Abstract
Background
The TolC protein from Sinorhizobium meliloti has previously been demonstrated to be required for establishing successful biological nitrogen fixation symbiosis with Medicago sativa. It is also needed in protein and exopolysaccharide secretion and for protection against osmotic and oxidative stresses. Here, the transcriptional profile of free-living S. meliloti 1021 tolC mutant is described as a step toward understanding its role in the physiology of the cell.
Results
Comparison of tolC mutant and wild-type strains transcriptomes showed 1177 genes with significantly increased expression while 325 had significantly decreased expression levels. The genes with an increased expression suggest the activation of a cytoplasmic and extracytoplasmic stress responses possibly mediated by the sigma factor RpoH1 and protein homologues of the CpxRA two-component regulatory system of Enterobacteria, respectively. Stress conditions are probably caused by perturbation of the cell envelope. Consistent with gene expression data, biochemical analysis indicates that the tolC mutant suffers from oxidative stress. This is illustrated by the elevated enzyme activity levels detected for catalase, superoxide dismutase and glutathione reductase. The observed increase in the expression of genes encoding products involved in central metabolism and transporters for nutrient uptake suggests a higher metabolic rate of the tolC mutant. We also demonstrated increased swarming motility in the tolC mutant strain. Absence of functional TolC caused decreased expression mainly of genes encoding products involved in nitrogen metabolism and transport.
Conclusion
This work shows how a mutation in the outer membrane protein TolC, common to many bacterial transport systems, affects expression of a large number of genes that act in concert to restore cell homeostasis. This finding further underlines the fundamental role of this protein in Sinorhizobium meliloti biology.
Background
The outer membrane protein TolC belongs to a family of envelope proteins found in Gram-negative bacteria [1] and is essential for the export of a wide range of toxic substances such as antibiotics, dyes, disinfectants and natural substances produced by the hosts, including bile, hormones and defense molecules [2,3]. TolC is also required for export of a range of extracellular proteins such as metalloproteases, α-hemolysins, lipases, enterotoxin II [4], the siderophore enterobactin [5], colicin uptake and secretion [6] and bacteriophage adsorption [7]. The TolC protein from Escherichia coli was also suggested as possibly involved in the efflux of not yet determined cellular metabolites [8]. Intracellular metabolite accumulation caused upregulation of several transcription factors including MarA, SoxS and Rob. These in turn upregulate TolC, leading to a decrease in metabolite concentration and restoration of cell homeostasis [8]. TolC family members are also required for colonization and persistence of bacteria in their host organisms. For example, Erwinia chrysanthemi [9] and Xylella fastidiosa [10]tolC mutants were unable to grow in planta and their virulence was severely compromised. TolC-deficient strains of Brucella suis [11] and Vibrio cholerae [12] also displayed an attenuation of infection or colonization in animal models, respectively. The TolC protein of Salmonella enterica was shown to be required for efficient adhesion and invasion of epithelial cells and macrophages and to colonize poultry [13,14]. Webber and collaborators [13] demonstrated that S. enterica mutants lacking acrA, acrB, or tolC genes encoding an efflux pump showed repression of operons involved in pathogenesis. Operons included chemotaxis, motility and type III secretion system genes, offering a possible explanation for the attenuated pathogenesis of these strains [13].
TolC protein of Sinorhizobium meliloti, the symbiotic partner of the leguminous plant Medicago sativa was recently characterised [15]. A S. meliloti tolC insertion mutant induced none or only very few nodules in M. sativa roots. Any nodules formed were brownish-white, non-nitrogen fixing, in contrast to the pink elongated nitrogen fixing nodules formed by wild-type S. meliloti 1021. The tolC gene mutation strongly affected the resistance phenotype to antimicrobial agents of plant origin and induced higher susceptibility to osmotic and oxidative stresses. Analysis of extracellular proteins showed that calcium-binding protein WgeA (formerly ExpE1), endoglycanase ExsH and the putative hemolysin-type calcium-binding protein SMc04171 were secreted in a TolC dependent manner. Another phenotype shown by the S. meliloti tolC mutant was absence of exopolysaccharides succinoglycan and galactoglucan from the culture supernatant [15]. Absence of galactoglucan in the tolC mutant is explained by the lack of WgeA protein secretion [16], but the contribution of TolC to succinoglycan production is so far not understood. Several phenotypes displayed by the S. meliloti tolC mutant strain illustrated the wide importance of this outer membrane protein to cellular functions. To better understand the contribution of TolC protein to S. meliloti cell physiology under free-living conditions, we investigated the effect of its inactivation on the transcriptome. Our data point towards an increased expression of genes encoding products involved in stress response, central metabolic pathways, and nutrient uptake transporters in the tolC mutant. Genes encoding products involved in nitrogen metabolism, transport and cell division displayed decreased expression.
Results and Discussion
Global changes in gene expression associated to a mutation in the tolC gene
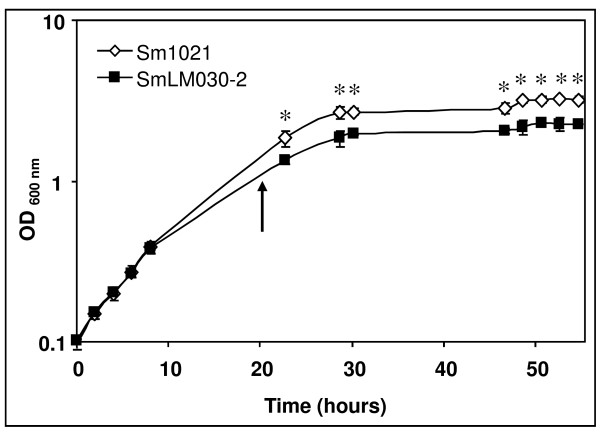
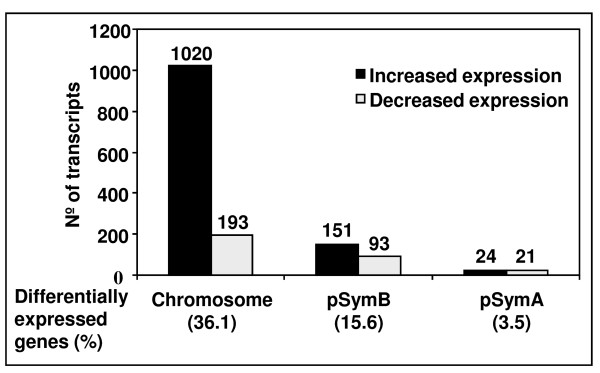
Cosme et al. [15] disrupted the S. meliloti 1021 tolC gene by inserting plasmid pK19mob2ΏHMB into its coding sequence, eliminating the last 102 nucleotides. This mutant, potentially expressing a truncated protein, displayed several phenotypes such as impaired symbiosis with Medicago, higher sensitivity to osmotic and oxidative stresses and absence of some extracellular proteins and exopolysaccharides [15]. Here, growth rates of wild-type and the tolC gene insertion mutant SmLM030-2 grown in GMS medium were determined (Fig. 1). During the first 8 hours the growth rate was comparable for both strains; subsequently the tolC mutant showed a lower growth rate and reduced biomass formation. To gain insight into what underlies these differences, transcriptomes of the wild-type and the tolC mutant strains cultured in GMS medium for 20 hours were compared. Microarray data analyzed using dChip (≥1.2-fold change lower confidence bound and a ≤0.4% FDR as cutoffs) and Partek Genomics Suite (FDR ≤ 5%; p-value ≤ 0.017) identified 2067 probe sets in common as being differentially expressed. From this list, we removed duplicated probes for the same genes and those covering intergenic regions, giving a subset of 1809 genes with differential expression (See Additional file 1: Table S1 and Additional file 2: Table S2). Clusters of Orthologous Groups (COGs) could be attributed to 1502 of these according to predicted gene functions (See Additional file 1: Table S1 and Additional file 2: Table S2). Replicon distribution of the 1502 tolC-dependent differentially expressed genes indicated that 1213 (80%) were chromosomal, 244 (16%) from pSymB and 45 (3%) from pSymA (Fig. 2). The annotated genome of S. meliloti 1021 has 54% of genes located in the chromosome, 25% on pSymB and 21% on pSymA. The distribution of tolC-dependently expressed genes shows a replicon bias with 1.50-fold higher impact on the chromosome encoded transcripts. Contrastingly, genes from pSymB and pSymA were under-represented with 0.65- and 0.14-fold, respectively.
Figure 1.
Effect of tolC mutation on growth of S. meliloti 1021. Growth curves of S. meliloti 1021 (◊) and SmLM030-2 tolC mutant (■) were obtained in GMS medium. Optical density values are the means of three independent experiments. The arrow indicates the time point where cells were collected for total RNA extraction. Error bars show standard deviations. Asterisks represent data points with significantly different means (p-value < 0.01).
Figure 2.
Distribution of differentially expressed genes in function of the S. meliloti 1021 replicons. The histogram shows the number of differentially expressed genes obtained when the tolC mutant transcriptome was compared to the wild-type strain and their distribution on the chromosome and the two megaplasmids pSymA and pSymB.
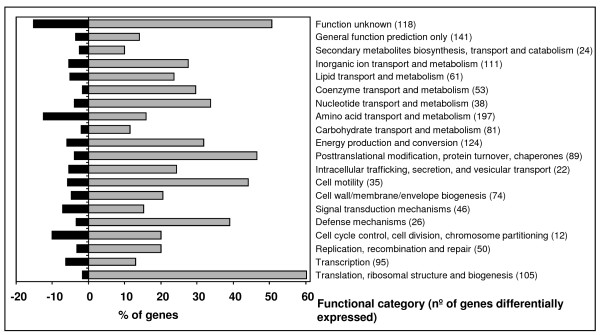
A total of 1177 genes (Table 1 and Additional file 1: Table S1) had significantly increased expression in the tolC mutant. These could be classified in 20 functional categories. Fig. 3 summarizes the percentages of differentially expressed genes in comparison to genes of the same category represented on the microarray. The largest categories, with more than 30% of the genes with significantly increased expression, included genes involved into protein synthesis, defense, cell motility, protein modification and turnover, energy production, nucleotide metabolism, and genes of unknown function (Fig. 3, grey bars). Microarray analysis revealed that expression of 325 genes was significantly decreased in the tolC mutant (Table 2 and Additional file 2: Table S2). Largest categories, with more than 10% of the genes with a significantly decreased expression include the genes involved in cell division, amino acid transport and metabolism, and of unknown function (Fig. 3, black bars).
Table 1.
Genes with more than 8-fold increased expression in the tolC mutant strain.
| Gene identifier | Annotation or description | Fold change1 (tolC vs. wild-type) |
|---|---|---|
| Signal transduction | ||
| SMb21560 | Putative two-component sensor histidine kinase | 14.7 |
| SMb21561 | Putative two-component response regulator | 27.1 |
| Translation | ||
| SMc00320 | rbfA probable ribosome-binding factor A, rRNA processing protein | 8.9 |
| SMc00323 | rpsO robable 30 S ribosomal protein S15 | 8.7 |
| SMc00324 | pnp probable polyribonucleotide nucleotidyltransferase | 10.1 |
| SMc00335 | rpsA 30 S ribosomal protein S1 | 10.2 |
| SMc00485 | rpsD probable 30 S ribosomal subunit protein S4 | 9.2/8.8 |
| SMc00522 | rhlE1 putative ATP-dependent RNA helicase | 8.5 |
| SMc00565 | rplI probable 50 S ribosomal protein L9 | 13.4 |
| SMc00567 | rpsR putative 30 S ribosomal protein S18 | 21.9 |
| SMc00568 | rpsF putative 30 S ribosomal protein S6 | 25.9 |
| SMc01287 | rpsM probable 30 S ribosomal protein S13 | 8.5 |
| SMc01290 | rplO probable 50 S ribosomal protein L15 | 10.5 |
| SMc01291 | rpmD probable 50 S ribosomal protein L30 | 12.9 |
| SMc01292 | rpsE probable 30 S ribosomal protein S5 | 15.9 |
| SMc01293 | rplR probable 50 S ribosomal protein L18 | 24.7/12.5 |
| SMc01294 | rplF probable 50 S ribosomal protein L6 | 12.3 |
| SMc01295 | rpsH probable 30 S ribosomal protein S8 | 12.9 |
| SMc01296 | rpsN probable 30 S ribosomal protein S14 | 13.3 |
| SMc01297 | rplE probable 50 S ribosomal protein L5 | 15.4 |
| SMc01298 | rplX probable 50 S ribosomal protein L24 | 13.1 |
| SMc01299 | rplN probable 50 S ribosomal protein L14 | 16.1/13.2 |
| SMc01300 | rpsQ probable 30 S ribosomal protein S17 | 20.8/12.0 |
| SMc01301 | rpmC probable 50 S ribosomal protein L29 | 13.1 |
| SMc01302 | rplP probable 50 S ribosomal protein L16 | 12.4 |
| SMc01303 | rpsC probable 30 S ribosomal protein S3 | 17.5/10.6 |
| SMc01304 | rplV probable 50 S ribosomal protein L22 | 13.2 |
| SMc01305 | rpsS probable 30 S ribosomal protein S19 | 15.2 |
| SMc01306 | rplB probable 50 S ribosomal protein L2 | 20.5/18.1 |
| SMc01307 | rplW probable 50 S ribosomal protein L23 | 31.9 |
| SMc01308 | rplD probable 50 S ribosomal protein L4 | 24.1 |
| SMc01309 | rplC probable 50 S ribosomal protein L3 | 22.4/16.5 |
| SMc01310 | rpsJ probable 30 S ribosomal protein S10 | 25.6/19.7 |
| SMc01312 | fusA1 probable elongation factor G | 29.6/21.0 |
| SMc01313 | rpsG probable 30 S ribosomal protein S7 | 30.4 |
| SMc01314 | rpsL probable 30 S ribosomal protein S12 | 19.5 |
| SMc01326 | tuf probable elongation factor TU protein | 10.2/10.1 |
| SMc02050 | tig probable trigger factor | 9.1 |
| SMc02053 | trmFO methylenetetrahydrofolate-tRNA-(uracil-5-)-methyltransferase | 10.4 |
| SMc02100 | tsf probable elongation factor TS (EF-TS) protein | 10.8 |
| SMc02101 | rpsB probable 30 S ribosomal protein S2 | 13.7 |
| SMc03242 | typA predicted membrane GTPase | 14.4 |
| SMc03859 | rpsP probable 30 S ribosomal protein S16 | 8.2 |
| Metabolism | ||
| SMa0680 | Decarboxylase (lysine, ornithine, arginine) | 11.2 |
| SMa0682 | Decarboxylase (lysine, ornithine, arginine) | 8.3 |
| SMa0765 | fixN2 cytochrome c oxidase subunit I | 9.8 |
| SMa0767 | fixQ2 nitrogen fixation protein | 11.5 |
| SMa1179 | nosR regulatory protein | 13.8 |
| SMa1182 | nosZ nitrous oxide reductase | 24.3 |
| SMa1183 | nosD nitrous oxidase accessory protein | 12.4 |
| SMa1188 | nosX accesory protein | 10.7 |
| SMa1208 | fixS1 nitrogen fixation protein | 10.6 |
| SMa1209 | fixI1 ATPase | 24.4 |
| SMa1210 | fixH nitrogen fixation protein | 10.1 |
| SMa1213 | fixP1 di-heme c-type cytochrome | 28.2 |
| SMa1214 | fixQ1 nitrogen fixation protein | 37.2 |
| SMa1216 | fixO1 cytochrome C oxidase subunit | 18.5 |
| SMa1243 | azu1 pseudoazurin | 9.6 |
| SMb21487 | cyoA putative cytochrome o ubiquinol oxidase chain II | 14.2 |
| SMb21488 | cyoB putative cytochrome o ubiquinol oxidase chain I | 22.2 |
| SMb21489 | cyoC putative cytochrome o ubiquinol oxidase chain III | 13.6 |
| SMc00090 | cyoN putative sulfate adenylate transferase cysteine biosynthesis protein | 37.5 |
| SMc00091 | cysD putative sulfate adenylate transferase subunit 2 cysteine biosynthesis protein | 21.1 |
| SMc00092 | cysH phosphoadenosine phosphosulfate reductase | 13.4 |
| SMc00595 | ndk probable nucleoside diphosphate kinase | 8.6 |
| SMc00868 | atpF probable ATP synthase B chain transmembrane protein | 8.1/8.0 |
| SMc00869 | atpF2 probable ATP synthase subunit B' transmembrane protein | 8.7 |
| SMc00871 | atpB probable ATP synthase A chain transmembrane protein | 8.3 |
| SMc01053 | cysG probable siroheme synthase | 13.9 |
| SMc01169 | ald probable alanine dehydrogenase oxidoreductase | 26.2 |
| SMc01923 | nuoJ probable NADH dehydrogenase I chain J transmembrane protein | 9.1 |
| SMc01925 | nuoL probable NADH dehydrogenase I chain L transmembrane protein | 10.0 |
| SMc02123 | Sulfate or sulfite assimilation protein | 12.6 |
| SMc02124 | cysI putative sulfite reductase | 20.2 |
| SMc02479 | mdh probable malate dehydrogenase | 9.9 |
| SMc02480 | sucC probable succinyl-CoA synthetase beta chain | 9.4 |
| SMc02481 | sucD probable succinyl-CoA synthetase alpha chain | 9.3 |
| SMc02499 | atpA probable ATP synthase subunit alpha | 8.2 |
| SMc02500 | atpG probable ATP synthase gamma chain | 16.2/11.1 |
| SMc02502 | atpC probable ATP synthase epsilon chain | 9.8 |
| SMc03858 | pheA putative chorismate mutase | 8.4 |
| Transport | ||
| SMa1185 | nosY permease | 8.5 |
| SMb20346 | Putative efflux transmembrane protein | 8.3 |
| SMc00873 | kup1 probable KUP system potassium uptake transmembrane protein | 11.4 |
| SMc02509 | sitA manganese ABC transporter periplasmic substrate binding protein | 9.4 |
| SMc03157 | metQ probable D-methionine -binding lipoprotein MetQ | 8.7/14.9 |
| SMc03158 | metI probable D-methionine transport system permease protein MetI | 12.3 |
| SMc03167 | MFS-type transport protein | 41.1 |
| SMc03168 | Multidrug resistance efflux system | 41.5 |
| Stress related | ||
| SMa0744 | groEL2 chaperonin | 18.3/13.7 |
| SMa0745 | groES2 chaperonin | 19.3 |
| SMa1126 | Putative protease, transmembrane protein | 16.4 |
| SMb21549 | thtR putative exported sulfurtransferase, Rhodanese protein | 29.3 |
| SMb21562 | Hypothetical membrane-anchored protein | 69.6 |
| SMc00913 | groEL1 60 KD chaperonin A | 17.5 |
| SMc02365 | degP1 probable serine protease | 20.4/18.5 |
| Motility | ||
| SMc03014 | fliF flagellar M-ring transmembrane protein | 8.3 |
| SMc03022 | motA chemotaxis (motility protein A) transmembrane | 16.2 |
| SMc03024 | flgF lagellar basal-body rod protein | 15.6 |
| SMc03027 | flgB flagellar basal-body rod protein | 9.3 |
| SMc03028 | flgC flagellar basal-body rod protein | 12.9 |
| SMc03030 | flgG flagellar basal-body rod protein | 11.0 |
| SMc03047 | flgE flagellar hook protein | 8.1 |
| SMc03054 | flhA probable flagellar biosynthesis transmembrane protein | 9.7 |
1 Some S. meliloti genes have more than one probe set represented on the array. In these cases, more than one fold change value is shown.
Figure 3.
Distribution of genes with differentially altered expression into COGs. Effect of the tolC gene mutation on the S. meliloti transcriptome analyzed according to the distribution of the genes with altered expression into 20 functional categories (COGs) as predicted using NCBI database. The black and grey bars represent the percentage of genes in each functional category whose transcription was decreased and increased, respectively, in the tolC mutant SmLM030-2 by comparison to the wild-type strain 1021.
Table 2.
Genes with more than 5-fold decreased expression in the tolC mutant strain.
| Gene identifier | Annotation or description | Fold change1 (tolC vs. wild-type) |
|---|---|---|
| Transcription and signal transduction | ||
| SMa0402 | Transcriptional regulator, GntR family | -8.4 |
| SMb21115 | Putative response regulator | -20.2 |
| SMc01042 | ntrB nitrogen assimilation regulatory protein | -8.0 |
| SMc01043 | ntrC nitrogen assimilation regulatory protein | -6.9 |
| SMc01504 | Receiver domain | -7.2 |
| SMc01819 | Transcription regulator TetR family | -10.0 |
| SMc03806 | glnK probable nitrogen regulatory protein PII 2 | -9.1 |
| Metabolism | ||
| SMa0387 | hisC3 histidinol-phosphate aminotransferase | -11.4 |
| SMa0398 | hisD2 histidinol dehydrogenase | -10.6 |
| SMa1683 | Arylsulfatase | -5.0 |
| SMb20984 | nirB nitrite reductase NAD(P)H | -22.7 |
| SMb20985 | nirD nitrite reductase NAD(P)H | -26.6 |
| SMb20986 | narB putative nitrate reductase, large subunit | -14.1 |
| SMb20987 | Putative uroporphiryn-III C-methyltransferase | -7.6 |
| SMb21094 | argH2 argininosuccinate lyase | -20.7 |
| SMb21163 | hutU urocanate hydratase (urocanase) | -10.3 |
| SMb21164 | hutG Putative formiminoglutamase | -11.5 |
| SMb21165 | hutH Putative histidine ammonia-lyase histidase | -7.7 |
| SMc01041 | dusB tRNA-dihydrouridine synthase B | -9.5 |
| SMc01814 | Probable glutamate synthase small chain | -12.5 |
| SMc01820 | Putative N-carbamyl-L-amino acid amidohydrolase | -12.7 |
| SMc01967 | speB2 putative agmatinase | -18.7 |
| SMc03208 | hmgA homogentisate 1,2-dioxygenase | -5.5 |
| SMc04026 | gltD probable glutamate synthase small chain | -9.2 |
| SMc04028 | gltB probable glutamate synthase NADPH large chain | -11.7 |
| SMc04153 | Putative aminomethyltransferase | -8.7 |
| SMc04323 | Probable aminotransferase | -7.8 |
| Transport | ||
| SMa0391 | ABC transporter, ATP-binding protein | -15.6 |
| SMa0392 | ABC transporter, periplasmic solute-binding protein | -8.3/-23.5 |
| SMa0394 | ABC transporter, permease | -10.5 |
| SMa0396 | ABC transporter, permease | -10.1 |
| SMa0581 | nrtC nitrate transporter, ATP binding protein | -24.8 |
| SMa0583 | nrtB nitrate transporter, permease | -33.0 |
| SMa0585 | nrtA nitrate ABC transporter, periplasmic nitrate binding protein | -34.8 |
| SMb20436 | Probable nitrate transporter | -62.2/-63.5 |
| SMb20602 | ABC transporter, ATP-binding protein | -12.0 |
| SMb20603 | ABC transporter, permease | -15.7 |
| SMb20604 | ABC transporter, permease | -25.0 |
| SMb20605 | ABC transporter, periplasmic solute-binding protein | -22.4 |
| SMb21095 | ABC transporter, permease | -10.3 |
| SMb21096 | ABC transporter, permease | -10.7 |
| SMb21097 | ABC transporter periplasmic solute-binding protein | -17.5 |
| SMb21114 | Putative nitrate transport protein | -10.3 |
| SMb21707 | ABC transporter, ATP-binding protein | -14.4 |
| SMc01597 | Putative amino acid permease | -8.1 |
| SMc01963 | Spermidine/putrescine transport system permease | -5.2 |
| SMc01964 | Putative spermidine/putrescine transport system permease ABC transporter | -5.8 |
| SMc01965 | Spermidine/putrescine ABC transporter ATP-binding subunit | -7.4 |
| SMc01966 | Putative spermidine/putrescine-binding periplasmic ABC transporter | -12.4 |
| SMc03807 | amtB probable ammonium transporter | -8.1 |
| SMc04147 | Putative amino acid permease | -10.7 |
1 Some S. meliloti genes have more than one probe set represented on the array. In these cases, more than one fold change value is shown.
To confirm data obtained by microarray analysis, we examined expression of ten representative genes of the functional categories of signal transduction, secondary metabolism, carbohydrate, amino acids and inorganic ion transport and metabolism, and protein turnover by real-time RT-PCR (Table 3). Gene glnA was chosen because its expression was moderately decreased in the tolC mutant background; genes smoG and rem had a moderately increased expression; genes gltB, argH2 and nrtA showed greatly decreased expression; SMc03167, cysN and degP1 gene expression was highly increased. wgeA was included as a control, since microarray analysis showed that its expression was not significantly altered. Results obtained by quantitative real-time PCR were in agreement with microarray data (Table 3). Below we discuss a selected subgroup of differentially expressed genes.
Table 3.
Quantitative real-time RT-PCR analysis performed in S. meliloti 1021 wild-type and tolC mutant cells.
| Gene | Microarray Fold change | Real-time PCR Fold change ± SD |
|---|---|---|
| rem | 4.2 | 4.6 ± 1.4 |
| wgeA | 1.0 | 1.0 ± 0.2 |
| SMoG | 3.9 | 2.9 ± 0.8 |
| SMc03167 | 41.1 | 58.2 ± 7.2 |
| glnA | -3.8 | -2.3 ± 0.1 |
| gltB | -11.7 | -11.0 ± 1.6 |
| argH2 | -20.7 | -4.5 ± 1.7 |
| nrtA | -34.8 | -19.4 ± 2.4 |
| cysN | 37.5 | 19.4 ± 0.9 |
| degP | 18.5 | 31.2 ± 1.1 |
Oxidative stress is induced in the tolC mutant
Bacteria have developed several different strategies to cope with fatal stress conditions. One is mediated by alternative sigma-32 factor (RpoH) that, besides high temperature, is activated by conditions that destabilize folded proteins or make correct nascent protein folding more difficult [17]. S. meliloti, as well as some other rhizobia, has two rpoH genes named rpoH1 and rpoH2. Comparison of the transcriptome of S. meliloti tolC mutant with the one of the wild-type strain, revealed a 3.4-fold increase in rpoH1 gene expression. rpoH2 gene expression was not altered. Both RpoH1 and RpoH2 sigma factors were studied in Rhizobium etli, with the results suggesting that they operate under different stress conditions: RpoH1 in heat-shock and oxidative stress, and RpoH2 in osmotic tolerance [18]. The increase of rpoH1 gene expression in the tolC mutant is probably a consequence of the stress conditions caused by accumulation of intracellular proteins or of other unknown cell metabolites and probably due to a higher metabolic rate. Similarly to the increase of rpoH1 gene expression, significantly increased expression of many genes that in other organisms are known to belong to the rpoH regulon was observed. These encompass genes encoding chaperones DnaJ, DnaK, ClpB, GroESL1, GroESL2, GroESL3, GroEL5, GrpE, HslO, HtpG, and IbpA, involved in the folding of newly synthesized proteins or in refolding of denatured proteins to maintain homeostasis. Under free-living conditions, S. meliloti RpoH1 seems to control the expression of groESL5 but not other chaperone encoding genes [19]. Here, beside groESL5, more than 17-fold increase of groESL1 and groESL2 operons expression was detected (Table 1), suggesting they may be regulated by another transcription factor or by RpoH1 in a stress condition dissimilar from the heat-shock tested by Bittner and Oke [19]. Several genes encoding proteases and protein modification enzymes such as ClpP1, ClpP2, ClpX, Lon, HslUV, HflCKX, FtsH, HtpX and Dcp also showed significantly increased expression in the tolC mutant. In addition to protecting proteins from destruction or degradation of the denatured ones the rpoH regulon also protects other macromolecules like DNA and RNA [17]. In the tolC mutant we observed increased expression of the gene encoding Mfd which recruits the DNA repair machinery to lesions, as well as genes such as mutM, recJ, topA and xerD encoding products known to maintain genomic integrity [20].
Reinforcing the idea of the tolC mutant strain being under stress, the expression of many transcripts encoding enzymes involved in detoxification and protection against oxidative stress was increased. Examples include gst1, gst4, gst7 and gst11, all of which encode glutathione S-transferases. Glutathione transferase proteins catalyze nucleophilic attack by the tripeptide glutathione (GSH) on a wide range of hydrophobic toxic compounds. They are also capable of non-catalytically binding a large number of endogenous compounds, playing an active role in protection against oxidative stress and detoxification of harmful xenobiotics [21]. Other genes with increased expression were katA (3.7-fold) encoding a catalase, sodB (2.4-fold) encoding a superoxide dismutase, cpo (2.5-fold) encoding a chloride peroxidase, and gor (1.8-fold) encoding a glutathione reductase. Gene thtR showed the greatest expression in this functional class with a 29.3-fold increase (Table 1). thtR encodes a protein homologous to tiosulphate sulfurtransferases of the Rhodanese family, which catalyze the transfer of the sulphate atom of thiosulphate to cyanide, to form sulphite and thyocianate. Several studies indicate that these proteins may function as antioxidants capable of scavenging oxidative species that would otherwise lead to inactivation of enzymes such as those containing Fe-S clusters [22].
To confirm microarray data and demonstrate that the tolC mutant is under oxidative stress, enzymatic activities of catalase, superoxide dismutase and glutathione reductase were determined in cells grown in GMS medium for 20 hours (Fig. 4). Results showed that the specific activity of glutathione reductase in the total protein extract of the tolC mutant was twice that of the wild-type strain (Fig. 4a). In-gel activity staining was used to visualize catalase activity. Despite increased expression of the katA gene and decreased katB expression compared to the wild-type strain, increased catalase activity was detected in the tolC mutant (Fig. 4b). SOD activity was also higher in the tolC mutant (Fig. 4c). The active SodB protein is a dimer [23] and corresponds probably to the lower band, while the upper band must be a multimeric form. Taken together, the increase of the three enzyme activities analyzed provides evidence that under conditions where the outer membrane protein TolC is non functional, cells suffer internal oxidative stress. Sikora et al. [24] recently demonstrated that mutants of Vibrio cholerae with compromised membrane phenotypes showed higher concentrations of radical oxygen species (ROS), induction of oxidative stress and changes in iron physiology. It is possible that the observed oxidative stress response of the S. meliloti tolC mutant is mainly caused by a compromised cell envelope, although a higher metabolic rate and accumulation of proteins and metabolites which can not be secreted may also contribute to stress.
Figure 4.
Activity of enzymes combating oxidative stress. Enzymatic activities of (a) glutathione reductase as measured spectrophotometrically at 412 nm; (b) catalase and (c) superoxidase dismutase in native gel after staining. Total protein extracts were obtained after growing the wild-type strain Sm1021 and the tolC mutant strain SmLM030-2 for 20 hours in GMS medium. 20 μg of crude extract were loaded in each lane. Arrows indicate the position of bands obtained.
In both Vibrio cholerae and E. coli, cell envelope perturbations resulted in induction of the extracytoplasmic stress factor RpoE, which directs transcription of genes involved in envelope maintenance [25]. We observed decreased expression of rpoE2, as well SMc01505 which is co-transcribed with rpoE2 and encoding an anti-sigma factor, suggesting that the lack of a functional TolC protein does not trigger RpoE-dependent stress response. Instead, by comparing the expression profile of the S. meliloti tolC mutant with that of the wild-type strain, we observed 69-, 27-, and 14-fold increased expression in genes SMb21562, SMb21561, and SMb21560, respectively (Table 1). Amino acid sequence of SMb21562 shows identity with the periplasmic protein CpxP from several Enterobacteria, displaying two characteristic LTxxQ motifs (data not shown). SMb21560 encodes a putative sensor histidine kinase homologous to CpxA. SMb21561 encodes a putative response regulator homologous to CpxR. The Cpx two-component regulator is a well characterized system to sense misfolded proteins in the periplasm and other perturbations in the cell envelope [26,27]. In Cpx signaling, unfolded proteins are recognized by CpxP, a periplasmically located inhibitor of the signaling sensor kinase CpxA, preventing CpxA to autophosphorylate. Nonphosphorylated CpxA is then unable to phosphorylate the cytoplasmic response regulator CpxR. The Cpx regulon of E. coli strain MC4100 contains at least 50 genes, some directly involved in maintenance of cell envelope proteins. These include periplasmic serine endoprotease DegP, disulfide oxidoreductase Dsb, periplasmic peptidyl-prolyl isomerase PpiA, phosphatidyl serine decarboxylase Psd, YccA, a modulator of FtsH proteolysis, periplasmic protein CpxP, and the two-component regulator CpxAR [28]. In addition, outer membrane protein OmpF, shikimate kinase AroK, and sigma-E regulator RpoE-RseABC are under negative control by Cpx [28]. Targets of the CpxR homologue in S. meliloti are completely unknown, but expression of genes encoding DegP proteases (degP1P3P4) and peptidyl-prolyl isomerase Ppi (ppiABD) were significantly increased in tolC mutant. A search for the E. coli CpxR binding site GTAAAN5GTAAA consensus sequence in the upstream coding regions of S. meliloti using the RSA-tools web interface revealed that this sequence matched the putative promoter region upstream of the predicted operon SMb21562/SMb21561/SMb21560. In a recent study, the CpxR protein from Yersinia enterocolitica was shown to negatively affect transcription of gene rpoE, coding for the extracytoplasmic sigma-E factor [29]. We also observed decreased expression of rpoE2 and rpoE8 genes. Our data suggest that in the absence of a functional TolC, cells trigger a Cpx instead of an RpoE-mediated response. A very different situation was observed in wild-type S. meliloti cells grown under different stress conditions such as osmotic shock [30,31], high metal ion concentration [32], acidic pH [33], heat shock and entry in stationary phase [34] where an rpoE2-mediated response was induced. This seems to indicate that the external stress imposed on the cells triggers a well defined extracytoplasmic response. When perturbations to the cell envelope, such as the absence of a functional outer membrane protein occur, cells seem to activate a distinct stress response pathway.
Genes involved in transcription and translation
It is possible that under the cytoplasmic and extracytoplasmic stress conditions experienced by the tolC mutant, many proteins and cofactors become inactive and need to be synthesized de novo or protected from denaturation. It is then not surprising that many genes encoding proteins involved in transcription and translation were found to have significantly increased expression in the tolC mutant strain. This was the case for genes encoding all RNA polymerase subunits (rpoABCZ), genes nusA and nusG involved in transcriptional pausing, termination, and antitermination, and the gene encoding transcription termination factor Rho. RNA degradation is mediated by the RNA degradosome, a multiprotein complex involving RNase E, polynucleotide phosphorylase (PNPase), helicase RhlB, and enolase [35]. In S. meliloti, those components are encoded by the genes rne, pnp, deaD, and eno, respectively, all of them showing increased expression in tolC mutant suggesting that, besides increased expression of genes encoding products involved in transcription, the mutant also increases expression of genes encoding products participating in RNA degradation.
Of the 105 genes differentially expressed and involved in translation and ribosome biogenesis only three had a decreased expression in the tolC mutant. Genes with increased expression encode 53 ribosomal proteins, along with initiation (infA, infB and infC), elongation (tsf, fusA1, efp) and release factors (prfA, prfB and prfC) (Table 1). In the tolC mutant we observed an increased expression of rbfA and rimM, coding for a ribosome binding factor and an rRNA-processing protein, respectively. Both gene products are essential for efficient processing of 16 S rRNA in E. coli [36]. The rrmJ gene encoding a ribosomal RNA large subunit methyltransferase and genes ksgA and hemK1 encoding two methylases involved in quality control by the small subunit of the ribosome [37] and methylation of release factors [38], respectively, also showed increased expression in the tolC mutant. Concerning amino acyl-tRNA modification we observed increased expression of the trmFO gene encoding a folate-dependent tRNA methyltransferase in the tolC mutant (Table 1). Maturation of tRNA precursors into functional tRNA molecules requires trimming of the primary transcript at both the 5'and 3'ends and is catalyzed by RNase P and RNase PH. Expression of genes encoding RNase P (rnpA) and RNase PH (rph), and genes encoding Rnase D (rnd1 and rnd2) which contribute to the 3'maturation of several stable RNAs also displayed increased expression levels in the tolC mutant. In contrast to S. meliloti cells exposed to osmotic stress which showed decreased expression of genes involved in protein metabolism [30,31], tolC mutant cells showed increased expression of these genes. As mentioned previously, a plausible explanation would be the need for new proteins to replace denatured ones due to oxidative stress conditions and the higher levels of metabolic enzymes needed for the cell to produce energy.
Genes involved in energy and central intermediary metabolism
We found increased expression of multiple genes involved in central metabolism and energy production in the tolC mutant (Fig. 5), suggesting a higher metabolic rate in response to tolC gene mutation. For instance, genes encoding 11 out of 12 of the enzymes involved in the tricarboxylic acid cycle (TCA) (acnA, icd, sucABCD, lpdA1A2, sdhABCD, fumC and mdh), along with genes encoding many enzymes of the Calvin-Benson-Bassham reductive pentose phosphate pathway (rbcL, pgk, fbaB, cbbF, tkt2, cbbT, rpiA and rpe) and most genes encoding enzymes for the glycolysis and gluconeogenesis pathways (cbbF, fbaB, tpiA1, gap, pgk, eno, pdhA) had significantly increased expression (Fig. 5). Alongside the increased expression of the genes encoding TCA enzymes, all genes encoding different protein complexes in the respiratory chain had also an increased expression. Genes include nuoA1B1C1D1E1F1G1HIJK1LMN and ndh forming NADH dehydrogenase (complex I); sdhABCD from fumarate reductase (complex II); fbcBCF from cytochrome c reductase (complex III); ctaCDEG and SMc01800 from cytochrome c oxidase (complex IV); and atpCDGABEF2FH from ATP synthase (complex V) (Table 1). cbb3-type cytochromes encoded by genes fixN2O2Q2P2 and fixP1Q1O1N1, and a low O2 affinity oxidase encoded by the cyoABC operon also showed increased expression in the tolC mutant (Table 1). Increased expression of all genes listed suggests that the tolC mutant strain may be metabolically more active. Nevertheless, the tolC mutant forms less biomass as seen in Fig. 1. This apparent contradiction can be explained if stress inflicted by cell envelope perturbations due to the absence of functional TolC protein results in a higher ATP turnover. Additional ATP would be consumed to maintain cell homeostasis and not to form biomass. It is also a formal possibility that perturbations to the cell envelope may reduce the proton electrochemical gradient, negatively affecting ATP synthesis and therefore creating the need to increase the expression of genes related to energy metabolism.
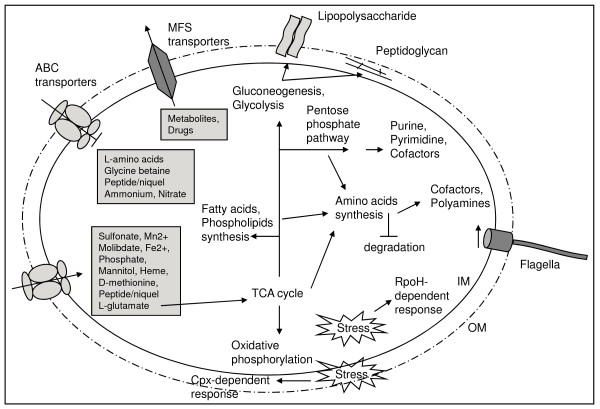
Figure 5.
Altered pathways and phenotypes on the dependence of tolC mutation in S. meliloti as depicted from the expression data. Arrows represent processes/pathways whose genes displayed increased expression and blocked arrows decreased expression in absence of a functional TolC protein. IM, inner membrane; OM, outer membrane.
Due to the general increase in expression of genes involved in translation, it was not surprising to see increased expression of genes encoding proteins involved in amino acid and cofactors biosynthesis in the tolC mutant (Fig. 5). Regarding cofactor biosynthesis we observed an increased expression in the tolC mutant of genes encoding enzymes for thiamine (thiE2, nifS), folate (folBCE, exsC), riboflavin (ribADEH), nicotinate and nicotimanide metabolism (nadABC, pntBAaAb), as well as genes panBC, coaAD, ilvCD2HI and acpS encoding enzymes required for pantothenate and CoA biosynthesis. Regarding amino acid metabolism by the tolC mutant there was an increased expression of genes encoding enzymes involved in the biosynthesis of the majority of them. These included serAB, glyA, SMc04029, lysC, asd, thrABC1, metAZHK, sda and metK1K2 for L-serine, L-glycine, L-threonine, L-methionine and L-cysteine biosynthesis; the genes leuBD, ilvCD2E1HI and pdhAaAb encoding enzymes for the synthesis of L-isoleucine, L-valine and L-leucine; the gene ald (Table 1) encoding an alanine dehydrogenase oxidoreductase synthesizing L-alanine from ammonia and pyruvate; the genes aroABCEFKQ, pheAAa, trpABDEF, tatA, tyrC, and aatAB encoding enzymes for biosynthesis of aromatic amino acids L-phenylalanine, L-tyrosine and L-tryptophan and genes hisABC1C2DEFGHIZ for the biosynthesis of L-histidine. Contrastingly, hutGHH2U genes involved in L-histidine degradation had more than 7-fold decreased expression (Table 2). Genes encoding enzymes for the biosynthesis of amino acid lysine (lysAC, asd, dapAA3BDF) had increased expression and those for degradation reduced expression levels (SMb21181, fadAB, phbA). Genes encoding urea cycle enzymes are argBDEJ, arcA1A2B and argF1GH1H2. With the exception of arcA2 and argH2 which encode a second copy of arginine deiminase and argininosuccinate lyase, respectively, all of them showed increased expression levels in tolC mutant. Commencing from ornithine or arginine it is possible to obtain the polyamines putrescine and agmatine. SMa0680 and SMa0682 (Table 1) encoding putative amino acid decarboxylases and the putative agmatinase encoded by gene speB were induced in the tolC mutant (Table 1). Polyamines are polycationic molecules that have important functions in cell physiology, contributing to stabilization of nucleic acids, production and function of outer membrane porins or are free radical scavengers when cells are exposed to oxidative stress [39]. Polyamine biosynthesis can therefore be another strategy used by the tolC mutant when under stress conditions.
In accordance with the hypothetical higher availability of metabolic intermediary compounds in the tolC mutant, fabBFGHIZ and accABCD encoding the enzymes for fatty acid biosynthesis; gpsA, plsC, cdsA, pgsA, pssA, and pcs involved in phospholipid biosynthesis; pyrBCDEFGH, cmk and ndk involved in pyrimidine nucleotides biosynthesis, and purBCDEFHKLMNQS and guaAB for purine nucleotides all had an increased expression in this mutant.
We observed 7-fold decreased expression of the genes ntrBC encoding the two-component regulatory system NtrBC in the tolC mutant, and decreased expression of NtrC-dependent genes encoding glutamine synthetases (glnII, glnA), regulatory PII proteins (glnB, glnK), and the AmtB transporter (amtB) (Table 2). A possible explanation could be intracellular differences in the C/N ratio between the two strains studied. Patriarca et al. [40] showed in Rhizobium etli cells grown in the presence of glutamine as single carbon and nitrogen source that the intracellular α-ketoglutarate/glutamine ratio influence NtrC activity.
Genes involved in transport
In keeping with the hypothesis of a higher metabolic rate in the tolC mutant, many genes related to nutrient uptake and assimilation showed increased expression in this strain including cysA2P2, SMb21132 and SMb21133 putatively involved in sulfate transport and cysDHIK1K2N encoding products involved in sulfate assimilation (Table 1). SMc04049 encoding a putative sulfite oxidase that converts sulfite back to sulfate had a decreased expression, possibly ensuring that in the tolC mutant sulfur flows in the direction of assimilation only. Other genes with increased expression in the tolC mutant were genes modABC encoding a putative molybdate ABC transporter; genes sitABCD encoding a manganese transporter; the genes pstABS and phoCDT encoding putative phosphate transporters; genes associated to biotin uptake (bioMN); kup1 and kup2 and corA2 putatively involved in K+ and Mg2+/Co2+ uptake, respectively; many genes related to iron (SMb21429, SMb21430, SMb21431 and SMb21432) and Fe3+-siderophore uptake (SMa1741, SMa1742, SMa1745, SMa1746 and exbBD); and genes encoding heme compound transporters (hmuTUV and ccmBC) (Fig. 5). An increase in the tolC mutant of the expression of smoEFGK genes involved in the uptake of mannitol, a carbon source provided in our experiments, was also observed. As regards amino acid transport, the genes metINQ, encoding an ABC transporter putatively involved in the transport of D-methionine (Table 1) also showed increased expression in the tolC mutant. We observed a strong decrease in the expression of genes involved nitrate, ammonium and amino acids transport in the tolC mutant (Fig. 5). For example, nitrate transporters encoded by nrtABC, SMb21114 and SMb20436 showed in excess of 10-fold decreased expression while the ammonium transporter encoded by the amtB gene showed 8-fold decreased expression. Genes associated with general amino acid transport (aapJMPQ) and branched-chain amino acids transport (SMb20602, SMb20603, SMb20604, SMb20605 and SMb21707) also displayed more than 12-fold decreased expression (Table 2). Genes encoding another ABC-type transporter putatively involved in the transport of spermidine/putrescine (SMc01963, SMc01964, SMc01965 and SMc01966) had 5-fold decreases expression while two putative ABC-type transporter systems of unknown function (SMb21095, SMb21096, SMb21097 and SMa0391, SMa0392, SMa0394 and SMa0396) had 10-fold decreased expression in the tolC mutant (Table 2). The decreased expression of genes involved in nitrogen-rich compound transport is probably an effect of decreased NtrC expression and is maybe a way to prevent a futile export and import cycle of these compounds.
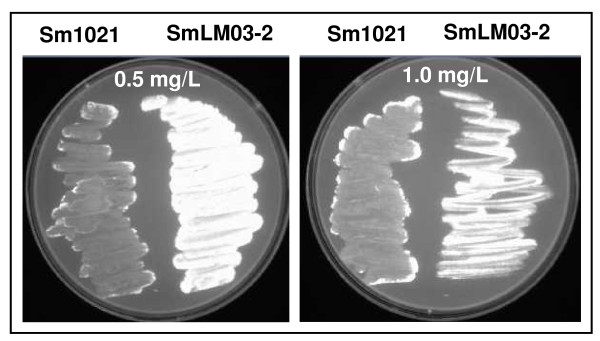
The tolC mutant exhibits an envelope defect, typified by its sensitivity to membrane-disrupting agents such as sodium dodecyl sulfate and deoxycholate [15]. When wild-type S. meliloti and tolC mutant strains were grown in solid GMS media supplemented with ethidium bromide it was observed that tolC mutant cells were fluorescent whilst wild-type cells were not (Fig. 6). This fluorescence results from the accumulation of ethidium bromide inside the tolC mutant cells, probably caused by their inability to pump this toxic compound out. This result suggests impairment of transport functions, most probably caused by the absence of the functional outer membrane protein TolC. Even when the tolC mutant is grown in GMS medium in the absence of toxic extracellular compounds, it is possible that unknown metabolites can not be secreted and accumulate in the cells, causing toxicity. To relieve that negative effect, cells would increase the expression of genes encoding certain transporters. This could explain the 5- and 41-fold increase in the expression of genes SMb20345/SMb20346 and SMc03167/SMc03168, respectively, which encode two putative transporters from the major facilitator superfamily, and the 1.4-fold increase in expression of truncated tolC gene. Similar reasoning was suggested by Rosner and Martin [8] in the case of E. coli TolC protein (together with other transport proteins) regarding the secretion of unknown cellular metabolites.
Figure 6.
Evaluation of efflux activity. S. meliloti 1021 (left side of the plate) and the tolC mutant SmLM03-2 (right side of the plate) were swabbed in GMS plates containing 0.5 and 1.0 mg/ml of ethidium bromide, following incubation at 30°C for 48 hours and detection under UV light.
Genes with a role in cell division and envelope biogenesis
In our data set the dnaA gene encoding a protein controlling chromosome replication initiation had increased expression in the tolC mutant. In C. crescentus DnaA controls expression of approximately 40 genes involved in, amongst others, DNA replication, recombination and repair, cell division and cell envelope biogenesis [41]. Expression profiles of genes putatively regulated by DnaA and involved in DNA replication, such as genes encoding subunits of DNA polymerase III, DnaB helicase, single strand DNA binding protein Ssb, RNase H and DNA polymerase I; DNA recombination (recJ, recN, recR, ruvC); and DNA repair (mutS, mutT, mutM, uvrA, uvrB, uvrC, uvrD, mfd) showed an increased expression in the tolC mutant. ctrA, encoding a member of the two-component signal transduction family involved in silencing replication initiation showed significantly decreased expression in the tolC mutant. We also observed increased expression of two genes encoding Maf-like proteins (SMc02311 and SMc02792). Expression of a maf-like gene was also increased in S. meliloti after NaCl osmotic shock [30]. In Bacillus subtilis, overexpression of maf results in inhibition of septation, leading to extensive filamentation [42]. To evaluate whether the tolC mutant cells showed morphological changes, microscopic analysis after staining of cells with crystal violet was performed at 17, 24 and 48 hours of growth. No significant differences were seen concerning size or shape of the two cell types at any time point (data not shown). Increased expression of maf-like genes could suggest inhibition of cell division in the tolC mutant in accordance to the lower optical density observed in the growth curve (Fig. 1). On the other hand, we observed an increased expression of genes involved in chromosomal replication. This apparent contradiction could be explained if, at the time of cell collection and total RNA extraction, the wild-type cells were growing less quickly than the tolC mutant cells, due to entry into stationary phase.
Expression profiling of genes encoding enzymes needed for lipopolysaccharide synthesis (LPS), such as the lpxABDKL genes involved in lipid A biosynthesis, and lpsBCDES, kdsA, kdsB and kdtA encoding enzymes for the biosynthesis of the LPS core showed a significantly increased expression in the tolC mutant. Regarding peptidoglycan biosynthesis we observed increased expression in the tolC mutant of murACEFG genes, the undecaprenyl pyrophosphate phosphatase uppP and synthase uppS. Three penicillin-binding protein encoding genes (mrcA1, mrcB and dac) and several putative lytic murein transglycosylases (SMc04411, mltB1, mltB2, SMc02785) also displayed increased expression. Multiple genes involved in capsular polysaccharide biosynthesis, including the rkpUAGHIJ and kpsF3 genes located on the chromosome and previously demonstrated to be involved in symbiotic capsular polysaccharide biosynthesis [43] and four genes from pSymB (rkpT2, rkpZ2, SMb20824, SMb20825) possibly also related to capsular polysaccharide biosynthesis showed increased expression in the tolC mutant. Increased expression of genes encoding products for synthesis of LPS, peptidoglycan and capsular polysaccharide may be linked to extracytoplasmic stress response activation to neutralize the compromised cell envelope.
We had previously shown that the tolC mutant strain is unable to produce succinoglycan in GMS medium [15]. Whether that was related to differences at transcriptional level or to post-transcriptional regulation was unknown. exo gene expression is positively regulated by the regulator MucR [44] and negatively by ExoR [45]. Here mucR gene expression was significantly increased whilst exoR was decreased when the transcription profile of the tolC mutant was compared to that of the wild-type strain. This could suggest increased expression of the exo genes directing succinoglycan biosynthesis in the tolC mutant. However, none of the exo genes had significant changes at the level of expression, with the exception of exoN encoding UDP-glucose pyrophosphorylase, which showed decreased expression, and the gene exoU encoding a glycosyltransferase the expression of which was increased. Apparently the absence of succinoglycan from the tolC mutant is not caused by differences at the transcription level. It appears more probable that, due to cell envelope perturbations, the exopolysaccharide polymerization and secretion multienzyme complex does not assemble properly or is inactive and therefore no exopolysaccharide is secreted. Also no difference was observed in the expression of genes involved in galactoglucan biosynthesis, with the exception of the transcriptional activator encoding gene wggR [46] that showed a decreased expression. Our results contrast with those obtained for S. meliloti cells stressed with salt or acid pH, where genes encoding proteins for exopolysaccharide biosynthesis showed increased expression [30,33].
Genes involved in motility and chemotaxis
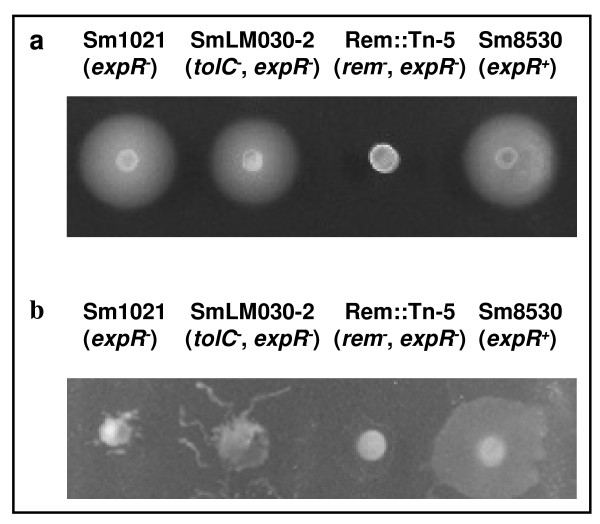
Analysis of gene expression levels in the flagellar regulon indicated an approximately 2-fold increased expression in the tolC mutant of cheABDRW1W2XY1Y2 and mcpU genes, whose products are involved in chemotaxis. Most of the fli, flh, mot, flg and fla genes encoding proteins for the basal body, L and P rings, hook filament, motor switch and flagellum also displayed increased expression in the tolC mutant (Table 1). To test whether differences in the expression of motility genes leads to a phenotype in GMS semi-solid media, swimming and swarming tests were performed using the two strains. Two further strains used in this test were an S. meliloti transposon mutant (Rem::Tn-5) as a negative control, unable to swim or swarm due to inactivation of the transcription regulator Rem, and strain Sm8530 (expR+) as positive control for the two types of motility. Representative images are shown in Fig. 7. Despite increased expression in the tolC mutant of several fli, flh, mot, flg and fla genes, we observed no difference between swimming motility of the tolC mutant and the wild-type strains, with both strains being able to swim (Fig. 7a). Regarding swarming motility, we found that after 24 hours of incubation the tolC mutant displayed a higher surface motility than the wild-type strain (Fig. 7b), consistent with our gene expression data. The swarming behavior of wild-type and tolC mutant strains was markedly different from the expR+ positive control strain Sm8530, which spread over the agar uniformly in all directions whilst the two first strains had a growth branching out from the center of the colony (Fig. 7b). S. meliloti cells stressed with acidic pH or increased osmotic pressure due to salt or sucrose showed decreased expression of genes involved in chemotaxis and motility, consistent with the cell needing to conserve energy [30,31,33]. Why the tolC mutant has increased swarming motility is not known.
Figure 7.
Swimming (a) and swarming (b) tests. Swimming and swarming plates containing 0.3% and 0.6% purified agar, respectively, were spotted with 5 μl of late exponential S. meliloti cultures grown overnight in GMS medium. The photographs were taken after 1 day of incubation for swarming and 3 days for swimming at 30°C.
Conclusions
The transcriptomic data presented here indicate that the absence of functional TolC protein in S. meliloti compromises cell homeostasis as reflected by the concomitant increase in expression levels of many genes putatively involved in cytoplasmic and extracytoplasmic stress responses. Intracellular stress can possibly be caused by accumulation of proteins and metabolites that can not be secreted combined with oxidative stress. To ameliorate adverse effects, a RpoH-dependent response is triggered with an increase in the expression of many genes encoding products protecting macromolecules like DNA, RNA and proteins and helping their turnover. Perturbations in the cell envelope caused by a potential accumulation of proteins such as the truncated TolC in the periplasm may have triggered a Cpx-dependent stress response with a set of genes encoding periplasmic proteases, chaperones and protein modifying enzymes having increased expression. Increased protein synthesis causes increased expression of the genes responsible for transcription, translation and energy producing pathways. The hypothetical higher metabolic demand was mirrored by increased expression of genes encoding nutrient uptake transport systems. Further support for our observations that cell envelope perturbation leads to extracytoplasmic and to oxidative stress comes from recent studies in Vibrio cholerae type II secretion mutants [24]. Sikora et al. [24] showed that type II secretion mutants having compromised membrane integrity, suffer from internal oxidative stress and increased levels of intracellular ferrous iron. Nevertheless, they observed the induction of an RpoE-mediated stress response, whilst we observed a Cpx-mediated stress response, emphasising the differences between the two types of mutations/organisms. Responses to stress caused by S. meliloti lack of functional TolC are distinct from other stress conditions such as osmotic shock and acid pH [30,33]. In the latter two there is general shut-down of the expression of genes involved in central metabolism, protein metabolism, iron uptake and chemotaxis. In contrast, the tolC mutant shows an increased expression of genes involved in all of these pathways. One possible explanation could be the higher need for energy and reducing power to combat oxidative stress and the possible accumulation of proteins that can not be secreted. Another possibility is related to an eventually compromised electrochemical proton gradient across the membrane. Since TolC is the outer membrane component of many transport systems [1], its inactivation may affect both proton transport and ATP synthesis and possibly the cell responds by increasing expression of genes involved in central metabolism to synthesize more ATP. Although many questions remain unanswered, our results highlight the mechanisms by which a large number of genes act together to restore cell homeostasis and, in particular, points to TolC protein as being fundamental in the biology of this microorganism.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this study were wild-type S. meliloti 1021 (Sm1021) [47], SmLM030-2 (Sm1021, pLS378 integrated into the tolC gene region) [15], Sm8530 (Sm1021, expR+) [48], and Rem::Tn-5 (Sm1021, rem-) [49]. For gene expression profiling, overnight cultures of S. meliloti 1021 and tolC mutant strain SmLM030-2 grown in TY complex medium [50] were diluted to an initial OD600 = 0.1 in GMS medium (Zevenhuizen, 1986). Triplicate flasks of each strain were cultured at 30°C in GMS medium at 180 rpm for 20 hours.
Isolation and processing of RNA samples
Cells were harvested, resuspended in RNAprotect bacteria reagent (Qiagen), and total RNA extraction was carried out using the RNeasy MiniKit (Qiagen) with DNase treatment following manufacturer's recommendations. Once absence of residual DNA was confirmed, concentration and purity were determined using a Nanodrop ND-1000 UV-visible spectrophotometer. RNA integrity was checked with an Agilent 2100 Bioanalyser using a RNA Nano assay (Agilent Technologies).
RNA was processed for use on Affymetrix (Santa Clara, CA, USA) GeneChip Medicago/Sinorhizobium Genome Arrays, according to the manufacturer's Prokaryotic Target Preparation Assay. Briefly, 10 μg of total RNA containing spiked in Poly-A RNA controls (GeneChip Expression GeneChip Eukaryotic Poly-A RNA Control Kit; Affymetrix) were used in a reverse transcription reaction with random primers (Invitrogen Life Technologies) to generate first-strand cDNA. After removal of RNA, 2 μg of cDNA was fragmented with DNase and end-labeled (GeneChip® WT Terminal Labeling Kit; Affymetrix). Size distribution of the fragmented and end-labeled cDNA, was assessed using an Agilent 2100 Bioanalyzer. 2 μg of end-labeled fragmented cDNA was used in a 200-μl hybridization cocktail containing added hybridization controls and hybridized on arrays for 16 hours at 48°C. Standard post hybridization wash and double-stain protocols (FS450_0001; GeneChip HWS kit, Affymetrix) were used on an Affymetrix GeneChip Fluidics Station 450. Arrays were scanned on an Affymetrix GeneChip scanner 3000 7G.
Microarray analysis
Scanned arrays were first analyzed using Affymetrix Expression Console software to obtain Absent/Present calls and assure that all quality parameters were in the recommended range. Subsequent analysis was carried out with DNA-Chip Analyzer 2008. First a digital mask was applied, leaving for analysis only the 8305 probe sets on the array representing Sinorhizobium meliloti transcripts. Then the 6 arrays were normalized to a baseline array with median CEL intensity by applying an Invariant Set Normalization Method [51]. Normalized CEL intensities of the arrays were used to obtain model-based gene expression indices based on a PM (Perfect Match)-only model [52]. Replicate data (triplicates) for each of the wild-type and tolC mutant strains were weighted gene-wise by using inverse squared standard error as weights. Genes compared were considered to be differentially expressed if the 90% lower confidence bound of the fold change between experiment and baseline was above 1.2, resulting in 3155 differentially expressed transcripts with a median False Discovery Rate (FDR) of 0.4%. The lower confidence bound criterion means that we can be 90% confident that the fold change is a value between the lower confidence bound and a variable upper confidence bound. Li and Wong [52] have shown that the lower confidence bound is a conservative estimate of the fold change and therefore more reliable as a ranking statistic for changes in gene expression. For a second analysis Partek Genomics Suite 6.4 was used. Here the 6 arrays were normalized and modeled using Robust Multichip Averaging (RMA). After RMA, probe sets analyzing expression of transcripts of Medicago truncatula and Medicago sativa, were filtered out. For the remaining S. meliloti probe sets differential expression was determined using 1-way Analysis of Variance (ANOVA). FDR analysis with a cut-off of 5% determined 2842 transcripts as differentially expressed, corresponding to an ANOVA p-value cut-off of <0.017. A set of 2067 differentially expressed transcripts was identified in the two independent analyses performed. All further analyses focused on this core set. Fold change values presented in Tables 1 and 2 and in the additional files 1 and 2 were obtained using Partek Genomics Suite 6.4.
Quantitative real-time RT-PCR
DNA microarray data were validated by quantitative real-time RT-PCR. For reverse transcription 1 μg of total RNA from S. meliloti 1021 and tolC mutant strains, derived from three independent samples, was used. cDNA was synthesized using TaqManR Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer's instructions. Primers used to amplify selected S. meliloti genes (See Additional file 3: Table S3) were designed using Primer Express 3.0 software (Applied Biosystems). RT-PCR amplification mixtures used 400 ng of template cDNA, 2× SYBR Green PCR Master Mix and 0.4 mM of reverse and forward primers for each gene in a total volume of 25 μl. Reactions containing nuclease-free water instead of the reverse transcriptase were included as negative control. Reactions were performed using a model 7500 thermocycler (Applied Biosystems). The expression ratio of the target genes was determined relative to reference gene hemA, which showed no variation in the transcript abundance under the experimental conditions used here. Relative quantification of gene expression by real-time RT-PCR was determined by applying the ΔΔCt method [53].
Preparation of cell lysates and measuring enzymatic activities
S. meliloti wild-type and tolC mutant cells were grown in GMS medium for 20 hours. Cells were harvested, washed and disrupted by sonication. The total protein concentration was measured by the Bradford method [54]. Catalase and superoxide dismutase activities were determined using the method of Clare et al. [55]. Crude extract (20 μg) of each sample was loaded on a standard nondenaturing polyacrylamide gel and samples electrophoresed for 6 hours at 70 V. To measure catalase activity, the gel was soaked in 50 mg/ml of horseradish peroxidase in 50 mM potassium phosphate, pH 7.0, at room temperature for 45 min and rinsed twice with phosphate buffer. The gel was then incubated with 5.0 mM H2O2 for 10 min then stained with 0.5 mg/ml diaminobenzidine in phosphate buffer. For superoxide dismutase measurement, the gel was soaked in the dark in 2.5 mM nitro blue tetrazolium with 3 mM H2O2 supplementation for 20 minutes. Gels were then incubated with 0.028 mM riboflavin and 2.8 mM TEMED in 36 mM phosphate buffer, pH 7.8 for 20 minutes, followed by irradiation with visible light until achromatic bands appeared. Glutathione reductase (GR) activity was measured as described by Smith et al. [56] following the disappearance of NADPH spectrophotometrically at 340 nm (E = 6.2 mM-1 cm-1). The reaction mixture contained 400 mM phosphate buffer (pH 7.5), 10 mM oxidized glutathione, 1 mM NADPH, 10 mM EDTA, 3 mM Dithionitrobenzoic acid and crude extract.
Assessment of cells efflux activity
Efflux activity was assayed by ethidium bromide agar screening [57]. Briefly, each S. meliloti culture was swabbed onto GMS plates containing ethidium bromide concentrations of 0.5 and 1.0 mg/L. Plates were incubated at 30°C for 48 hours, after which fluorescence under UV light associated with the bacterial mass was recorded.
Motility assays
Motility assays were carried out as described by Soto et al. [58]. Swimming plates with 0.3% bacto agar (Difco) and swarming plates with 0.6% Noble agar (Difco) were prepared using GMS medium. For estimation of motility, overnight GMS cultures (5 μl) were inoculated on the surface of the agar and incubated at 30°C for 1 and 3 days to measure swarming and swimming motility, respectively. Three separate experiments, each containing two technical replicates were performed.
Microarray data accession number
The microarray data were deposited in the Array Express database under accession number E-MEXP-2561.
Authors' contributions
LFM and JDB designed the work, supervised the research study, and prepared the manuscript. MRS, AMC, JMCM and MFM performed all experimental work. All authors read and approved the final manuscript.
Supplementary Material
Genes with increased expression in the S. meliloti tolC mutant. Table S1. Complete list of all S. meliloti SmLM030-2 genes with increased expression (>1.2-fold change; p < 0.017) compared to the expression in the wild-type S. meliloti 1021. Genes classified into COGs are the ones analyzed.
Genes with decreased expression in the S. meliloti tolC mutant. Table S2. Complete list of all S. meliloti SmLM030-2 genes with decreased expression (>1.2-fold change; p < 0.017) compared to expression in the wild-type S. meliloti 1021. Genes classified into COGs are the ones analyzed.
Primer sequences used in this study. Table S3. Gene-specific primers used for real-time RT-PCR.
Contributor Information
Mário R Santos, Email: mario.r.santos@ist.utl.pt.
Ana M Cosme, Email: ana.cosme@ist.utl.pt.
Jörg D Becker, Email: jdbecker@igc.gulbenkian.pt.
João MC Medeiros, Email: joao.medeiros@ist.utl.pt.
Márcia F Mata, Email: marcia.mata@ist.utl.pt.
Leonilde M Moreira, Email: lmoreira@ist.utl.pt.
Acknowledgements
Dr Anke Becker from University of Freiburg, Germany, is acknowledged for providing strains Sm8530 and Rem::Tn-5. This work was supported by FEDER and Fundação para a Ciência e a Tecnologia, Portugal (contracts PTDC/AGR-AAM/66977/2006, PTDC/AGR-GPL/70592/2006, and Ph.D. grants to A.M.C. and M.R.S.).
References
- Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuel C, Grosse C, Taudte N, Scherer J, Wesenberg D, Krauss GJ, Nies DH, Grass G. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J Bacteriol. 2005;187:6701–6707. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- German GJ, Misra R. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J Mol Biol. 2001;308:579–585. doi: 10.1006/jmbi.2001.4578. [DOI] [PubMed] [Google Scholar]
- Rosner JL, Martin RG. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol. 2009;191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabote RD, Johnson OL, Zetina E, San Francisco SK, Fralick JA, San Francisco MJ. Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J Bacteriol. 2003;185:5772–5778. doi: 10.1128/JB.185.19.5772-5778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JD, Reddy SL, Hopkins DL, Gabriel DW. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol Plant Microbe Interact. 2007;20:403–410. doi: 10.1094/MPMI-20-4-0403. [DOI] [PubMed] [Google Scholar]
- Posadas DM, Martin FA, Sabio y Garcia JV, Spera JM, Delpino MV, Baldi P, Campos E, Cravero SL, Zorreguieta A. The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect Immun. 2007;75:379–389. doi: 10.1128/IAI.01349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber MA, Bailey AM, Blair JM, Morgan E, Stevens MP, Hinton JC, Ivens A, Wain J, Piddock LJ. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol. 2009;191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006;8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- Cosme AM, Becker A, Santos MR, Sharypova LA, Santos PM, Moreira LM. The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol Plant Microbe Interact. 2008;21:947–957. doi: 10.1094/MPMI-21-7-0947. [DOI] [PubMed] [Google Scholar]
- Moreira LM, Becker JD, Puhler A, Becker A. The Sinorhizobium meliloti ExpE1 protein secreted by a type I secretion system involving ExpD1 and ExpD2 is required for biosynthesis or secretion of the exopolysaccharide galactoglucan. Microbiology. 2000;146:2237–2248. doi: 10.1099/00221287-146-9-2237. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salazar JM, Sandoval-Calderon M, Guo X, Castillo-Ramirez S, Reyes A, Loza MG, Rivera J, Alvarado-Affantranger X, Sanchez F, Gonzalez V. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology. 2009;155:386–397. doi: 10.1099/mic.0.021428-0. [DOI] [PubMed] [Google Scholar]
- Bittner AN, Foltz A, Oke V. Only one of five groEL genes is required for viability and successful symbiosis in Sinorhizobium meliloti. J Bacteriol. 2007;189:1884–1889. doi: 10.1128/JB.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. Febs J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- Cereda A, Carpen A, Picariello G, Tedeschi G, Pagani S. The lack of rhodanese RhdA affects the sensitivity of Azotobacter vinelandii to oxidative events. Biochem J. 2009;418:135–143. doi: 10.1042/BJ20081218. [DOI] [PubMed] [Google Scholar]
- Santos R, Bocquet S, Puppo A, Touati D. Characterization of an atypical superoxide dismutase from Sinorhizobium meliloti. J Bacteriol. 1999;181:4509–4516. doi: 10.1128/jb.181.15.4509-4516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora AE, Beyhan S, Bagdasarian M, Yildiz FH, Sandkvist M. Cell envelope perturbation induces oxidative stress and changes in iron homeostasis in Vibrio cholerae. J Bacteriol. 2009;191:5398–5408. doi: 10.1128/JB.00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora AE, Lybarger SR, Sandkvist M. Compromised outer membrane integrity in Vibrio cholerae Type II secretion mutants. J Bacteriol. 2007;189:8484–8495. doi: 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Ann Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnebaumer K, Sander G, Shutinoski B, Schmidt MA, Heusipp G. Controlled activation of the Cpx system is essential for growth of Yersinia enterocolitica. FEMS Microbiol Lett. 2009. [DOI] [PubMed]
- Dominguez-Ferreras A, Perez-Arnedo R, Becker A, Olivares J, Soto MJ, Sanjuan J. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J Bacteriol. 2006;188:7617–7625. doi: 10.1128/JB.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberg S, Tian ZX, Krol E, Linke B, Meyer F, Wang Y, Puhler A, Weidner S, Becker A. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J Biotechnol. 2003;106:255–268. doi: 10.1016/j.jbiotec.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Rossbach S, Mai DJ, Carter EL, Sauviac L, Capela D, Bruand C, de Bruijn FJ. Response of Sinorhizobium meliloti to elevated concentrations of cadmium and zinc. Appl Environ Microbiol. 2008;74:4218–4221. doi: 10.1128/AEM.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellweg C, Puhler A, Weidner S. The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol. 2009;9:37. doi: 10.1186/1471-2180-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauviac L, Philippe H, Phok K, Bruand C. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J Bacteriol. 2007;189:4204–4216. doi: 10.1128/JB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Ann Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Bylund GO, Wipemo LC, Lundberg LA, Wikstrom PM. RimM and RbfA are essential for efficient processing of 16 S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. RNA folding and ribosome assembly. Curr Opin Chem Biol. 2008;12:667–673. doi: 10.1016/j.cbpa.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Shipman L, Zhang M, Anton BP, Roberts RJ, Cheng X. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J Mol Biol. 2004;340:695–706. doi: 10.1016/j.jmb.2004.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Romero DG, Swiatlo E. Role of polyamine transport in Streptococcus pneumoniae response to physiological stress and murine septicemia. Microb Pathog. 2008;45(3):167–172. doi: 10.1016/j.micpath.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Patriarca EJ, Tate R, Iaccarino M. Key role of bacterial NH(4)(+) metabolism in Rhizobium-plant symbiosis. Microbiol Mol Biol Rev. 2002;66:203–222. doi: 10.1128/MMBR.66.2.203-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- Butler YX, Abhayawardhane Y, Stewart GC. Amplification of the Bacillus subtilis maf gene results in arrested septum formation. J Bacteriol. 1993;175:3139–3145. doi: 10.1128/jb.175.10.3139-3145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Kiss E, Reuhs BL, Carlson RW, Kondorosi A, Putnoky P. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J Bacteriol. 1998;180:5426–5431. doi: 10.1128/jb.180.20.5426-5431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram-Drogatz PA, Quester I, Becker A, Puhler A. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol Gen Genet. 1998;257:433–441. doi: 10.1007/s004380050667. [DOI] [PubMed] [Google Scholar]
- Yao SY, Luo L, Har KJ, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J Bacteriol. 2004;186:6042–6049. doi: 10.1128/JB.186.18.6042-6049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlawane C, Baumgarth B, Serrania J, Ruberg S, Becker A. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J Bacteriol. 2008;190:3456–3466. doi: 10.1128/JB.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellock BJ, Teplitski M, Boinay RP, Bauer WD, Walker GC. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J Bacteriol. 2002;184:5067–5076. doi: 10.1128/JB.184.18.5067-5076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobigaylo N, Wetter D, Szymczak S, Schiller U, Kurtz S, Meyer F, Nattkemper TW, Becker A. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl Environ Microbiol. 2006;72:4329–4337. doi: 10.1128/AEM.03072-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clare DA, Duong MN, Darr D, Archibald F, Fridovich I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal Biochem. 1984;140:532–537. doi: 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- Smith IK, Vierheller TL, Thorne CA. Assay of glutathione reductase in crude tissue homogenates using 5,5'-dithiobis(2-nitrobenzoic acid) Anal Biochem. 1988;175:408–413. doi: 10.1016/0003-2697(88)90564-7. [DOI] [PubMed] [Google Scholar]
- Couto I, Costa SS, Viveiros M, Martins M, Amaral L. Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. J Antimicrob Chemother. 2008;62:504–513. doi: 10.1093/jac/dkn217. [DOI] [PubMed] [Google Scholar]
- Soto MJ, Fernandez-Pascual M, Sanjuan J, Olivares J. A fadD mutant of Sinorhizobium meliloti shows multicellular swarming migration and is impaired in nodulation efficiency on alfalfa roots. Mol Microbiol. 2002;43:371–382. doi: 10.1046/j.1365-2958.2002.02749.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes with increased expression in the S. meliloti tolC mutant. Table S1. Complete list of all S. meliloti SmLM030-2 genes with increased expression (>1.2-fold change; p < 0.017) compared to the expression in the wild-type S. meliloti 1021. Genes classified into COGs are the ones analyzed.
Genes with decreased expression in the S. meliloti tolC mutant. Table S2. Complete list of all S. meliloti SmLM030-2 genes with decreased expression (>1.2-fold change; p < 0.017) compared to expression in the wild-type S. meliloti 1021. Genes classified into COGs are the ones analyzed.
Primer sequences used in this study. Table S3. Gene-specific primers used for real-time RT-PCR.