Abstract
Almost three decades ago, we presented a model where the extracellular matrix (ECM) was postulated to influence gene expression and tissue-specificity through the action of ECM receptors and the cytoskeleton. This hypothesis implied that ECM molecules could signal to the nucleus and that the unit of function in higher organisms was not the cell alone, but the cell plus its microenvironment. We now know that ECM invokes changes in tissue and organ architecture and that tissue, cell, nuclear, and chromatin structure are changed profoundly as a result of and during malignant progression. Whereas some evidence has been generated for a link between ECM-induced alterations in tissue architecture and changes in both nuclear and chromatin organization, the manner by which these changes actively induce or repress gene expression in normal and malignant cells is a topic in need of further attention. Here, we will discuss some key findings that may provide insights into mechanisms through which ECM could influence gene transcription and how tumor cells acquire the ability to overcome these levels of control.
I. INTRODUCTION
No man is an island, entire of itself; every man is a piece of the continent, a part of the main.
John Donne (1573–1631)
Over the years, studies of transcription, chromatin structure, and nuclear organization, mostly performed in lower organisms or in cells isolated from higher organisms and cultivated on tissue culture plastic, have provided important and valuable evidence for elucidating intracellular-signaling events important for gene expression. But much still remains to be elucidated in order to understand the intricate complexity of gene expression within a three-dimensional (3D) tissue context. Vertebrate cells exist as part of an integrated unit of multiple cell types, that is, an organ, in a milieu rich in growth factors, hormones, and an assortment of extracellular matrix (ECM) molecules, the combination of which is organ-specific. Similarly, the nucleus within each cell is not an isolated vessel. It is connected to its external milieu and the ECM through a dynamic intracellular network that includes the nuclear matrix, the nuclear envelope, and the cytoskeleton. In the past few decades, a number of studies have provided evidence that the physical and biochemical signals transmitted from the ECM to the nucleus are indeed dynamic and reciprocal (Bissell et al., 1982; Lelievre et al., 1998; Plachot and Lelievre, 2004; Roskclley et al., 1994; Zoubiane et al., 2004). More specifically, the nature of ECM receptors and their signaling, and the nature of the connections between these receptors and intracellular components such as the cytoskeleton and its interacting proteins, promote changes in nuclear events which influence gene expression and initiate the production of specific signaling molecules that are fed back into the extracellular environment which, in turn, influences the cell and the nucleus (Bissell et al., 1982) (Fig. 1). Understanding how signaling by the ECM and its receptors are integrated into messages that alter chromatin structure and nuclear function is essential for knowing how a normal cell functions in the context of a tissue and an organ, and for identifying aberrations that lead to the development of diseases such as cancer. Here, we will discuss the influence of ECM on cellular and nuclear functions, as well as chromatin structure, and relate these changes to alterations in transcriptional events essential for functional differentiation and transformation.
Fig. 1.
A schematic diagram illustrating the basic principles of dynamic reciprocity between neighboring cells and their extracellular environment. Mechanical and biochemical signals received at points of cell–cell or cell–ECM contact are transduced to the nucleus by transmembrane receptors, signaling molecules, and cytoskeletal components where they initiate nuclear events resulting in the expression of specific gene products that are excreted back into the extracellular milieu. Green arrows represent the bidirectional flow of mechanical and biochemical signals between the ECM and the nucleus. RTKs represent receptor tyrosine kinase. (Modified, with permission, from Bissell et al., 2005.) (See Color Insert.)
II. THE ECM
There are two broad subtypes of ECM: interstitial/stromal ECM and basement membrane (BM) (Guo and Giancotti, 2004). The interstitial matrix surrounds cells in the connective tissue (Guo and Giancotti, 2004), while the BM is present at the basolateral surface of different cell types in many tissues (Kalluri, 2003). The BM is composed mainly of laminin, type IV collagen, entactin/nidogen, and proteoglycans such as heparin sulfate that are deposited by many different cell types (Kalluri, 2003).
In order to fully appreciate how profoundly a cell and its nucleus are affected by the ECM, one must consider that the composition of the ECM is constantly influenced by physiological effectors such as growth factors, cytokines, and hormones, and, thus, is continuously changing throughout development, aging, tissue repair, and tumor progression (Guo and Giancotti, 2004; Labat-Robert, 2003; Mott and Werb, 2004). There are now multiple types of ECM receptors that have been identified including integrins, syndecans, glypicans, and dystroglycan (Guo and Giancotti, 2004; Rizki and Bissell, 2004; Weir and Muschler, 2003). Different integrins are known to activate distinct signaling pathways but the same integrin receptor can stimulate multiple signaling pathways (DeMali et al., 2003; Guo and Giancotti, 2004).
Exposure to ECM promotes changes in cell shape, initiates the clustering of neighboring integrin receptors at cell-matrix contact sites, and promotes the recruitment of kinases and adaptor proteins that link these receptor molecules to the actin cytoskeleton (Schatzmann et al., 2003). These changes initiate a series of mechanical and biochemical signals within the cell that ultimately change the program of gene expression and influence cellular processes such as survival, transcription factor activation, and apoptosis (Schatzmann et al., 2003). In the case of mammary epithelial cells, changes in cell shape that are mediated by exposure to a laminin-rich ECM gel (lrECM) activate expression of the milk protein lactoferrin (Close et al., 1997; Roskelley et al., 1994) (Fig. 2). When these cell shape changes are combined with further biochemical and mechanical signals generated from the prolactin receptor and laminin 1-induced activation of integrin receptors, the transcription of additional milk proteins such as β-casein is initiated, but the expression of growth factors such as TGFβ1 is inhibited concomitantly (Streuli et al., 1991, 1993, 1995). Following initial exposure to lrECM, morphogenic events take place that lead to the development of spherical, acinar structures with hollow lumen (Barcellos-Hoff et al., 1989). The formation of these structures is accompanied by growth arrest, enhancement of milk protein production, down-regulation of growth factors such as TGFα, and the expression of new milk proteins such as whey acidic protein (WAP) (Barcellos-Hoff et al., 1989; Chen and Bissell, 1989; Lin et al., 1995; Petersen et al., 1992). As a result, the ability of lrECM to promote the functional differentiation of normal mammary epithelial cells into polar, acinar structures that produce and secrete milk requires several levels of control (Bissell et al., 1999; Lin and Bissell, 1993; Roskelley et al. 1995).
Fig. 2.
An illustration of the different levels through which ECM controls gene expression and tissue function. As cells transition from a 2D monolayer to a 3D environment, they undergo changes in cell shape that influence the expression of certain genes. Exposure to ECM engages specific cell surface receptors and initiates the transduction of biochemical and mechanical signals through the cell to the nucleus, where they further influence gene expression. As the duration of exposure time to ECM increases, cells undergo morphogenic events involving the formation of acinar structures and once again exhibit changes in their gene expression profile. Thus, tissue structure influences gene expression and, therefore, dictates tissue function. (Modified, with permission, from Bissell et al., 1999, 2005; Roskelley et al., 1995.) (See Color Insert.)
III. ECM-RESPONSE DNA ELEMENTS
One of the most convincing pieces of evidence linking the ECM to gene expression was the identification of an ECM response element in the promoter region of the bovine β-casein gene. Deletion analysis of the bovine β-casein promoter revealed that the transcriptional activation of this promoter was completely dependent on a 160-bp BCE-1 transcriptional enhancer located approximately 1.5-kb upstream of the transcription start site (Schmidhauser et al., 1992). Since this discovery, ECM response elements have been identified in several other genes from different species (DiPersio et al., 1991; Novaro et al., 2004; Schmidhauser et al., 1992, 1994) (Table I). However, exactly how the ECM influences gene expression through these response elements is poorly understood.
Table I.
ECM-Response Elements Identified to Date
| Element | Gene | Type of ECM | Cell type | Organism | References |
|---|---|---|---|---|---|
| BCE-I enhancer | β-casein | IrECM | Mammary epithelial |
Cow | Schmidhauser et al., 1992 |
| Promoter | Erα | Laminin | Mammary epithelial |
Mouse | Novaro et al., 2004 |
| Enhancer | Albumin | Collagen | Hepatocytes | Mouse | DiPersio et al., 1991 |
| Proximal G-string |
LpSl | Collagen | Aboral ectoderm |
Sea urchin | Seid et al., 1997 |
| Promoter | MMTV | lrECM | Mammary epithelial |
Mouse | Schmidhauser et al., 1994 |
| Enhancer | SV40a | lrECM | Mammary epithelial |
Mouse | Schmidhauser et al., 1994 |
| Enhancer | CMVb | lrECM | Mammary epithelial |
Mouse | Schmidhauser et al., 1994 |
SV4O represents Simian vacuolating virus 40.
CMV represents cytomegalovirus enhancer.
IV. POTENTIAL MECHANISMS FOR THE TRANSCRIPTIONAL ACTIVATION OF ECM-RESPONSE DNA ELEMENTS
A. Exposure to ECM Influences the Nuclear Translocation and DNA-Binding Properties of Specific Transcription Factors That Bind to ECM-Response Elements
One mechanism through which exposure to ECM may influence the activation of response elements is by altering the levels of specific transcription factors residing in the nucleus. Cognate DNA-binding elements for STAT5, and C/EBPβ, reside in both the bovine β-casein and mouse ERα ECM-response elements (Myers et al., 1998; Novaro et al., 2004). Mutation analysis of these binding sites revealed that their requirement for activation of the β-casein promoter is absolute (Myers et al., 1998). In mammary epithelial cells, exposure to lrECM and prolactin promotes the phosphorylation and translocation of STAT5 into the nucleus (Edwards et al., 1998; Gouilleux et al., 1994; Groner and Gouilleux, 1995; Ihle, 1996). Chromatin-binding proteins locate their cognate sites by scanning the 3D space of the nucleus and bind transiently to chromatin with a residence time on the order of seconds (Phair et al., 2004). An increase in STAT5 levels within the nucleus likely increases the frequency with which this factor interacts with its target DNA sites. In support of this, we have observed that exposure of mouse mammary epithelial cells to lrECM and prolactin induces the recruitment of STAT5 to the β-casein promoter (Xu et al., 2005). Whether lrECM treatment alters the nuclear levels of other factors important for β-casein expression such as C/EBPβ or GR remains to be determined. In a previous study, the culturing of primary rabbit mammary epithelial cells on collagen increased the expression levels of C/EBPβ (Jolivet et al., 2001). In addition, we have also observed an increase in the binding of C/EBPβ to the endogenous mouse β-casein promoter in response to lrECM and prolactin treatment (Xu et al., 2005). Such evidence supports the possibility that lrECM affects gene transcription by influencing transcription factors levels within the nucleus.
B. Exposure to ECM May Initiate Mechanical Signals That Alter the Organization of Nuclear Factors in a Manner That Promotes Activation of ECM-Response Elements
However, the possibility remains that the expression Level of some nuclear proteins does not change in response to ECM treatment. Exposure to ECM has profound effects on cell shape, and cytoskeletal organization (Roskelley et al., 1994; Zoubiane et al., 2004) and the integrity of the cytoskeleton is essential for both STAT5 nuclear translocation and β-casein transcription (Phung-Koskas et al., 2005; Roskelley et al., 1994; Zoubiane et al., 2004). Furthermore, the cytoskeleton bridges the nucleus with the ECM by interacting with both nuclear envelope proteins and integrin receptors on the plasma membrane (Crisp et al., 2006; D’Angelo and Hetzer, 2006; Fey et al., 1984a,b; Janmey, 1998; Wilhelmsen et al., 2005). Lamins help constitute the nuclear envelope and organize DNA into loop domains (Davie, 1995). Alterations in the organization of cytoskeletal filaments, distortions in nuclear shape, and a redistribution of nucleoli along an axis of applied tension have been observed in bovine capillary epithelial cells in response to the mechanical tug of an integrin receptor lining the cell surface (Maniotis et al., 1997, 2005). Thus, it is possible that the ECM promotes transcriptional activation of response elements by changing cytoskeletal organization, which may, in turn, alter the arrangement of nuclear components to promote a localized concentration of transcription factors to areas of the nucleus containing target DNA sequences. In addition, exposure to ECM may also stimulate cellular events that posttranslationally modify and, therefore, alter the properties of transcription factors and chromatin-remodeling enzymes.
C. ECM-Induced Activation of DNA-Response Elements Involves Mechanisms That Invoke Changes in Chromatin Structure
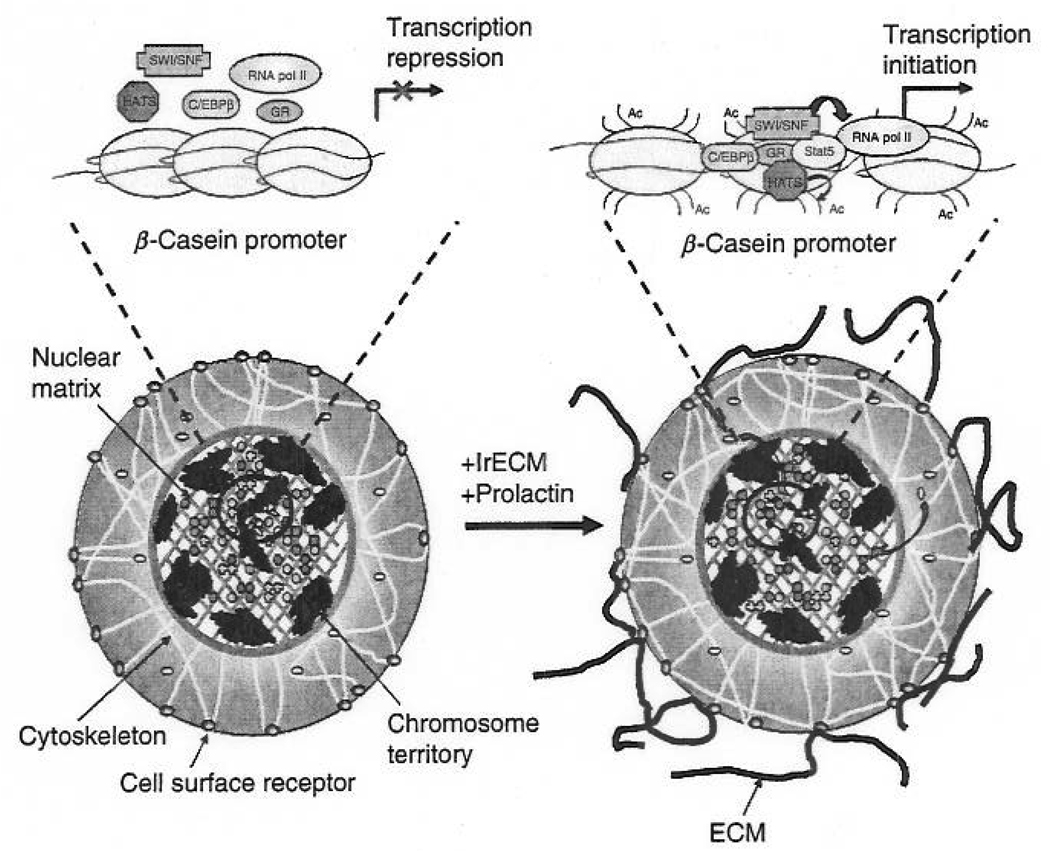
Regardless of whether the ECM mediates an increase in the nuclear levels of transcription factors and/or an increase in the proximity of these factors to their target DNA-binding sites, the fact remains that exposure of mammary epithelial cells to lrECM induces STAT5 and C/EBPβ binding to DNA sites that are within close proximity of one another within the BCE-1 element and the mouse β-casein promoter (Myers et al., 1998; Rosen et al., 1999; Xu et al., 2005). We have shown previously that the treatment of stably transfected mouse mammary epithelial cells with a general histone deacetylase Inhibitor increases BCE-1 activity, albeit not to the same extent as cells treated with inhibitor and lrECM together (Myers et al., 1998). In a study, we also observed an increase in the association of acetylated histones with the endogenous mouse β-casein promoter after lrECM and prolactin treatment (Xu et al., 2005). Further examination of the effects of lrECM on transcription factor recruitment revealed that exposure to lrECM induced also the binding of Brg1 (Xu et al., 2005), an ATPase subunit of the SWI/SNF chromatin-remodeling complex (de la Serna et al., 2006). Overexpression of a dominant-negative Brg1 isoform significantly decreased β-casein RNA levels (Xu et al., 2005). These results indicate that lrECM-mediated induction of β-casein gene transcription requires both histone acetylation and ATP-dependent chromatin remodeling.
STAT5, GR, and C/EBPβ have been shown to interact with histone acetyl-transferases (Lee et al., 1985; Li et al., 2003; Litterst et al., 2003; Pfitzner et al., 1998) and the transactivation potential of GR depends on its ability to recruit the chromatin-remodeling factor Brg1 to target genes (Nagaich et al., 2004; Trotter and Archer, 2004). We have observed by coimmunoprecipitation analyses that STAT5, C/EBPβ, and GR are associated with the SWI/SNF complex in response to lrECM and prolactin treatment (Xu et al., 2005). In addition, this treatment promotes the binding of RNA. polymerase II to the endogenous mouse β-casein promoter (Xu et al., 2005). Therefore, besides altering the nuclear localization and DNA-binding properties of transcription factors, exposure to lrECM promotes transcription factor-mediated recruitment of chromatin-remodeling factors to the bovine BCE-1 enhancer and the endogenous mouse β-casein promoter. A localized decondensation in chromatin structure would allow additional factors such as RNA polymerase II to access their target DNA sites and potentiate further rounds of transcription (Fig. 3). Such events support a popular paradigm for transcriptional activation that describes the recruitment of specific transcription factors to their cognate DNA sequences and their subsequent recruitment of chromatin-remodeling and DNA-modifying enzymes followed by the general transcriptional machinery (Kosak and Groudine, 2004).
Fig. 3.
Schematic representation of lrECM- and prolactin-induced changes in nuclear organization and transcription factor binding in mammary epithelial cells that lead to the initiation of β-casein gene transcription. In the absence of prolactin and lrECM, STAT5 is predominantly cytoplasmic in primary cells. Exposure to lrECM and prolactin induces STAT5 phosphorylation, nuclear translocation, and binding to its cognate DNA sequence in the β-casein promoter. In addition, this treatment increases the association of acetylated histones and promotes the binding of additional transcription factors including C/EBPβ, SWI/SNF, GR, and RNA polymerase (pol) II to the β-casein promoter. Ac represents acetylation at lysine residues along histone N-terminal tails. (See Color Insert.)
Interestingly, mouse mammary epithelial cells transiently transfected with a construct containing the BCE-1 element positioned upstream of a minimal bovine β-casein promoter cloned adjacent to a CAT reporter gene did not display an increase in BCE-1 activity when exposed to lrECM and prolactin, while cells stably transfected with the same construct displayed a significantly large induction (Myers et al., 1998). The fact that BCE-1 required integration into the genome to become transcriptionally active suggests that the proximity of this sequence to its target transcription factors, chromatin-remodeling factors, and the basal transcriptional machinery likely plays an important role in its activation. Thus, to accurately depict the factors and events involved in ECM-mediated gene expression, we must consider how the ECM could influence the general organization of the nucleus.
V. POTENTIAL MECHANISMS THROUGH WHICH ECM INFLUENCES THE GENERAL ORGANIZATION OF NUCLEAR FACTORS AND OVERALL TRANSCRIPTIONAL ACTIVITY
The nucleus has a 10-micron diameter and contains approximately 2 m of DNA (Getzenberg et al., 1991; Hager et al., 2004; Misteli, 2005). In order for this vast amount of DNA to fit within the confines of the nuclear envelope, it must be assembled into chromatin that undergoes several orders of compaction, resulting in a DNA-protein assembly that differentially impedes the access of transcription factors to their cognate sites. Residing alongside chromatin is a large number of nuclear proteins involved in diverse nuclear processes such as transcription, RNA splicing, and DNA repair. Add to this the presence of RNA and proteins required for maintaining structural integrity of the nucleus and it becomes clear that the nucleus is a very crowded organelle indeed.
Past studies have shown that transcription factors are located in discrete bodies within the nucleus (Handwerger and Gall, 2006; Lamond and Spector, 2003; Zimber et al., 2004). A number of investigators have speculated that these bodies are either protein storage sites or centers for the recruitment of specific regulatory cofactors in response to extracellular and intracellular signals (Boisvert et al., 2000; Borden, 2002; Pearson and Pelicci, 2001). Factor recruitment to nuclear bodies is important for events such as proliferation, senescence, and apoptosis. It is thus clear that understanding the factors that affect nuclear organization and the role of nuclear organization in gene expression is essential for understanding cell development and ECM-induced differentiation.
A. Of Mouse and Women: Application to Human Breast Epithelial Cells
The principles learned from the phenotypic and functional behavior of mouse mammary epithelial cells in 3D culture have been applied to human cells with an added advantage: the 3D assay provides a rapid and robust assay with which to distinguish between nonmalignant and malignant breast cells (Bissell et al., 1999, 2005; Petersen et al., 1992; Schmeichel and Bissell, 2003). To examine the principle of ECM signaling to the nucleus in human cells, we showed that culturing nonmalignant HMT-3522 (S1) human mammary epithelial cells on lrECM gels induced acinar morphogenesis and caused the nuclear mitotic apparatus (NuMA) and the serine/arginine repeat-related nuclear matrix protein of 160 kDa (SRm160) to coalesce into discrete foci (Knowles et al., 2006; Lelievre et al., 1998). SRm160 is a splicing factor found in interchromatin granule clusters (IGCs). IGCs are considered to be storage sites for splicing factors (Huang et al., 1994; Misteli et al., 1997; Spector, 1993). Importantly, disturbing the integrity of NuMA foci with an antibody to its C-terminal end reversed lrECM-mediated changes in chromatin organization, disrupted the BM integrity, and activated one or more matrix metalloproteinases (MMPs). Treatment of cells on lrECM with trichostatin A, a histone deacetylase inhibitor, also resulted in the disruption of NuMA foci, and the loss of BM. Plachot and Lelievre (2004) subsequently showed that treatment of acini formed by S1-HMT3522 cells with 5-Aza-2′-deoxycytidine, an inhibitor of DNA methylation, induced DNA hypomethylation and prevented the establishment of apical polarity (Plachot and Lelievre, 2004). These results lead further credence to the concept of a dynamically reciprocal interaction between the nucleus and the ECM, and show that DNA organization, nuclear matrix organization, and the BM are all intimately connected. While the principles appear to hold true, the exact events through which ECM influences nuclear and chromatin organization remain to be resolved.
B. ECM-Induced Differentiation May Involve the Selective Activation of Particular Tissue-Specific Genes, But an Overall Decrease in Gene Activity
Exposure of mouse mammary epithelial cells to lrECM decreased total levels of acetylated histones (Pujuguet et al., 2001) and the assembly of human mammary epithelial cells into functionally differentiated acinar structures on lrECM (Barcellos-Hoff et al., 1989; Petersen et al., 1992) is accompanied by a decrease in overall acetylated H4 levels, an increase in HP1γ, and an increase in the expression levels of MeCP2 (Plachot and Lelievre, 2004). These events promote and/or are a consequence of gene silencing, and, therefore, suggest strongly that lrECM-induced differentiation of human mammary epithelial cells may involve changes in chromatin structure that are conducive to selective activation of particular tissue-specific genes, but an overall decrease in gene activity.
Exposure to lrECM promotes cell cycle arrest (Fournier et al., 2006; Petersen et al., 1992) and this effect may play an important role in its ability to decrease overall transcriptional activity. For instance, in the nuclei of cells in G0/G1, the retinoblastoma (Rb) protein is completed with c-abl, a nonreceptor tyrosine kinase (Welch and Wang, 1993, 1995). When associated with Rb, nuclear c-abl is inactive; in cycling cells, however, cyclin-dependent kinases phosphorylate Rb and disrupt this complex, causing activation of nuclear c-abl which, in turn, activates RNA polymerase II by phosphorylating its C-terminal-repeated domain (Baskaran et al., 1993, 1996). In a study, the number of active RNA polymerase II molecules and the number of active transcription sites significantly decreased in mouse F9 teratocarcinoma and totipotent mouse embryonic stem cells when they were induced to differentiate (Faro-Trindade and Cook, 2006). Thus, exposure to lrECM may decrease total levels of transcriptional activity by indirectly decreasing RNA polymerase II activity.
A decrease in the number of transcription sites would eliminate the need for certain transcription factors and create a temporary surplus of these factors that could be redistributed to discrete nuclear domains such as promyelocytic leukemia (PML) nuclear bodies for storage and/or eventual degradation (Borden, 2002; Kosak and Groudine, 2004; Zimber et al., 2004). In support of this possibility, increasing the levels of a particular transcription factor by transient or stable expression causes excess factors to localize into PML bodies (Tsukamoto et al., 2000).
A decrease in demand for transcription factors may also reduce the need for splicing factors. The C-terminal tail of the largest subunit of RNA polymerase II recruits splicing factors from nearby IGCs during transcription (Misteli and Spector, 1999). Treatment with α-amanitin, a specific inhibitor of RNA polyermase II (Lindell et al., 1970), causes these clusters to round up and prevents the movement of splicing factors from IGCs (Misteli et al., 1997). Thus, a decrease in overall transcription levels resulting from exposure to lrECM could be responsible for the formation of SRm160 foci we observed in mammary epithelial cells (Lelievre et al., 1998). In support of this, hnRNP RNA-processing proteins were shown to accumulate into HERDS, heterogeneous clusters, within the interchromatin space in response to transcriptional arrest (Biggiogera et al., 2004).
C. ECM-Induced Changes in Overall Chromatin Structure May Have Profound Implications on Nuclear Organization and Gene Expression
ECM-induced foci formation may be explained also by the ability of lrECM to promote an overall decrease in total histone acetylation levels. In proliferating monkey CV-1 cells, the formation of a nuclear body containing classes I and II histone deacetylases, as well as SMRT and NCoR nuclear receptor corepressors were dependent on histone deacetylase activity (Downes et al., 2000). The integrity of these domains was disrupted by treatment with histone deacetylase inhibitors, substantiating again that the act of histone deacetylation plays an important role in nuclear organization. A function for DNA-mediated organization of nuclear factors is supported also by the findings of Kaminker et al., (2005), which showed that lrECM-induced cessation of growth in epithelial breast cell lines led to the formation of TIN2 foci. However, whereas treatment with DNase I eliminated the foci, treatment with RNase did not. One possible explanation for histone deacetylase-mediated formation of nuclear domains could be that histone deacetylation promotes the formation of highly condensed chromatin structures that would restrict the access of transcription factors to their target DNA sequences, thereby causing them to either localize to more accessible regions of the nucleus or amalgamate with nearby domains enriched in transcription factors. Such an event would also promote HERD formation since it would contribute to a decrease in transcription and RNA synthesis.
VI. ADVANCING TOWARD A DEEPER UNDERSTANDING OF THE MALIGNANT PHENOTYPE
On the basis of the discussion presented so far, our understanding of the effects of ECM on cellular, and, in particular, nuclear events important for normal cell differentiation is in its infancy. Furthermore, much remains to be learned of the effect of ECM on tumor cell development. We have shown previously that nonmalignant S1 and malignant T4-2 mammary epithelial cells express similar levels of β1-integrin and the coxsackievirus and adeno-virus receptor (CAR) when cultured on a 2D plastic surface (Anders et al., 2003; Weaver et al., 1997) (Table II). However, when cultured in 3D lrECM, the expression levels of these receptors decline dramatically in nonmalignant cells, but remain unaltered in malignant cells (Anders et al., 2003; Weaver et al., 1997). In addition, unlike S1 cells, T4-2 cells cultured on lrECM express a profile of cell surface receptors that favors increased proliferation through pathways involving the epidermal growth factor receptor (EGFR) and phosphatidylinositol-3′-kinase (PI3K) (Liu et al., 2004; Wang et al., 1998; Weaver et al., 1997).
Table II.
Expression Profile of Cell Surface and Signaling Proteins in Nonmalignant (S1) HMT-3522 Cells and Malignant T4-2 Cells Cultured on 2D Plastic and 3D lrECM.
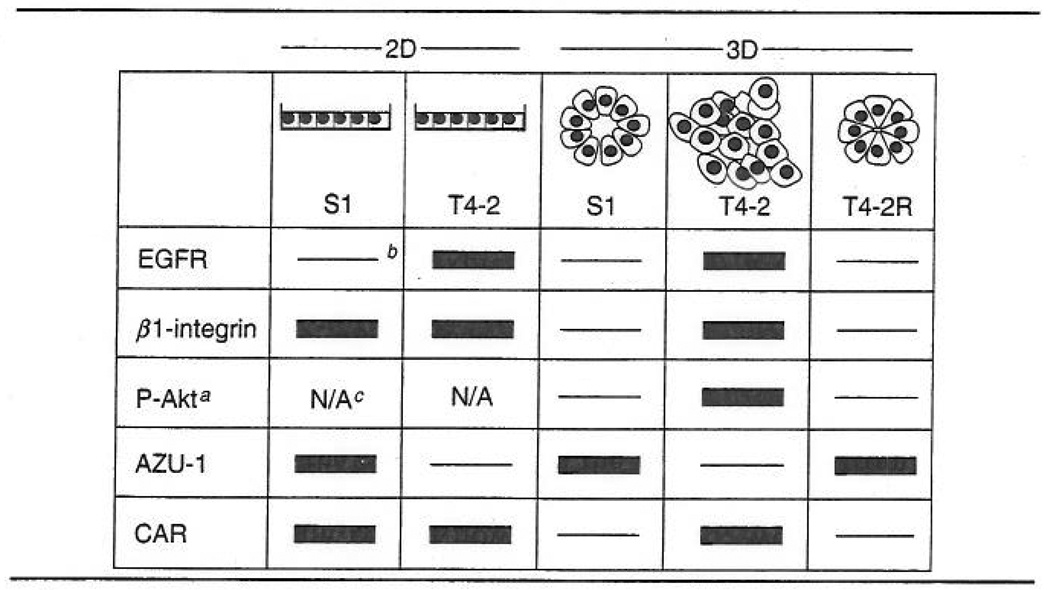 |
P-Akt represents phosphorylated Akt.
Thin lines represent negative expression; Solid boxes represent positive expression.
N/A represent not available.
Interestingly, the levels of β1-integrin, CAR, phosphorylated Akt, and EGFR dramatically decline in T4-2 cells when cultured on 3D lrECM in the presence of a reverting agent that promotes the assembly of these cells into organized, polar structures closely resembling those formed by non-malignant cells (Anders et al., 2003; Liu et al., 2004; Wang et al., 1998; Weaver et al., 1997). In addition, exposure to reverting agents induces T4-2 cells to express the tumor suppressor protein, anti-zuai-1 (AZU-1), to levels observed in S1 cells when cultured on either 2D plastic or 3D lrECM (Chen et al., 2000). In a study, lrECM-induced morphogenesis of normal breast epithelial cells was shown to cause a drastic change in NuMA nuclear organization without inducing morphogenesis or changes in the distribution of NuMA in malignant cells (Knowles et al., 2006; Weaver et al., 1997). Thus, the nature with which a nonmalignant cell communicates with its 3D environment at both the cell surface and the nucleus differs from that of a malignant cell.
The ability of reverting agents to reverse the expression of T4-2 cell surface receptors and signaling proteins shows that the acquisition of a malignant phenotype is accompanied by changes in tissue architecture and reversible changes in protein expression that allow a transformed cell to bypass the strict hierarchical events inherent to normal cell differentiation. With this said, additional studies on normal and malignant mammary epithelial cells need to be performed to further understand and identify the cellular and nuclear events that allow a cancer cell to flourish uncontrollably in an environment that promotes the differentiation and growth cessation of normal cells.
VII. A 3D RECONSTRUCTION FOR THE FUTURE OF CANCER RESEARCH
The union between a cell and its surrounding environment has profound implications on cellular processes such as proliferation, differentiation, and apoptosis. However, during tumorigenesis, the communication between a cancer cell and its environment becomes altered. Mechanisms once enacted to provoke controlled growth and tissue-specific gene expression become obsolete, conferring a tumor cell with a growth advantage nonexistent to a normal cell. While efforts have been made to elucidate the mechanisms through which ECM influences normal and tumor cell behavior and development, the underlying complexity with which a cell communicates with its external environment has thwarted advancements toward obtaining a deeper and truer understanding.
The fact that only a handful of studies have looked at the relationship between ECM and the nucleus is surprising since it is now accepted that ECM signals to gene expression and almost every cellular component outside of the nucleus is affected by ECM. One can only imagine what intriguing discoveries remain to be uncovered in the nucleus by future investigations. Such efforts will not only provide valuable basic information about signaling regulation but also may lead to identification of novel targets for drug therapy.
ACKNOWLEDGMENTS
Our work was supported by grants from the Office of Biological and Environmental Research of the Department of Energy (DE-AC03-76SF00098), the National Cancer Institute (CA057621-13 to Zena Werb and M.J.B.), the Department of Defense Breast Cancer Research Program (Innovator Award DAMD17-02-1-438 to M.J.B., and postdoctoral fellowships W81XWH0410581 to V.A.S. and DAMD17-02-1-0441 to R.X.), and a postdoctoral fellowship from the Canadian Institute for Health Research to V.A.S.
REFERENCES
- Anders M, Hansen R, Ding RX, Rauen KA, Bissell MJ, Korn WM. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the cox-sackievirus and adenovirus receptor. Proc. Natl. Acad. Sci. USA. 2003;100:1943–1948. doi: 10.1073/pnas.0337599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran R, Chiang GG, Wang JY. Identification of a binding site in c-Ab1 tyrosine kinase for the C-terminal repeated domain of RNA polymerase II. Mol. Cell. Biol. 1996;16:3361–3369. doi: 10.1128/mcb.16.7.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera M, Bottone MG, Scovassi AI, Soldani C, Vecchio L, Pellicciari C. Rearrangement of nuclear ribonucleoprotein (RNP)-containing structures during apoptosis and transcriptional arrest. Biol. Cell. 2004;96:603–615. doi: 10.1016/j.biolcel.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757s–1763s. discussion 1763s-1764s. [PubMed] [Google Scholar]
- Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: The role of extracellular matrix and its degrading enzymes. Cold Spring Harb. Symp. Quant. Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: Possible functions for PML nuclear bodies. Mol. Cell Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Schmeichel KL, Mian IS, Lelievre S, Petersen OW, Bissell MJ. AZU-1: A candidate breast tumor suppressor and biomarker for tumor progression. Mol. Biol. Cell. 2000;11:1357–1367. doi: 10.1091/mbc.11.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LH, Bissell MJ. A novel regulatory mechanism for whey acidic protein gene expression. Cell Regul. 1989;1:45–54. doi: 10.1091/mbc.1.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close MJ, Howlett AR, Roskelley CD, Desprez PY, Bailey N, Rowning B, Teng CT, Stampfer MR, Yaswen P. Lactoferrin expression in mammary epithelial cells is mediated by changes in cell shape and actin cytoskeleton. J. Cell Sci. 1997;110(Pt. 22):2861–2871. doi: 10.1242/jcs.110.22.2861. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell. Mol. Life Sci. 2006;63:316–332. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JR. The nuclear matrix and the regulation of chromatin organization and function. Int. Rev. Cytol. 1995;162A:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- DeMali KA, Wennerberg K, Burridge K. Integrin signaling to the actin cytoskeleton. Curr. Opin. Cell Biol. 2003;15:572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- DiPersio CM, Jackson DA, Zaret KS. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol. Cell. Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc. Natl. Acad. Sci. USA. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GM, Wilford FH, Liu X, Hennighausen L, Djiane J, Streuli CH. Regulation of mammary differentiation by extracellular matrix involves protein-tyrosine phosphatases. J. Biol. Chem. 1998;273:9495–9500. doi: 10.1074/jbc.273.16.9495. [DOI] [PubMed] [Google Scholar]
- Faro-Trindade I, Cook PR. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value. Mol. Biol. Cell. 2006;17:2910–2920. doi: 10.1091/mbc.E05-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey EG, Capco DG, Krochmalnic G, Penman S. Epithelial structure revealed by chemical dissection and unembedded electron microscopy. J. Cell Biol. 1984a;99:203s–208s. doi: 10.1083/jcb.99.1.203s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey EG, Wan KM, Penman S. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: Three-dimensional organization and protein composition. J. Cell Biol. 1984b;98:1973–1984. doi: 10.1083/jcb.98.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MV, Martin KJ, Kenny PA, Xhaja K, Bosch I, Yaswen P, Bissell MJ. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006;66:7095–7102. doi: 10.1158/0008-5472.CAN-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzenberg RH, Pienta KJ, Ward WS, Coffey DS. Nuclear structure and the three-dimensional organization of DNA. J. Cell. Biochem. 1991;47:289–299. doi: 10.1002/jcb.240470402. [DOI] [PubMed] [Google Scholar]
- Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner B, Gouilleux F. Prolactin-mediated gene activation in mammary epithelial cells. Curr. Opin. Genet. Dev. 1995;5:587–594. doi: 10.1016/0959-437x(95)80027-1. [DOI] [PubMed] [Google Scholar]
- Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell. Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Hager GL, Nagaich AK, Johnson TA, Walker DA, John S. Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta. 2004;1677:46–51. doi: 10.1016/j.bbaexp.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: New insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN. STATs: Signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- Janmey PA. The cytoskeleton and cell signaling: Component localization and mechanical coupling. Physiol. Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- Jolivet G, Meusnier C, Chaumaz G, Houdebine LM. Extracellular matrix regulates alpha s1-casein gene expression in rabbit primary mammary cells and CCAAT enhancer binding protein (C/EBP) binding activity. J. Cell. Biochem. 2001;82:371–386. doi: 10.1002/jcb.1166. [DOI] [PubMed] [Google Scholar]
- Kalluri R. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, Bissell MJ, Campisi J, Lelievre SA. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: A novel role for telomere-associated protein TIN2. J. Cell Sci. 2005;118:1321–1330. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DW, Sudar D, Bator-Kelly C, Bissell MJ, Lelievre SA. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proc. Natl. Acad. Sci. USA. 2006;103:4445–4450. doi: 10.1073/pnas.0509944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Labat-Robert J. Age-dependent remodeling of connective tissue: Role of fibronectin and laminin. Pathol. Biol. (Paris) 2003;51:563–568. doi: 10.1016/j.patbio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lee EY, Lee WH, Kaetzel CS, Parry G, Bissell MJ. Interaction of mouse mammary epithelial cells with collagen substrata: Regulation of casein gene expression and secretion. Proc. Natl. Acad. Sci. USA. 1985;82:1419–1423. doi: 10.1073/pnas.82.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc. Natl. Acad. Sci. USA. 1998;95:14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CQ, Bissell MJ. Multi-faceted regulation of cell differentiation by extracellular matrix. FASEB J. 1993;7:737–743. doi: 10.1096/fasebj.7.9.8330681. [DOI] [PubMed] [Google Scholar]
- Lin CQ, Dempsey PJ, Coffey RJ, Bissell MJ. Extracellular matrix regulates whey acidic protein gene expression by suppression of TGF-alpha in mouse mammary epithelial cells: Studies in culture and in transgenic mice. J. Cell Biol. 1995;129:1115–1126. doi: 10.1083/jcb.129.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Kliem S, Marilley D, Pfitzner E. NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain. J. Biol. Chem. 2003;278:45340–45351. doi: 10.1074/jbc.M303644200. [DOI] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J. Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Valyi-Nagy K, Karavitis J, Moses J, Boddipali V, Wang Y, Nunez R, Setty S, Arbieva Z, Bissell MJ, Folberg R. Chromatin organization measured by AluI restriction enzyme changes with malignancy and is regulated by the extracellular matrix and the cytoskeleton. Am. J. Pathol. 2005;166:1187–1203. doi: 10.1016/S0002-9440(10)62338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr. Opin. Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol. Cell. Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Novaro V, Radisky DC, Ramos Castro NE, Weisz A, Bissell MJ. Malignant mammary cells acquire independence from extracellular context for regulation of estrogen receptor alpha. Clin. Cancer Res. 2004;10:402S–409S. doi: 10.1158/1078-0432.ccr-031209. [DOI] [PubMed] [Google Scholar]
- Pearson M, Pelicci PG. PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene. 2001;20:7250–7256. doi: 10.1038/sj.onc.1204856. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung-Koskas T, Pilon A, Pous C, Betzina C, Sturm M, Bourguet-Kondracki ML, Durand G, Drechou A. STAT5B-mediaced growth hormone signaling is organized by highly dynamic microtubules in hepatic cells. J. Biol. Chem. 2005;280:1123–1131. doi: 10.1074/jbc.M409918200. [DOI] [PubMed] [Google Scholar]
- Plachot C, Lelievre SA. DNA methylation control of tissue polarity and cellular differentiation in the mammary epithelium. Exp. Cell Res. 2004;298:122–132. doi: 10.1016/j.yexcr.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Pujuguet P, Radisky D, Levy D, Lacza C, Bissell MJ. Trichostatin A inhibits beta-casein expression in mammary epithelial cells. J. Cell. Biochem. 2001;83:660–670. doi: 10.1002/jcb.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki A, Bissell MJ. Extracellular matrix: Tissue-specific regulator of cell proliferation. In: Stein GS, Pardee AB, editors. Cell Cycle and Growth Control: Biomolecular Regulation and Cancer. New Jersey: John Wiley & Sons, Inc.; 2004. p. 25. [Google Scholar]
- Rosen JM, Wyszomierski SL, Hadsell D. Regulation of milk protein gene expression. Annu. Rev. Nutr. 1999;19:407–436. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc. Natl. Acad. Sci. USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr. Opin. Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzmann F, Marlow R, Streuli CH. Integrin signaling and mammary cell function. J. Mammary Gland Biol. Neoptasia. 2003;8:395–408. doi: 10.1023/B:JOMG.0000017427.14751.8c. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J. Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Myers CA, Sanzo KT, Bolten S, Bissell MJ. A novel transcriptional enhancer is involved in the prolactin- and extracellular matrix-dependent regulation of beta-casein gene expression. Mol. Biol Cell. 1992;3:699–709. doi: 10.1091/mbc.3.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Bissell MJ. Transcriptional activation by viral enhancers: Critical dependence on extracellular matrix-cell interactions in mammary epithelial cells. Mol. Carcinog. 1994;10:66–71. doi: 10.1002/mc.2940100203. [DOI] [PubMed] [Google Scholar]
- Seid CA, Ramachandran RK, George JM, Govindarajan V, Gonzalez-Rimbau MF, Flytzanis CN, Tomlinson CR. An extracellular matrix response element in the promoter of the LpS1 genes of the sea urchin Lytechinus pictus. Nucleic Acids Res. 1997;25:3175–3182. doi: 10.1093/nar/25.15.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Nuclear organization of pre-mRNA processing. Curr. Opin. Cell Biol. 1993;5:442–447. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: Basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J. Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH, Schmidhauser C, Kobrin M, Bissell MJ, Derynck R. Extracellular matrix regulates expression of the TGF-beta 1 gene. J. Cell Biol. 1993;120:253–260. doi: 10.1083/jcb.120.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J. Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol. Cell. Biol. 2004;24:3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc. Natl. Acad. Sci. USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir ML, Muschler J. Dystroglycan: Emerging roles in mammary gland function. J. Mammary Gland Biol. Neoplasia. 2003;8:409–419. doi: 10.1023/B:JOMG.0000017428.38034.a7. [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JY. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JY. Abrogation of retinoblastoma protein function by c-Abl through tyrosine kinase-dependent and -independent mechanisms. Mol. Cell. Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Spencer VA, Bissell MJ. SWI/SNF complex and histone acetylation cooperate with STAT5 and C/EBPbeta to regulate beta-casein transcription. Mol. Biol. Cell. Suppl. 2005;16 Abstract no. 500. [Google Scholar]
- Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: Functional roles and cellular signalling in health and disease. Cell. Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Zoubiane GS, Valentijn A, Lowe ET, Akhtar N, Bagley S, Gilmore AP, Streuli CH. A role for the cytoskeleton in prolactin-dependent mammary epithelial cell differentiation. J. Cell Sci. 2004;117:271–280. doi: 10.1242/jcs.00855. [DOI] [PubMed] [Google Scholar]