Using a targeted genetic deletion, we show that the S100A4 metastasis factor is required for macrophage recruitment to sites of inflammation in vivo. S100A4−/− primary macrophages display defects in chemotaxis due to myosin-IIA overassembly and altered CSF-1 receptor signaling. These studies establish S100A4 as a regulator of macrophage motility.
Abstract
S100A4, a member of the S100 family of Ca2+-binding proteins, is directly involved in tumor metastasis. In addition to its expression in tumor cells, S100A4 is expressed in normal cells and tissues, including fibroblasts and cells of the immune system. To examine the contribution of S100A4 to normal physiology, we established S100A4-deficient mice by gene targeting. Homozygous S100A4−/− mice are fertile, grow normally and exhibit no overt abnormalities; however, the loss of S100A4 results in impaired recruitment of macrophages to sites of inflammation in vivo. Consistent with these observations, primary bone marrow macrophages (BMMs) derived from S100A4−/− mice display defects in chemotactic motility in vitro. S100A4−/− BMMs form unstable protrusions, overassemble myosin-IIA, and exhibit altered colony-stimulating factor-1 receptor signaling. These studies establish S100A4 as a regulator of physiological macrophage motility and demonstrate that S100A4 mediates macrophage recruitment and chemotaxis in vivo.
INTRODUCTION
S100A4, also known as mts1, fibroblast-specific protein (FSP1), 18A2, pEL98, p9Ka, 42A, calcium protein placental homologue, and calvasculin (Garrett et al., 2006), is a member of the S100 family of dimeric, EF-hand Ca2+-binding proteins that are found exclusively in vertebrates. The S100 protein family comprises >20 members, which exhibit 22–57% sequence identity (Marenholz et al., 2004). Functional diversity of the S100 family is achieved by the Ca2+-dependent recognition of distinct protein ligands by the individual S100 proteins, their ability to form homo- and heterodimers, and their tissue-specific expression patterns (Zimmer et al., 1995; Donato, 2001; Santamaria-Kisiel et al., 2006). The interaction of S100 proteins with a diverse range of protein targets impacts numerous cellular processes, such as protein phosphorylation, cell growth and survival, cell motility, and differentiation (Donato, 2001).
The biological function of S100A4 has been investigated most extensively with respect to its role in promoting tumor metastasis. Elevated S100A4 expression levels correlate with several metastatic cancers, including breast (Rudland et al., 2000), colorectal (Gongoll et al., 2002), bladder (Davies et al., 2002), esophageal (Ninomiya et al., 2001), non–small-cell lung (Kimura et al., 2000), gastric (Cho et al., 2003), medulloblastoma (Hernan et al., 2003), pancreatic (Rosty et al., 2002), prostate (Saleem et al., 2005), and thyroid (Zou et al., 2005). Mouse and rat models of breast cancer have demonstrated a causal role for S100A4 overexpression in promoting metastasis (Davies et al., 1993; Ambartsumian et al., 1996; Davies et al., 1996). Consistent with these observations, inhibition of S100A4 expression in tumor cells suppresses metastatic potential (Maelandsmo et al., 1996; Takenaga et al., 1997; Xue et al., 2003).
In addition to its expression in tumor cells, S100A4 is expressed in normal cells and tissues, including fibroblasts, macrophages, lymphocytes, and bone marrow-derived cells (Grigorian et al., 1994; Takenaga et al., 1994c,b). Although the normal physiological function of S100A4 is not well established, several studies suggest that S100A4 expression promotes a migratory phenotype, and in particular, promotes the epithelial-mesenchymal transition. For example, S100A4 expression in fibroblasts and tumor cells is associated with mesenchymal cell morphology and increased motility (Takenaga et al., 1994a; Strutz et al., 1995; Jenkinson et al., 2004; Chen et al., 2007), whereas reduction or loss of S100A4 expression correlates with decreased migration and inhibition of epithelial transformation (Okada et al., 1997; Takenaga et al., 1997; Bjornland et al., 1999; Grum-Schwensen et al., 2005; Stein et al., 2006).
At the molecular level, S100A4 functions as a Ca2+-activated switch. Calcium-binding drives the adoption of an “open” conformation, which allows S100A4 to bind and regulate the activity of its protein targets (Vallely et al., 2002; Malashkevich et al., 2008). At present, the best-characterized S100A4 target is nonmuscle myosin-IIA (Kriajevska et al., 1994; Ford et al., 1997; Li et al., 2003). p53 has also been reported as an S100A4 target (Grigorian et al., 2001; Fernandez-Fernandez et al., 2005), but recent studies demonstrate that S100A4 does not bind the full-length p53 monomer and only exhibits very weak interactions with p53 tetramers (van Dieck et al., 2009). These observations suggest that the proposed regulation of p53 by S100A4 requires reexamination. Other potential cytoskeletal and signaling targets include tropomyosin (Takenaga et al., 1994d), and liprin β1, an interacting protein of the LAR family of transmembrane tyrosine phosphatases (Kriajevska et al., 2002), but these interactions are not well characterized.
To examine the role of S100A4 in macrophages, we established S100A4-deficient mice by gene targeting. Macrophages derived from these mice display reduced recruitment to sites of inflammation in vivo and consistent with this defect, S100A4−/− bone marrow macrophages (BMMs) exhibit significant defects in chemotactic motility. The motility defects associated with the loss of S100A4 expression in macrophages are the consequence of the dysregulation of myosin-IIA assembly as well as increased Pyk2 activation, enhanced paxillin phosphorylation, and decreased Rac activity. These data provide, for the first time, genetic evidence that S100A4 contributes to the normal physiological motility of macrophages, and they demonstrate a role for S100A4 in regulating macrophage recruitment and chemotaxis in vivo.
MATERIALS AND METHODS
Generation of S100A4 Knockout Mice
S100A4−/− mice were generated through a research agreement with Ozgene Pty Ltd (Bentley DC, WA, Australia). A genomic clone of the S100A4 gene was isolated from a mouse C57BL/6 genomic DNA library. The targeting vector introduced a loxP site 305 base pairs upstream of the start methionine, which is present in the second exon of the S100A4 gene. A phosphoglycerine kinase (PGK) Neo cassette flanked by loxP sites was inserted 426 base pairs downstream of the stop codon in exon 3 (Figure 1A). The entire targeting vector was sequenced because the 5′ and 3′ homology arms contain exonic and intronic sequences from upstream and downstream S100 family members. The targeting vector was electroporated into C57BL/6-derived Bruce4 embryonic stem (ES) cells (Kontgen et al., 1993). ES cells that integrated the targeting vector were identified by Southern blot, and the PGK Neo cassette was removed by treatment with Cre recombinase. S100A4-targeted ES cells were microinjected into C57BL/6 blastocysts and chimeric mice were bred to generate heterozygous F1 mice on an inbred C57BL/6 genetic background. Floxed mice were crossed with Cre-deleter C57BL/6 mice (Schwenk et al., 1995), which resulted in the complete removal of the S100A4 coding exons on one chromosome. Mice were bred to homozygosity and to remove Cre recombinase. Mice were maintained and inbred in a pathogen-free barrier facility. All experiments were performed according to protocols approved by the Animal Welfare Committee at the Albert Einstein College of Medicine (Bronx, NY).
Figure 1.
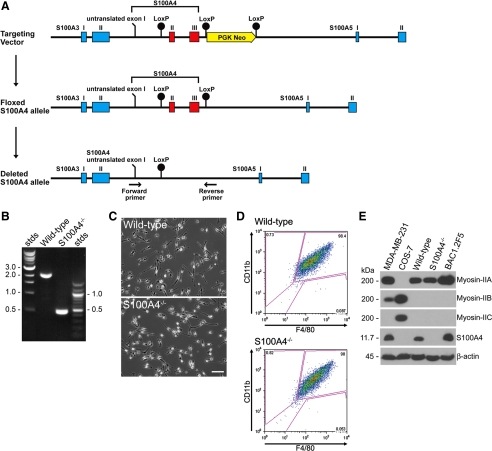
Generation of S100A4 knockout mice. (A) Map of the S100A4 genomic locus with the S100A4 coding exons indicated in red. The targeting vector introduced loxP sites (black circles) that flank the S100A4 gene and a PGK Neo cassette for ES cell selection. (B) PCR of tail DNA to distinguish between wild-type (2.1 kb) and S100A4−/− (0.5 kb) mice. (C) Morphology of wild-type and S100A4−/− BMMs plated on glass in standard growth medium. Bar, 50 μm. (D) Characterization of cell surface markers on isolated BMMs. Ninety-eight percent of the wild-type and S100A4−/− BMMs stain positively for CD11b and F4/80. (E) Immunoblot analysis of S100A4 and myosin-II expression in BMMs isolated from wild-type and S100A4−/− mice, and from BAC1.2F5, a cultured macrophage line. MDA-MB-231 and COS7 cells are controls for myosin-IIB and myosin-IIC expression, respectively. β-Actin was used as a loading control.
Mouse Genotyping
Wild-type and S100A4-null animals were genotyped from tail DNA by polymerase chain reaction (PCR) analysis. The following primers were used for PCR of the S100A4 locus: forward primer (5′-AGCTGGGGTTTTTCCACTTT-3′) and reverse primer (5′-ATCCAACCCTTCATGGACAG-3′). Expected PCR products are 2.1- and 0.5-kb fragments for wild-type and S100A4−/− mice, respectively.
Isolation of Bone Marrow Macrophages
BMMs were isolated from 6- to 12-wk-old mice as described previously (Stanley, 1997). In brief, bone marrow cells from femurs and tibias were seeded onto T-75 flasks, containing α-minimal essential medium (MEM) Plus (Invitrogen, Carlsbad, CA) supplemented with 15% fetal calf serum (FCS) and 10% WEHI conditioned medium. After 3 d, contaminating red blood cells, fibroblasts, and mature macrophages were removed by subtractive adherence to the plates, and nonadherent cells were transferred to fresh α-MEM Plus containing 15% FCS and 1000 U/ml human recombinant colony-stimulating factor-1 (CSF-1) (kindly provided by Dr. E. Richard Stanley, Albert Einstein College of Medicine). After an additional 2–3 d, adherent cells were cultured in α-MEM Plus containing 15% fetal bovine serum (FBS) and 10,000 U/ml human recombinant CSF-1 (standard growth medium). Day 5–14 BMMs were used for all experiments. For CSF-1 stimulation experiments, BMMs were CSF-1-starved for 16–20 h before the start of the experiment.
For BMM transduction, the human S100A4 and TurboRFP lentiviral expression plasmids were purchased from Open Biosystems (Huntsville, AL). To generate infectious lentiviral particles, human embryonic kidney (HEK)293T cells were cotransfected with the second generation packaging plasmids pCMV-dR8.2 dvpr and pCMV-VSVG (Addgene, Cambridge, MA) and the S100A4 or TurboRFP LentiORF plasmids. Virus was collected after 3 d and used to infect BMMs at day 8 of culture for 24 h in the presence of 5 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO). Infected cells were cultured in BMM standard growth medium containing 1 μg/ml blasticidin S (MP Biomedicals, Solon, OH) for an additional 3–5 d and then used for experiments.
Antibodies and Reagents
The mouse monoclonal β-actin antibody was from Sigma-Aldrich. Antibodies to S100A4 were described previously (Li and Bresnick, 2006). Antibodies to the C termini of human nonmuscle myosin-IIA and myosin-IIB, which react with the human, rat, and mouse myosins (Choi et al., 1996; Lo et al., 2004; Dulyaninova et al., 2007), were used for blots against whole cell lysates. The antibody against human nonmuscle myosin-IIC, which reacts with monkey and mouse myosin-IIC (Buxton et al., 2003; Golomb et al., 2004), was a kind gift from Dr. Robert Adelstein (National Heart, Lung, and Blood Institute, Bethesda, MD). The BT561 human NMHC-IIA polyclonal antibody (Biomedical Technologies, Stoughton, MA) and antibodies to full-length platelet myosin-IIA (kind gift from Dr. Robert Adelstein) were used for Triton-insoluble fractions. Phospho-regulatory light chain (RLC) antibodies (T18/S19) were purchased from Cell Signaling Technology (Danvers, MA). Paxillin antibodies were from Millipore Bioscience Research Reagents (Temecula, CA). The paxillin pY118 and Pyk2 pY402 antibodies were from BioSource International (Camarillo, CA). Human fibronectin and the Pyk2, CD16/32, and phycoerythrin (PE)-Cy7–conjugated CD11b antibodies were from BD Biosciences (San Jose, CA). The fluorescein isothiocyanate (FITC)-conjugated GR-1, Alexa 488-conjugated F4/80, and allophycocyanin (APC)-conjugated CD11b antibodies were from Invitrogen. Recombinant mouse CSF-1 was purchased from R&D Systems (Minneapolis, MN). (−)-Blebbistatin was from Toronto Research Chemicals (North York, ON, Canada).
Thioglycollate-induced Peritonitis
Eight- to 12-wk-old mice were injected intraperitoneally with 1 ml of sterile 3.8% of aged thioglycollate (Sigma-Aldrich). Seventy-two hours after injection mice were carbon dioxide asphyxiated, and cells were collected by intraperitoneal lavage using a total of 9 ml of ice-cold phosphate-buffered saline (PBS) supplemented with 0.5% bovine serum albumin (BSA) and 5 mM EDTA. Trace contaminating erythrocytes were selectively lysed by the addition of 10 ml of 1× RBC lysis buffer (150 mM NH4Cl, 0.1 mM EDTA, and 10 mM KHCO3) and incubated on ice for 15 min. Cells were counted with a hemocytometer and adjusted to 5 × 106/ml with cold PBS containing 0.5%BSA. In total, six to 11 mice were used for each time point in three independent experiments.
Flow Cytometry
Cells derived from peritoneal exudates and BMMs were resuspended in PBS containing 0.5% BSA, and Fc receptors were blocked with CD16/32 antibodies. Peritoneal cells (5 × 105) were stained with FITC-Gr-1 or Alexa 488-F4/80 and PE-Cy7-CD11b antibodies. BMMs were reacted with Alexa 488-F4/80 and APC-CD11b antibodies. After staining, cells were washed twice with PBS and then resuspended and fixed in 1% paraformaldehyde/PBS. For flow cytometric analysis, forward and side scatter gates were set to exclude dead cells and aggregates. Data were collected using a FACScan cytometer (BD Biosciences) and analyzed with FlowJo version 8.7 software (TreeStar, (Ashland, OR).
Cell Culture
The BAC1.2F5 macrophage cell line was cultured in α-MEM containing 10% FBS and 3000 U/ml human recombinant CSF-1. MDA-MB-231 and COS-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum.
Intracellular S100A4 Concentration
To determine the total intracellular S100A4 protein concentration, wild-type BMMs or MDA-MB-231 cells were trypsinized and imaged with an UPlanFl 10×, 0.3 numerical aperture (NA) objective. The cell diameter was measured for 100 cells, and the total cell volume was calculated using the equation for the volume of a sphere, 4/3(πr3). Known cell numbers of BMMs or MDA-MB-231 cells were lysed directly in 2× Laemmli sample buffer and loaded onto a 12% Tricine SDS-polyacrylamide gel along with a standard curve of purified S100A4. S100A4 blots were processed as described below. The relative amount of S100A4 was estimated by densitometry using ImageQuant version 5.0 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Triton Cytoskeleton Assay
Triton-insoluble fractions of CSF-1–stimulated (20 ng/ml) wild-type and S100A4−/− macrophages were prepared by lysis in ice-cold Triton X (Tx)-100 buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5% Triton X-100, 5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 5 μg/ml each of chymostatin, leupeptin, and pepstatin A). After 5 min of incubation on ice, the supernatant, which contains Triton X-100–soluble proteins, was removed. The Triton-insoluble fraction was washed twice with ice-cold PBS, resuspended in 2× Laemmli sample buffer, boiled, and separated on 8% SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting with antibodies to myosin-IIA or β-actin. For whole cell lysates, cells were washed with cold PBS and lysed in ice-cold Tx-100 buffer containing 1% SDS, 1 mM EDTA, and 1 mM DTT. Protein concentrations were determined with the DC protein assay (Bio-Rad Laboratories, Hercules, CA) using BSA as a standard. The relative amounts of cytoskeletal myosin-IIA, actin, and paxillin were determined by densitometry using ImageQuant version 5.2.
For experiments using blebbistatin, CSF-1–starved cells were treated with 5 μM active (−)-blebbistatin or with vehicle alone (0.05% dimethyl sulfoxide [DMSO]) for 15 min. Cells were stimulated with 20 ng/ml CSF-1 for 15 min, lysed with ice-cold Tx-100 buffer, and processed as described above.
Immunoblots
To prepare whole cell extracts, BMMs were lysed in a buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 50 mM KCl, 0.5 mM DTT, 1 mM PMSF, and 5 μg/ml each of chymostatin, leupeptin, and pepstatin) supplemented with 2% SDS. For S100A4 or β-actin immunoblots, lysates were separated on a 12% Tricine SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. For myosin-II immunoblots, lysates were separated on a 6% glycine SDS-polyacrylamide gel and transferred to a nitrocellulose membrane.
For phospho-paxillin and phospho-Pyk2 immunoblots, BMMs were lysed in the buffer described above supplemented with 20 mM NaF and a 1:100 dilution of phosphatase inhibitor cocktails I and II (Sigma-Aldrich). Lysates were separated on an 8% Tricine SDS-polyacrylamide gel. Immunoreactive proteins were detected using the SuperSignal West Pico chemiluminescent detection system (Pierce Chemical, Rockford, IL). The relative amounts of pY118 paxillin/total paxillin and pY402 Pyk2/total Pyk2 were estimated by densitometry. β-Actin immunoblots were used as a loading control. Phosphorylation was expressed as the fold change relative to unstimulated cells.
For phospho-RLC antibody (T18/S19) immunoblots, total cell lysates from CSF-1–stimulated cells were prepared by direct addition of ice-cold 10% trichloroacetic acid supplemented with 10 mM DTT to the cell culture dish. Cell samples were collected by scraping followed by microcentrifugation. Cell pellets were washed twice with ice-cold acetone containing 10 mM DTT and resuspended in 2× Laemmli sample buffer and separated on 15% SDS-PAGE. Relative phosphorylation of the RLC was estimated by densitometry as described above.
Rac Activation Assay
At specific times after CSF-1 stimulation, dishes were placed on ice and BMMs were washed with ice-cold PBS containing 0.5 mM sodium vanadate. Cells were lysed in a buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 1 mM DTT, 5% glycerol, 0.5 mM sodium vanadate, 1 mM PMSF, and 5 μg/ml each of chymostatin, leupeptin, and pepstatin. Total protein concentrations were determined using the DC protein assay. Active guanosine triphosphate (GTP)-bound Rac1 pull-down assays were performed with a Rac1 activation kit (Pierce Chemical). One-twelfth of the total lysate was used for the detection of total Rac1. The remaining lysate (0.8 mg of total protein) was used to pull down GTP-Rac1 as per the manufacturer's instruction. Total cell lysates and Rac1 pull-downs were resolved by 12% Tricine SDS-PAGE. Proteins were detected with the Rac1 antibody supplied in the kit. The relative amounts of active and total Rac1 were estimated by densitometry. Relative Rac1 activation was determined by dividing the values obtained by densitometry of Rac1-GTP pull downs by the values obtained for total Rac1. Activation was expressed as the fold change relative to unstimulated cells.
Boyden Chamber Assays
CSF-1–starved BMMs were seeded into the top chamber of an 8-μm transwell in CSF-1–free medium or in medium containing 20 ng/ml recombinant mouse CSF-1 for chemotaxis and chemokinesis experiments, respectively. The transwells were placed into wells containing medium supplemented with 20 ng/ml CSF-1, and the cells were allowed to migrate at 37°C for 3 h or for 5 h (rescue experiments). For controls, CSF-1–free medium was placed into the lower well. Membranes were fixed with 3.7% formaldehyde and stained with 4,6-diamidino-2-phenylindole (DAPI). From nine to 20 random fields of cells on the underside of the membrane were counted using an UPlanFl 10×, 0.3 NA objective. Migration toward CSF-1 was expressed as the fold change relative to migration in the absence of CSF-1 in the lower well.
Random Motility
BMMs (8 × 104) were plated on 35-mm bacterial plastic Petri dishes in standard macrophage growth medium. For imaging, the cells were maintained at 37°C using a PDMI-2 stage microincubator (Harvard Apparatus, Holliston, MA), and the pH was maintained by perfusing the medium with 5% CO2. Evaporation was prevented by overlaying the medium with mineral oil. Phase images were acquired with an UPlanFl 10×, 0.3 NA objective every 5 min for 5–6 h.
For kymography of randomly migrating cells, 8 × 104 BMMs were plated on 35-mm glass-bottomed dishes (MatTek, Ashland, MA) coated with 3 μg/cm2 (13.2 μg/ml) fibronectin. Before imaging, the medium was replaced with prewarmed growth medium containing 20 mM HEPES. The temperature was maintained at 37°C using a heated stage (WPI, Sarasota, FL). Images were recorded every 20 s over a 20-min period using a UPlanFl 20×, 0.5 NA objective.
Micropipette Assay
BMMs (8 × 104) were plated on 35-mm glass-bottomed dishes coated with fibronectin as described above. A Femtojet micromanipulator and pump (Eppendorf-Brinkmann Instruments, Westbury, NY) were used to control the position and the flow from the micropipette. A micropipette was filled with 120 ng/ml recombinant mouse CSF-1 and 0.5 mg/ml 10-kDa rhodamine-dextran and placed ∼60 μm from the edge of a quiescent cell. Images were recorded every 20 s over a 30-min period using a UPlanFl 20×, 0.5 NA objective with the magnification selection knob set to 1.5. Cell perimeters were traced and the coordinates of the cell centroid for each frame of the film were determined using ImageJ (National Institutes of Health, Bethesda, MD; Rasband, 1997–2007). Cell motility parameters and difference pictures were calculated using macros developed by the Analytical Imaging Facility and the Gruss-Lipper Biophotonics Center (Albert Einstein College of Medicine). Directionality is defined as the net path divided by the total path length. The angle θ describes the motility path of a cell and is defined by two reference lines; the line from the cell centroid to the tip of the micropipette at t = 0, and a line from the cell centroid at t = 0 to the cell centroid at t = final.
For assays performed in the presence of blebbistatin, CSF-1–starved macrophages were treated with 5 μM (−)-blebbistatin or DMSO (0.05%) for 15 min before directional stimulation with 120 ng/ml CSF-1.
Cells directionally stimulated with CSF-1 for 15 min were fixed by the addition of an equal volume of 8% paraformaldehyde to the cell culture medium (final concentration, 4%). Fixed cells were permeabilized with 0.2% Triton X-100 for 5 min, blocked with 10% horse serum and 1% BSA in Tris-buffered saline, and stained with the pY118 paxillin antibody. For the analysis of paxillin pY118 immunofluorescence in directionally stimulated cells, cell perimeters were traced and the coordinates of the cell centroid were determined. A 90° angle was drawn from the cell centroid to the cell edge along the side of the cell facing the micropipette. The mean pixel intensity was determined for the cellular region delineated by the 90° angle (cell front) and for the entire cell. The data are expressed as the ratio of the mean pixel intensity of the cell front (region facing the micropipette) to the entire cell.
Microscopy
All images were acquired using IPLab Spectrum software and a CoolSNAP HQ interline 12-bit, cooled charge-coupled device camera (Roper Scientific, Trenton, NJ) mounted on an IX70 microscope (Olympus, Melville, NY).
Kymography
For kymographs of randomly migrating cells, cells were overlaid with a template of eight equally spaced lines radiating from the cell nucleus to the cell periphery (Miller et al., 2004). Kymographs were generated and analyzed with ImageJ and Excel software (Microsoft, Redmond, WA) at all eight positions for each cell. For directionally stimulated cells, kymographs were generated by drawing a line from the tip of the micropipette to the cell centroid before CSF-1 stimulation. The frequency, persistence, and velocity of lamellipodial protrusions and retractions were quantified as described previously (Hinz et al., 1999).
Statistical Analysis
Unpaired Student's t tests were performed to assess the statistical significance of all assays.
RESULTS
Generation of S100A4-deficient Mice
C57BL/6 mice heterozygous for floxed S100A4, in which loxP sites flank the two S100A4 coding exons, were generated and crossed with Cre-deleter mice (Schwenk et al., 1995). Recombination resulted in complete removal of the S100A4 coding exons on one chromosome (Figure 1A). S100A4+/− mice were bred to homozygosity (Figure 1B) and the resulting S100A4−/− mice were fertile, grew normally, and exhibited no overt abnormalities as compared with wild-type mice. To examine S100A4 expression in cells of the mononuclear phagocyte system, bone marrow-derived macrophages were isolated by selecting for CSF-1–responsive macrophage lineage precursors (Stanley, 1997). On the basis of morphology and surface expression of CD11b and F4/80 (Figure 1, C and D, and Supplemental Figure S1), 98% of the cells obtained with this protocol were macrophages. Both Bac1.2F5 cells, a cultured CSF-1–dependent macrophage line, and wild-type BMMs expressed high levels of S100A4 (Figure 1E). For MDA-MB-231 cells and wild-type BMMs, we estimated intracellular concentrations of 4.7 and 3.4 μM S100A4, respectively, whereas S100A4 protein expression in S100A4−/− BMMs was undetectable. Wild-type and S100A4−/− BMMs expressed equivalent levels of nonmuscle myosin-IIA and none of the macrophage lines examined express the B or C isoforms of nonmuscle myosin-II (Figure 1E). Based on quantitative reverse transcription (qRT)-PCR, we did not detect any changes in the mRNA expression of S100 family members known to be expressed in macrophages with the exception of S100A8, which was expressed at 100-fold lower levels in S100A4−/− BMMs (Supplemental Figure S2, A and B). However, S100A8 protein expression was not detected in either wild-type or S100A4−/− BMMs (Supplemental Figure S2C).
S100A4 Mediates the Recruitment of Macrophages to Sites of Inflammation In Vivo
To evaluate how the loss of S100A4 affects macrophage recruitment to sites of inflammation in vivo, we used a thioglycollate-induced model of peritonitis. Seventy-two hours after thioglycollate (TG) injection, peritoneal exudates were analyzed by flow cytometry using side scatter and cell surface marker expression to distinguish the cell populations present in the peritoneal cavity (Chan et al., 1998; Cook et al., 2003; Xia et al., 2009; Supplemental Figure S3). There was no difference in the total number of peritoneal cells in exudates from untreated and TG-treated wild-type and S100A4−/− mice (data not shown); however, there was a 36% decrease in resident peritoneal macrophages in S100A4−/− animals (Table 1). In TG-injected animals, resident macrophages constituted <1% of the total cells recovered from peritoneal exudates (Supplemental Figure S3). At 72 h after TG treatment, there was a 38% reduction in the recruitment of inflammatory macrophages to the peritoneal cavity in S100A4−/− mice (Table 1). Interestingly, in 72-h TG exudates from S100A4−/− animals, there was a 31% increase in eosinophils.
Table 1.
Percentage of cells in peritoneal exudatesa
| Resident |
72-h TG |
|||
|---|---|---|---|---|
| Wild type | S100A4−/− | Wild type | S100A4−/− | |
| Macrophages | 36.8 ± 4.4 | 23.5 ± 2.6* | 52.0 ± 3.2 | 32.1 ± 4.0** |
| Neutrophils | n.d.b | n.d. | 3.7 ± 0.7 | 4.2 ± 0.9 |
| Eosinophils | n.d. | n.d. | 35.5 ± 2.6 | 46.7 ± 3.5*** |
a n = 6–11 mice per condition; p values: *, 0.031; **, 0.0009; and ***, 0.018.
b n.d., not determined.
S100A4−/− BMMs Exhibit Defects in CSF-1–stimulated Motility
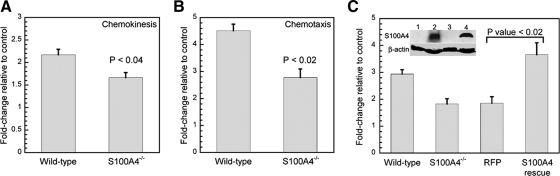
Because the loss of S100A4 resulted in impaired recruitment of macrophages in vivo, we examined the motility properties of S100A4−/− macrophages in vitro. Deletion of S100A4 had no significant effect on the migration speed, directionality, or net path length of randomly migrating macrophages (Table 2). Nor did we detect any alterations in the dynamics of lamellipodial protrusions of randomly migrating cells (Supplemental Figure S4). To investigate whether S100A4 regulates directed cell migration, wild-type and S100A4−/− macrophages were examined for their ability to migrate toward CSF-1 in a Boyden chamber assay. In chemokinesis assays, the loss of S100A4 had a modest effect on migration with a 24% reduction in the motility of S100A4−/− BMMs (Figure 2A). In chemotaxis assays, wild-type BMMs showed a 4.5-fold increase in migration in response to CSF-1, whereas S100A4−/− BMMs displayed only a 2.7-fold increase (60% of the wild-type response) in their CSF-1–stimulated motility (Figure 2B). Lentiviral transduction of S100A4−/− BMMs with human S100A4 rescued the defect in CSF-1–mediated chemotaxis, whereas expression of Turbo RFP had no effect on the chemotactic capabilities of S100A4-null macrophages (Figure 2C).
Table 2.
Random motility of BMMs
| Wild type | S100A4−/− | |
|---|---|---|
| Total path length (μm) | 313 ± 16a | 309 ± 15 |
| Net path length (μm) | 122 ± 15 | 95 ± 10 |
| Directionality (net/total distance) | 0.40 ± 0.04 | 0.31 ± 0.03 |
| Velocity (μm/min) | 1.04 ± 0.05 | 1.03 ± 0.03 |
a Values represent the mean ± SEM for 32 wild-type and S100A4−/− BMMs. Migration was monitored over 5 h. No statistical differences were observed for any of the measured parameters.
Figure 2.
S100A4−/− BMMs exhibit reduced migration toward CSF-1. BMMs were CSF-1 starved for 16 h before seeding in an 8-μm transwell. The CSF-1–stimulated migration of each cell population is expressed as the fold change relative to unstimulated cells. Data represent the mean ± SEM for three independent experiments performed in triplicate. (A) Chemokinesis (3 h) of wild-type and S100A4−/− BMMs; 20 ng/ml CSF-1 was present in both the upper and lower chambers. (B) Chemotaxis (3 h) of wild-type and S100A4−/− BMMs; 20 ng/ml CSF-1 was present in only the lower chamber. (C) Chemotaxis (5 h) of wild-type BMMs, S100A4−/− BMMs, and S100A4−/− BMMs expressing either TurboRFP or human S100A4; 20 ng/ml CSF-1 was present in only the lower chamber. Inset, expression of S100A4 in primary BMMs. β-Actin was used as a loading control. 1, S100A4−/− BMMs transduced with a TurboRFP lentivirus; 2, S100A4−/− BMMs transduced with the human S100A4 lentivirus; 3, S100A4−/− BMMs; and 4, wild-type BMMs.
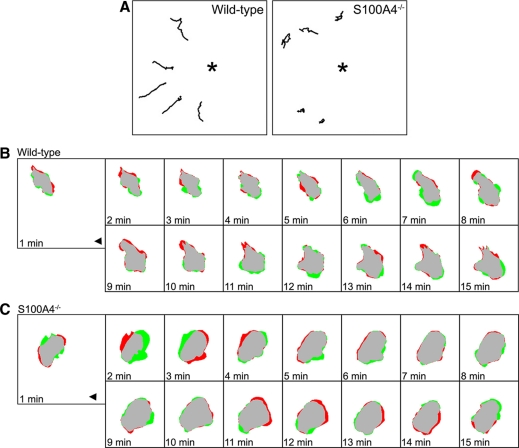
To quantitatively assess how the loss of S100A4 affects CSF-1–stimulated motility, we used a micropipette chemotaxis assay (Mouneimne et al., 2004; Li and Bresnick, 2006). A CSF-1–filled pipette was placed ∼60 μm away from the edge of a quiescent cell, and migration toward the source of CSF-1 was monitored by time-lapse microscopy (Supplemental Video 1). Centroid plot analysis, which shows the position of cell centroids traced at consecutive time intervals, demonstrated that S100A4−/− BMMs exhibited a random walking path or moved in tight circles, compared with wild-type macrophages, which displayed more linear movement during chemotaxis toward CSF-1 (Figure 3A and Supplemental Video 1). Although wild-type and S100A4−/− cells migrated at the same velocities, S100A4−/− BMMs showed a significant reduction in the net path length. As a consequence, directionality, which is the ratio of the net to the total path, was lower in S100A4-deficient BMMs (Table 3). An examination of the direction change, which relates the cell trajectory to the position of the micropipette and reflects the frequency of turning during migration, revealed that S100A4−/− BMMs have a 2.8-fold higher turning frequency compared with wild-type cells (Table 3), which is consistent with the reduced directionality measured for S100A4−/− cells. Although not statistically significant (p = 0.052), we observed a consistent reduction in the chemotactic index of S100A4−/− BMMs. Difference pictures of a representative wild-type BMM show that most protrusions localized to the anterior half of the cell that faces the micropipette (Figure 3B). In contrast, corresponding difference pictures of a representative S100A4−/− cell indicate that protrusions occur along the anterior, as well as the sides and posterior of the cell (Figure 3C).
Figure 3.
S100A4−/− BMMs show impaired persistence during chemotaxis. (A) Trace of cell centroids for five representative cells of each genotype during chemotaxis toward a point source of CSF-1. The stars indicate the position of the micropipette. (B) Difference pictures of representative wild-type and S100A4−/− BMMs responding to a gradient of CSF-1. Protrusions are shaded green and retractions are shaded red. The arrowhead indicates the position of the micropipette. A micropipette filled with 120 ng/ml CSF-1 was placed ∼60 μm away from the cell, and the cell response was monitored over 30 min, with images collected every 20 s. Cells were plated on 13.2 μg/ml fibronectin.
Table 3.
CSF-1–stimulated motility of BMMs
| Wild type | S100A4−/− | p value | |
|---|---|---|---|
| Total path length (μm) | 28.63 ± 1.67a | 29.65 ± 1.93 | N.S.b |
| Net path length (μm) | 19.11 ± 1.70 | 11.73 ± 2.02 | 0.009 |
| Directionality (net/total distance) | 0.66 ± 0.04 | 0.40 ± 0.06 | 0.002 |
| Velocity (μm/min) | 1.43 ± 0.08 | 1.48 ± 0.10 | N.S. |
| Direction change (°) | 14.66 ± 2.79 | 40.91 ± 10.69 | 0.027 |
| Chemotactic index (cos θ) | 0.95 ± 0.02 | 0.66 ± 0.14 | 0.052 |
a Values represent the mean ± SEM for 18 wild-type and 19 S100A4−/− BMMs. Chemotaxis towards a point source of CSF-1 was monitored over 30 min.
b N.S. represents p > 0.05.
Loss of S100A4 Increases the Frequency but Decreases the Persistence and Size of Protrusions Due to Actomyosin-IIA Overassembly
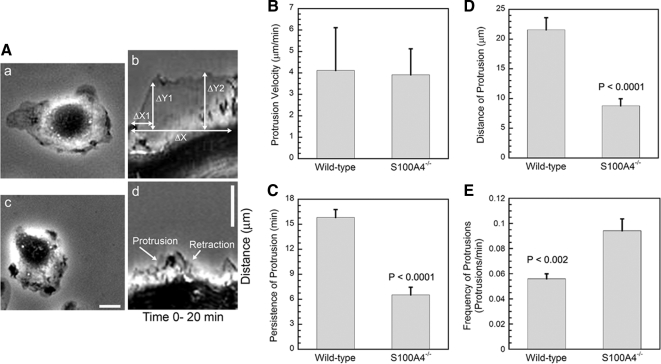
To examine the basis for the migratory defects observed with S100A4−/− macrophages, we used kymograph analysis to examine protrusion dynamics in cells responding to CSF-1 from a micropipette (Figure 4A). S100A4−/− BMMs extended protrusions with the same velocities as wild-type cells (Figure 4B); however, the protrusions of S100A4−/− macrophages were 2.4-fold less persistent and 2.5-fold smaller than those of wild-type macrophages (Figure 4, C and D). In addition, S100A4−/− cells had more frequent protrusions (1.6-fold) than wild-type BMMs (Figure 4E).
Figure 4.
S100A4−/− BMMs form small, highly unstable protrusions. (A) Representative kymographs from wild-type (a and b) and S100A4−/− (c and d) BMMs after directional stimulation with CSF-1. BMMs were plated on 13.2 μg/ml fibronectin. The phase-contrast micrographs (a and c) show wild-type and S100A4−/− BMMs before stimulation. For the kymographs (b and d), the time and distance are noted on the horizontal and vertical axes, respectively. ΔX, protrusion persistence; ΔY2, protrusion distance; and ΔY1/ΔX1, protrusion velocity. Bars, 10 μm. Quantification of protrusion velocity (B), persistence (C), distance (D), and frequency (E). Values represent the mean ± SEM for 18 wild-type and 19 S100A4−/− cells.
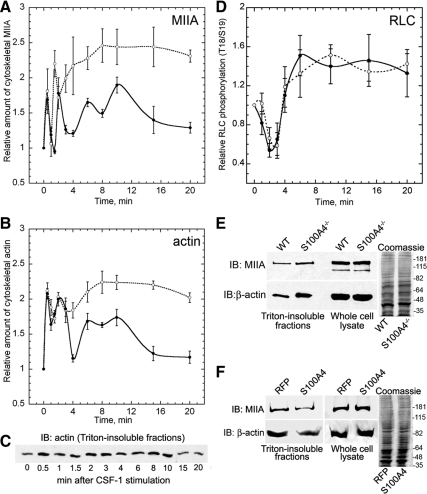
Given that in vitro and in vivo studies demonstrate a role for S100A4 in regulating myosin-IIA assembly (Ford et al., 1997; Li et al., 2003; Li and Bresnick, 2006), we examined the kinetics of myosin-IIA assembly in CSF-1–stimulated wild-type and S100A4−/− BMMs. In wild-type cells, we observed four peaks of Triton-insoluble (e.g., assembled) myosin-IIA at 0.5, 2, 6, and 10 min after CSF-1 stimulation (Figure 5A). Coincident with the peaks of myosin-IIA assembly, we also detected cross-linked β-actin in wild-type cells (Figure 5, B and C). In S100A4−/− BMMs, the first peak of myosin-IIA assembly occurred at 0.5 min; however, the second peak was shifted to a slightly earlier time (1.5 min). The third and fourth peaks merged into a single broad peak that began 3–4 min after CSF-1 stimulation and remained high until 20 min. At these later times, there was a 1.5- to 1.8-fold increase in the relative amount of myosin-IIA in Triton cytoskeletons compared with wild-type BMMs (Figure 5, A and E). A similar increase in cytoskeletal actin was observed in S100A4−/− BMMs, which paralleled the change in myosin-IIA assembly (Figure 5B). Reexpression of human S100A4 in S100A4−/− BMMs reduced the amount of cytoskeletal myosin-IIA by 37 ± 8.5% (mean ± SD from two independent experiments; Figure 5F), which is comparable with wild-type levels. We also examined the phosphorylation status of total RLC in CSF-1–stimulated cells. In contrast to the alterations observed in myosin-IIA filament assembly for S100A4-null cells, the kinetics of RLC monophosphorylation (data not shown) and diphosphorylation (Figure 5D) were the same for wild-type and S100A4−/− cells.
Figure 5.
Loss of S100A4 promotes myosin-IIA and β-actin overassembly but does not affect RLC phosphorylation. Myosin-IIA (A) and β-actin (B) assembly were assessed by isolating Triton X-100–resistant cytoskeletons from CSF-1–stimulated wild-type (solid lines) and S100A4−/− (dotted lines) BMMs. For all curves, values represent the mean ± SEM for three to four independent experiments. (C) Representative immunoblot of β-actin in Triton-insoluble fractions from wild-type BMMs at different times after stimulation with CSF-1. (D) Kinetics of total T18/S19 RLC phosphorylation in stimulated wild-type (solid lines) and S100A4−/− (dotted lines) BMMs. (E) Relative amount of myosin-IIA heavy chain and β-actin in Triton-insoluble fractions and whole cell lysates from wild-type and S100A4−/− BMMs stimulated with CSF-1 for 20 min. Right, Coomassie-stained whole cell lysates used for the immunoblots. (F) Relative amount of myosin-IIA heavy chain and β-actin in Triton-insoluble fractions from S100A4−/− BMMs expressing either TurboRFP or human S100A4 after stimulation with CSF-1 for 20 min. Right, Coomassie-stained whole cell lysates used for the immunoblots.
To modulate actomyosin overassembly, we treated S100A4−/− BMMs with low concentrations of the myosin-II ATPase inhibitor blebbistatin. Blebbistatin preferentially binds to myosin-ADP·Pi, thus stabilizing myosin-II in a weak actin binding state (Kovacs et al., 2004; Limouze et al., 2004), resulting in an overall decrease in actomyosin. Treatment of CSF-1–stimulated S100A4−/− macrophages with 5 μM blebbistatin decreased assembled actin and myosin-IIA (Figure 6A), consistent with a reduction in myosin-IIA actin cross-linking activity. Next, we used the micropipette chemotaxis assay to examine protrusion dynamics in blebbistatin-treated S100A4−/− BMMs. Low blebbistatin concentrations did not affect protrusion persistence in wild-type cells but did restore protrusion persistence in S100A4−/− cells (Figure 6B). However, other protrusion parameters such as distance, velocity, and frequency were not affected by treatment with blebbistatin under our experimental conditions (data not shown). Time-lapse microscopy of blebbistatin-treated S100A4−/− BMMs showed that even though tail retraction is somewhat impaired, the cells polarize and exhibit more linear movement toward the source of CSF-1 (Supplemental Video 2).
Figure 6.
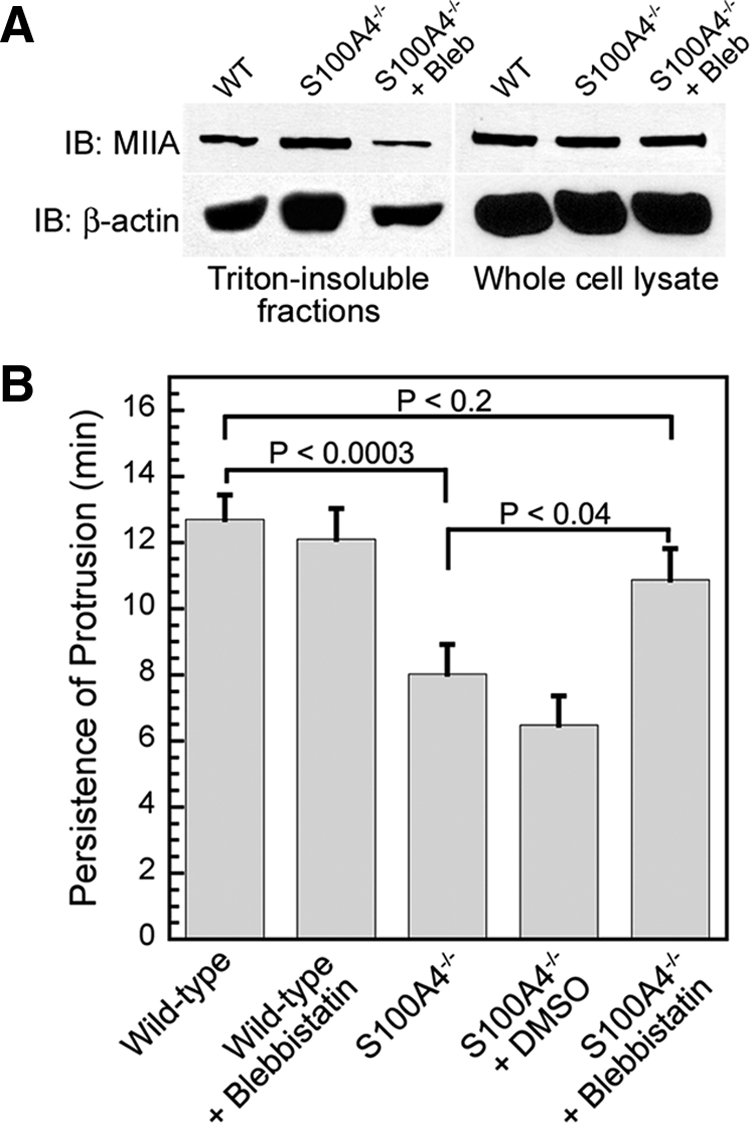
Blebbistatin treatment restores protrusion persistence in CSF-1–stimulated S100A4−/− BMMs. (A) Relative amount of myosin-IIA and β-actin from Triton-insoluble cytoskeletons from S100A4−/− BMMs stimulated with CSF-1 for 15 min in the presence of 5 μM blebbistatin. (B) Protrusion persistence of S100A4−/− BMMs after directional stimulation with CSF-1 in the presence of 0.05% DMSO or 5 μM blebbistatin. Values represent the mean ± SEM for 15–17 cells of each genotype and treatment. There is no statistically significant difference in the protrusion persistence of untreated and DMSO-treated S100A4−/− cells.
Responsiveness of the Cytoskeleton to CSF-1 Receptor Signaling Is Altered in S100A4−/− Macrophages
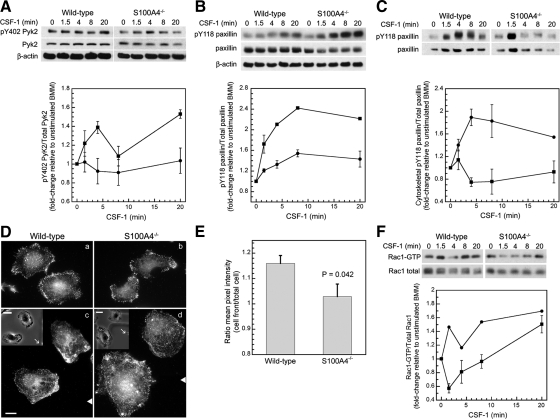
Based on our observations that S100A4−/− BMMs rapidly extend small, short-lived protrusions, we considered that loss of S100A4 may also affect focal adhesions through these myosin-II–associated perturbations. First, we examined the activation of Pyk2, a nonreceptor protein tyrosine kinase closely related to FAK that is expressed in macrophages (Okigaki et al., 2003). In wild-type BMMs, phosphorylation on Y402 remained relatively unchanged after CSF-1 stimulation. This contrasted with S100A4−/− BMMs, which exhibited a 1.4-fold increase in the relative amount of pY402 Pyk2 by 4 min after CSF-1 stimulation (Figures 7A). However, basal Pyk2 phosphorylation was higher in wild-type than in S100A4−/− BMMs (compare 0 min in Figure 7A).
Figure 7.
S100A4−/− BMMs exhibit an altered response to CSF-1 receptor signaling. Analysis of cell signaling in CSF-1–stimulated wild-type (circles) and S100A4−/− (squares) BMMs. Phosphorylation or activation is expressed as the fold change relative to unstimulated cells. Data represent the mean for two to four independent experiments. SEs are shown only for data points from three or four independent experiments. (A) Top, representative immunoblot of pY402 Pyk2, total Pyk2, and β-actin. Bottom, kinetics of Pyk2 Y402 phosphorylation. (B) Top, representative immunoblot of pY118 paxillin, total paxillin, and β-actin. Bottom, kinetics of paxillin Y118 phosphorylation. (C) Top, representative immunoblot of pY118 paxillin and total paxillin from Triton-insoluble cytoskeletons of stimulated cells. Bottom, kinetics of pY118 paxillin associated with Triton cytoskeletons. (D) Paxillin pY118 localization in globally (a and b) and directionally (c and d) CSF-1–stimulated wild-type and S100A4−/− cells. (c and d) White triangles indicate the position of the micropipette relative to the cells (not the actual distance). Bar, 10 μm. Inset, phase images of cells at the start of CSF-1 stimulation. White arrows indicate the position of the micropipette. Bar, 20 μm. (E) Quantification of pY118 paxillin in directionally stimulated cells. The cell front is defined as a 90° wedge facing the micropipette. Values represent the ratio ± SEM for 10 wild-type and nine S100A4−/− BMMs. (F) Top, representative immunoblot of GTP-bound and total Rac1. Bottom, kinetics of Rac1 activation.
In macrophages, Pyk2 phosphorylates paxillin (Gismondi et al., 1997; Hiregowdara et al., 1997). Because phosphorylation on paxillin Y118 is associated with adhesion assembly (Zaidel-Bar et al., 2007), we used this as a marker for the formation of focal complexes. Similar to Pyk2, we observed alterations in total paxillin phosphorylation levels in S100A4-deficient cells. Phosphorylation on paxillin Y118 increased 1.5-fold by 8 min after CSF-1 stimulation in wild-type macrophages. In S100A4−/− BMMs, paxillin Y118 phosphorylation increased rapidly after CSF-1 stimulation, with maximal changes of 2.4-fold detected by 8 min (Figure 7B). Because CSF-1 induced robust phosphorylation of paxillin on Y118 in both wild-type and S100A4−/− BMMs, we examined the redistribution of pY118 paxillin to Triton-insoluble cytoskeletons after CSF-1 addition. In wild-type macrophages, CSF-1 promoted the rapid redistribution of pY118 paxillin, with a 1.9-fold increase in phosphorylated paxillin associated with the cytoskeletal fraction by 4 min after stimulation followed by a gradual decline in pY118 paxillin associated with the cytoskeleton (Figure 7C). Although S100A4−/− BMMs displayed an overall increase in total pY118 paxillin, the amount of phospho-paxillin associated with Triton cytoskeletons was reduced significantly compared with wild-type cells. There was a slight increase in pY118 paxillin at 1.5 min after stimulation followed by a decrease in cytoskeleton-associated pY118 paxillin (Figure 7C); however, in unstimulated cells more pY118 paxillin was associated with the cytoskeleton in S100A4−/− than wild-type BMMs. Immunofluorescence studies demonstrated that for cells globally stimulated with CSF-1, pY118 paxillin uniformly distributed along the cell periphery in both wild-type and S100A4−/− cells, with fewer focal complexes detected in S100A4−/− BMMs (Figure 7D, a and b), which is consistent with biochemical assays. In wild-type cells that were stimulated with CSF-1 from a micropipette, pY118 paxillin was enriched in protrusions that face the source of chemoattractant by 13%, whereas in S100A4−/− BMMs phosphorylated paxillin localized principally along the cell perimeter (Figure 7D, c and d; and E). Consistent with these alterations in signaling to the adhesion apparatus, S100A4−/− BMMs exhibited a 10–20% reduction at early times of cell attachment compared with wild-type cells (Supplemental Figure S5). By 15 min, no differences in attachment could be detected between wild-type and S100A4-null cells.
Because Rac1 is involved in the formation and turnover of adhesions (del Pozo et al., 2000; Wells et al., 2004; Wheeler et al., 2006), we examined the kinetics of Rac activation in CSF-1–stimulated wild-type and S100A4−/− BMMs. In wild-type cells, a sharp rise in Rac activity (1.5-fold over unstimulated cells) was observed at 1.5 min after CSF-1 stimulation, followed by a slow increase in Rac activation (Figure 7F). This profile contrasted sharply with the kinetics observed in S100A4−/− BMMs, where Rac1 activity rapidly decreased followed by a gradual increase in activity. As a consequence, 1.5 min after CSF-1 stimulation, Rac activity was 2.6-fold lower in S100A4−/− macrophages as compared with wild-type cells.
DISCUSSION
In this study, we demonstrate that macrophages express high levels of S100A4 and that S100A4 regulates the motile and invasive capabilities of these cells. Based on the criteria of morphology, CSF-1–dependent growth, and expression of the cell surface markers CD11b and F4/80, these studies were performed on a highly enriched population of bone marrow-derived macrophages (98%) that is free of contaminating fibroblast-like cells. Despite early studies demonstrating S100A4 expression in monocytes, macrophages, and lymphocytes by immunoblotting techniques (Takenaga et al., 1994b,c), the detection of S100A4-expressing immune cells by immunohistochemical methods has been controversial due to the possible lack of selectivity of S100A4 antibodies. Thus, it has been suggested that S100A4 (also known as FSP1) is exclusively expressed in fibroblasts (Strutz et al., 1995; Inoue et al., 2005a; Inoue et al., 2005b; Le Hir and Kaissling, 2005). As a consequence, S100A4 is used frequently as a marker to identify fibroblasts, within the tumor microenvironment (Egeblad et al., 2005; Sugimoto et al., 2006; Zeisberg et al., 2007; Joyce and Pollard, 2009) and under other pathological states, including fibrosis, rheumatoid arthritis, and pulmonary disease (Schneider et al., 2008). BMMs and mouse embryonic fibroblasts derived from wild-type mice express comparable levels of S100A4 (Supplemental Figure S6); thus, histological studies with S100A4 antibodies are unlikely to distinguish between fibroblasts and macrophages. Our results indicate that the identification of fibroblasts in mouse tissues based solely on immunohistochemical analysis of S100A4 expression requires careful reexamination.
Although wound healing and transwell assays suggested a role for S100A4 in promoting the motility of fibroblasts and cancer cells (Bjornland et al., 1999; Grum-Schwensen et al., 2005; Stein et al., 2006; Chen et al., 2007), a detailed analysis of S100A4-dependent motility has not been performed in any cell type. Results presented here unequivocally demonstrate the involvement of S100A4 in regulating cell migration both in vivo and in vitro. Interestingly, the loss of S100A4 has no affect on random motility in vitro, whereas S100A4-deficient BMMs exhibit a significant impairment in chemotactic motility. During CSF-1–stimulated chemotaxis, S100A4−/− BMMs rapidly extend small, highly unstable protrusions with high frequency. Due to the reduction in lamellipodial stability, S100A4−/− macrophages turn more frequently and translocate with less persistence than wild-type cells. These data support a role for S100A4 in maintaining cell polarization during chemotactic migration, and they are consistent with previous studies demonstrating that interactions between myosin-IIA and S100A4 mediate directional protrusions in tumor cells (Li and Bresnick, 2006). Whether the maintenance of polar directionality during chemotactic migration will be a general feature of S100A4 function in all cell types will require a detailed comparative analysis of additional S100A4-expressing and S100A4-deficient cell lines.
S100A4 binds in a Ca2+-dependent manner to the C-terminal coiled-coil of the myosin-IIA heavy chain to promote filament depolymerization and increase the amount of soluble myosin-IIA (Li et al., 2003; Garrett et al., 2008; Malashkevich et al., 2008). In addition, S100A4 increases the apparent critical monomer concentration for myosin-IIA assembly (Li et al., 2003). Consistent with these in vitro analyses, studies presented here demonstrate that the loss of S100A4 results in increased myosin-IIA filament assembly in vivo. These changes in myosin-IIA assembly are mediated through S100A4's interaction with the heavy chain because the extent and kinetics of RLC phosphorylation are the same in wild-type and S100A4-deficient cells. Although RLC phosphorylation is unchanged, our data show that the increase in myosin-IIA filament assembly in S100A4−/− cells allows for enhanced actin filament cross-linking. In contrast to muscle myosin-II, nonmuscle myosin-II assembles into small filaments (Niederman and Pollard, 1975; Verkhovsky et al., 1995), thus limiting the number of myosin-II heads that can interact with F-actin relative to the more extensive assemblies present in muscle. However, the enhanced myosin-II assembly in S100A4-null BMMs would increase the number of myosin heads within filaments as well as potential interactions with actin. Augmented interactions with actin would promote cortical stability and may increase contractile force and retrograde flow in the lamellum, thus limiting actin polymerization in the lamellipodium and decreasing protrusion (Vicente-Manzanares et al., 2009). Consistent with this idea, blebbistatin treatment, which would have a disruptive effect on myosin-II's actin cross-linking activity, partially restores the protrusive defects observed in S100A4−/− BMMs. Although we measured global effects of blebbistatin on actomyosin cross-linking, the observed rescue in protrusion persistence could be the consequence of local alterations in the activity of specific myosin-II pools.
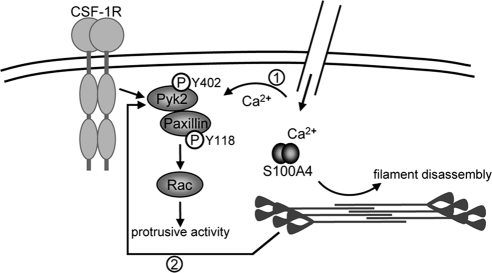
In addition to affecting myosin-IIA assembly directly, our data show that the loss of S100A4 also impacts signaling downstream of the CSF-1 receptor. CSF-1 ligation in quiescent macrophages stimulates lamellipodial protrusions and the formation of focal complexes via a signaling pathway that involves Pyk2 activation (Hatch et al., 1998), paxillin phosphorylation, and translocation to the plasma membrane (Nakamura et al., 2000; Pixley et al., 2001; Zaidel-Bar et al., 2007), the recruitment of SH2 containing proteins that promote Rac activity (Vedham et al., 2005; Sakai et al., 2006; Lee et al., 2007; Frank and Hansen, 2008) and the formation of a stable lamellipodium (del Pozo et al., 2000; Nishiya et al., 2005; Sakai et al., 2006; Figure 8). Cytoskeletal integrity and calcium signaling have been shown to mediate Pyk2 activation, and we propose that alterations in both contribute to the elevated Pyk2 activation observed in S100A4-null cells.
Figure 8.
Model showing how S100A4 impacts CSF-1 signaling in macrophages. Pyk2 activation is mediated by calcium signaling (1) and cytoskeletal integrity (2). Activated Pyk2 promotes paxillin phosphorylation on pY118, and the recruitment of intermediary signaling molecules that recruit and activate Rac, which modulates the formation of adhesions as well as protrusive activity. The loss of S100A4 results in actomyosin overassembly and may alter local calcium dynamics near the plasma membrane, resulting in hyperactivation of Pyk2 and associated dysregulation of downstream signaling events.
Pyk2 activation is mediated, in part, by an intact cytoskeleton (Hiregowdara et al., 1997; Brinson et al., 1998; Wu et al., 2006), and we propose that stabilization of the actomyosin cytoskeleton observed in S100A4-deficient cells contributes to the increase in Pyk2 activation detected in these cells. Augmented Pyk2 activation would also drive enhanced phosphorylation on paxillin Y118. In addition, recent studies suggest that myosin-II contractility mediates phosphorylation on paxillin Y118 (Schneider et al., 2009; Pasapera et al., 2010); thus, myosin-II overassembly in S100A4-null cells may also directly modulate paxillin phosphorylation. Although these cells exhibit increased pY118 paxillin, phosphorylated paxillin fails to translocate to the Triton-cytoskeleton, which will affect the recruitment of intermediate signaling complexes and subsequent Rac activation. Because Rac1 regulates protrusive activity in macrophages, as well as the formation and turnover of adhesions (del Pozo et al., 2000; Wells et al., 2004; Wheeler et al., 2006), an inability to form new adhesions at the leading edge after directional stimulation with CSF-1 would contribute to the reduced lamellipodial stability observed for S100A4−/− BMMs.
In addition, to its regulation by the cytoskeleton, Pyk2 is activated by increased intracellular calcium (Sanjay et al., 2001; Faccio et al., 2003; Yamamoto et al., 2008); thus, it could be argued that the loss of S100A4 could activate Pyk2 via the disruption of calcium homeostasis. Although originally considered to function as calcium buffering proteins, recent studies demonstrate that S100 proteins act as calcium sensors to modulate cellular responses to specific biological stimuli (Xiong et al., 2000; McNeill et al., 2007; Prosser et al., 2008). Based on these observations we propose that S100A4 regulates local calcium dynamics at the leading edge. In vivo studies with a biosensor that reports on the Ca2+-bound activated form of S100A4 supports the idea that S100A4 functions as a regiospecific calcium sensor rather than as a buffer of global increases in cytosolic calcium. In lysophosphatidic acid-stimulated and randomly migrating fibroblasts, localized and transient S100A4 activation is observed in cell extensions and areas of the leading edge undergoing dynamic remodeling (Garrett et al., 2008). Similarly, in fibroblasts and human breast cancer cells migrating into a wound, S100A4 activation is highest at the leading edge (unpublished observations). Moreover, recent studies have identified short-lived calcium microdomains in polarized migrating cells that are required to maintain the structure and activity of the leading edge (Evans and Falke, 2007; Wei et al., 2009). Such microdomains might be responsible for the regiospecific regulation of both Pyk2 and S100A4.
Perhaps most notably our studies demonstrate that S100A4 expression is important for the recruitment of myeloid cells to sites of inflammation, which is consistent with the observed defects in chemotactic motility. One surprising finding was that increased numbers of eosinophils were detected in TG-stimulated S100A4−/− animals. At this time, there is no information as to whether S100A4 is expressed in eosinophils; thus, the biological significance of these observations will require further investigation. However, given that S100A4 is important for neutrophil and macrophage recruitment to sites of inflammation in vivo, S100A4 may be make a widespread contribution to immune function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Adelstein for antibodies to human myosin-IIC and full-length platelet myosin-IIA and Dr. E. Richard Stanley for recombinant human CSF-1. We are grateful to Dr. Dianne Cox (Albert Einstein College of Medicine) and Dr. Anne Davidson (Feinstein Institute for Medical Research, Manhasset, NY) for helpful discussions. We gratefully thank Dr. Violetta Leschenko (Albert Einstein College of Medicine) for assistance with qRT-PCR analysis. This work was supported by National Institutes of Health grants CA-129598 (to A.R.B.) and AI-07289 (to S.C.A. and Stan Nathenson) and Department of Defense grant BC061082 (to R.P.H.).
Abbreviations used:
- BMM
bone marrow macrophage
- CSF-1
colony stimulating factor-1
- ES
embryonic stem
- PGK
phosphoglycerine kinase
- Pyk2
proline-rich tyrosine kinase 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0609) on June 2, 2010.
REFERENCES
- Ambartsumian N. S., Grigorian M. S., Larsen I. F., Karlstrom O., Sidenius N., Rygaard J., Georgiev G., Lukanidin E. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–1630. [PubMed] [Google Scholar]
- Bjornland K., Winberg J. O., Odegaard O. T., Hovig E., Loennechen T., Aasen A. O., Fodstad O., Maelandsmo G. M. S100A4 involvement in metastasis: deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res. 1999;59:4702–4708. [PubMed] [Google Scholar]
- Brinson A. E., Harding T., Diliberto P. A., He Y., Li X., Hunter D., Herman B., Earp H. S., Graves L. M. Regulation of a calcium-dependent tyrosine kinase in vascular smooth muscle cells by angiotensin II and platelet-derived growth factor. Dependence on calcium and the actin cytoskeleton. J. Biol. Chem. 1998;273:1711–1718. doi: 10.1074/jbc.273.3.1711. [DOI] [PubMed] [Google Scholar]
- Buxton D. B., Golomb E., Adelstein R. S. Induction of nonmuscle myosin heavy chain II-C by butyrate in RAW 264.7 mouse macrophages. J. Biol. Chem. 2003;278:15449–15455. doi: 10.1074/jbc.M210145200. [DOI] [PubMed] [Google Scholar]
- Chan J., Leenen P. J., Bertoncello I., Nishikawa S. I., Hamilton J. A. Macrophage lineage cells in inflammation: characterization by colony-stimulating factor-1 (CSF-1) receptor (c-Fms), ER-MP58, and ER-MP20 (Ly-6C) expression. Blood. 1998;92:1423–1431. [PubMed] [Google Scholar]
- Chen P. S., et al. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J. Cell Sci. 2007;120:2053–2065. doi: 10.1242/jcs.03460. [DOI] [PubMed] [Google Scholar]
- Cho Y. G., et al. Overexpression of S100A4 is closely related to the aggressiveness of gastric cancer. APMIS. 2003;111:539–545. doi: 10.1034/j.1600-0463.2003.1110502.x. [DOI] [PubMed] [Google Scholar]
- Choi O. H., Park C. S., Itoh K., Adelstein R. S., Beaven M. A. Cloning of the cDNA encoding rat myosin heavy chain-A and evidence for the absence of myosin heavy chain-B in cultured rat mast (RBL-2H3) cells. J. Muscle Res. Cell Motil. 1996;17:69–77. doi: 10.1007/BF00140325. [DOI] [PubMed] [Google Scholar]
- Cook A. D., Braine E. L., Hamilton J. A. The phenotype of inflammatory macrophages is stimulus dependent: implications for the nature of the inflammatory response. J. Immunol. 2003;171:4816–4823. doi: 10.4049/jimmunol.171.9.4816. [DOI] [PubMed] [Google Scholar]
- Davies B. R., Davies M. P., Gibbs F. E., Barraclough R., Rudland P. S. Induction of the metastatic phenotype by transfection of a benign rat mammary epithelial cell line with the gene for p9Ka, a rat calcium-binding protein, but not with the oncogene EJ-ras-1. Oncogene. 1993;8:999–1008. [PubMed] [Google Scholar]
- Davies B. R., O'Donnell M., Durkan G. C., Rudland P. S., Barraclough R., Neal D. E., Mellon J. K. Expression of S100A4 protein is associated with metastasis and reduced survival in human bladder cancer. J. Pathol. 2002;196:292–299. doi: 10.1002/path.1051. [DOI] [PubMed] [Google Scholar]
- Davies M. P., Rudland P. S., Robertson L., Parry E. W., Jolicoeur P., Barraclough R. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–1637. [PubMed] [Google Scholar]
- del Pozo M. A., Price L. S., Alderson N. B., Ren X. D., Schwartz M. A. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R. S 100, a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Dulyaninova N. G., House R. P., Betapudi V., Bresnick A. R. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol. Biol. Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M., Littlepage L. E., Werb Z. The fibroblastic coconspirator in cancer progression. Cold Spring Harb. Symp. Quant. Biol. 2005;70:383–388. doi: 10.1101/sqb.2005.70.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. H., Falke J. J. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc. Natl. Acad. Sci. USA. 2007;104:16176–16181. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccio R., Novack D. V., Zallone A., Ross F. P., Teitelbaum S. L. Dynamic changes in the osteoclast cytoskeleton in response to growth factors and cell attachment are controlled by beta3 integrin. J. Cell Biol. 2003;162:499–509. doi: 10.1083/jcb.200212082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez M. R., Veprintsev D. B., Fersht A. R. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl. Acad. Sci. USA. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford H. L., Silver D. L., Kachar B., Sellers J. R., Zain S. B. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–16327. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- Frank S. R., Hansen S. H. The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Semin. Cell. Dev. Biol. 2008;19:234–244. doi: 10.1016/j.semcdb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S. C., Hodgson L., Rybin A., Toutchkine A., Hahn K. M., Lawrence D. S., Bresnick A. R. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S. C., Varney K. M., Weber D. J., Bresnick A. R. S100A4, a mediator of metastasis. J. Biol. Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- Gismondi A., Bisogno L., Mainiero F., Palmieri G., Piccoli M., Frati L., Santoni A. Proline-rich tyrosine kinase-2 activation by beta 1 integrin fibronectin receptor cross-linking and association with paxillin in human natural killer cells. J. Immunol. 1997;159:4729–4736. [PubMed] [Google Scholar]
- Golomb E., Ma X., Jana S. S., Preston Y. A., Kawamoto S., Shoham N. G., Goldin E., Conti M. A., Sellers J. R., Adelstein R. S. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J. Biol. Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- Gongoll S., Peters G., Mengel M., Piso P., Klempnauer J., Kreipe H., von Wasielewski R. Prognostic significance of calcium-binding protein S100A4 in colorectal cancer. Gastroenterology. 2002;123:1478–1484. doi: 10.1053/gast.2002.36606. [DOI] [PubMed] [Google Scholar]
- Grigorian M., et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: functional consequences of their interaction. J. Biol. Chem. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- Grigorian M., Tulchinsky E., Burrone O., Tarabykina S., Georgiev G., Lukanidin E. Modulation of mts1 expression in mouse and human normal and tumor cells. Electrophoresis. 1994;15:463–468. doi: 10.1002/elps.1150150163. [DOI] [PubMed] [Google Scholar]
- Grum-Schwensen B., Klingelhofer J., Berg C. H., El-Naaman C., Grigorian M., Lukanidin E., Ambartsumian N. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- Hatch W. C., Ganju R. K., Hiregowdara D., Avraham S., Groopman J. E. The related adhesion focal tyrosine kinase (RAFTK) is tyrosine phosphorylated and participates in colony-stimulating factor-1/macrophage colony-stimulating factor signaling in monocyte-macrophages. Blood. 1998;91:3967–3973. [PubMed] [Google Scholar]
- Hernan R., Fasheh R., Calabrese C., Frank A. J., Maclean K. H., Allard D., Barraclough R., Gilbertson R. J. ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 2003;63:140–148. [PubMed] [Google Scholar]
- Hinz B., Alt W., Johnen C., Herzog V., Kaiser H. W. Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp. Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- Hiregowdara D., Avraham H., Fu Y., London R., Avraham S. Tyrosine phosphorylation of the related adhesion focal tyrosine kinase in megakaryocytes upon stem cell factor and phorbol myristate acetate stimulation and its association with paxillin. J. Biol. Chem. 1997;272:10804–10810. doi: 10.1074/jbc.272.16.10804. [DOI] [PubMed] [Google Scholar]
- Inoue T., Plieth D., Neilson E. G. Antibodies against macrophages that overlap in specificity with fibroblasts. Kidney Int. 2005a;68:2400. doi: 10.1111/j.1523-1755.2005.00358.x. author reply 2400–2401. [DOI] [PubMed] [Google Scholar]
- Inoue T., Plieth D., Venkov C. D., Xu C., Neilson E. G. Antibodies against macrophages that overlap in specificity with fibroblasts. Kidney Int. 2005b;67:2488–2493. doi: 10.1111/j.1523-1755.2005.00358.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson S. R., Barraclough R., West C. R., Rudland P. S. S100A4 regulates cell motility and invasion in an in vitro model for breast cancer metastasis. Br. J. Cancer. 2004;90:253–262. doi: 10.1038/sj.bjc.6601483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce J. A., Pollard J. W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Endo Y., Yonemura Y., Heizmann C. W., Schafer B. W., Watanabe Y., Sasaki T. Clinical significance of S100A4 and E-cadherin-related adhesion molecules in non-small cell lung cancer. Int. J. Oncol. 2000;16:1125–1131. doi: 10.3892/ijo.16.6.1125. [DOI] [PubMed] [Google Scholar]
- Kontgen F., Suss G., Stewart C., Steinmetz M., Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int. Immunol. 1993;5:957–964. doi: 10.1093/intimm/5.8.957. [DOI] [PubMed] [Google Scholar]
- Kovacs M., Toth J., Hetenyi C., Malnasi-Csizmadia A., Sellers J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Kriajevska M., Fischer-Larsen M., Moertz E., Vorm O., Tulchinsky E., Grigorian M., Ambartsumian N., Lukanidin E. Liprin beta 1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1) J. Biol. Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- Kriajevska M. V., Cardenas M. N., Grigorian M. S., Ambartsumian N. S., Georgiev G. P., Lukanidin E. M. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J. Biol. Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- Le Hir M., Kaissling B. Antibodies against macrophages that overlap in specificity with fibroblasts. Kidney Int. 2005;68:2400. doi: 10.1111/j.1523-1755.2005.00704_1.x. author reply 2400–2401. [DOI] [PubMed] [Google Scholar]
- Lee W. L., Cosio G., Ireton K., Grinstein S. Role of CrkII in Fcgamma receptor-mediated phagocytosis. J. Biol. Chem. 2007;282:11135–11143. doi: 10.1074/jbc.M700823200. [DOI] [PubMed] [Google Scholar]
- Li Z. H., Bresnick A. R. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- Li Z. H., Spektor A., Varlamova O., Bresnick A. R. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- Limouze J., Straight A. F., Mitchison T., Sellers J. R. Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- Lo C. M., Buxton D. B., Chua G. C., Dembo M., Adelstein R. S., Wang Y. L. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelandsmo G. M., Hovig E., Skrede M., Engebraaten O., Florenes V. A., Myklebost O., Grigorian M., Lukanidin E., Scanlon K. J., Fodstad O. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res. 1996;56:5490–5498. [PubMed] [Google Scholar]
- Malashkevich V. N., Varney K. M., Garrett S. C., Wilder P. T., Knight D., Charpentier T. H., Ramagopal U. A., Almo S. C., Weber D. J., Bresnick A. R. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenholz I., Heizmann C. W., Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem. Biophys. Res. Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- McNeill E., Conway S. J., Roderick H. L., Bootman M. D., Hogg N. Defective chemoattractant-induced calcium signalling in S100A9 null neutrophils. Cell Calcium. 2007;41:107–121. doi: 10.1016/j.ceca.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Yano H., Uchida H., Hashimoto S., Schaefer E., Sabe H. Tyrosine phosphorylation of paxillin alpha is involved in temporospatial regulation of paxillin-containing focal adhesion formation and F-actin organization in motile cells. J. Biol. Chem. 2000;275:27155–27164. doi: 10.1074/jbc.M000679200. [DOI] [PubMed] [Google Scholar]
- Niederman R., Pollard T. D. Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J. Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya I., et al. Increased expression of S100A4 and its prognostic significance in esophageal squamous cell carcinoma. Int. J. Oncol. 2001;18:715–720. doi: 10.3892/ijo.18.4.715. [DOI] [PubMed] [Google Scholar]
- Nishiya N., Kiosses W. B., Han J., Ginsberg M. H. An alpha4 integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat. Cell Biol. 2005;7:343–352. doi: 10.1038/ncb1234. [DOI] [PubMed] [Google Scholar]
- Okada H., Danoff T. M., Kalluri R., Neilson E. G. Early role of Fsp1 in epithelial-mesenchymal transformation. Am. J. Physiol. 1997;273:F563–F574. doi: 10.1152/ajprenal.1997.273.4.F563. [DOI] [PubMed] [Google Scholar]
- Okigaki M., Davis C., Falasca M., Harroch S., Felsenfeld D. P., Sheetz M. P., Schlessinger J. Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc. Natl. Acad. Sci. USA. 2003;100:10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera A. M., Schneider I. C., Rericha E., Schlaepfer D. D., Waterman C. M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixley F. J., Lee P. S., Condeelis J. S., Stanley E. R. Protein tyrosine phosphatase phi regulates paxillin tyrosine phosphorylation and mediates colony-stimulating factor 1-induced morphological changes in macrophages. Mol. Cell. Biol. 2001;21:1795–1809. doi: 10.1128/MCB.21.5.1795-1809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser B. L., Wright N. T., Hernandez-Ochoa E. O., Varney K. M., Liu Y., Olojo R. O., Zimmer D. B., Weber D. J., Schneider M. F. S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. J. Biol. Chem. 2008;283:5046–5057. doi: 10.1074/jbc.M709231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. S. ImageJ. Bethesda, MD: National Institutes of Health; 1997–2007. ImageJ. [Google Scholar]
- Rosty C., Ueki T., Argani P., Jansen M., Yeo C. J., Cameron J. L., Hruban R. H., Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am. J. Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudland P. S., Platt-Higgins A., Renshaw C., West C. R., Winstanley J. H., Robertson L., Barraclough R. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595–1603. [PubMed] [Google Scholar]
- Sakai H., Chen Y., Itokawa T., Yu K. P., Zhu M. L., Insogna K. Activated c-Fms recruits Vav and Rac during CSF-1-induced cytoskeletal remodeling and spreading in osteoclasts. Bone. 2006;39:1290–1301. doi: 10.1016/j.bone.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Saleem M., Adhami V. M., Ahmad N., Gupta S., Mukhtar H. Prognostic significance of metastasis-associated protein S100A4 (Mts1) in prostate cancer progression and chemoprevention regimens in an autochthonous mouse model. Clin. Cancer Res. 2005;11:147–153. [PubMed] [Google Scholar]
- Sanjay A., et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria-Kisiel L., Rintala-Dempsey A. C., Shaw G. S. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. C., Hays C. K., Waterman C. M. EGF-induced contraction regulates paxillin phosphorylation to temporally separate traction generation from de-adhesion. Mol. Biol. Cell. 2009 doi: 10.1091/mbc.E09-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Hansen J. L., Sheikh S. P. S100A4: a common mediator of epithelial-mesenchymal transition, fibrosis and regeneration in diseases? J. Mol. Med. 2008;86:507–522. doi: 10.1007/s00109-007-0301-3. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E. R. Murine bone marrow-derived macrophages. Methods Mol. Biol. 1997;75:301–304. doi: 10.1385/0-89603-441-0:301. [DOI] [PubMed] [Google Scholar]
- Stein U., et al. The metastasis-associated gene S100A4 is a novel target of beta-catenin/T-cell factor signaling in colon cancer. Gastroenterology. 2006;131:1486–1500. doi: 10.1053/j.gastro.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Strutz F., Okada H., Lo C. W., Danoff T., Carone R. L., Tomaszewski J. E., Neilson E. G. Identification and characterization of a fibroblast marker: FSP1. J. Cell Biol. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Mundel T. M., Kieran M. W., Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Endo H., Sakiyama S. Involvement of S100-related calcium-binding protein pEL98 (or mts1) in cell motility and tumor cell invasion. Jpn. J. Cancer Res. 1994a;85:831–839. doi: 10.1111/j.1349-7006.1994.tb02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Sakiyama S. Cellular localization of pEL98 protein, an S100-related calcium binding protein, in fibroblasts and its tissue distribution analyzed by monoclonal antibodies. Cell Struct. Funct. 1994b;19:133–141. doi: 10.1247/csf.19.133. [DOI] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Sakiyama S. Expression of a calcium binding protein pEL98 (mts1) during differentiation of human promyelocytic leukemia HL-60 cells. Biochem. Biophys. Res. Commun. 1994c;202:94–101. doi: 10.1006/bbrc.1994.1898. [DOI] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Sakiyama S. Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene. 1997;14:331–337. doi: 10.1038/sj.onc.1200820. [DOI] [PubMed] [Google Scholar]
- Takenaga K., Nakamura Y., Sakiyama S., Hasegawa Y., Sato K., Endo H. Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J. Cell Biol. 1994d;124:757–768. doi: 10.1083/jcb.124.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallely K. M., Rustandi R. R., Ellis K. C., Varlamova O., Bresnick A. R., Weber D. J. Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41:12670–12680. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]
- van Dieck J., Fernandez-Fernandez M. R., Veprintsev D. B., Fersht A. R. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J. Biol. Chem. 2009;284:13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedham V., Phee H., Coggeshall K. M. Vav activation and function as a rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol. Cell. Biol. 2005;25:4211–4220. doi: 10.1128/MCB.25.10.4211-4220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky A. B., Svitkina T. M., Borisy G. G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wang X., Chen M., Ouyang K., Song L. S., Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C. M., Walmsley M., Ooi S., Tybulewicz V., Ridley A. J. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 2004;117:1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- Wheeler A. P., Wells C. M., Smith S. D., Vega F. M., Henderson R. B., Tybulewicz V. L., Ridley A. J. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J. Cell Sci. 2006;119:2749–2757. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- Wu S. S., Jacamo R. O., Vong S. K., Rozengurt E. Differential regulation of Pyk2 phosphorylation at Tyr-402 and Tyr-580 in intestinal epithelial cells: roles of calcium, Src, Rho kinase, and the cytoskeleton. Cell Signal. 2006;18:1932–1940. doi: 10.1016/j.cellsig.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Xia W., Hilgenbrink A. R., Matteson E. L., Lockwood M. B., Cheng J. X., Low P. S. A functional folate receptor is induced during macrophage activation and can be used to target drugs to activated macrophages. Blood. 2009;113:438–446. doi: 10.1182/blood-2008-04-150789. [DOI] [PubMed] [Google Scholar]
- Xiong Z., O'Hanlon D., Becker L. E., Roder J., MacDonald J. F., Marks A. Enhanced calcium transients in glial cells in neonatal cerebellar cultures derived from S100B null mice. Exp. Cell Res. 2000;257:281–289. doi: 10.1006/excr.2000.4902. [DOI] [PubMed] [Google Scholar]
- Xue C., Plieth D., Venkov C., Xu C., Neilson E. G. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- Yamamoto S., et al. TRPM2-mediated Ca2+influx induces chemokine production in monocytes that aggravates inflammatory neutrophil infiltration. Nat. Med. 2008;14:738–747. doi: 10.1038/nm1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- Zeisberg E. M., Potenta S., Xie L., Zeisberg M., Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- Zimmer D. B., Cornwall E. H., Landar A., Song W. The S100 protein family: history, function, and expression. Brain Res. Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- Zou M., Al-Baradie R. S., Al-Hindi H., Farid N. R., Shi Y. S100A4 (Mts1) gene overexpression is associated with invasion and metastasis of papillary thyroid carcinoma. Br. J. Cancer. 2005;93:1277–1284. doi: 10.1038/sj.bjc.6602856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.