CSPP and CSPP-L are centrosomal proteins of known mitotic function. Here, we identify CSPP proteins as ciliary proteins and place them into a NPHP protein network crucial for normal cilia-dependent renal and retinal tissue architecture. Importantly, CSPP-L is found to be required for ciliogenesis and shown to be a cilia length modulator.
Abstract
We described previously the cell cycle- and microtubule-related functions of two splice isoforms of the centrosome spindle pole-associated protein (CSPP and CSPP-L). Here, we show that endogenous CSPP isoforms not only localize to centrosomes and the midbody in cycling cells but also extend to the cilia axoneme in postmitotic resting cells. They are required for ciliogenesis in hTERT-RPE1 cells in vitro and are expressed in ciliated renal, retinal, and respiratory cells in vivo. We report that CSPP isoforms require their common C-terminal domain to interact with Nephrocystin 8 (NPHP8/RPGRIP1L) and to form a ternary complex with NPHP8 and NPHP4. We find CSPP-L to be required for the efficient localization of NPHP8 but not NPHP4 to the basal body. The ciliogenesis defect in hTERT-RPE1 cells is, however, not mediated through loss of NPHP8. Similar to the effects of ectopical expression of CSPP-L, cilia length increased in NPHP8-depleted cells. Our results thus suggest that CSPP proteins may be involved in further cytoskeletal organization of the basal body and its primary cilium. To conclude, we have identified a novel, nonmitotic function of CSPP proteins placing them into a ciliary protein network crucial for normal renal and retinal tissue architecture and physiology.
INTRODUCTION
Centrosomes are well known for their role in the organization of the microtubule (MT) cytoskeleton in interphase and mitosis, thus impacting intracellular trafficking, intracellular signaling, cell motility, adhesion, and polarity. Structurally, the centrosome is a nonmembranous organelle that consists of a tethered unequal pair of centrioles embedded within an electron-dense matrix termed pericentriolar material. The older mother centriole is distinguished from the younger daughter centriole by the presence of distal and subdistal appendages. These appendages are important for multiple functions, including MT anchoring and docking of the mother centriole to the cell membrane upon cilia formation (for a recent review, see Bettencourt-Dias and Glover, 2007). The centrosome is duplicated once during the cell cycle to allow bipolar spindle assembly in mitosis. During duplication, a new daughter centriole arises at each centriole. The duplicated centrosome pair is untethered before spindle assembly where both centrosomes acquire MT nucleation activity. The maturation of the older centriole of the daughter centrosome that includes the acquisition of distal and subdistal appendages is not concluded until completion of mitosis and reentry into interphase (Azimzadeh and Bornens, 2007).
During the cell division cycle DNA and centrosome duplication have to be coordinated. The detailed examination of the structural organization and the dynamic protein composition of centrosomes revealed many molecular links to diverse cell biological processes (Doxsey et al., 2005a,b; Schatten, 2008). For example, several proteins involved in DNA replication and DNA damage response (DDR) localize to the centrosome (Zhang et al., 2007), and centrosomal proteins have been identified as target of the DDR (Smith et al., 2009). Furthermore, loss of centrosome integrity can cause cell cycle arrest (Srsen et al., 2006; Mikule et al., 2007). These examples emphasize the interrelation of centrosomes and DNA in terms of duplication and integrity surveillance. In addition, several centrosomal proteins localize to the midbody and are involved in the regulation of cytokinesis (Doxsey, 2005; Barr and Gruneberg, 2007). Taken together, the centrosome is not only a follower of the cell division cycle but is actively monitored to ensure the balanced segregation of the duplicated genome in mitosis.
In cells that withdraw from proliferation the centrosome can be docked via the distal appendages of the mother centriole to the cell membrane. Undergoing a process of morphogenesis to form a basal body, the mother centriole can template the assembly of an MT axoneme at its distal end to form either a motile or nonmotile cilium (for reviews, see Dawe et al., 2007; Nigg and Raff, 2009). Almost all cells of the human body can display primary cilia (Wheatley, 1995; Wheatley et al., 1996). Primary cilia functions are cell type specific and range from locomotion to acting as environmental sensors participating in cell signaling (Shah et al., 2009; Veland et al., 2009). It is now well known that genetic defects affecting cilia or basal bodies result in a myriad of pathologies, collectively termed ciliopathies (Cardenas-Rodriguez and Badano, 2009). They are either specific to one or do involve multiple organs. These pathologies include infertility, obesity, blindness, deafness, respiratory disease, cystic kidney disease, hydrocephalus, situs inversitus, skeletal malformations, and other developmental defects (Fliegauf et al., 2007).
Notably, several causative mutations have been identified in genes whose gene products have been localized to centrosomes/basal bodies (Marshall, 2008). Mutation of these proteins not only might affect cilia-associated functions but also might extend to abnormalities in cell polarity, motility, and adhesion, thereby disrupting normal tissue architecture and function. This is well studied in renal cystic diseases such as polycystic kidney disease (PKD) for which two major disease genes, PKD1 and PKD2, have been identified. Autosomal dominant polycystic kidney disease (ADPKD) is the most frequent life-threatening genetic disease, with an incident rate of ∼1 in 500 (Harris and Torres, 2009). Although less frequent than ADPKD, the autosomal recessive cystic renal disorder Nephronophthisis (NPHP) is the most frequent genetic cause of end-stage renal failure in children and young adults. To date, causative mutations in 10 genes (NPHP1–9 and NPHP11) have been identified for this monogenic autosomal recessive cystic kidney disease (Otto et al., 2009; Hildebrandt et al., 2009a,b). In contrast to polycystic kidney disease, in NPHP the kidney size is not increased despite the development of predominantly corticomedullary tubular cysts. Individual NPHP proteins have been functionally associated with centrosomes, cilia, and, in some cases cell–cell junctions. It is thus thought that cyst formation in NPHP is a result of the deregulation of ciliary/centrosome associated signaling pathways that affect the planar cell polarity of the tubular epithelia cells. Furthermore, the loss of cilia associated functions of NPHP proteins is frequently associated with extrarenal involvement of other tissues that require cilia integrity, the most frequent pathologies being retinitis pigmentosa (Senior–Løken syndrome), liver fibrosis, and cerebellar vermis aplasia (Joubert syndrome; Hildebrandt and Zhou, 2007; Salomon et al., 2009).
We originally identified the centrosomal protein encoding centrosome spindle pole associated protein (CSPP1) gene as overexpressed in a mRNA expression screen aimed at the identification of genes involved in progression of human B cell lymphoma (Patzke et al., 2005). Cloning of two splice isoforms, CSPP and CSPP-L, and their characterization by ectopic expression and small interfering RNA (siRNA) depletion linked CSPP isoforms functionally to centrosomes, microtubule organization, and cell cycle progression (Patzke et al., 2005, 2006). CSPP isoforms localized to centrosomes throughout the cell division cycle. The larger isoform decorated kinetochore MTs in metaphase, concentrated at the midspindle in anaphase and localized to the midbody during cytokinesis. Overexpression of either isoform promoted aneuploidy through the generation of abnormal spindles. This finding is supportive for an oncogenic role of CSPP overexpression. A recent study addressing the mitotic function of CSPP isoform showed that CSPP proteins were required for the recruitment of the myosin GTPase exchange factor MyoGEF to the midspindle, thus facilitating cytokinesis by aiding actomyosin ring formation and constriction (Asiedu et al., 2009).
We have now generated a monoclonal antibody (mAb) directed against the common C-terminal domain of CSPP and CSPP-L to further examine their expression and function in cells and tissues. The tissue-specific expression, subcellular localization, and protein interactions described in the presented study link CSPP to cilia related functions of renal and retinal cells. Thus we hypothesize that loss or malfunction of CSPP isoforms could contribute to ciliary disease.
MATERIALS AND METHODS
Generation of a CSPP-specific Antibody
Bacterial expression constructs of CSPP fragments were obtained by subcloning of polymerase chain reaction (PCR)-derived cDNA fragments into pGEX-6-P3 (glutathione transferase [GST]-tag; GE Healthcare, Pittsburgh, PA) and pQE-32 (His-tag; QIAGEN, Hilden, Germany). Tagged proteins were overexpressed in Escherichia coli BL21(DE3) and purified by affinity chromatography. Hybridomas expressing CSPP-specific antibodies were generated from mice immunized with GST-CSPP and identified by affinity for HIS-CSPP (BioGenes, Berlin, Germany).
Antibodies, Plasmids, and siRNA Duplexes
The following primary antibodies were use in this study: mouse anti-β-catenin (610154; BD Biosciences Pharmingen, San Diego, CA), rabbit anti-CSPP1 (11931-1-AP; Proteintech Europe, Manchester, United Kingdom), mouse anti-c-myc, rabbit anti-FLAG, mouse anti-γ-tubulin, mouse anti-acetylated tubulin (all Sigma Chemical, Poole, Dorset, United Kingdom), goat anti-NPHP4 clone N-17 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-NPHP1 and rabbit anti-IFT88 (kind gifts from Greg Pazour, University of Massachusetts Medical School, Worcester, MA), and mouse anti-glutamylated tubulin (a kind gift from Carsten Janke, Institut Curie, Orsay, Cedex, France). Secondary antibodies for immunofluorescence and Western blot analysis were purchased from Invitrogen (Carlsbad, CA) and Jackson ImmunoResearch Laboratories (West Grove, PA).
Plasmids encoding for hemagglutinin (HA)- and Myc-tagged CSPP/CSPP-L expression constructs were described previously (Patzke et al., 2005, 2006). The FLAG-NPHP8 expression construct was a kind gift from Ronald Roepman (Nijmegen Center for Molecular Life Sciences, Nijmegen, The Netherlands) and has been described previously (Arts et al., 2007). The FLAG-NPHP4 expression construct was kindly provided by Thomas Benzing (Uniklinik Köln, Cologne, Germany).
The following siRNA duplexes were obtained from Dharmacon RNA Technologies (Lafayette, CO): ON-TARGET plus SMARTpool siRNAs targeting CSPP1 and a single siRNA targeting a shared sequence of CSPP and CSPP-L mRNAs: 5′-GAAGATTTGCGCAGTGGAC-3′ and a signal siRNA targeting a CSPP-L mRNA specific sequence: 5′-ACAUGGAAAUGAAGGGAAA-3′ (Patzke et al., 2005), siGenome SMARTpool siRNAs targeting RPGRIP1L/NPHP8, green fluorescent protein (GFP), and pericentrin (PCNT) targeting siRNAs described previously (Mikule et al., 2007).
Immunoprecipitations and Immunoblotting
For immunoprecipitation and immunoblot analysis, cells were washed twice in phosphate-buffered saline (PBS) and lysed in lysis buffer [50 mM Tris-Cl, pH 7.6, 20 mM Na2HPO4, 1 mM EDTA, 150 mM NaCl, and 1% (vol/vol) NP-40] supplemented with inhibitor cocktails targeting phosphatases (Sigma Chemical) and proteases (Roche Diagnostics, Basel, Switzerland). Crude cell lysates were clarified by centrifugation (20,000 × g at 4°C for 15 min). Immunoprecipitations were carried out overnight at 4°C, and immunocomplexes were purified using protein A/G-Sepharose (Santa Cruz Biotechnology) and three consecutive washes in lysis buffer. Immunocomplexes or samples of total cell lysates were resolved by SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane (Millipore, Billerica, MA) for Western blot analysis. For coimmunoprecipitation (IP) from retinal extracts, bovine retinal extracts (∼300 μg) were subjected to IP using CSPP-L or NPHP8 antibodies essentially as described previously (Khanna et al., 2005, 2009).
Cell Culture and Transfection Procedures
Human embryonic kidney (HEK)293T and HeLa cells were grown in DMEM. hTERT-RPE1 cells were grown in DMEM/F-12. Both media were supplemented with 10% heat-inactivated fetal calf serum, penicillin, and streptomycin at 37°C in a humidified atmosphere with 5% CO2.
HEK293T and HeLa cells were transfected with plasmids or siRNAs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. hTERT-RPE1 cells were transfected with siRNA duplexes using Oligofectamine (Invitrogen) as described in Mikule et al. (2007).
Immunofluorescence Imaging
For immunofluorescence microscopy, cells were grown on sterilized no. 1.5 glass coverslips (Glasswarenfabrik Karl Hecht KG, Sondheim, Germany) and fixed in ice-cold (−20°C) methanol. For immunostaining, cells were washed three times with PBS and blocked 15 min in PBS containing 1% bovine serum albumin and 0.5% Triton X-100 (PBSAT) or ImageiT FX (Invitrogen) before incubation with primary antibodies in PBSAT. Cells were washed three times with PBSAT before incubation with fluorescently labeled secondary antibodies, counterstained for DNA using 4,6-diamidino-2-phenylindole (DAPI) or DRAQ5, washed, and mounted slides using Prolong Gold (Invitrogen). All confocal images were obtained using either an AxioVert 200 microscope (Carl Zeiss, Jena, Germany), a 100×/1.4 PlanApo differential interference contrast (DIC) lens, an Ultraview spinning disk (PerkinElmer Life and Analytical Sciences, Boston, MA), and an ORCA-ER camera (Hamamatsu, Bridgewater, NJ) or with an AxioImager Z1 ApoTome microscope system (Carl Zeiss), a 100×/1.4 PlanApo DIC lens, and a AxioCam MRm camera. To display the entire cell volume, these images are presented as maximal projections of z-stacks using MetaMorph software (Molecular Devices, MDS Analytical Technologies, Sunnyvale, CA) or ImageJ (public domain National Institutes of Health Image software, National Institutes of Health, Bethesda, MD). Live cell imaging of transiently transfected, serum-starved hTERT-RPE1 cells grown in 35-mm ibiTreat μ-culture dishes (Ibidi, Munich, Germany) was performed using a CellObserver microscope system (Carl Zeiss) equipped with a 40×/1.3 PlanApo phase 3 lens and an AxioCam MRm camera.
Immunohistochemistry
Expression of CSPP proteins was studied by immunohistochemistry using the EnVision + peroxidase system (Dako Denmark, Glostrup, Denmark). CSPP antibodies were carefully titrated, and secondary antibodies and detection reagents were tested to exclude detection of false positives. Monoclonal and polyclonal antibodies were used at 1:50 and 1:250 dilution, respectively. Formalin-fixed 4-μm sections from paraffin-embedded blocks were dewaxed using xylene and serial ethanol dilutions and subsequently pretreated by heat in a microwave oven for antigen retrieval (750 W; 4 × 5 min) in Tris-EDTA, pH 9.0. This was followed by sequential incubations with primary antibodies for 30 min, secondary antibodies for 30 min, and incubation with DAB for 5 min. Sections were counterstained with hematoxylin. Stained sections were dehydrated by successive washes in 70, 95, and 100% ethanol; transferred to xylene; and mounted using glycerol vinyl alcohol aqueous mounting solution. Tissue sections were imaged using a Uplan 40×/0.75 phase 2 or Uplan FL 100×/1.30 lens on a motorized AX-70 microscope (Olympus Optical, Hamburg, Germany) equipped with a syncroCOOL 435 charge-coupled device camera (Syncroscopy, Cambridge, United Kingdom).
RESULTS
Generation of a CSPP-specific mAb
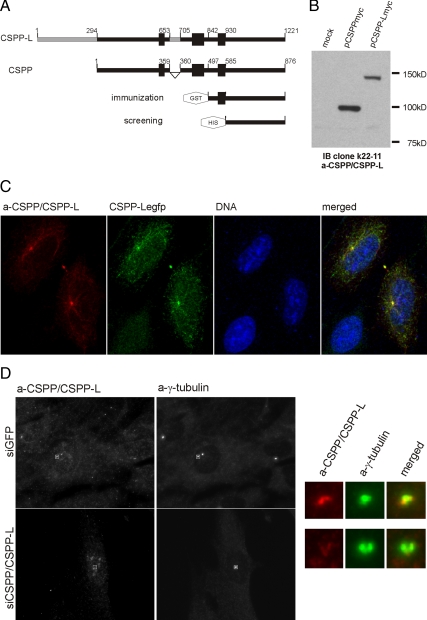
A mAb directed against the common C terminus of CSPP and CSPP-L was generated by immunization of mice with a bacterially expressed GST-fusion protein that made up the C-terminal 379 aa of CSPP and CSPP-L. Individual clones were screened for reactivity against a bacterially expressed His-tagged fusion protein making up the common C-terminal 291 aa and further tested for reactivity against both CSPP isoforms ectopically expressed in HEK293T cells (Figure 1A). A clone, k22-11, was identified whose antibodies specifically detected both ectopically expressed CSPP isoforms by means of Western blot analysis and immunofluorescence (Figure 1, B and C). The sensitivity of this antibody was too low to detect endogenous CSPP isoforms by Western blot of total cell lysates (Figure 1B) but sufficient to detect a centrosomal antigen by immunofluorescence analysis of HeLa cells (Figure 1C). The immunofluorescence staining consistently showed a two barrel like staining pattern embedded within γ-tubulin containing foci. Importantly, the mAb staining was lost in cells depleted for CSPP isoforms by transfection with an individual siRNA against a sequence common to CSPP and CSPP-L mRNAs as well as siRNA pool targeting both isoforms (Figure 1D and Supplemental Figure 1).
Figure 1.
Generation of a CSPP/CSPP-L–specific antibody. (A) Schematic representation of CSPP and CSPP-L proteins and bacterially expressed constructs used for antibody generation. Bold bars indicate coiled-coil regions. (B) Western blot using the generated monoclonal CSPP/CSPP-L–specific antibody for detection of CSPP isoforms in total cell lysates of HEK293T cells transfected with indicated plasmids. (C) Immunofluorescence detection of CSPP-Legfp (green) in transiently transfected HeLa cells using the generated monoclonal CSPP/CSPP-L–specific antibody (red). (D) Immunofluorescence staining against CSPP/CSPP-L (red) and γ-tubulin (green) of HeLa cells 72 h posttransfection with either GFP or CSPP/CSPP-L mRNA targeting siRNA.
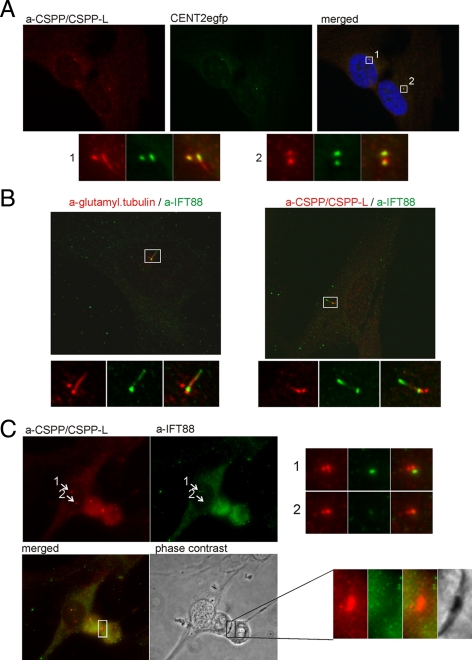
Cspp Isoforms Localize to Centrioles and the Midbody in Cycling Cells and to Centrioles and the Cilia Axoneme in Resting Cells
The observed centrosomal staining pattern suggested that CSPP isoforms localized to the centrioles. We therefore analyzed the localization of CSPP isoforms in hTERT-RPE1 cells that stably express GFP-centrin 2 (CETN2) fusion protein, a structural component of the centrioles. CSPP isoforms colocalized with the centriolar marker. However, frequently extracentriolar staining, extending from one of the centrioles reminiscent of cilia was observed (Figure 2A). We therefore investigated the localization of CSPP isoforms in hTERT-RPE1 cells that had been serum starved for 48 h to induce primary cilia formation. CSPP isoforms not only localized to centrioles but also extended into the lower cilia axoneme as shown by costaining for the intraflagellar transport protein 88 (IFT88), which decorates the cilia axoneme and the transition zone. In contrast to IFT88, CSPP proteins were only detected at the lower part of the cilia axoneme (Figure 2B).
Figure 2.
CSPP proteins localize to centrioles, cilia, and the midbody. (A) Immunofluorescence images of hTERT-RPE1 cells stably expressing the GFP tagged centriolar protein centrin 2 (green) stained for CSPP/CSPP-L (red). Panels below show magnified montages of centrosomes indicated in the merged image. CSPP proteins colocalize with CENT2gfp at centrioles but show additional staining at the tip of one centriole. (B) Immunofluorescence images of hTERT-RPE1 cells 72 h after serum starvation stained for the ciliary marker IFT88 (green) and glutamylated tubulin (red; left) or CSPP/CSPP-L (red; right). Bottom panels show magnified images of indicated cilia in the merged image. CSPP proteins are detected at centrioles and along the lower cilia axoneme. (C) Immunofluorescence images of hTERT-RPE1 cells in G2 phase (left cell) and telophase/cytokinesis (right cell) stained for CSPP/CSPP-L (red) and the interflagellar transport protein IFT88 (green). Phase-contrast imaging reveals localization of CSPP/CSPP-L to the phase-dense midbody. Panels to the right show magnified images of indicated centrosomes.
In cycling cells, IFT88 localizes to the distal appendages of the mother centriole (Figure 2C). In Figure 2C, one interphase and one mitotic (telophase/cytokinesis) cell are depicted. The interphase cell displays duplicated and separated centriole pairs, which both are stained by the CSPP antibody although only one centriole stains at the distal end for IFT88. During telophase, CSPP isoforms are still localized to centrioles but also stained midbodies that were identified by phase-contrast imaging. The expression and localization of CSPP-L to centrosomes and midbodies also was observed in unciliated cells such as human B lymphocytes (Supplemental Figure 2).
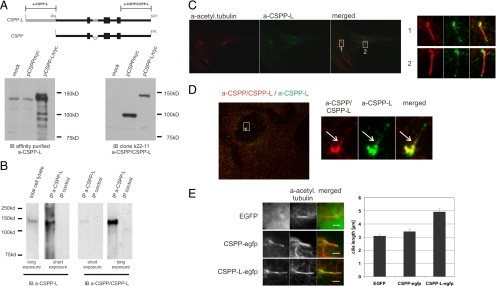
CSPP-L Localizes to Basal Bodies and the Cilia Axoneme
During the course of our study, a polyclonal antibody against the CSPP-L specific N terminus became available. This polyclonal antibody specifically detects Myc-tagged CSPP-L but not CSPP in immuno blots of total cell lysates of HEK293T transfectants, and it detects an endogenous protein of the size of CSPP-L in total cell lysates of untransfected HEK293T cells (Figure 3A). Furthermore, this polyclonal antibody immunoprecipitated an endogenous protein of approximately 150 kDa from total cell lysates of hTERT-RPE1 cells. The immunoprecipitated protein was stained by the mAb that is directed against the common C-terminal end of CSPP and CSPP-L (Figure 3B). We conclude that the polyclonal antibody is specific to CSPP-L. Immunofluorescence staining of serum-starved hTERT-RPE1 cells for CSPP-L and acetylated tubulin, a tubulin modification found on centrioles and the cilia axoneme, showed that CSPP-L localized to the centrosome and the cilia axoneme (Figure 3C). Notably, the cilia staining seemed not to be limited to the lower axoneme as observed with the mAb. CSPP-L decorated patches along the whole axoneme. Furthermore, costaining with the mAb against the common C-terminal domain of both isoforms and the CSPP-L specific antibody not only showed colocalization at centrioles and the lower axoneme but also additional staining of CSPP-L around the centrosome and along the upper cilia axoneme (Figure 3D).
Figure 3.
Identification of CSPP-L as a ciliary protein. (A) Schematic presentation of CSPP and CSPP-L isoforms with labeling of regions used for generation of the monoclonal CSPP/CSPP-L–specific and the polyclonal CSPP-L–specific antibodies. The affinity-purified CSPP-L–specific antibody detects Myc-tagged CSPP-L and an endogenous protein of equivalent molecular weight but not Myc-tagged CSPP in Western blots of total cell lysates of transiently transfected HEK293T cells. Immunoblotting of the same lysates with the monoclonal CSPP/CSPP-L–specific antibody is shown as control. (B) The polyclonal CSPP-L–specific antibody detects and immunoprecipitates an endogenous protein of 150 kDa in total cell lysates of hTERT-RPE1 cells that is also detected by the monoclonal CSPP/CSPP-L–specific antibody. (C) Immunofluorescence images of serum starved hTERT-RPE1 cells stained for CSPP-L (green) and acetylated tubulin (red) showing localization of CSPP-L to the basal body and the cilia axoneme. Panels to the right show magnified images of indicated cilia. (D) Immunofluorescence images of serum-starved hTERT-RPE1 cells stained with the polyclonal CSPP-L–specific (green) and the CSPP/CSPP-L–specific mAb (red). Colocalization is observed at the basal body and in patches at the lower cilia axoneme (arrow). (E) Costaining for the ciliary marker acetylated tubulin in serum starved hTERT-RPE1 cells transiently expressing either EGFP, CSPP-egfp, or CSPP-L-egfp (green) show both EGFP-fusion proteins specifically enriched at basal bodies and along cilia axonemes, including the ciliary tip structure (see Supplemental Figure 3). CSPP-L-egfp transfectants show statistically significant longer cilia axonemes than CSPP-egfp and EGFP transfectants (error bars depict SE of the mean cilia length measured in three independent experiments).
We next examined the localization of ectopically expressed enhanced green fluorescent protein (egfp)-tagged CSPP isoforms in transiently transfected, serum-starved hTERT-RPE1 cells. Both isoforms specifically concentrated around the basal body and within the cilia axoneme (Figure 3E and Supplemental Figure 3). Notably, the axonemes of in particular CSPP-Legfp transfectants seemed elongated in live transfectants. This increase in cilia length was confirmed in fixed cells by costaining for the ciliary marker acetylated tubulin. The average length of the cilia axoneme in CSPP-Legfp transfectants (4.9 μm; SEM = 0.2 μm) was found to be significantly longer than in CSPP-egfp (3.4 μm; SEM = 0.2 μm) and EGFP (3.1 μm; SEM = 0.1 μm) control transfectants (Figure 3E). Expression of truncated CSPP/CSPP-L constructs identified the common central coiled-coil (amino acids 295–708 in CSPP-L, 1–503 in CSPP) as being required for ciliary localization, whereas the common C-terminal domain of both isoforms only enriched around centrosomes. The CSPP-L specific N-terminal domain (amino acids 1-295 in CSPP-L) neither showed ciliary localization but decorated stress fibers in live cells and colocalized with actin fibers in fixed cells (Supplemental Figure 3). Collectively, these data identify CSPP proteins as novel ciliary proteins and identify a CSPP-L–specific actin or stress fiber targeting protein domain.
CSPP and CSPP-L Are Expressed in Ciliated Cells of Renal, Retinal, and Bronchial Tissues
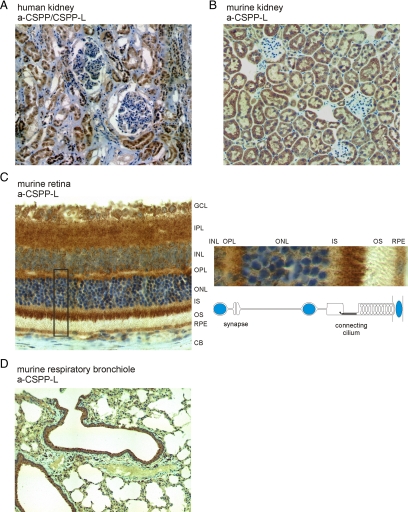
Supportive for a cilary function, we observed that CSPP1 mRNA is predominantly expressed in ciliated tissues during mouse embryogenesis (Supplemental Figure 4). We therefore examined the expression of CSPP proteins in three independent biopsies of normal human kidney using the monoclonal CSPP antibody. CSPP protein expression was specifically detected in tubular epithelia cells of the nephron, whereas mesenchymal cells and cells of the blood filtrating glomeruli did not express CSPP proteins (Figure 4A). An identical staining pattern was obtained using the polyclonal CSPP-L–specific antibody (data not shown). This antibody also showed cross-reactivity to murine CSPP-L and resulted in a similar staining pattern in kidney sections of BALBc nu/nu mice (Figure 4B).
Figure 4.
CSPP proteins are expressed in ciliated renal, retinal, and respiratory epithelia cells. Sections of formalin fixed tissues were stained by immunohistochemistry with either the mouse monoclonal CSPP/CSPP-L–specific antibody (A) or the rabbit polyclonal CSPP-L–specific antibody (B–D) (brown). Nuclei were detected by counterstaining with hematoxylin (blue). (A) Staining of human renal tissue section with the CSPP/CSPP-L–specific mAb by immunohistochemistry shows expression of CSPP isoforms (brown) in tubular epithelia cells, whereas cells of glomeruli and the mesenchymal cells showed no staining. (B) Detection of CSPP-L in renal tissue sections from male BALB/C nu/nu mice shows identical staining pattern to that observed in human kidney sections. (C) CSPP-L shows a distinct expression pattern in murine retina sections. Photoreceptor cells of the outer nuclear layer show subcellular accumulation of CSPP-L mainly at the connecting cilium comprising transition zone between inner and outer segment and partially at regions of synapses to the bipolar cells of the inner nuclear layer. The outer segment is negative for CSPP-L. Right, magnification of indicated area and schematic view of cellular organization. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; IS, inner segment; OS, outer segment; RPE, retina pigment epithelium; CB, choroidal border. (D) High CSPP-L expression is detected in bronchiole lining ciliated respiratory epithelia cells. Also see Supplemental Figure 4.
Next, we analyzed CSPP-L expression in the murine retina (Figure 4B). CSPP-L expression could be detected in cells of the pigmented epithelium, the sensory photoreceptor cells of the outer nuclear layer, and the signal transducing cells of the inner nuclear layer, whereas the mesenchyme of the choroid (underlying the pigmented epithelium) showed no CSPP-L expression. Interestingly, the strongest CSPP-L expression was detected in cells of the outer nuclear layer and showed a characteristic subcellular localization to the connecting cilium that composes the transition zone between the inner segment and the outer segment (Figure 4C). A second zone of concentrated CSPP-L expression is detected in the apical part of the outer plexiform layer adjacent to the outer nuclear layer where synapses between bipolar cells of the inner nuclear layer and the photoreceptor cells of the outer nuclear layer are formed. Notably, both neuronal cell types, the bipolar cells and the ganglia cells, also showed CSPP-L expression. This localization is very similar to what is reported for NPHP8 and NPHP4 (Arts et al., 2007). NPHP8 and NPHP4 belong to a group of eleven genes (NPHP1–11) identified by positional cloning to harbor causative mutations in patients suffering from NPHP. Individual NPHP proteins have been functionally associated with centrosomes, cilia, and cell–cell junctions. It is thus thought that NPHP is a result of the deregulation of ciliary/centrosome–associated signaling pathways in tubular epithelia cells, leading to cyst formation. The loss of cilia associated functions of NPHP4 and NPHP8 proteins is frequently associated with an extra-renal phenotype, primarily retinal degeneration and neurological disorders.
Finally, gene expression in the mouse embryo (Supplemental Figure 4) and transcriptional profiling of mucociliary differentiation in human airway epithelial cells indicated CSPP expression in multiciliary epithelium (Ross et al., 2007). Immunohistochemistry staining of tissue sections of murine respiratory bronchioles identified the highly polarized, multiciliated bronchial epithelia cells to express CSPP-L (Figure 4D). This result was further confirmed by immunofluorescence staining of isolated murine trachea epithelia cells (Supplemental Figure 4).
CSPP proteins are thus expressed in multiciliated airway epithelia cells and in monociliated renal and retinal cells. The subcellular localization observed in the highly polarized retinal photoreceptor cells suggests a concentration of CSPP-L at the connecting cilium and to lower degree at the synapse to the bipolar cells. Expression of CSPP-L also was detected in neuronal cells of the retina.
CSPP Isoforms Are Required for Ciliogenesis
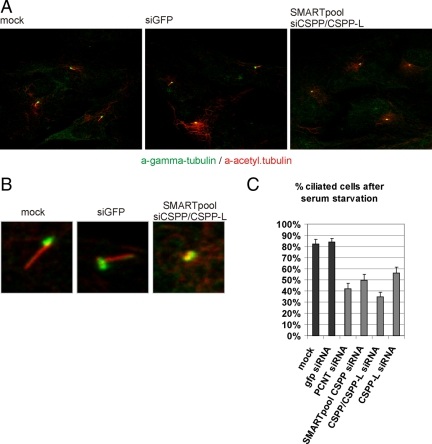
To explore the putative ciliary function of CSPP proteins, we tested their requirement for cilia formation in hTERT-RPE1 cells in response to serum starvation. hTERT-RPE1 cells were consecutively transfected with siRNA targeting both CSPP isoforms or targeting the CSPP-L isoform alone (Figure 5 and Supplemental Figure 5). To achieve an efficient knockdown, cells were allowed to grow in serum containing medium for 48h after a first siRNA transfection before being subjected to a second transfection that was followed by serum starvation for 48 h to induce ciliogenesis. As controls, cells were transfected with siRNA targeting GFP or the centrosomal protein PCNT. PCNT has been shown previously to be required for ciliogenesis in hTERT-RPE1 cells (Mikule et al., 2007). The knockdown of CSPP isoforms inhibited ciliogenesis to a similar degree as the knockdown of pericentrin, whereas ciliogenesis was unimpaired in GFP siRNA transfectants (Figure 5, A and B). Fifty percent (SD = 5%) of the cells transfected with SMARTpool siRNAs targeting CSPP1 transcripts displayed either no cilia or cilia with severely shortened axonemes (Figure 5C). A similar loss of ciliogenesis was observed in hTERT-RPE1 cells transfected with the CSPP and CSPP-L targeting single siRNA used in previous studies (Patzke et al., 2005), thus excluding potential unspecificity of the siRNA mix. Interestingly, a single siRNA specific to the mRNA of the larger CSPP-L isoform also impaired ciliogenesis (Figure 5C and Supplemental Figure 5A), indicating that this isoform alone is a major contributor to ciliogenesis in hTERT-RPE1 cells.
Figure 5.
Knockdown of CSPP proteins impairs serum starvation induced ciliogenesis in hTERT-RPE1 cells. Immunofluorescence images of siRNA-transfected hTERT-RPE1 cells stained for the centrosomal marker γ-tubulin (green) and the cilia marker acetylated tubulin (red). CSPP/CSPP-L mRNA targeting siRNA transfectants but not control transfectants show defects in ciliogenesis. Images of larger field (A) and close-up view on representative primary cilia (B) show severely shortened cilia axonemes in CSPP/CSPP-L siRNA transfectants. (C) Quantification of the efficacy of ciliogenesis in response to serum starvation after siRNA transfection with indicated siRNAs. Depletion of pericentrin and CSPP/CSPP-L efficiently inhibit cilia formation compared with control cells or GFP siRNA transfected cells. Individual siRNA molecules targeting both CSPP isoforms or CSPP-L alone result in similar ciliogenesis defects. At least 300 cells were scored in three independent transfections of each siRNA. Error bars indicate SD. Also see Supplemental Figure 4.
It could be argued that the knockdown of CSPP/CSPP-L before the serum starvation resulted in an arrest in other phases of the cell cycle than G0/G1 similar to what was observed in p53-deficient HEK293T cells (Patzke et al., 2005) and that this could be the explanation for the reduced cilia formation. However, hTERT-RPE1 cells arrested in G0/G1 after CSPP/CSPP-L knockdown (Supplemental Figure 5, B and C), in agreement with the results obtained after depletion of other centrosomal proteins (Srsen et al., 2006; Mikule et al., 2007). It is suggested that the impaired cilia formation is a result of the lack of CSPP/CSPP-L, rather than a result of arrest in a cell cycle phase that is not permissive for cilia formation.
CSPP Proteins Colocalize and Interact with NPHP Proteins of the Transition Zone
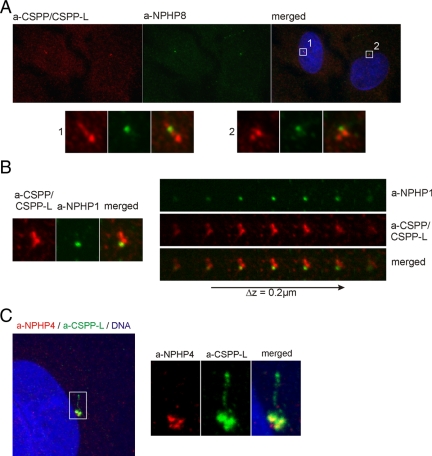
The cell type-specific expression, the requirement for ciliogenesis and the intriguing localization to the lower cilia axoneme throughout the transition zone suggested a possible involvement of CSPP isoforms in human ciliopathies. We therefore tested known ciliopathy related proteins that localize to the basal body and the transition zone for colocalization and interaction with CSPP or CSPP-L. These include NPHP1 (Fliegauf et al., 2006), NPHP4 (interacts with NPHP1; Mollet et al., 2002. 2005; Otto et al., 2002), and the retinitis pigmentosa GTPase regulator-interacting protein 1-like protein NPHP8 (interacts with NPHP4; Delous et al., 2007; Arts et al., 2007). NPHP8 and NPHP1 localized specifically to the transition zone that is formed at the tip of the mother centriole/basal body, colocalizing with CSPP proteins (Figure 6, A and B). NPHP4 showed a less focused localization being detected around both centrioles and to minor degree at the transition zone as shown by costaining with the CSPP-L–specific antibody (Figure 6C).
Figure 6.
CSPP and CSPP-L colocalize with NPHP proteins at the transition zone and at the basal body. Immunofluorescence images of hTERT-RPE1 cells 72 h after serum starvation. (A) Cells were stained for NPHP8 (green) and CSPP/CSPP-L (red). Boxes indicate magnified areas displayed in bottom panels. (B) The centrosome/basal body of a ciliated hTERT-RPE1 cell stained for the marker of the transition zone, NPHP1 (green), and CSPP/CSPP-L. CSPP/CSPP-L is detected at centrioles and the lower cilia axoneme showing colocalization with NPHP1 in individual confocal planes (bottom). (C) Immunofluorescence images of serum-starved hTERT-RPE1 cells stained for DNA (blue), CSPP-L (green), and NPHP4 (red) showing colocalization of both proteins at the basal body.
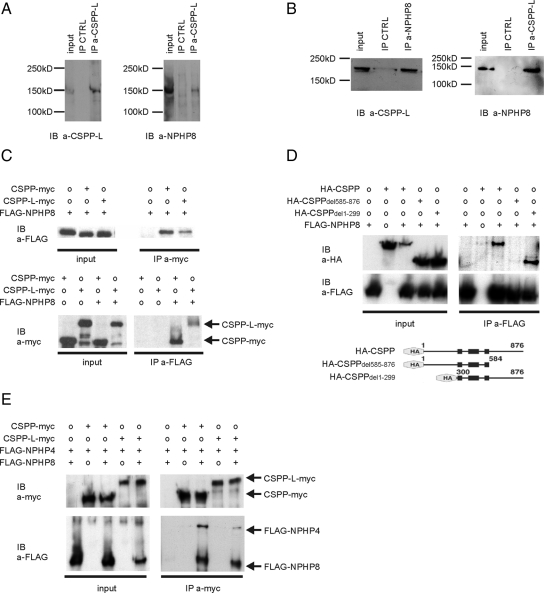
We screened for possible protein–protein interactions in hTERT-RPE1 cells and identified NPHP8 as a CSPP-L–interacting protein by means of coimmunoprecipitation with the CSPP-L specific antibody (Figure 7A). This interaction was further confirmed in bovine retina extracts by reciprocal coimmunoprecipitation experiments by using NPHP8-specific and CSPP-L–specific antibodies, respectively (Figure 7B). Notably, bovine CSPP-L migrated at a slightly higher molecular weight, which may indicate posttranslational modification or the existence of further splice variants. Next, differentially tagged proteins were transiently overexpressed in HEK293T cells and tested for interaction by coimmunoprecipitation from total cell lysates to further characterize the nature of the interaction of NPHP8 and CSPP proteins. FLAG-NPHP8 was confirmed as a CSPP-myc– and CSPP-L-myc–interacting protein. Immunoprecipitation of either myc-tagged CSPP isoform coimmunoprecipitated a FLAG-tagged construct of the C-terminal part of NPHP8 (amino acid residues 411-1055; Arts et al., 2007) and vice versa (Figure 7C). Furthermore, this result indicated that CSPP and CSPP-L share a NPHP8 binding domain. To identify this domain, the FLAG-NPHP8 construct was transiently coexpressed with either full-length or truncated HA-tagged variants of CSPP lacking either the N- or the C-terminal domain (Patzke et al., 2005) and subjected to immunoprecipitation using a FLAG-tag–specific antibody. Immunoprecipitated FLAG-NPHP8 copurified both full-length HA-CSPP and the construct lacking the N-terminal domain, whereas the interaction was lost upon deletion of the C-terminal domain. This result supports the specificity of the interaction, because both HA-tagged and Myc-tagged CSPP can be coprecipitated, and identifies the common C-terminal 292 aa of CSPP and CSPP-L to be required for the interaction with NPHP8 (Figure 7D).
Figure 7.
CSPP and CSPP-L interact with NPHP proteins of the basal body/transition zone. (A) Western blots of immunoprecipitates from total cell lysates of hTERT-RPE1 cells. NPHP8 is identified as a CSPP-L interacting protein by immunoblotting for NPHP8-specific antibody (right; NPHP8 antibody as in Arts et al., 2007). Left, control immunoblotting for CSPP-L. (B) The interaction of CSPP-L and NPHP8 proteins also is detected by reciprocal immunoprecipitations from bovine retinal extracts (NPHP8 antibody as in Khanna et al., 2009). (C) Western blot of immunoprecipitates from total cell lysates of HEK293T transfected with indicated expression plasmids. Immunoprecipitation (IP) of Myc-tagged CSPP isoforms with a Myc-tag–specific antibody coimmunoprecipitated FLAG-tagged NPHP8 as detected by immunoblotting (IB) with a FLAG-tag–specific antibody (top). Inversely, immunoprecipitation of FLAG-NPHP8 with a FLAG-tag–specific antibody coimmunoprecipitated Myc-tagged CSPP isoforms as detected by immunoblotting with a Myc-tag–specific antibody (bottom). Neither the Myc nor the FLAG antibody showed cross-reactivity to the other tag. (D) Western blot of immunoprecipitates with a FLAG-tag–specific antibody from total cell lysates of HEK293T transfected with indicated expression plasmids. Immunoblotting with either FLAG- or Myc-tag–specific antibodies reveals that coimmunoprecipitation of HA-CSPP by FLAG-NPHP8 is lost when the common C-terminal domain of CSPP and CSPP-L is deleted (HA-CSPPdel585-876). The FLAG-tag–specific antibody shows no cross-reactivity for the HA-tag. (E) Western blot of immunoprecipitates with a Myc-tag–specific antibody from total cell lysates of HEK293T transfected with indicated expression plasmids. Efficient coprecipitation of FLAG-NPHP4 by Myc-tagged CSPP isoforms is dependent on the coexpression of FLAG-NPHP8, suggesting ternary complex formation.
NPHP8 is a NPHP4-interacting protein (Delous et al., 2007; Arts et al., 2007). It is therefore possible that CSPP could complex with NPHP4 directly or could form a ternary complex with NPHP8. To test this hypothesis, individual Myc-tagged CSPP isoforms were transiently expressed in HEK293T cells together with either FLAG-tagged NPHP4 (FLAG-NPHP4) alone or with a combination of FLAG-NPHP4 and FLAG-NPHP8 (Figure 7E). CSPP isoforms were immunoprecipitated from total lysates with a Myc-tag–specific antibody and tested for coprecipitation of FLAG-tagged NPHP4 or NPHP8. CSPP and CSPP-L poorly copurified FLAG-NPHP4 in the absence of FLAG-NPHP8, whereas coexpression of FLAG-NPHP8 increased significantly the efficacy of FLAG-NPHP4 copurification. This result indicated that CSPP and CSPP-L can form a ternary complex with NPHP4 via its binding to NPHP8.
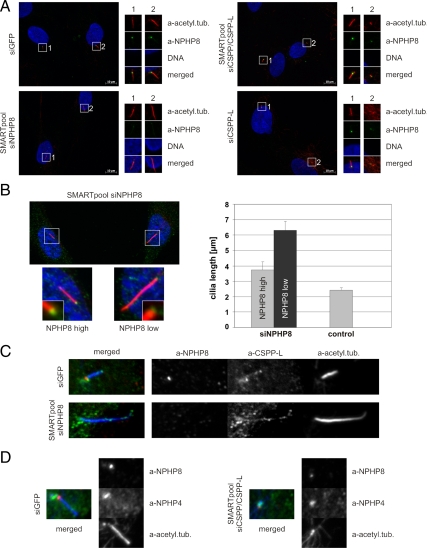
CSPP Proteins Stabilize NPHP8 at the Transition Zone but Loss of NPHP8 Is Not the Cause of Ciliogenesis Impairment
NPHP8 is a known regulator of cilia-mediated signaling, and the NPHP8 knockout mouse displays decreased cilia numbers in tissues undergoing morphogenic transitions, although primary mouse embryonic fibroblasts derived from this mouse are competent in ciliogenesis (Vierkotten et al., 2007). We therefore investigated whether loss of CSPP/CSPP-L would perturb the localization of NPHP8. Whereas control siRNA transfectants displayed normal cilia and normal localization of NPHP8 to the base of the primary cilium (the area between centrioles and the cilia axoneme that is devoid of staining for acetylated tubulin), NPHP8 staining is severely diminished in the cilia defective CSPP/CSPP-L SMARTpool siRNA or CSPP-L siRNA transfectants (Figure 8A). Thus, CSPP-L either recruits or maintains NPHP8 at the transition zone. Yet, interestingly, knockdown of NPHP8 did not impair ciliogenesis in serum-starved hTERT-RPE1, and cilia seemed longer than in control transfectants. We therefore measured cilia length in control and NPHP8 siRNA transfectants (Figure 8B). The average cilia length of presumably untransfected cells of the NPHP8 siRNA transfection, which displayed high mean NPHP8 signal intensity, was measured to 3.7 μm (SEM = 0.5 μm). Although longer than cilia length of control cells, 2.4 μm (SEM = 0.1 μm), this difference was not statistically significant. In contrast, the average cilia length of cells that lost NPHP8 signal in the NPHP8 siRNA transfection was 6.3 μm (SEM = 0.3 μm) and showed a statistically significant difference to control transfectants and to cells displaying near normal NPHP8 signal intensities in the NPHP8 siRNA transfection. Notably, CSPP-L still decorated the prolonged cilia axonemes of NPHP8-depleted cells (Figure 8C).
Figure 8.
CSPP proteins are required for stabilization of NPHP8 but not NPHP4 at the basal body but NPHP8 is not required for ciliogenesis. Immunofluorescence images of serum-starved hTERT-RPE1 cells transfected with siRNAs targeting GFP (control; A, top left), NPHP8 (SMARTpool; A, bottom left), CSPP/CSPP-L (SMARTpool; A, top right) or CSPP-L alone (A, bottom right) stained for the centriole and ciliary marker acetylated tubulin (red), NPHP8 (green), and DNA (blue). Magnifications of indicated areas are presented to the right (A) or below (B) of each merged overlay image. Centrioles are not stained by the acetylated tubulin-specific antibody due to formaldehyde fixation. The concentration of NPHP8 proteins at the basal body is diminished in cilia-defective CSPP or CSPP-L siRNA-targeting transfectants (right). In contrast, ciliogenesis itself can occur independently of NPHP8 protein expression (A, bottom left; and B) and leads to statistically significant longer average cilia length (B). Examples of the NPHP8 concentration-dependent cilia length increase are shown to the left, and a quantification of this defect is shown in the bar diagram (error bars depict SE of the mean cilia length measured in three independent experiments). (C) NPHP8 (red) is not required for the ciliary localization of CSPP-L (green) to the cilia axoneme (acetylated tubulin, blue) because NPHP8 (red)-depleted cells showed similar axonemal localization of CSPP-L (green) as in control transfectants. (D) CSPP-L is required for the recruitment or maintenance of NPHP8 (red) but not NPHP4 (green) at the basal body (acetylated tubulin, blue). Also see Supplemental Figure 6.
Finally, recombinant CSPP proteins were able to form a ternary complex with NPHP4 in the presence of NPHP8 (Figure 7). We therefore also investigated whether the localization of NPHP4 to basal bodies would be affected in CSPP/CSPP-L–depleted cells (Figure 8D and Supplemental Figure 6). The localization of NPHP4 seemed unchanged in cilia-defective basal bodies of CSPP/CSPP-L siRNA transfectants displaying decreased NPHP8 levels (Figure 8D). Similarly, the localization of NPHP4 was found unchanged in NPHP8-depleted cells (Supplemental Figure 6). These results suggest that CSPP-L specifically recruits or maintains NPHP8 at the basal body.
DISCUSSION
All reported studies on CSPP isoforms and their centrosome- and MT-associated functions mainly examined their role in facilitating mitosis and cytokinesis (Patzke et al., 2005, 2006; Asiedu et al., 2009). Our present investigation is therefore the first to shed light on a postmitotic (cytoskeleton associated) function of CSPP proteins in both cells and tissues. Four different experimental strategies used to study CSPP proteins in vitro and in vivo link them functionally to the primary cilium: 1) the analysis of subcellular localization of endogenous and ectopic CSPP proteins in hTERT-RPE1 and primary trachea epithelia cells; 2) the analysis of cell type specific expression in human renal and murine renal, retinal, and bronchial tissue sections in conjunction with the tissue-specific mRNA expression reported during mouse embryogenesis; 3) the analysis of the effects of CSPP overexpression and depletion on cilia length and the efficiency of ciliogenesis of hTERT-RPE1 cells, respectively; and 4) the identification and characterization of the interaction and complex formation with known ciliary proteins NPHP8 and NPHP4.
The immunofluorescence staining suggests that CSPP proteins can occur in different protein pools localized to 1) centrosomes, 2) midspindel/midbodies, and 3) the cilia axoneme. Both antibodies used in this study detected endogenous CSPP isoforms at the centrosome, the midbody and along the cilia axoneme. However, not all CSPP proteins stained by the affinity-purified polyclonal CSPP-L–specific antibody also were stained by the mAb that is directed against the common C-terminal domain of both CSPP isoforms (Figure 3). This discrepancy may be explained by either the lower affinity of the mAb compared with the CSPP-L–specific polyclonal antibody or epitope masking of the C-terminal domain by the formation of protein complexes such as with NPHP8. Alternatively, because we have shown previously that CSPP proteins can be phosphorylated at serine residues (Patzke et al., 2005), posttranslational modification could mask the epitope detected by the mAb.
Our results show that CSPP proteins not only are expressed in NPHP protein-expressing cell types in renal, retinal, and respiratory tissue sections but also that they interact directly with NPHP8 and indirectly with NPHP4. Furthermore, the interaction of endogenous CSPP proteins with NPHP8 is not only detected in cell lines but also confirmed in bovine retina extracts. Our results suggest that CSPP proteins are required for correct localization of NPHP8 to the tip of the basal body that is forming the transition zone. In hTERT-RPE1 cells the targeted knockdown of CSPP-L alone is sufficient to impair ciliogenesis and to decrease the recruitment of NPHP8 to centrosomes, indicating its sole significance in this process in this cell line (Figures 5C and 8A). Our finding that ciliogenesis is not impaired by NPHP8 knockdown indicates that loss of NPHP8 from the transition zone alone is not sufficient to explain the CSPP-L siRNA-mediated ciliogenesis defect. This finding is consistent with the observation of primary cilia in primary mouse embryonic fibroblasts derived from the NPHP8 knockout mouse (Vierkotten et al., 2007). The observed increase in cilia length might reflect a homeostatic response in which hTERT-RPE1 cells try to compensate for lack of signaling through the cilium as NPHP8 has been shown to be required for Shh signaling. Alternatively, it could be the consequence of imbalanced sorting of cilia length determining proteins (e.g., Rab8; Nachury et al., 2007) over the transition zone, because the NPHP8 interactor NPHP4 and its interactor NPHP1 also are suggested to regulate transport through the transition zone (Winkelbauer et al., 2005; Jauregui et al., 2008). However, NPHP8 seems to be expendable for the localization of NPHP4 and CSPP-L to the primary cilium. Thus, NPHP8 might act antagonistically on CSPP-L, which we found not only to be required for ciliogenesis but also to positively regulate cilia length upon ectopic expression. Interestingly, NPHP8 has been found to interact with RPGR (Khanna et al., 2009), which in turn is associated with NPHP6 (CEP290; Chang et al., 2006) that has been shown to regulate ciliogenesis via recruitment of Rab8 (Kim et al., 2008; Tsang et al., 2008). Although our study did not unravel the mechanism of CSPP/CSPP-L dependence in ciliogenesis, we identified a second arm of the NPHP network feeding into the control of ciliogenesis or eventually cilia homeostasis.
Depletion of several different proteins localizing to distinct structural moieties of the centrosome have been shown to result in ciliogenesis defects similar to CSPP/CSPP-L depletion reported here. These studies highlight the importance of the general structural integrity of the centrosome (Mikule et al., 2007; Graser et al., 2007). However, based on the fact that we found CSPP-L to contain an actin- or stress fiber-targeting domain in its isoform specific N-terminal domain (Supplemental Figure 3), one may speculate that CSPP-L helps to position the centrosome during its maturation/morphogenesis to the basal body in the progress of ciliogenesis. Indeed, actin remodeling is required for ciliogenesis and cilia positioning. Contractile proteins such as myosin are constituents of basal bodies (Gordon et al., 1980; Gordon and Lane, 1984; Dawe et al., 2007, 2009) and a role of actin modulation in ciliogenesis, maintenance, and function is starting to emerge (Kim et al., 2010; Molla-Herman et al., 2010). Interestingly, CSPP-L has been shown to recruit a myosin GTPase exchange factor MyoGEF to facilitate actomyosin ring contraction during cell division (Asiedu et al., 2009). Hence, localized organization or activation of the actomyosin network might be the common functional denominator for CSPP-L at the centrosome during ciliogenesis and at the midspindle/midbody during cell division that is also found in nonciliated lymphocytes (Supplemental Figure 2). In this context, it is interesting to note that the C-terminal domain of CSPP and CSPP-L that is required for interaction with NPHP8 was found previously to be required to restrict the MT organizing activity of CSPP proteins to mitosis (Patzke et al., 2006). NPHP8 could be a modulator of this MT-organizing activity and could thereby contribute to cilia assembly/maintenance.
The interaction of NPHP8 with CSPP proteins still allowed ternary complex formation with NPHP4, which in turn is a known NPHP1 interacting protein (Mollet et al., 2005). The partial colocalization of CSPP-L and NPHP4 as well as NPHP1 at the basal body/transition zone (Figure 6E) may be supportive for the existence of CSPP-L–NPHP8–NPHP4 (–NPHP1) complex formation in cells. Furthermore, CSPP proteins are enriched at the connecting cilium and at synapses of rod and cone cells in the outer nuclear layer (Figure 4), thus closely resembling reported staining pattern of NPHP8 and NPHP4 (see supplemental figure 5 in Arts et al., 2007). Also, NPHP1 is a known constituent of the retinal connecting cilium as well as of the transition zone of the renal and respiratory cilia (Fliegauf et al., 2006; Jiang et al., 2009). Notably, in addition to their ciliary localization, these proteins also have been shown to localize to cell–cell and cell–matrix contacts where they complex with proteins involved in cytoskeleton organization to facilitate epithelial morphogenesis (Donaldson et al., 2000; Mollet et al., 2005; Delous et al., 2009); and importantly, NPHP associated mutations in NPHP8 weaken or abrogate their interaction with NPHP4 (Arts et al., 2007; Delous et al., 2007). Although little is known about the dynamic behavior of these proteins, in a unifying concept NPHP8 might modulate cilia growth and polarization on two levels: indirectly through modulation of transport across the transition zone thereby affecting planar-cell-polarity signaling pathways and directly through interactions with cytoskeleton affecting proteins such as NPHP4, NPHP6, and Rab8 via RPGR, and CSPP proteins identified here (Supplemental Figure 7). Our results may suggest that CSPP-L and NPHP8 can form a functional unit of which (in hTERT-RPE1 cells) CSPP-L is required for cilia formation and NPHP8 recruitment or maintenance. Once the cilium is formed, NPHP8 might antagonistically act on CSPP-L to regulate cilia length. This might involve further interactions with other proteins such as NPHP4. However, additional work is required to investigate whether, when, and where these proteins can occur as multiprotein complexes together with CSPP in vivo. Furthermore the identification of putative binding partners of CSPP-L within the ciliary axoneme and its tip region will be a prerequisite to understand its effect on axoneme length control.
Finally, like many NPHP proteins, CSPP is highly conserved in vertebrate evolution (Patzke et al., 2005). Although unlike NPHP1 and NPHP4, no CSPP homologues were identifiable in genomes of the flagellated chlorophyte Chlamydomonas rheinhardtii or in genomes of nematodes, we identified a homologue in the choanoflagellate Monosiga brevicollis (Supplemental Figure 8), one of the closest unicellular relatives of animals. This phylogenetic profile is reminiscent of that of the here identified CSPP-interacting protein NPHP8 (Vierkotten et al., 2007). The vertebrate-specific coevolution may strengthen the hypothesis of a cell-type–specific functional relation of both proteins.
To conclude, our investigation of CSPP expression at cellular and tissue levels identify a novel, nonmitotic function for CSPP isoforms and place these proteins into a NPHP protein containing network at the primary cilium. The activity of this network is crucial for normal renal and retinal tissue architecture and function. Importantly, the larger isoform CSPP-L is found to be required for cilia formation and found to promote cilia prolongation. To date, 10 disease genes have been identified accounting collectively for only ∼30% of the nephronophthisis patients (Hildebrandt et al., 2009a,b; Otto et al., 2009). Our results define CSPP1 as a novel candidate gene for multiorgan phenotype ciliopathies. The putative role of CSPP proteins in human ciliopathies, in particular nephronophthisis-associated ciliopathies, should therefore be investigated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Hans-Christian Aasheim, David Warren, and Kjell Nustad for help and advice in bacterial expression and purification of GST- and HIS-tagged proteins; Dr. Jostein Dahle and Anne Kristine Hjelmerud for preparation of mouse tissues; Dr. Jan Delabie for evaluation of immunohistology; Dr. Steinar Funderud for critical reading of the manuscript; and all members of the Doxsey laboratory for help and discussion. We also thank the reviewers for valuable criticism and suggestions. Furthermore, we thank Drs. Ronald Roepman, Thomas Benzing, and Gregory Pazour for sharing reagents used in this study. S. P. acknowledges financial support by The Norwegian Cancer Society and Radiumhospitalets legater. H. K. is supported by National Institutes of Health grant EY-007961, Foundation Fighting Blindness, and Midwest Eye Banks and Transplantation Center. S.J.D. acknowledges support by National Institutes of Health grant 2 R01 051994-15.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0503) on June 2, 2010.
REFERENCES
- Arts H. H., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- Asiedu M., Wu D., Matsumura F., Wei Q. Centrosome/spindle pole-associated protein regulates cytokinesis via promoting the recruitment of MyoGEF to the central spindle. Mol. Biol. Cell. 2009;20:1428–1440. doi: 10.1091/mbc.E08-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D. M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Cardenas-Rodriguez M., Badano J. L. Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- Chang B., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Adams M., Wheway G., Szymanska K., Logan C. V., Noegel A. A., Gull K., Johnson C. A. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J. Cell Sci. 2009;122:2716–2726. doi: 10.1242/jcs.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Delous M., et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- Delous M., Hellman N. E., Gaude H. M., Silbermann F., Le B. A., Salomon R., Antignac C., Saunier S. Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum. Mol. Genet. 2009;18:4711–4723. doi: 10.1093/hmg/ddp434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. C., Dempsey P. J., Reddy S., Bouton A. H., Coffey R. J., Hanks S. K. Crk-associated substrate p130(Cas) interacts with nephrocystin and both proteins localize to cell-cell contacts of polarized epithelial cells. Exp. Cell Res. 2000;256:168–178. doi: 10.1006/excr.2000.4822. [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005a;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Doxsey S. J. Molecular links between centrosome and midbody. Mol. Cell. 2005;20:170–172. doi: 10.1016/j.molcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005b;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 2006;17:2424–2433. doi: 10.1681/ASN.2005121351. [DOI] [PubMed] [Google Scholar]
- Gordon R. E., Lane B. P. Immunolocalization of myosin and tropomyosin in cells undergoing ciliogenesis during regeneration of rat tracheal epithelium. Tissue Cell. 1984;16:337–343. doi: 10.1016/0040-8166(84)90053-3. [DOI] [PubMed] [Google Scholar]
- Gordon R. E., Lane B. P., Miller F. Identification of contractile proteins in basal bodies of ciliated tracheal epithelial cells. J. Histochem. Cytochem. 1980;28:1189–1197. doi: 10.1177/28.11.7000888. [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., Le C. M., Nigg E. A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. C., Torres V. E. Polycystic kidney disease. Annu. Rev. Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Attanasio M., Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J. Am. Soc. Nephrol. 2009a;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009b;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Zhou W. Nephronophthisis-associated ciliopathies. J. Am. Soc. Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- Jauregui A. R., Nguyen K. C., Hall D. H., Barr M. M. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. T., Chiou Y. Y., Wang E., Chien Y. L., Ho H. H., Tsai F. J., Lin C. Y., Tsai S. P., Li H. Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum. Mol. Genet. 2009;18:1566–1577. doi: 10.1093/hmg/ddp068. [DOI] [PubMed] [Google Scholar]
- Khanna H., et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H., et al. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J. Biol. Chem. 2005;280:33580–33587. doi: 10.1074/jbc.M505827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S. R., Gleeson J. G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J. E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Za-Blanc P., Gleeson J. G. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. Basal bodies platforms for building cilia. Curr. Top. Dev. Biol. 2008;85:1–22. doi: 10.1016/S0070-2153(08)00801-6. [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A., et al. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J. Cell Sci. 2010;123:1785–1795. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- Mollet G., et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat. Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum. Mol. Genet. 2005;14:645–656. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Otto E., et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am. J. Hum. Genet. 2002;71:1161–1167. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E. A., et al. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11) J. Med. Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- Patzke S., Hauge H., Sioud M., Finne E. F., Sivertsen E. A., Delabie J., Stokke T., Aasheim H. C. Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene. 2005;24:1159–1173. doi: 10.1038/sj.onc.1208267. [DOI] [PubMed] [Google Scholar]
- Patzke S., Stokke T., Aasheim H. C. CSPP and CSPP-L associate with centrosomes and microtubules and differently affect microtubule organization. J. Cell. Physiol. 2006;209:199–210. doi: 10.1002/jcp.20725. [DOI] [PubMed] [Google Scholar]
- Ross A. J., Dailey L. A., Brighton L. E., Devlin R. B. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell. Mol. Biol. 2007;37:169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- Salomon R., Saunier S., Niaudet P. Nephronophthisis. Pediatr. Nephrol. 2009;24:2333–2344. doi: 10.1007/s00467-008-0840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H. The mammalian centrosome and its functional significance. Histochem. Cell Biol. 2008;129:667–686. doi: 10.1007/s00418-008-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. S., Ben-Shahar Y., Moninger T. O., Kline J. N., Welsh M. J. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E., Dejsuphong D., Balestrini A., Hampel M., Lenz C., Takeda S., Vindigni A., Costanzo V. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat. Cell Biol. 2009;11:278–285. doi: 10.1038/ncb1835. [DOI] [PubMed] [Google Scholar]
- Srsen V., Gnadt N., Dammermann A., Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 2006;174:625–630. doi: 10.1083/jcb.200606051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W. Y., Bossard C., Khanna H., Peranen J., Swaroop A., Malhotra V., Dynlacht B. D. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veland I. R., Awan A., Pedersen L. B., Yoder B. K., Christensen S. T. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 2009;111:39–53. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierkotten J., Dildrop R., Peters T., Wang B., Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- Wheatley D. N. Primary cilia in normal and pathological tissues. Pathobiology. 1995;63:222–238. doi: 10.1159/000163955. [DOI] [PubMed] [Google Scholar]
- Wheatley D. N., Wang A. M., Strugnell G. E. Expression of primary cilia in mammalian cells. Cell Biol. Int. 1996;20:73–81. doi: 10.1006/cbir.1996.0011. [DOI] [PubMed] [Google Scholar]
- Winkelbauer M. E., Schafer J. C., Haycraft C. J., Swoboda P., Yoder B. K. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- Zhang S., Hemmerich P., Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J. Cell. Biochem. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.