattle are considered to be the most important animal reservoirs of E. coli O157:H7 [6-12]. Transmission of E. coli O157:H7 by fecal contaminated water [13,14] is thought to be a major source of infection. Some person-to-person transmission has been also reported [15,16], but the main source of human infection with E. coli O157:H7 is contamination of food products.
The infective dose of E. coli O157:H7 is low for both calves and humans, in some cases approximately only 102 organisms are required to cause infection [17]. Neonatal calves are particularly susceptible to E. coli O157:H7, but adult cattle do not generally exhibit clinical signs following experimental or natural infection. Adult cattle typically continue to shed bacteria in their feces for weeks to months, or for the lifetime of the animal. Carcasses of non-colonized cattle have sometimes been found to contain E. coli O157:H7 in the abattoir, suggesting that cross-contamination during meat processing can be a major source of contamination of beef products and subsequent infection of humans [9].
One of the important virulence factors of E. coli O157:H7 is the eaeA gene that encodes the 97 kDa surface protein intimin. Intimin is required for E. coli O157:H7 colonization, the development of attaching and effacing epithelial lesions, and disease in neonatal calves, pigs, and mice [18]. Intimin-specific antiserum can block adherence of E. coli O157:H7 to HEp-2 cells in tissue culture [19]. Calves challenged with intimin-deficient mutant bacteria do not develop diarrhea or attaching/effacing lesions, nor are colonized to the same extent as animals infected with wild type or complemented mutant strains [20]. Earlier studies have proposed that mucosal IgA directed against intimin might serve an analogous function in vivo [21]. However, experimental challenge of cattle previously infected with E. coli O157:H7 has failed to demonstrate protective immune responses [22], perhaps because E. coli O157:H7 generate very low titers of specific mucosal IgA responses directed against intimin or other E. coli O157:H7 antigens [23]. E. coli O157:H7 colonization of mice can be reduced when the animals are fed recombinant tobacco expressing intimin [24]. It is suggested that intimin on the surface of EHEC would bind to nucleolin [25]. The present study was undertaken to test the hypothesis that a specific adaptive mucosal immune response directed against the surface antigen intimin might prevent or reduce the colonization of E. coli O157:H7 in cattle.
Methods
Cloning the eaeA gene into pRB3
The eaeA gene was amplified from pEB310 using primers SW20H3: 5'-CGCCCAAGCTTCGTTGTTAAGTCAATGG-3' and EaeA 3': 5'-CGCGGATCCAGTAGTAGATTTGATTATAAGAGG-3' by PCR and cloned into the HindIII/SmaI site of pRB3. Plasmid DNA was introduced into S. Dublin aroA::tet by electroporation. His-tagged EaeA was produced by cloning the coding region of eaeA into pET16b (Novagen, Gibbstown, NJ). Expression and purification of His-tagged EaeA on NTA-Nickel resin (Qiagen, Valencia, CA) was performed according to the manufacturer's instructions. His-tagged EaeA was concentrated and stored in 50 mM Tris-HCL 250 mM NaCl, 0.1 mM EDTA and 1 mM DTT.
Identification of Salmonella- and E. coli O157:H7-free calves
Clinically healthy male Holstein/Friesian calves, aged 1 to 2 weeks, were obtained from a local supplier. The weight of the calves ranged between 40 and 45 kg. Animals were cared for according to the Association for Assessment and Accreditation of Laboratory Animal Care guidelines under the oversight of the Texas A&M University Institutional Animal Care and Use Committee AUP 2000-252. Calves were fed 2 liters of antibiotic-free whey-based milk replacer twice daily and given water ad libitum. Before being used for experiments, calves were clinically evaluated for fever and infection with Salmonella and E. coli 0157:H7. The presence of Salmonella was evaluated by incubation of fecal samples in tetrathionate broth (Difco), followed by enrichment in Rappaport-Vassiliadis R10 broth (Difco), then by plating onto XLT-4 plates (BBL). All calves were free of Salmonella and E. coli O157:H7.
Calves were divided into 3 treatment groups. The control group consisted of 3 calves that were fed only milk replacer (950 ml) and 50 ml of inoculum buffer (a suspension of 5% magnesium trisilicate, 5% sodium bicarbonate, and 5% magnesium carbonate) on the inoculation days. The second group consisted of the vector group (4 animals). The calves in this group were inoculated with 1010 colony forming units (CFU) of the S. Dublin aroA strain with the empty pRB3 vector suspended in 50 ml of inoculum buffer in milk replacer. The third group (hereafter called the vaccinated group) consisted of 5 calves inoculated with 109-1010 CFU S. Dublin aroA pRB3::eaeA suspended in 50 ml of inoculum buffer in milk replacer.
Vaccination of calves with S. Dublin strains
Overnight cultures were grown in LB broth, and the optical density at 600 nm was determined. A volume containing the desired quantity of bacteria was added to 50 ml inoculum buffer. The inoculum was added to 950 ml of milk replacer and used to orally inoculate calves on days 0, 14 and 28. For all experiments, the bacterial titer of the inoculum was determined by plating serial dilutions onto LB agar plates, incubating plates overnight at 37°C, and enumerating the colonies.
Challenge of calves with virulent E. coli O157:H7 strain 86-24
All calves were challenged orally with 1010 CFU E. coli O15:H7 strain 86-24 [26] (kindly provided by Dr. Rod Moxley from the University of Nebraska, Lincoln-Nebraska) on day 98. Animals were inoculated with one liter of milk replacer containing 1010 CFU E. coli O15:H7 strain 86-24 that had been suspended in 50 ml of 5% magnesium trisilicate, 5% sodium bicarbonate, and 5% magnesium carbonate inoculum buffer.
Collection of fecal samples
Fecal samples from all the calves were collected daily from day 0 to day 42 post-inoculation to determine the level of vaccine strain shedding. After 42 days, fecal samples were collected weekly to determine the presence of S. Dublin vaccine strain shedding. Fecal samples were collected daily for 30 days to determine the amount of E. coli shedding following oral challenge with E. coli O157:H7 strain 86-24 on day 98.
Collection of peripheral blood for serum IgA measurement and enumeration of intimin-specific IgA-secreting cells
Peripheral blood was collected weekly for serum IgA and ELISpot assays. Blood was collected in serum separator tubes, kept at 37°C for 6 hours, and centrifuged at 2000 rpm for 30 min. Clarified serum was collected and stored at -20°C for serum IgA detection.
Serum IgA antibody measurement
An Immulon 2-HB flat bottom 96-well microtiter plate (Thermo Labsystems, Franklin, MA) was coated overnight at 4°C with 100 μl/well (5 μg/ml) His-tagged intimin. The plate was washed 3 times with wash buffer (PBS with 0.05% Tween 20), then blocked for 1 h at 37°C with 250 μL/well PBS containing 3% (wt/vol) dried nonfat milk powder (Carnation, Nestle, Glendale, CA). Bovine serum was diluted to 1:1000 with blocking buffer, and 100 μl/well volume of diluted serum was added to the plate and incubated for 3 h at 37°C. Wells were washed 3 times with wash buffer. One-hundred μl rabbit of anti-bovine IgA-horseradish peroxidase conjugate (Bethyl, Montgomery, TX) diluted 1:1000 in wash buffer with 3% (wt/vol) dried nonfat milk powder were added to each well. The plates were then incubated for 1 h at 37°C. Following incubation, the plates were washed 3 times with wash buffer, then incubated with 100 μL/well of a 1:1 mixture of 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) peroxidase substrate and peroxidase solution B (KPL, Gaithersburg, MD) for 1 h at 37°C. After color development (5 min at RT), absorbance was measured at 410 nm on a microplate reader (FLUOstar Optima, BMG Labtechnologies, INC, Durham, NC).
Mucosal (fecal) IgA antibody measurement
For the detection of mucosal IgA, feces were collected in individual sterile 50 ml centrifuge tubes from all calves. Fecal samples were weighed and liquefied by the addition of ice-cold 0.1 M sodium acetate buffer pH 4.5 (ratio 1:2). This fecal sample-buffer mixture was incubated at 56°C for 30 min. To inactivate proteolytic enzymes, a cocktail of soybean trypsin inhibitor, aprotinin, and phenylmethylsulphonyl chloride (PMSF) was added to this mixture and incubated for 30 min on ice [27]. After incubation, the fecal suspension was centrifuged at 15,000 g at 4°C for 30 min. Clarified supernatant was filtered through a low binding sterile filter (0.45 mM). Titers of intimin-specific IgA in the fecal sample were detected essentially the same way as described above for the intimin-specific IgA in serum, using fecal supernatant instead of serum samples.
ELISpot assay to detect intimin-specific IgA-secreting cells in peripheral blood
Peripheral blood samples were collected directly into 8 ml BD Vacutainer CPT (Becton Dickinson Vacutainer systems, Franklin lakes, NJ) that contained 1.0 ml of 0.1 M sodium citrate, 1 ml of Ficoll-Hypaque and a gel barrier. Peripheral blood mononuclear cells (PBMC) were isolated as described earlier [28]. Tubes were centrifuged at 3000 RPM for 30 min. The buffy coat containing white blood cells was collected and washed in PBS-citrate. Red blood cells were lysed by incubating the cell suspension in RBC lysis buffer (Analytical Genetic Testing Central, INC. Denver, CO) for 15 min. Cells were washed 2 times with PBS-citrate and finally resuspended in RPMI medium (Gibco BRL, Life Technologies, Inc., Grand Island, NY) supplemented with 15% fetal bovine serum, L-glutamine and sodium pyruvate. Viable cells were counted using trypan blue exclusion dye and a hemacytometer. Cells were kept on ice until they were aliquoted for the ELISpot assay. For the detection of intimin-specfic IgA-secreting cells, individual wells of an ELISpot plate (Millipore Cooperation, 290 Concord Road Billerica, Massachusetts) were incubated at 4°C overnight with 0.5 ug affinity purified bovine IgA diluted in coating buffer (50 mM carbonate buffer, pH 9.6). The coated plate was emptied and rinsed once with supplemented RPMI medium, then blocked with supplemented RPMI media at RT for 3 hrs. For ELISpot assays, 105 PBMC were plated in duplicate wells. Cells were stimulated with His-tagged intimin (200 ng per well) or PHA (100 ng/well from the Sigma Chemical Company, St. Louis, MO) and incubated for 18 hrs at 37°C in a humidified incubator containing 10% CO2. After incubation, cells were rinsed once with distilled water, then washed 3 times with PBS containing 0.05% Tween 20 (PBS-T). Anti-IgA antibodies conjugated to HRPO (100 ul of 1:1000 dilution in PBS-T) were added to each well, and the plate was further incubated for 3 hrs at 37°C. The plate was washed again with PBS-T and spots developed using 100 ul of substrate solution (3-amino-9-ethylcarbazol tablet from Sigma Chemicals, St. Louis, MO, reconstituted as per the manufacturer's recommendation). After development, the plate was emptied and rinsed ten times with distilled water. Antigen stimulated spots were reported by subtracting the number of spots obtained from wells without stimulant from the number of spots obtained in stimulant-added wells.
Qualitative Salmonella fecal culture
Shedding of Salmonella was monitored by collecting daily fecal swabs, followed by enrichment in tetrathionate broth (Beckton Dickinson and Company, Franklin Lakes, NJ), and in Rappaport-Vassiliadis R10 broth (Beckton Dickinson and Company, Franklin Lakes, NJ). Bacteria were enumerated by plating serial dilutions onto XLT-4 plates (Beckton Dickinson and Company, Franklin Lakes, NJ).
Quantitative E. coli O157:H7 fecal culture
Ten g samples of feces were immediately processed in a Stomacher, serially diluted in sterile phosphate-buffered saline, and plated in triplicate onto Sorbitol-MacConkey agar. The sensitivity of the direct plating was 50 CFU/g. A 10 g fecal sample was also added to enrichment broth (Tryptic Soy Broth with 0.15% bile salts), incubated overnight at 37°C, and plated onto selective medium. Colonies isolated on selective medium were confirmed as E. coli O157:H7 following the instructions of latex agglutination kit (BD Difco™E. Coli Antisera kit from Becton, Dickinson and Company, Cockeysville, MD).
Necropsy
Calves were euthanized by captive bolt, and a complete necropsy was performed. At necropsy, tissue samples for bacteriology and histopathology were collected from abomasum, omasum, duodenum, jejunum, ileum, cecum spiral colon, distal colon, rectum and mesenteric lymph node. Homogenates of each tissue were prepared in the Stomacher by mincing two 6 mm biopsy punches of each sample in phosphate-buffered saline. The tissue homogenates were then plated and incubated overnight at 37°C for enumeration of bacteria.
Statistical analysis
Data were analyzed using SAS version 9.1 (SAS Institute, Cary, NC). Statistical analysis was performed by repeated measures analysis test for between-subject effects (TRT), within-subject effects (Time), and within-subject-by-between-subject interaction effect (TRT*Time). Interaction effects are the joint effects of pairs, triplets, or higher-order combinations of the independent variables, different from what would be predicted from any of the independent variables acting alone. When an interaction is present, the effect of an independent on a dependent varies according to the values of another independent. If the probability of F is less than 0.05 for any such combination, we conclude that the interaction of the combination has an effect on the dependent. Note that the concept of interaction between two independents is not related to the issue of whether the two variables are correlated.
Results
Construction of attenuated Salmonella strains expressing cloned intimin
The eaeA gene including the upstream promoter region from E. coli O157:H7 86-24 was amplified by PCR from pEB310 and cloned into the low copy, RK2-based plasmid pRB3 [29]. This plasmid contains the par (partition) locus from RK2 and insures plasmid segregation and stable maintenance even in the absence of selection. This plasmid has been previously used for in vivo complementation of S. Typhimurium mutations in mice [30,31]. Western blot analysis revealed the production of full length EaeA as well as a smaller ~50-60 kD protein in E. coli K12 carrying pEB310. An immunoreactive protein corresponding to the smaller protein was also present in S. Dublin aroA with pRB3::eaeA, but the full length 96 kD EaeA protein was not seen (Figure 1). The presence of the smaller protein in both E. coli and S. Dublin suggests that EaeA is subject to proteolytic cleavage. A significant quantity of the smaller immunoreactive protein was expressed in S. Dublin aroA pRB3::eaeA, and this strain was used for subsequent vaccine trials.
Figure 1.
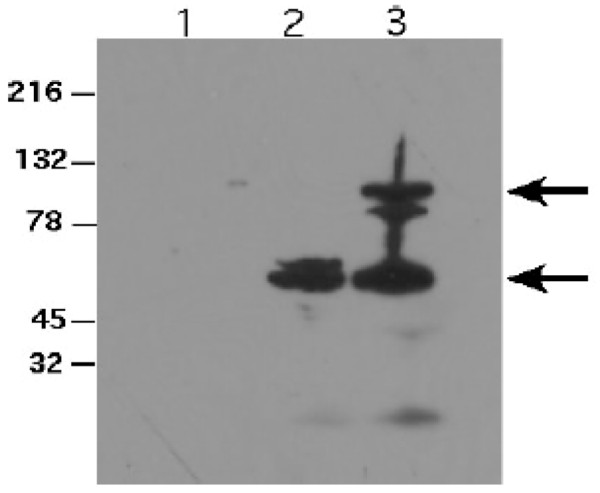
Expression of Intimin in aroA mutant S. Dublin. Protein from overnight cultures of S. Dublin aroA, S. Dublin aroA with pRB3::eaeA, or E. coli K12 with pEB310 were separated on 4-20% SDS-PAGE and expression of intimin determined by western blot. Lane 1, S. Dublin aroA, lane 2 S. Dublin aroA with pRB3::eaeA, and lane 3, E. coli K12 with pEB310. The full-length intimin protein is 96 kD (upper arrow) present in E. coli K12 with pEB310 but absent in S. Dublin aroA with pRB3::eaeA. A smaller immunoreactive protein (lower arrow) is present in both S. Dublin with pRB3::eaeA and E. coli K12 with pEB310.
Vaccination of calves with S. Dublin vaccine strains
All calves were orally inoculated with the non-recombinant S. Dublin aroA vector or with intimin-expressing S. Dublin aroA on days 0, 14 and 28. A detailed time line for the experiment is provided in Figure 2.
Figure 2.
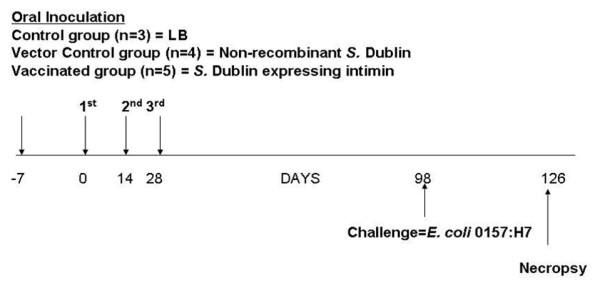
Time frame of experiment and collection of samples. Calves were divided into 3 treatment groups. Calves (n = 3) in the control group were fed only milk on days 0, 14 and 28. Calves (n = 4) in the vector group were orally inoculated with the live attenuated non-recombinant S. Dublin strain on days 0, 14 and 28. Calves (n = 5) in the vaccinated group were inoculated orally with live attenuated recombinant S. Dublin expressing E. coli O157:H7 intimin on days 0, 14 and 28. All calves were challenged orally with E. coli O157:H7 on day 98 post-vaccination. Fecal samples from calves were collected daily from all calves from day 0 to day 42 post-inoculation for Salmonella culture, and fecal samples were collected weekly thereafter. After oral E. coli O157:H7 challenge, fecal samples were collected daily to monitor E. coli O157:H7 shedding. Peripheral blood samples were collected weekly after inoculation to measure levels of serum IgA and to detect intimin-specific IgA-secreting cells by ELISpot assay.
Calves that received S. Dublin strains shed for variable amounts of time following vaccination. Most calves shed the vaccine strains intermittently until the end of the experiment (126 days post-immunization), indicating the establishment of the vector/vaccine strain in the host. Figure 3 depicts fecal shedding of S. Dublin strains until 98 days post-immunization. Following the first immunization, both vector and vaccinated groups had similar percentages of calves positive for shedding of S. Dublin aroA (vector 33%, vaccinated 40%). However, after the third immunization, the vector group contained significantly higher numbers of calves positive for S. Dublin shedding as compared to animals immunized with intimin-expressing S. Dublin. In order to normalize short-term fluctuations and highlight longer-term trends, we calculated the moving average for the shedding of Salmonella after immunization (Figure 3). By the end of the experiment, the frequency of positive shedders in both the groups was similar (~44%).
Figure 3.
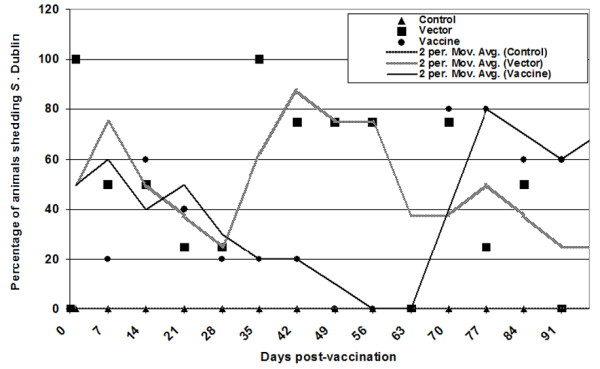
Duration (x axis) and percentage (y axis) of S. Dublin positive calves as measured by qualitative evaluation of fecal shedding. Calves were inoculated orally as indicated on days 0, 14 and 28. Solid markers (triangle, square or circle) indicate the percentage of calves shedding S. Dublin. A moving average trendline was calculated to normalize fluctuations in shedding. The moving average was calculated by setting the period as 2, with the average of the first two data points used as the first point in the moving average trendline. The average of the second and third data points was used as the second point in the trendline, and subsequent trendline points were calculated accordingly.
Serum IgA antibody response
Intimin-specific IgA was measured weekly in the serum of all calves throughout the experiment (Figure 4). We observed an increase in intimin-specific IgA in calves receiving either the vector or the intimin-expressing strain. Statistical analyses revealed a significant time effect (P = 0.036), but no significant treatment effect (P = 0.87) or treatment × time interaction (P = 0.4). Also, prior to the E. coli O157:H7 challenge, there was a significant time effect (P = 0.0009), but no significant treatment effect (P = 0.25) or treatment × time interaction (P = 0.26). A sporadic increase in intimin-specific IgA levels was observed in the control group. However, we did not notice any symptoms of clinical infection in these animals. Mucosal (fecal) IgA responses with intimin-specific IgA antibody were observed in all animals (Figure 5). There was a significant time effect (P < 0.001), but no significant treatment effect (P = 0.2) or treatment × time effect (P = 0.5). Fecal anti-intimin IgA levels prior to the E. coli O157:H7 challenge showed a significant time effect (P < 0.001) and treatment × time interaction (P = 0.0009), but no significant treatment effect (P = 0.2).
Figure 4.
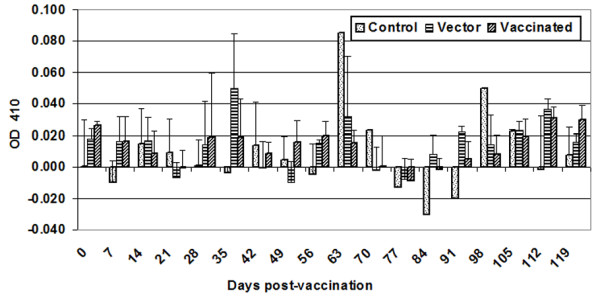
Serum IgA antibody response. Peripheral blood was collected weekly from all calves during the length of the vaccination study. Intimin-specific serum IgA antibody concentrations were measured by ELISA. The bar graph depicts the mean (± standard deviation) of animals in each group. Normalization of data was performed by subtracting the OD410 of diluent-only wells from experimental values.
Figure 5.
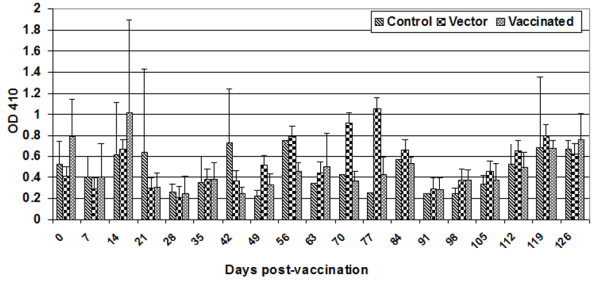
Mucosal (fecal) IgA antibody response. Intimin-specific fecal IgA antibody concentrations were measured by ELISA. The bar graph depicts the mean (± standard deviation) of calves in each group. Normalization of data was performed by subtracting the OD410 of diluent-only wells from experimental values.
Intimin-specific IgA secreting cells in peripheral blood mononuclear cells
An increase in intimin-specific IgA secreting cells was measured by the ELISpot assay (Figure 6). There was a significant treatment effect (P = 0.04) and time effect (P = 0.001), but no significant treatment × time interaction (P = 0.19). Using multiple comparisons, there was a significant difference between control and vaccinated animals, and between control and vector animals. However, before the E. coli O157:H7 challenge, there was a significant time effect (P < 0.03) and treatment × time interaction (P = 0.03), but no significant treatment effect (P = 0.6).
Figure 6.
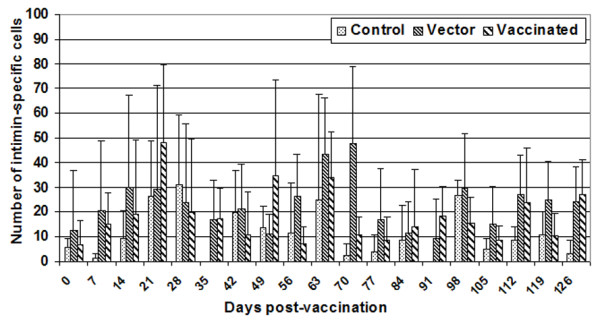
Intimin specific IgA secreting cells in peripheral blood mononuclear cells. 105 PBMC were plated on bovine IgA-coated wells. Cells were stimulated with intimin (200 ng per well) and incubated for 18 hrs at 37°C. After incubation, cells were incubated with anti-IgA antibody conjugated to HRPO, and spots were developed using substrate solution. Intimin-specific IgA-secreting cells were determined by subtracting the number of spots obtained in wells without any stimulant from the number of spots obtained in wells to which intimin was added. The bar graph depicts the mean (± standard deviation) of animals in each group.
Shedding of E. coli O157:H7
All calves were challenged orally with E. coli O157:H7 on day 98 post-vaccination. E. coli O157:H7 shedding was measured after the challenge (Figure 7). There was a significant time effect (P < 0.001) and treatment × time interaction (P = < 0.001), but no significant treatment effect (P = 0.27). However, when the trend of shedding E. coli O157:H7 was calculated as a polynomial trendline, the trend revealed that the vaccinated calves descended into the "valley" of the trendline earlier than those who received the vector, whereas the control group never reached the valley of the trendline during the entire study period. Moreover, levels of shedding were lower in animals receiving the intimin-expressing vaccine strain. This indicates an early, albeit transient, clearance of the challenge strain in vaccinated calves.
Figure 7.
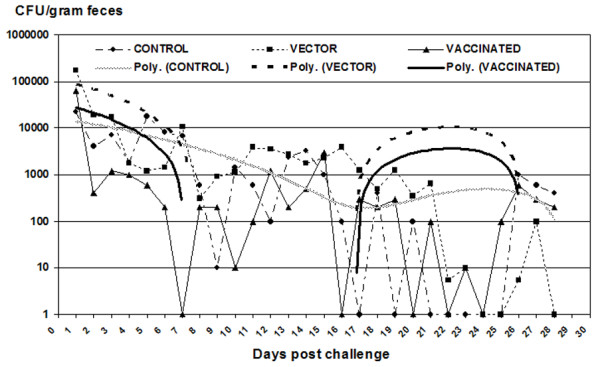
Magnitude and duration of fecal shedding of E. coli O157:H7. Magnitude (median of each group, CFU/gram of feces) and duration of fecal shedding post-challenge with E. coli O157:H7 were calculated quantitatively by direct plating as well as by enrichment culture following oral challenge with E. coli O157:H7 during the 30 days post challenge. Specimens containing less than the detection limit (E. coli O157:H7 found only by enrichment) were assigned a value of 10. Negative specimens were assigned a value of 1. Samples too numerous to count (TNTC) were considered to contain the maximum number counted.
Bacteriology and Histopathology
None of the examined tissue was positive for the colonization of bacteria. No notable differences were detected in the histopathology among the treatment groups.
Discussion
Enterohemorrhagic Escherichia coli (EHEC) such as strain O157:H7 is an etiologic agent of acute enteric diseases in both humans and neonatal calves [32]; however, mature cattle are not affected. E. coli O157:H7 can enter the human food supply from cattle via fecal contamination of beef carcasses at slaughter [33]. Intimin is an outer membrane protein expressed by several human and animal enteric pathogens, including enteropathogenic E. coli and EHEC [34-38]. Antibodies to intimin may prevent the initial steps of EHEC colonization in the gastrointestinal tract [39-41]. Anti-intimin immune responses can modulate the outcome of experimental infection with the EPEC-like bacterium Citrobacter rodentium in rabbits and supports the inclusion of intimin as a component of an EPEC or EHEC vaccine [42]. Vaccination of cattle has significant potential as a pre-harvest intervention strategy to reduce E. coli O157:H7 shedding. However, the ability of intimin to elicit protective immune responses in the bovine intestinal tract has not previously been demonstrated. We hypothesized that the mucosal immune response elicited by live attenuated Salmonella enterica serovar Dublin expressing the intimin protein of E. coli O157:H7 would reduce the magnitude and duration of E. coli O157:H7 colonization and fecal shedding.
In the present study, orally-administered S. Dublin vector or S. Dublin expressing E. coli O157:H7 intimin were recovered for up to 98 days post-inoculation in the feces of calves, confirmed the establishment of intestinal carriage. The frequency of positive S. Dublin shedders in both vaccinated groups at the end of experiment were similar as observed in other studies in which vaccine strains administered to cattle were shed for a considerable period of time [31]. From 35-70 days post-inoculation, the S. Dublin vector control strain was detectable in a significantly higher proportion of calves than the intimin-expressing S. Dublin vaccine strain. Levels of intimin-specific IgA in serum and feces were not significantly higher in calves receiving intimin-expressing S. Dublin. However, cattle immunized with the intimin-expressing strain group exhibited a reduced magnitude and duration of E. coli O157:H7 shedding following oral challenge.
Earlier studies reported that infection of seropositive adult cattle with E. coli O157:H7 increases serum antibody titers to intimin and to the translocated intimin receptor (Tir) [23]. Intimin interacts not only with Tir, but also with host cell intimin receptor(s) on the luminal surface of intestinal epithelia, including integrin and nucleolin [43,44]. These receptors are potentially accessible as binding sites for intimin during vaccination with recombinant S. Dublin. While antibodies directed against either Tir or intimin might impede intimin-Tir interactions, antibodies to intimin might be anticipated to inhibit EHEC binding to alternative host receptors as well. Recently, it has been shown that vaccination with a combination of antigens associated with type III secretion system-mediated adherence; the translocon filament protein, EspA, the extracellular region of the outer membrane adhesin, intimin, and Tir significantly reduced shedding of EHEC O157 from experimentally infected animals [45].
In our study, initial shedding of E. coli O157:H7 after challenge was comparable in the groups receiving either vector or intimin-expressing S. Dublin, and significantly lower than in the control group. Earlier studies have indicated that E. coli persists (a challenge dose of 109) for days to weeks in the bovine intestinal tract before being cleared [46]. Similar results were observed in this study; however, it is important to note that the challenge dose in the present study was higher (1010). The polynomial trendline revealed that E. coli O157:H7 was cleared more rapidly from vaccinated calves than from control or vector-vaccinated animals. This provides evidence in support of the principle that potentiation of immune responses to intimin at the mucosal surface can reduce shedding of the pathogenic E. coli O157:H7 strain. Also, the possibility of interaction of various components of adaptive immunity due to initial Salmonella (vector) infection could not be ruled out [47]. We originally hypothesized that the protective responses would be related to fecal concentrations of intimin-specific IgA. However, the mechanism of vaccine protection is clearly more complex, as anti-intimin fecal IgA levels did not correlate with fecal shedding. Enteric mucosal IgA responses against intimin and type III secreted proteins were identified in rectal mucus and in the rectal tissue respectively [48,49]. These studies definitely indicate the importance of other clinical samples (tissue and rectal mucus) for studying the mucosal immune response. Moreover, protection against enteric pathogens by immunization does not essentially require secretory IgA [45], and intestinal clearance of intimin-expressing Citrobacter rodentium has been shown to require B cells and IgG antibodies, but not secretory IgA [45]. Importantly, intimin-specific antibody titers in colostrum and serum of dams were found to be increased after parenteral vaccination with intimin [50]. In another study, immunization of calves with the cell-binding domain of intimin subtypes beta or gamma via the intramuscular route induced antigen-specific serum IgG1 and, in some cases salivary IgA responses, but did not reduce the magnitude or duration of faecal excretion of EHEC upon subsequent experimental challenge [51]. The role of IgG in intimin-expressing vaccine induced protection of calves is worthy of further investigation.
Of note, reduction in colonization and shedding was obtained in this study by oral vaccination without a preceding parenteral inoculation. However, oral vaccines can significantly boost mucosal immune responses when primed by parenteral vaccine administration [52,53]. Parenteral priming of the immune system facilitates the gut associated lymphoid tissue to react more rapidly to antigens delivered by oral immunization, and may decrease the likelihood of inducing oral immune tolerance [24]. Another possible explanation for the modest degree of reduction in colonization and shedding of E. coli O157:H7 may be that intimin-specific mucosal IgA was already present prior to immunization. Thus, this antibody may have interfered with ability of the intimin-expressing S. Dublin vaccine strain to effectively reach gut-associated lymphoid tissue and augment local immune responses. One limitation to the present study is the need for replication in outbred populations of cattle having a more defined immune status.
Conclusions
In summary, a live S. Dublin vaccine strain expressing the E. coli O157:H7 intimin protein effectively colonized the intestines of calves after vaccination. Immunization resulted in a transient clearance and subsequently reduced colonization and shedding of E. coli O157:H7 following challenge.
Authors' contributions
SK coordinated sampling of animals, completed immunological assays, analyzed data and drafted the manuscript. SZ cultured S. Dublin. FCF and SJL designed and constructed the S. Dublin vaccine strain. DH and RP isolated PBMC from blood and evaluated shedding of bacteria. WA performed statistical analyses. LGA, FCF, and SJL are funded co-applicants who designed the study and edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Sangeeta Khare, Email: skhare@cvm.tamu.edu.
Walid Alali, Email: walali@uga.edu.
Shuping Zhang, Email: zhang@cvm.msstate.edu.
Doris Hunter, Email: dhunter@cvm.tamu.edu.
Roberta Pugh, Email: rpugh@cvm.tamu.edu.
Ferric C Fang, Email: fcfang@u.washington.edu.
Stephen J Libby, Email: slibby@u.washington.edu.
L Garry Adams, Email: gadams@cvm.tamu.edu.
Acknowledgements
We acknowledge funding from USDA CSREES National Research Initiative number 9800488 and grant number 1999-35201-8578 to support the project. We are grateful for the active contribution of all student workers and the farm manager (Mr. Alan Patranella) who provided animal care.
References
- Armstrong GL, Hollingsworth J, Morris JG. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18(1):29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- Berkelman RL. Emerging infectious diseases in the United States, 1993. J Infect Dis. 1994;170(2):272–277. doi: 10.1093/infdis/170.2.272. [DOI] [PubMed] [Google Scholar]
- Lawson JM. Update on Escherichia coli O157:H7. Curr Gastroenterol Rep. 2004;6(4):297–301. doi: 10.1007/s11894-004-0081-7. [DOI] [PubMed] [Google Scholar]
- Mariani-Kurkdjian P, Bingen E. [Escherichia coli O157:H7, an emerging pathogen] Presse Med. 1999;28(37):2067–2074. [PubMed] [Google Scholar]
- Richards A, Goodship JA, Goodship TH. The genetics and pathogenesis of haemolytic uraemic syndrome and thrombotic thrombocytopenic purpura. Curr Opin Nephrol Hypertens. 2002;11(4):431–435. doi: 10.1097/00041552-200207000-00010. [DOI] [PubMed] [Google Scholar]
- Berry ED, Dorsa WJ, Siragusa GR, Koohmaraie M. Bacterial cross-contamination of meat during liquid nitrogen immersion freezing. J Food Prot. 1998;61(9):1103–1108. doi: 10.4315/0362-028x-61.9.1103. [DOI] [PubMed] [Google Scholar]
- Duffy G, O'Brien SB, Carney E, Sheridan JJ, McDowell DA, Blair IS. Characterisation of E. coli O157 isolates from bovine hide and beef trimming in Irish abattoirs by pulsed field gel electrophoresis. J Microbiol Methods. 2005;60(3):375–382. doi: 10.1016/j.mimet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Flores RA, Tamplin ML, Marmer BS, Phillips JG, Cooke PH. Transfer coefficient models for Escherichia coli O157:H7 on contacts between beef tissue and high-density polyethylene surfaces. J Food Prot. 2006;69(6):1248–1255. doi: 10.4315/0362-028x-69.6.1248. [DOI] [PubMed] [Google Scholar]
- Hepburn NF, MacRae M, Ogden ID. Survival of Escherichia coli O157 in abattoir waste products. Lett Appl Microbiol. 2002;35(3):233–236. doi: 10.1046/j.1472-765X.2002.01173.x. [DOI] [PubMed] [Google Scholar]
- Sharma M, Richards GM, Beuchat LR. Survival and growth of Escherichia coli O157:H7 in roast beef and salami after exposure to an alkaline cleaner. J Food Prot. 2004;67(10):2107–2116. doi: 10.4315/0362-028x-67.10.2107. [DOI] [PubMed] [Google Scholar]
- Uhitil S, Jaksic S, Petrak T, Botka-Petrak K. Presence of Escherichia coli O157:H7 in ground beef and ground baby beef meat. J Food Prot. 2001;64(6):862–864. doi: 10.4315/0362-028x-64.6.862. [DOI] [PubMed] [Google Scholar]
- Wachtel MR, McEvoy JL, Luo Y, Williams-Campbell AM, Solomon MB. Cross-contamination of lettuce (Lactuca sativa L.) with Escherichia coli O157:H7 via contaminated ground beef. J Food Prot. 2003;66(7):1176–1183. doi: 10.4315/0362-028x-66.7.1176. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Lisay CM, Yurkovetskiy AV, Sawan SP. Persistent silver disinfectant for the environmental control of pathogenic bacteria. Am J Infect Control. 2003;31(4):208–214. doi: 10.1067/mic.2003.23. [DOI] [PubMed] [Google Scholar]
- Mattick K, Durham K, Domingue G, Jorgensen F, Sen M, Schaffner DW, Humphrey T. The survival of foodborne pathogens during domestic washing-up and subsequent transfer onto washing-up sponges, kitchen surfaces and food. Int J Food Microbiol. 2003;85(3):213–226. doi: 10.1016/S0168-1605(02)00510-X. [DOI] [PubMed] [Google Scholar]
- Weber DJ, Rutala WA. The emerging nosocomial pathogens Cryptosporidium, Escherichia coli O157:H7, Helicobacter pylori, and hepatitis C: epidemiology, environmental survival, efficacy of disinfection, and control measures. Infect Control Hosp Epidemiol. 2001;22(5):306–315. doi: 10.1086/501907. [DOI] [PubMed] [Google Scholar]
- Williams RC, Isaacs S, Decou ML, Richardson EA, Buffett MC, Slinger RW, Brodsky MH, Ciebin BW, Ellis A, Hockin J. Illness outbreak associated with Escherichia coli O157:H7 in Genoa salami. E. coli O157:H7 Working Group. Cmaj. 2000;162(10):1409–1413. [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Richards BL, Rice DH, Hancock DD. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol Infect. 2001;127(3):555–560. doi: 10.1017/S095026880100615X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66(9):4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee ML, O'Brien AD. Truncated enterohemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to HEp-2 cells. Infect Immun. 1996;64(6):2225–2233. doi: 10.1128/iai.64.6.2225-2233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick NA, Booher SL, Moon HW. Intimin facilitates colonization by Escherichia coli O157:H7 in adult ruminants. Infect Immun. 2002;70(5):2704–2707. doi: 10.1128/IAI.70.5.2704-2707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima H, Takahashi I, Kiyono H. Mucosal immune network in the gut for the control of infectious diseases. Rev Med Virol. 2001;11(2):117–133. doi: 10.1002/rmv.307. [DOI] [PubMed] [Google Scholar]
- Cray WC, Moon HW. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61(4):1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider G, Berberov EM, Moxley RA. Isotype-specific antibody responses against Escherichia coli O157:H7 locus of enterocyte effacement proteins in adult beef cattle following experimental infection. Vet Immunol Immunopathol. 2007;118(3-4):229–238. doi: 10.1016/j.vetimm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Judge NA, Mason HS, O'Brien AD. Plant cell-based intimin vaccine given orally to mice primed with intimin reduces time of Escherichia coli O157:H7 shedding in feces. Infect Immun. 2004;72(1):168–175. doi: 10.1128/IAI.72.1.168-175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair JF, O'Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002;277(4):2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109(9):705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- VanCott JL, Brim TA, Simkins RA, Saif LJ. Isotype-specific antibody-secreting cells to transmissible gastroenteritis virus and porcine respiratory coronavirus in gut- and bronchus-associated lymphoid tissues of suckling pigs. J Immunol. 1993;150(9):3990–4000. [PubMed] [Google Scholar]
- Khare S, Hondalus MK, Nunes J, Bloom BR, Garry Adams L. Mycobacterium bovis DeltaleuD auxotroph-induced protective immunity against tissue colonization, burden and distribution in cattle intranasally challenged with Mycobacterium bovis Ravenel S. Vaccine. 2007;25(10):1743–1755. doi: 10.1016/j.vaccine.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Berggren RE, Wunderlich A, Ziegler E, Schleicher M, Duke RC, Looney D, Fang FC. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10(5):489–495. doi: 10.1097/00042560-199512050-00001. [DOI] [PubMed] [Google Scholar]
- Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72(2):1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol. 2002;43(3):771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- Dean-Ntstrom BTB Evelyn A, Moon Harley W, O'Brien Alison D. Bovine infection with Shiga Toxin-producing Escherichia coli. American Society for Microbiology, Washington, D.C; 1998. [Google Scholar]
- Russell JB, Diez-Gonzalez F, Jarvis GN. Invited review: effects of diet shifts on Escherichia coli in cattle. J Dairy Sci. 2000;83(4):863–873. doi: 10.3168/jds.S0022-0302(00)74950-2. [DOI] [PubMed] [Google Scholar]
- DeVinney R, Gauthier A, Abe A, Finlay BB. Enteropathogenic Escherichia coli: a pathogen that inserts its own receptor into host cells. Cell Mol Life Sci. 1999;55(6-7):961–976. doi: 10.1007/PL00013202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Ruschkowski S, Kenny B, Stein M, Reinscheid DJ, Stein MA, Rosenshine I. Enteropathogenic E. coli exploitation of host epithelial cells. Ann N Y Acad Sci. 1996;797:26–31. doi: 10.1111/j.1749-6632.1996.tb52946.x. [DOI] [PubMed] [Google Scholar]
- Frankel G, Phillips AD, Trabulsi LR, Knutton S, Dougan G, Matthews S. Intimin and the host cell--is it bound to end in Tir(s)? Trends Microbiol. 2001;9(5):214–218. doi: 10.1016/S0966-842X(01)02016-9. [DOI] [PubMed] [Google Scholar]
- Kaper JB, McDaniel TK, Jarvis KG, Gomez-Duarte O. Genetics of virulence of enteropathogenic E. coli. Adv Exp Med Biol. 1997;412:279–287. doi: 10.1007/978-1-4899-1828-4_47. [DOI] [PubMed] [Google Scholar]
- Ochoa TJ, Barletta F, Contreras C, Mercado E. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans R Soc Trop Med Hyg. 2008;102(9):852–856. doi: 10.1016/j.trstmh.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agin TS, Zhu C, Johnson LA, Thate TE, Yang Z, Boedeker EC. Protection against hemorrhagic colitis in an animal model by oral immunization with isogeneic rabbit enteropathogenic Escherichia coli attenuated by truncating intimin. Infect Immun. 2005;73(10):6608–6619. doi: 10.1128/IAI.73.10.6608-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro I, Frankel G, Adu-Bobie J, Dougan G, Trabulsi LR, Carneiro-Sampaio MM. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli virulence-associated proteins: intimin, BfpA, EspA, and EspB. J Pediatr Gastroenterol Nutr. 1998;27(2):166–171. doi: 10.1097/00005176-199808000-00007. [DOI] [PubMed] [Google Scholar]
- Zapata-Quintanilla LB, Tino-De-Franco M, Nunez SP, Carbonare CB, Vulcano AB, Carbonare SB. Seric and secretory antibodies reactive to alpha, beta and gamma intimins of Escherichia coli in healthy Brazilian adults. FEMS Immunol Med Microbiol. 2009;57(1):32–9. doi: 10.1111/j.1574-695X.2009.00576.x. [DOI] [PubMed] [Google Scholar]
- Ghaem-Maghami M, Simmons CP, Daniell S, Pizza M, Lewis D, Frankel G, Dougan G. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect Immun. 2001;69(9):5597–5605. doi: 10.1128/IAI.69.9.5597-5605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves NS, Hale C, Dougan G, Frankel G, MacDonald TT. Binding of intimin from enteropathogenic Escherichia coli to lymphocytes and its functional consequences. Infect Immun. 2003;71(5):2960–2965. doi: 10.1128/IAI.71.5.2960-2965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair JF, Dean-Nystrom EA, O'Brien AD. The established intimin receptor Tir and the putative eucaryotic intimin receptors nucleolin and beta1 integrin localize at or near the site of enterohemorrhagic Escherichia coli O157:H7 adherence to enterocytes in vivo. Infect Immun. 2006;74(2):1255–1265. doi: 10.1128/IAI.74.2.1255-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeilly TN, Mitchell MC, Rosser T, McAteer S, Low JC, Smith DG, Huntley JF, Mahajan A, Gally DL. Immunization of cattle with a combination of purified intimin-531, EspA and Tir significantly reduces shedding of Escherichia coli O157:H7 following oral challenge. Vaccine. 2010;28(5):1422–1428. doi: 10.1016/j.vaccine.2009.10.076. [DOI] [PubMed] [Google Scholar]
- Bretschneider G, Berberov EM, Moxley RA. Reduced intestinal colonization of adult beef cattle by Escherichia coli O157:H7 tir deletion and nalidixic-acid-resistant mutants lacking flagellar expression. Vet Microbiol. 2007;125(3-4):381–386. doi: 10.1016/j.vetmic.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Killar LM, Eisenstein TK. Immunity to Salmonella typhimurium infection in C3H/HeJ and C3H/HeNCrlBR mice: studies with an aromatic-dependent live S. typhimurium strain as a vaccine. Infect Immun. 1985;47(3):605–612. doi: 10.1128/iai.47.3.605-612.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider G, Berberov EM, Moxley RA. Enteric mucosal antibodies to Escherichia coli O157:H7 in adult cattle. Vet Rec. 2008;163(7):218–219. doi: 10.1136/vr.163.7.218. [DOI] [PubMed] [Google Scholar]
- Nart P, Holden N, McAteer SP, Wang D, Flockhart AF, Naylor SW, Low JC, Gally DL, Huntley JF. Mucosal antibody responses of colonized cattle to Escherichia coli O157-secreted proteins, flagellin, outer membrane proteins and lipopolysaccharide. FEMS Immunol Med Microbiol. 2008;52(1):59–68. doi: 10.1111/j.1574-695X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- Dean-Nystrom EA, Gansheroff LJ, Mills M, Moon HW, O'Brien AD. Vaccination of pregnant dams with intimin(O157) protects suckling piglets from Escherichia coli O157:H7 infection. Infect Immun. 2002;70(5):2414–2418. doi: 10.1128/IAI.70.5.2414-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diemen PM, Dziva F, Abu-Median A, Wallis TS, van den Bosch H, Dougan G, Chanter N, Frankel G, Stevens MP. Subunit vaccines based on intimin and Efa-1 polypeptides induce humoral immunity in cattle but do not protect against intestinal colonisation by enterohaemorrhagic Escherichia coli O157:H7 or O26:H. Vet Immunol Immunopathol. 2007;116(1-2):47–58. doi: 10.1016/j.vetimm.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterslager TG, Florack DE, van der Wal TJ, Molthoff JW, Langeveld JP, Bosch D, Boersma WJ, Hilgers LA. Oral immunisation of naive and primed animals with transgenic potato tubers expressing LT-B. Vaccine. 2001;19(17-19):2749–2755. doi: 10.1016/S0264-410X(00)00513-2. [DOI] [PubMed] [Google Scholar]
- Stok W, van der Heijden PJ, Bianchi AT. Conversion of orally induced suppression of the mucosal immune response to ovalbumin into stimulation by conjugating ovalbumin to cholera toxin or its B subunit. Vaccine. 1994;12(6):521–526. doi: 10.1016/0264-410X(94)90311-5. [DOI] [PubMed] [Google Scholar]


