Abstract
Pancreatic cancer shows very poor prognosis and is the fifth leading cause of cancer death in Japan. Previous studies indicated some genetic factors contributing to the development and progression of pancreatic cancer; however, there are limited reports for common genetic variants to be associated with this disease, especially in the Asian population. We have conducted a genome-wide association study (GWAS) using 991 invasive pancreatic ductal adenocarcinoma cases and 5,209 controls, and identified three loci showing significant association (P-value<5×10−7) with susceptibility to pancreatic cancer. The SNPs that showed significant association carried estimated odds ratios of 1.29, 1.32, and 3.73 with 95% confidence intervals of 1.17–1.43, 1.19–1.47, and 2.24–6.21; P-value of 3.30×10−7, 3.30×10−7, and 4.41×10−7; located on chromosomes 6p25.3, 12p11.21 and 7q36.2, respectively. These associated SNPs are located within linkage disequilibrium blocks containing genes that have been implicated some roles in the oncogenesis of pancreatic cancer.
Introduction
Pancreatic cancer is the fifth leading cause of cancer death with an estimated death of 24,634 patients in Japan in year 2007. Its 5-year survival rate is as low as 6.7% (http://www.fpcr.or.jp/publication/pdf/statistics2009/fig01.pdf and http://www.fpcr.or.jp/publication/pdf/statistics2009/fig20.pdf). Since no specific symptom is observed in the patients with pancreatic cancer at an early stage, most of the patients were diagnosed at their advanced stage with a very low possibility of cure for the disease [1], [2].
Previous reports indicated the involvement of both environmental and genetics factors in the etiology of this deleterious disease. Several case-control and cohort epidemiological studies have identified a number of possible risk factors such as smoking [3], diabetes [4], chronic pancreatitis [5], which are likely to predispose individual to the disease. In addition, familial aggregation of the disease has implied the possible involvement of genetic factors in pancreatic cancer [6]; approximately 10% of the patients were reported to have family history and individuals having first-degree relatives with pancreatic cancer revealed 2- to 4- fold higher risk of the disease [7]–[9]. These data indicated that genetic factors are likely to play some roles in the development of pancreatic cancer. In the last decade, the advancement of molecular biology improved the understanding of the pathogenesis of pancreatic cancer and characterized a number of genes that mutated in pancreatic cancers, such as somatic mutations in genes INK4A(CDKN2A), TP53, DPC4, BRCA1/2, STK11, APC, KRAS and ATM and PALB2 are found in pancreatic cancers [10]–[18].
Two recent GWAS studies for pancreatic cancer using Caucasian populations have identified associations with genome-wide significance on chromosomes 9p34.2 (ABO), 13q22.1, 1q32 (NR5A2) and 5p15.33 (CLPTM1L-TERT), and highlighted that accumulation of these common genetic risk variants with modest effects are likely to play an important role on this complex disease, either individually or in interaction with environmental factors [19]–[22]. As the ethnicity is one of the critical factors in the pathogenesis of the genetic diseases with complex gene-gene and gene-environmental interactions, we (Biobank Japan (BBJ) in The University of Tokyo and National Cancer Center (NCC) Japan) combined samples of 991 cases with pancreatic cancer and 5209 controls (Table S1), attempted to identify common genetic variations associated with susceptibility to pancreatic cancer in the Japanese population.
Results
After the standard quality control of the genotype results (Table S2), association analysis was performed for 420,236 SNPs using logistic regression analysis on the basis of allelic, dominant and recessive models after adjustment of age, sex and smoking status for each individual. The Q-Q plot for this GWAS based on allelic P-values by logistic regression revealed no significant population stratification with genomic inflation factor λ of 1.026 (Figure 1).
Figure 1. Q-Q plot for GWAS of pancreatic cancer in Japanese population.
This Q-Q plot is based on logistic regression allelic P-values after standard quality control. (genomic inflation factor λ = 1.026).
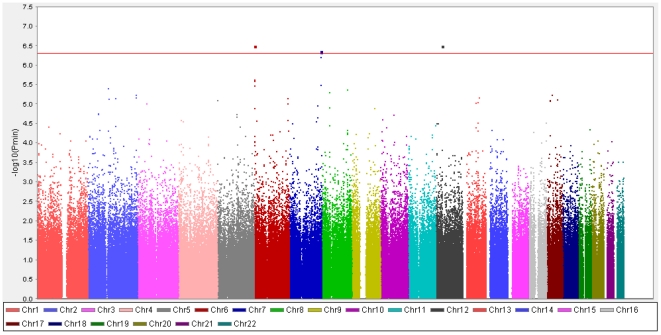
We successfully identified three genomic regions, 6p25.3, 12p11.21 and 7q36.2, shown to be significantly associated (P-value<5.0×10−7) with increased risk of pancreatic cancer in Japanese population as indicated in the Manhattan plot in Figure 2 (referred to ref. 23).
Figure 2. Manhattan plot for GWAS of pancreatic cancer in Japanese population.
The plot is based on logistic regression model after correction of age, sex and smoking status. The P min indicates the minimum P-value from logistic regression analysis for three models: allelic, dominant and recessive. Red line indicates genome-wide significant level (P-value = 5×10−7).
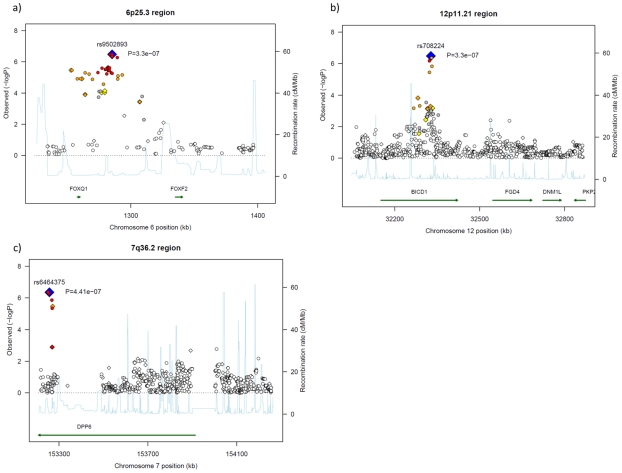
The most significantly-associated SNP, rs9502893 (P-value of 3.30×10−7, per-allele odds ratio (OR) of 1.29 with 95% confidence interval (CI) of 1.17–1.43), is located within a 75-kb linkage disequilibrium (LD) block on chromosome 6p25.3 (Table 1). This LD block includes FOXQ1 (forkhead box (Fox) Q1) gene, which is located 25 kb upstream to this marker SNP (Figure 3a). Imputation analysis also revealed modest association at SNPs located near to or on the FOXQ1 gene suggesting it to be one of the causative genes for pancreatic cancer (Figure 3a and Table S3).
Table 1. SNPs that show suggestive association with increase risk of pancreatic cancer in Japanese population.
| CHR* | SNP | Position* | Risk allele | RAF | Allelic | Dominant | Recessive | P min | Gene | Relativeloc* | ||||||||||
| Case | Control | P-value | OR | L95 | U95 | P-value | OR | L95 | U95 | P-value | OR | L95 | U95 | |||||||
| 6 | rs9502893 | 1285189 | G | 0.411 | 0.351 | 3.30E-07 | 1.29 | 1.17 | 1.43 | 2.97E-05 | 1.36 | 1.18 | 1.57 | 2.18E-05 | 1.50 | 1.24 | 1.80 | 3.30E-07 | FOXQ1 | 25196 |
| 12 | rs708224 | 32327676 | A | 0.718 | 0.656 | 3.30E-07 | 1.32 | 1.19 | 1.47 | 8.54E-07 | 1.42 | 1.23 | 1.63 | 2.09E-03 | 1.46 | 1.15 | 1.86 | 3.30E-07 | BICD1 | 0 |
| 7 | rs6464375 | 153256776 | A | 0.116 | 0.103 | 1.15E-01 | 1.13 | 0.97 | 1.32 | 7.36E-01 | 1.03 | 0.87 | 1.22 | 4.41E-07 | 3.73 | 2.24 | 6.21 | 4.41E-07 | DPP6 | 0 |
| 7 | rs7779540 | 153253595 | A | 0.116 | 0.103 | 1.08E-01 | 1.14 | 0.97 | 1.33 | 7.12E-01 | 1.03 | 0.87 | 1.23 | 4.58E-07 | 3.72 | 2.23 | 6.20 | 4.58E-07 | DPP6 | 0 |
| 7 | rs6973850 | 153269181 | A | 0.116 | 0.106 | 2.23E-01 | 1.10 | 0.94 | 1.29 | 9.76E-01 | 1.00 | 0.84 | 1.18 | 6.27E-07 | 3.64 | 2.19 | 6.04 | 6.27E-07 | DPP6 | 0 |
| 6 | rs11242679 | 1282311 | A | 0.366 | 0.311 | 2.40E-06 | 1.28 | 1.15 | 1.42 | 1.15E-05 | 1.37 | 1.19 | 1.58 | 2.07E-03 | 1.39 | 1.13 | 1.71 | 2.40E-06 | FOXQ1 | 22318 |
| 6 | rs7750826 | 1281867 | G | 0.365 | 0.311 | 2.57E-06 | 1.28 | 1.15 | 1.41 | 1.30E-05 | 1.37 | 1.19 | 1.57 | 1.98E-03 | 1.39 | 1.13 | 1.71 | 2.57E-06 | FOXQ1 | 21874 |
| 7 | rs10487687 | 153271407 | A | 0.150 | 0.136 | 8.99E-02 | 1.13 | 0.98 | 1.30 | 6.76E-01 | 1.03 | 0.88 | 1.21 | 3.35E-06 | 2.66 | 1.76 | 4.02 | 3.35E-06 | DPP6 | 0 |
| 6 | rs11242674 | 1252846 | A | 0.355 | 0.301 | 3.46E-06 | 1.28 | 1.15 | 1.41 | 9.64E-06 | 1.37 | 1.19 | 1.58 | 4.59E-03 | 1.37 | 1.10 | 1.69 | 3.46E-06 | FOXQ1 | −4829 |
| 2 | rs6711606 | 101288602 | A | 0.135 | 0.116 | 1.27E-02 | 1.20 | 1.04 | 1.39 | 1.86E-01 | 1.12 | 0.95 | 1.32 | 4.02E-06 | 2.81 | 1.81 | 4.37 | 4.02E-06 | RNF149 | 0 |
| 8 | rs10088262 | 124834883 | A | 0.374 | 0.341 | 3.42E-03 | 1.16 | 1.05 | 1.28 | 4.30E-06 | 1.40 | 1.21 | 1.61 | 3.98E-01 | 0.91 | 0.74 | 1.13 | 4.30E-06 | FAM91A1 | −15180 |
| 8 | rs7832232 | 38588460 | A | 0.483 | 0.454 | 1.43E-02 | 1.13 | 1.03 | 1.25 | 7.63E-01 | 0.98 | 0.84 | 1.14 | 5.10E-06 | 1.45 | 1.24 | 1.71 | 5.10E-06 | RNF5P1 | −10528 |
| 2 | rs6736997 | 235279936 | A | 0.372 | 0.328 | 2.95E-04 | 1.20 | 1.09 | 1.33 | 4.96E-02 | 1.15 | 1.00 | 1.33 | 5.85E-06 | 1.57 | 1.29 | 1.91 | 5.85E-06 | ARL4C | −209504 |
| 17 | rs225190 | 27901771 | G | 0.410 | 0.360 | 5.99E-06 | 1.26 | 1.14 | 1.39 | 1.92E-04 | 1.32 | 1.14 | 1.52 | 2.37E-04 | 1.43 | 1.18 | 1.72 | 5.99E-06 | MYO1D | 0 |
| 2 | rs4663158 | 235263691 | A | 0.397 | 0.352 | 1.39E-04 | 1.21 | 1.10 | 1.34 | 2.89E-02 | 1.17 | 1.02 | 1.35 | 6.91E-06 | 1.53 | 1.27 | 1.85 | 6.91E-06 | ARL4C | −193259 |
| 13 | rs2039553 | 79197723 | A | 0.291 | 0.268 | 1.32E-02 | 1.15 | 1.03 | 1.28 | 4.39E-01 | 1.06 | 0.92 | 1.21 | 7.01E-06 | 1.73 | 1.36 | 2.19 | 7.01E-06 | NDFIP2 | 171501 |
| 2 | rs1427593 | 137271694 | A | 0.110 | 0.080 | 1.55E-05 | 1.42 | 1.21 | 1.66 | 7.10E-06 | 1.49 | 1.25 | 1.77 | 4.30E-01 | 1.31 | 0.67 | 2.58 | 7.10E-06 | THSD7B | −193238 |
| 6 | rs3016539 | 162156065 | A | 0.903 | 0.871 | 1.67E-05 | 1.42 | 1.21 | 1.67 | 7.28E-06 | 1.50 | 1.26 | 1.79 | 2.90E-01 | 1.36 | 0.77 | 2.43 | 7.28E-06 | PARK2 | 0 |
| 2 | rs12615966 | 104745389 | A | 0.112 | 0.097 | 1.40E-02 | 1.22 | 1.04 | 1.42 | 1.59E-01 | 1.13 | 0.95 | 1.35 | 7.44E-06 | 3.15 | 1.91 | 5.21 | 7.44E-06 | LOC284998 | −6744 |
| 17 | rs2257205 | 53803296 | A | 0.378 | 0.327 | 1.58E-05 | 1.25 | 1.13 | 1.38 | 7.74E-06 | 1.38 | 1.20 | 1.59 | 2.97E-02 | 1.25 | 1.02 | 1.53 | 7.74E-06 | RNF43 | 0 |
| 5 | rs6879627 | 2162901 | G | 0.575 | 0.522 | 8.12E-06 | 1.25 | 1.14 | 1.39 | 4.66E-04 | 1.31 | 1.12 | 1.52 | 1.57E-04 | 1.42 | 1.18 | 1.69 | 8.12E-06 | LOC731559 | 225138 |
| 17 | rs4924935 | 18694595 | G | 0.269 | 0.228 | 8.80E-05 | 1.25 | 1.12 | 1.40 | 8.15E-06 | 1.37 | 1.19 | 1.58 | 5.06E-01 | 1.11 | 0.82 | 1.48 | 8.15E-06 | PRPSAP2 | (7622 |
| 17 | rs1737947 | 18772157 | G | 0.252 | 0.212 | 3.88E-05 | 1.27 | 1.13 | 1.43 | 8.49E-06 | 1.37 | 1.19 | 1.58 | 2.89E-01 | 1.19 | 0.87 | 1.62 | 8.49E-06 | PRPSAP2 | 0 |
| 13 | rs1886449 | 72830115 | A | 0.424 | 0.383 | 2.61E-04 | 1.21 | 1.09 | 1.33 | 5.62E-02 | 1.15 | 1.00 | 1.33 | 9.24E-06 | 1.51 | 1.26 | 1.80 | 9.24E-06 | LOC730242 | −206271 |
| 13 | rs1585440 | 65379816 | C | 0.761 | 0.713 | 9.28E-06 | 1.30 | 1.16 | 1.45 | 3.09E-05 | 1.35 | 1.17 | 1.55 | 7.36E-03 | 1.50 | 1.12 | 2.03 | 9.28E-06 | LOC387933 | −118820 |
| 3 | rs4683235 | 46476935 | A | 0.215 | 0.173 | 9.93E-06 | 1.31 | 1.16 | 1.48 | 7.53E-05 | 1.34 | 1.16 | 1.54 | 2.24E-03 | 1.70 | 1.21 | 2.38 | 9.93E-06 | LTF | 0 |
Odds ratios, 95% confidence limits and P-values were obtained using logistic regression analysis according to allelic, dominant and recessive model after adjustment of age, sex and smoking.
RAF, risk allele frequency; OR, odds ratio; L95, U95, lower and upper confidence limits; P min, minimum P-value among three genetic models.
*Position and relative loci (Relativeloc) are based on NCBI Human Genome Build 36.
Figure 3. Regional association plots for three pancreatic cancer risk loci.
(a) 6p25.3 region, SNP rs9502893 located 25 kb upstream to gene FOXQ1. (b) 12p11.21 region, SNP rs708224 is located at the second intron of gene BICD1. (c) 7q36.2 region, SNP rs6464375 is located at the first intron of gene DPP6 transcript variant 3. Each of the marker SNPs is marked by a blue diamond. SNPs that are genotyped in the Illumina platform are plotted as diamonds; Imputed SNPs are plotted as circles. The color intensity reflects the extent of LD with the marker SNP, red (r2≥0.8), orange (0.5≤r2<0.8), yellow (0.2≤r2<0.5) and white (r2<0.2). Light blue line indicated local recombination rate.
The second significantly-associated SNP, rs708224, located in the second intron of the gene BICD1 (Bicaudal-D homolog 1) on chromosome 12p11 (P-value of 3.30×10−7, per-allele OR of 1.32 with 95% CI of 1.19–1.47) (Table 1). The 80-kb LD block showing the association corresponds to the second intron of BICD1 as revealed by the imputation analysis shown in Figure 3b (Table S3).
The third locus is marked by rs6464375, rs7779540, rs6973850 and rs1048768 in the first intron of DPP6 gene. These SNPs indicated suggestive associations only under recessive model with minimum P-value of 4.41×10−7 (OR of 3.73 with 95%CI of 2.24–6.21) as shown in Table 1 and Figure 3c.
Discussion
Here we present results of GWAS analysis on 991 cases with pancreatic cancer and 5209 controls. Our study represents the first GWAS attempt to identify common variants associated with pancreatic cancer in Japanese population and successfully identified SNPs located on chromosomal loci of 6p25.3, 12p11.21 and 7q36.2 are significantly associated with increased risk of pancreatic cancer in Japanese population.
It is known that the development of the common disease is caused by the accumulation of common genetic variants, and each of this variant has a very modest effect on the risk (for example OR of <1.2). In order to detect such small fraction, GWAS involving much larger populations (5000–10000) should be required. Our study was expected to identify SNPs with moderate effects (i.e OR>1.4). Hence SNPs that show very modest effect might have failed to be identified through this study.
The most significantly associated SNP in this GWAS, rs9502893 (P-value = 3.30×10−7, OR = 1.29) is located within a 75 kb LD block which encompasses gene FOXQ1 on chromosome loci 6p25.3. FOXQ1 encodes for protein forkhead box (Fox) Q1. The Fox family of transcription factors consists of at least 43 members and mutations in Fox genes can cause significant effects on human common disease and cancers [24], [25]. A Fox member, FoxM1, is well-known to be associated with oncogenesis of pancreatic cancer. Down-regulation of this protein results in the inhibition of migration, invasion and angiogenesis in pancreatic cancer cells [26]. Furthermore, a recent study showed that FoxQ1 is overexpressed in pancreatic cancer, suggesting its role in pancreatic cancer tumorigenesis [27]. Although the SNP that we identified is approximately 25 kb downstream to this gene, the associated SNP may ‘tag’ the causative variant located on the expression regulatory region of the gene and subsequently alter expression of the gene. However, further study is needed to elucidate a precise biological role and mechanism of the gene function with regard to pancreatic carcinogenesis.
The second most significantly associated SNP, rs708224 (P-value = 3.30×10−7, OR = 1.32) is located within the BICD1 gene. This gene encodes a protein Bicaudal-D homolog 1, which plays a role in vacuolar trafficking. Previous studies reported substantial evidences indicating a link between vacuolar gene and shorter telomeres in yeast model [28]–[30]. In addition, Mangino et al. suggested that genetic variations within the BICD1 gene could alter its transcriptional levels and in turn influence telomere length in humans [31]. Several recent studies have documented reduced telomere length in pancreatic ductal adenocarnoma specimens, suggesting telomeric dysfunction in pancreatic cancer cells [32]–[34]. Thus, it is of importance to determine the functional consequences of rs708224 and/or variations linked to this SNP in the pathogenesis of pancreatic cancer.
Several SNPs located in the first intron of DPP6 indicated suggestive associations with an increased risk of pancreatic cancer in this study. DPP6 encodes protein dipeptidyl-peptidase 6, which binds to specific voltage-gated potassium channels and alters their expression and biophysical properties. A recent study on core signaling pathways in human pancreatic cancers found three somatic mutations in DPP6 among 24 pancreatic cancer samples examined by detailed sequence analyses. This report also suggested that DPP6 might play a crucial role in regulation of invasion of pancreatic cancer cells [35]. Hence, our study strengthens the risk of DPP6 in pancreatic cancer and warrants further screening on this gene to confirm its association with pancreatic cancer.
Recent GWAS reports have indicated several loci on chromosomes 9p34.2, 13q22.1, 1q32.1 and 5p15.33 to be associated with an increased risk of pancreatic cancer in Caucasian population [21], [22]. Among the significantly associated SNPs, rs9543325 on chromosome 13q22.1 showed moderate association in our study populations (P-value (allelic model) of 1.69×10−4; OR of 1.21 with 95%CI of 1.10–1.34) (Table S4). On the other hand, SNPs on chromosomes 9p34.2 (rs505922) and 1q32.1 (rs3790844) showed a weak association in our study populations (P-values of 3.69×10−2 and 1.24×10−2; ORs of 1.11 and 1.14 with 95% CI of 1.01–1.22 and 1.03–1.27, respectively) (Table S4). We were unable to replicate the remaining loci (SHH and two loci on chromosomes 5p15.33 and 15q14) in these reports, probably because most of these associated SNPs are either non-polymorphic or possess very low allelic frequencies (MAF = 0.01) in Japanese population. The power of our study was not sufficient enough to detect positive associations for these variants with the low allelic frequency. Such ethnic difference in genetic architecture of disease susceptibility is not rare. For example, two recent GWAS reported common variants on KCNQ1 gene associated with type 2 diabetes mellitus in Japanese population, but European GWAS were unable to identify the associations due to the low allelic frequency of these variants in the population [36], [37]. In addition, identification of susceptibility loci may be also influenced by the differences in the LD structure across different populations and by potential interaction with other genetic variants and environmental factors [38].
In summary, this study represents the first GWAS to identify common variants possibly associated with pancreatic cancer in Japanese population. Our study confirmed the association from the Caucasian GWAS studies and revealed several novel possible candidate associated loci that were not detected in the previous Caucasian GWAS studies. Nevertheless, further additional replications are required to confirm or exclude the current findings.
Materials and Methods
Case and control subjects
A total of 331 and 675 cases that were clinically and/or histologically diagnosed to have an invasive pancreatic ductal adenocarcinoma were obtained from Biobank Japan (http://biobankjp.org) at the Institute of Medical Science, The University of Tokyo as well as National Cancer Center Hospital, respectively. The control samples consisted of Japanese volunteers that were obtained from Osaka-Midosuji Rotary Club, Osaka, Japan (n = 906) as well as from staff members in Keio University, Japan, who participated in its health-check program (n = 677). In addition, individuals who were registered in Biobank Japan as subjects with various diseases except cancer (n = 3,728) (those having pulmonary tuberculosis, chronic hepatitis-B, keroid, drug-induced skin rash, peripheral artery disease, arrhythmia, stroke and myocardial infarction) were used as controls. All samples were obtained after obtaining the written informed consent. This project was approved by the ethics committee at The Institute of Medical Sciences, The University of Tokyo, National Cancer Center and Keio University. Individuals who had clinical history of diabetes mellitus (a possible confounding factor for pancreatic cancer) were excluded from these control sets. For sample quality control, we excluded five cases with call rate<0.98. After performing principal component analysis, we excluded outliers of 10 cases and 102 controls, who did not belong to the major Japanese cluster (Hondo cluster) (Figure S1) [39]. We eventually performed the association study based on 991 cases and 5209 controls (Table S1). Power calculation showed that our study would have over 90% power to detect a per-allele OR of 1.4 or greater for an allele with 30% frequency at the genome-wide significance level (α = 5×10−7).
SNP genotyping and quality control
All the individuals were genotyped using either Illumina Infinium HumanHap550v3 or Illumina Infinium Human610-Quad DNA Analysis Genotyping BeadChip. SNPs common in the two platforms were used for further analysis. We applied SNP quality control for all sets of samples as follows; SNP call rate should be >0.99 in both cases and controls, and P-value of Hardy-Weinberg equilibrium test should be >1.0×10−6 in controls. SNPs with minor allele frequency (MAF) of <0.01 in both case and control samples were excluded from the further analysis (Table S2).
Statistical analysis
We analyzed each SNP using logistic regression adjusted for age (continuous), sex and smoking status (current/former, never). P-values and OR with 95%CI were calculated for allelic, dominant and recessive models. We used the minimum P-values obtained from three models to evaluate the statistical significance of the association. All OR were reported with respect to the risk allele. All the statistical analyses were performed using R statistical environment version 2.9.0 (http://www.r-project.org/) or PLINK 1.06 (http://pngu.mgh.harvard.edu/~purcell/plink/). R statistical environment version 2.9.0 was employed to draw Q-Q plot and regional association plot.
Genotype Imputation
We performed genotype imputation analysis for each set of samples by utilizing a Hidden Markov model as programmed in MACH version 1.0 (http://www.sph.umich.edu/csg/abecasis/mach/index.html). To infer untyped and missing genotypes around the candidate chromosomal loci, we provided genotypes from our own samples together with haplotypes for reference samples (Japanese from Tokyo, JPT) from HapMap database (http://hapmap.ncbi.nlm.nih.gov/). SNPs with low genotyping rate (<99%), showing deviations from Hardy-Weinberg equilibrium (<1.0×10−6), or MAF (<0.01) were excluded from the analysis. MACH version 1.0 was used to estimate haplotypes, map crossover and error rates using 50 iterations of the Markov chain Monte Carlo algorithm. By utilizing the genotype information from the HapMap database, maximum likelihood genotypes were generated. For quality control, we retained imputed SNPs with the estimated r2 of >0.3. We also picked up a total of 17 SNPs (P-value<0.001) to verify the association using Invader and TaqMan genotyping methods (data not shown).
Supporting Information
Sample characteristic of this study.
(0.02 MB XLS)
Total number of SNPs excluded according to each quality control criteria.
(0.02 MB XLS)
Imputation analysis around significantly associated SNPs.
(0.04 MB XLS)
Association study of SNPs which shown to be significantly associated with increased risk of pancreatic cancer in Caucasian population in Japanese.
(0.02 MB XLS)
Principal component analysis for GWAS of pancreatic cancer in Japanese population. a) Principal component analysis for GWAS of pancreatic cancer in Japanese population refer to four HapMap population control subjects including CEU indicates Caucasians from Utah; YRI, Nigerians from Yoruba; CHB, Han Chinese from Beijing and JPT, Japanese from Tokyo. b) Principal component analysis of study subjects referred only to Asian populations. We utilized samples from the homogenous case-control (Hondo) cluster.
(9.43 MB TIF)
Acknowledgments
We express our heartfelt gratitude to all the patients who participate in this study. We would like to thank Dr Yoichiro Kamatani for his constructive comments and suggestions. Our thankfulness also goes to the member of The Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan for making this study possible. We thank Drs. Hideki Ueno, Masafumi Ikeda, Chigusa Morizane, Yoshihiro Sakamoto, Minoru Esaki, Tomoo Kosuge and Nobuyoshi Hiraoka for ascertainment of the patients and their clinico-pathological information at the National Cancer Center Hospital. We also would like to express our gratefulness to Miss Kumi Matsuda for her outstanding technical assistance.
Footnotes
Competing Interests: Although the affiliations of the following three authors (Akira Saito [Statistical Genetics Analysis Division, StaGen Co., Ltd.], Suenori Chiku [Science Solutions Division, Mizuho Information and Research Institute, Inc.] and Hirohiko Totsuka [Bioinfomatics Group, Research and Development Center, Solution Division]) are in the private sector, each of them worked for one of the corresponding authors, Teruhiko Yoshida, as a part of a contract research solely funded by an academic research fund granted from NiBio (http://www.nibio.go.jp/english/) to T. Yoshida. Therefore, this study neither received any funding from the companies to which the above three authors belong, nor did their affiliating companies per se play any role in this research except that they sent their staff (the above three authors) to the laboratory of Teruhiko Yoshida under a research contract paid by Yoshida's grant. There is no competing interests involved between Teruhiko Yoshida and these companies, including intellectual properties. The authors also confirm that they can adhere to all the PLoS ONE policies on sharing data and materials.
Funding: This work was conducted as a part of Biobank Japan Project that was supported by the Ministry of Education, Culture, Sports, Sciences and Technology from the Japanese Government and Center for Genomic Medicine, RIKEN, Japan. It was also supported in part by the program for promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kelsen DP, Portenoy R, Thaler H, Tao Y, Brennan M. Pain as a predictor of outcome in patients with operable pancreatic carcinoma. Surgery. 1997;122:53–59. doi: 10.1016/s0039-6060(97)90264-6. [DOI] [PubMed] [Google Scholar]
- 2.Catanzaro A, Richardson S, Veloso H, Isenberg GA, Wong RC, et al. Long-term follow-up of patients with clinically indeterminate suspicion of pancreatic cancer and normal EUS. Gastrointest Endosc. 2003;58:836–840. doi: 10.1016/s0016-5107(03)02301-0. [DOI] [PubMed] [Google Scholar]
- 3.Anderson KE, Mack T, Silverman D. Cancer of the pancreas. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. New York: Oxford University Press; 2006. pp. 721–762. [Google Scholar]
- 4.Stevens RJ, Roddam AW, Beral V. Pancreatic cancer in type 1 and young-onset diabetes: systematic review and meta-analysis. Br J Cancer. 2007;96:507–509. doi: 10.1038/sj.bjc.6603571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, et al. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 1993;328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 6.Del Chiaro M, Zerbi A, Falconi M, Bertacca L, Polese M, et al. Cancer risk among the relatives of patients with pancreatic ductal adenocarcinoma. Pancreatology. 2007;7:459–469. doi: 10.1159/000108963. [DOI] [PubMed] [Google Scholar]
- 7.McWilliams RR, Rabe KG, Olswold C, De Andrade M, Petersen GM. Risk of malignancy in first-degree relatives of patients with pancreatic carcinoma. Cancer. 2005;104:388–394. doi: 10.1002/cncr.21166. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez E, La Vecchia C, D'Avanzo B, Negri E, Franceschi S. Family history and the risk of liver, gallbladder, and pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 1994;3:209–212. [PubMed] [Google Scholar]
- 9.Tersmette AC, Petersen GM, Offerhaus GJ, Falatko FC, Brune KA, et al. Increased risk of incident pancreatic cancer among first-degree relatives of patients with familial pancreatic cancer. Clin Cancer Res. 2001;7:738–744. [PubMed] [Google Scholar]
- 10.Yan L, McFaul C, Howes N, Leslie J, Lancaster G, et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology. 2005;128:2124–2130. doi: 10.1053/j.gastro.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 12.Barton CM, Staddon SL, Hughes CM, Hall PA, O'Sullivan C, et al. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64:1076–1082. doi: 10.1038/bjc.1991.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berrozpe G, Schaeffer J, Peinado MA, Real FX, Perucho M. Comparative analysis of mutations in the p53 and K-ras genes in pancreatic cancer. Int J Cancer. 1994;58:185–191. doi: 10.1002/ijc.2910580207. [DOI] [PubMed] [Google Scholar]
- 14.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–3. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 15.Hruban RH, Iacobuzio-Donahue C, Wilentz RE, Goggins M, Kern SE. Molecular pathology of pancreatic cancer. Cancer J. 2001;7:251–258. [PubMed] [Google Scholar]
- 16.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 17.Wong T, Howes N, Threadgold J, Smart HL, Lombard MG, et al. Molecular diagnosis of early pancreatic ductal adenocarcinoma in high-risk patients. Pancreatology. 2001;1:486–509. doi: 10.1159/000055852. [DOI] [PubMed] [Google Scholar]
- 18.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLeod SL, Chowdhury P. The genetics of nicotine dependence: relationship to pancreatic cancer. World J Gastroenterol. 2006;12:7433–7439. doi: 10.3748/wjg.v12.i46.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milne RL, Greenhalf W, Murta-Nascimento C, Real FX, Malats N. The inherited genetic component of sporadic pancreatic adenocarcinoma. Pancreatology. 2009;9:206–214. doi: 10.1159/000210261. [DOI] [PubMed] [Google Scholar]
- 21.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 25.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 27.Cao D, Hustinx SR, Sui G, Bala P, Sato N, et al. Identification of novel highly expressed genes in pancreatic ductal adenocarcinomas through a bioinformatics analysis of expressed sequence tags. Cancer Biol Ther. 2004;3:1081–1089. doi: 10.4161/cbt.3.11.1175. [DOI] [PubMed] [Google Scholar]
- 28.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, et al. Telomere Length as a Quantitative Trait: Genome-Wide Survey and Genetic Mapping of Telomere Length-Control Genes in Yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rog O, Smolikov S, Krauskopf A, Kupiec M. The yeast VPS genes affect telomere length regulation. Curr Genet. 2005;47:18–28. doi: 10.1007/s00294-004-0548-y. [DOI] [PubMed] [Google Scholar]
- 31.Mangino M, Brouilette S, Braund P, Tirmizi N, Vasa-Nicotera M, et al. A regulatory SNP of the BICD1 gene contributes to telomere length variation in humans. Hum Mol Genet. 2008;17:2518–2523. doi: 10.1093/hmg/ddn152. [DOI] [PubMed] [Google Scholar]
- 32.Büchler P, Conejo-Garcia JR, Lehmann G, Müller M, Emrich T, et al. Real-time quantitative PCR of telomerase mRNA is useful for the differentiation of benign and malignant pancreatic disorders. Pancreas. 2001;22:331–340. doi: 10.1097/00006676-200105000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Kobitsu K, Tsutsumi M, Tsujiuchi T, Suzuki F, Kido A, et al. Shortened telomere length and increased telomerase activity in hamster pancreatic duct adenocarcinomas and cell lines. Mol Carcinog. 1997;18:153–159. doi: 10.1002/(sici)1098-2744(199703)18:3<153::aid-mc4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 34.van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 37.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–456. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample characteristic of this study.
(0.02 MB XLS)
Total number of SNPs excluded according to each quality control criteria.
(0.02 MB XLS)
Imputation analysis around significantly associated SNPs.
(0.04 MB XLS)
Association study of SNPs which shown to be significantly associated with increased risk of pancreatic cancer in Caucasian population in Japanese.
(0.02 MB XLS)
Principal component analysis for GWAS of pancreatic cancer in Japanese population. a) Principal component analysis for GWAS of pancreatic cancer in Japanese population refer to four HapMap population control subjects including CEU indicates Caucasians from Utah; YRI, Nigerians from Yoruba; CHB, Han Chinese from Beijing and JPT, Japanese from Tokyo. b) Principal component analysis of study subjects referred only to Asian populations. We utilized samples from the homogenous case-control (Hondo) cluster.
(9.43 MB TIF)