DYF-1 is a highly conserved protein. Our results demonstrate that DYF-1 is a canonical subunit of IFT particle complex B and strongly support the hypothesis that the IFT machinery has species- and tissue-specific variations with functional ramifications.
Abstract
DYF-1 is a highly conserved protein essential for ciliogenesis in several model organisms. In Caenorhabditis elegans, DYF-1 serves as an essential activator for an anterograde motor OSM-3 of intraflagellar transport (IFT), the ciliogenesis-required motility that mediates the transport of flagellar precursors and removal of turnover products. In zebrafish and Tetrahymena DYF-1 influences the cilia tubulin posttranslational modification and may have more ubiquitous function in ciliogenesis than OSM-3. Here we address how DYF-1 biochemically interacts with the IFT machinery by using the model organism Chlamydomonas reinhardtii, in which the anterograde IFT does not depend on OSM-3. Our results show that this protein is a stoichiometric component of the IFT particle complex B and interacts directly with complex B subunit IFT46. In concurrence with the established IFT protein nomenclature, DYF-1 is also named IFT70 after the apparent size of the protein. IFT70/CrDYF-1 is essential for the function of IFT in building the flagellum because the flagella of IFT70/CrDYF-1–depleted cells were greatly shortened. Together, these results demonstrate that IFT70/CrDYF-1 is a canonical subunit of IFT particle complex B and strongly support the hypothesis that the IFT machinery has species- and tissue-specific variations with functional ramifications.
INTRODUCTION
The assembly and maintenance of flagella and cilia depend on the microtubule-based transport system known as intraflagellar transport (IFT; Rosenbaum and Witman, 2002), a process characterized as a bidirectional movement of large protein particles between the flagellar base and tip (Kozminski et al., 1993). The IFT machinery includes three key components: the IFT particle, the anterograde motors, and the retrograde motor. IFT particles are composed of at least 18 polypeptides that are organized into complex A and complex B. These particles are also called IFT trains because they appear as repetitive arrays of variable numbers of the same unit (Pigino et al., 2009). In this report, these two terms are used interchangeably. IFT trains serve as adaptors to bridge the axonemal precursors required for flagellar assembly with the motors (Qin et al., 2004; Hou et al., 2007; Ahmed et al., 2008). In the anterograde direction from the flagellar base to the tip, IFT particles are transported by either the heterotrimetric kinesin-II motor alone or kinesin-II together with the homodimer OSM-3. In the retrograde direction, IFT is powered by the cytoplasmic dynein 1b. Presently, little is known concerning how the motor activity is choreographed with the directional movement of IFT particles or how IFT carries cargo. To gain insight into the regulation of IFT, we chose to understand the function of DYF-1, the only protein to have shown a clear role in regulating IFT by activating the motor OSM-3 in Caenorhabditis elegans (Ou et al., 2005).
In C. elegans, two sequential IFT pathways are essential for full assembly of the cilia. Kinesin-II and OSM-3 function collaboratively to assemble the proximal part of the cilium, whereas OSM-3 alone is responsible for building the remainder of the distal segment (Snow et al., 2004). The osm-3 mutant has shortened cilia missing the distal segment, and this partially shortened ciliary defect is also seen in the dyf-1 mutant. In the dyf-1 mutant, the OSM-3 kinesin is capable of entering the ciliary compartment, but cannot bind to the microtubule or move and thus is inactive. Therefore, DYF-1 was postulated to be an OSM-3 positive regulator, required for either mediating OSM-3 binding with the IFT particle or docking onto microtubules (Ou et al., 2005). The interaction of DYF-1 with the IFT particle is not mediated through OSM-3 because in the osm-3 mutant DYF-1 moves together with IFT particles along the remaining proximal part of the cilia (Ou et al., 2005). DYF-1 is predicted to be a complex B–associated protein because it moves together with IFT complex B in bbs7 and bbs8 mutants in which complex A and B move separately with different speeds (Ou et al., 2005, 2007).
Evidence obtained from zebrafish and Tetrahymena revealed that DYF-1 must have additional roles in ciliogenesis in addition to regulating the activity of OSM-3. The zebrafish DYF-1 homologue Fleer is required for systemic ciliogenesis (Pathak et al., 2007), whereas the OSM-3 homologue KIF17 is required for the formation of photoreceptor cilia but not pronephric cilia (Insinna and Besharse, 2008; Insinna et al., 2008). The shortened olfactory and pronephric cilia of zebrafish fleer mutant have an ultrastructural defect of the doublet microtubules in the axoneme. This defect likely results from a reduced level of glutamylated tubulin (Pathak et al., 2007), which is important in stabilizing the axonemes (Bobinnec et al., 1998; Redeker et al., 2005). These zebrafish results lead to two hypotheses for the role of Fleer in ciliogenesis. The first is that Fleer serves as a structural component of the ciliary axoneme, which is missing in the fleer mutant; the second is that Fleer functions as an IFT cargo adapter for a tubulin glutamic acid ligase, which is responsible for flagellar tubulin glutamylation (Pathak et al., 2007).
Consistent with the role of DYF-1 in ciliogenesis, a more recent study in Tetrahymena showed that cells lacking an orthologue of DYF-1, Dyf1p, fail to assemble axonemes or only assemble extremely short remnants that have diverse structural defects. The defects include the absence of a central pair and outer doublet microtubules, and incomplete or absent B tubules on the outer microtubules (Dave et al., 2009). However, in contrast to the results with the fleer zebrafish, the level of tubulin glutamylation was increased in the axonemal remnants of the DYF1p knockout Tetrahymena cells (Dave et al., 2009). Taken together, these studies indicate that although DYF-1 is a conserved and critical component required for the assembly of ciliary axoneme, the specific contributions of DYF-1 during ciliogenesis vary in these organisms.
The most prominent questions arising from previous studies are how DYF-1 biochemically interacts with the IFT machinery and whether DYF-1 functions independently of OSM-3 in ciliogenesis. In this study, we address the function of this protein in the model organism Chlamydomonas reinhardtii, which is well-suited for genetic and biochemical studies of flagella. In this organism, kinesin-II is believed to be the sole anterograde IFT motor (Kozminski et al., 1993, 1995; Cole et al., 1998). The C. reinhardtii dyf-1 homologue has been identified, and three peptides for DYF-1 protein were found in the flagellar proteomic analysis, suggesting it is a component of flagella (Pazour et al., 2005). We report here that this protein is an integral component of IFT particle complex B and that it is essential for flagella assembly in C. reinhardtii.
MATERIALS AND METHODS
Strains and Cultures
C. reinhardtii wild-type (wt) strain cc125, cell wall–deficient strain cw92, and the temperature-sensitive flagella assembly mutant fla10ts (fla10–1 allele, cc1919) were obtained from the Chlamydomonas center (http://www.chlamy.org). Cells were grown on Tris-acetate- phosphate (TAP) solid plates or in M1 liquid medium with constant aeration in a Conviron environmental cabinet (Asheville, NC) programmed at 18°C with a light-dark cycle of 14:10 h.
Phylogenetic Analysis
The sequences of IFT70/CrDYF-1 homologues were obtained from the National Center for Biotechnology Information databases. Gene accession numbers are listed in the legend to Figure 1 and in Table 1. The sequences were aligned with ClustalX 1.81 (UCD Conway Institute, University College Dublin, Dublin, Ireland). A neighbor-joining tree was calculated using the TreeView 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
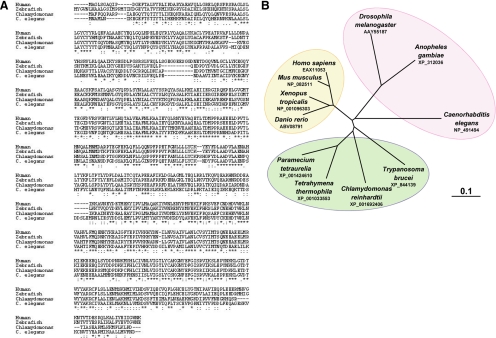
Figure 1.
IFT70 is a highly conserved protein. (A) Amino acid sequence alignment among the IFT70/DYF-1 othologues from invertebrate and vertebrate species including human (EAX11053), zebrafish (ABV08791), C. reinhardtii (XP_001692406), C. elegans (NP_491494), and Tetrahymena (XP_001033553). (B) The phylogenetic tree of IFT70/DYF-1 proteins. Branch lengths represent evolutionary relatedness.
Table 1.
IFT70/DYF-1 is the most conserved protein among IFT particle complex B subunits
| C. reinhardtii | T. brucei | C. elegans | D. melanogaster | Homo sapiens | |
|---|---|---|---|---|---|
| IFT70 | XP_001692406 | XP_844139 (75) | AAY55187 (63) | NP_491494 (65) | NP_689488 (73) |
| IFT46 | ABH06907 | XP_845431 (62) | AAL48848 (49) | NP_001076767 (66) | NP_064538 (72) |
| IFT52 | AAL12162 | XP_827974 (56) | NP_609045 (54) | NP_741633 (58) | NP_057088 (68) |
| IFT80 | ABQ96217 | XP_827975 (60) | NP_610064 (54) | NP_508106 (58) | NP_065851 (67) |
| IFT172 | XP_001691740 | XP_822375 (60) | NP_647700 (58) | NP_510681 (55) | NP_056477 (65) |
| IFT20 | AAM75748 | XP_845450 (58) | NP_724409 (51) | NP_740843 (55) | AAP50265 (64) |
| IFT88 | AAG37228 | XP_828263 (60) | ABG02143 (49) | NP_508511 (56) | NP_783195 (58) |
| IFT25 | ABU90455 | XP_829387 (47) | — | — | NP_057210 (57) |
| IFT27 | XP_001689745 | XP_844145 (51) | — | — | AAP36177 (57) |
| IFT57 | XP_001698648 | XP_823340 (52) | NP_608792 (49) | NP_492749 (53) | NP_060480 (57) |
| IFT72 | AAO92260 | XP_845960 (49) | — | NP_495359 (47) | NP_001092693 (49) |
| IFT81 | AAT99262 | XP_822517 (48) | — | NP_508900 (46) | NP_054774 (54) |
| IFT22 | XP_001689669 | XP_829740 (52) | — | NP_503073 (47) | NP_073614 (50) |
The percentages of positive conserved amino acids of IFT-B subunits between C. reinhardtii and other organisms are listed in parentheses after the GenBank accession numbers. —, there is no homologue of the IFT particle protein in the genome sequence of the organism.
Antibodies
Polyclonal anti-IFT70/CrDYF-1 antisera were raised against the IFT70/CrDYF-1 N-terminal amino acid residues 1–380. The cDNA fragment encoding the N-terminal amino acid residues 1–380 of IFT70/CrDYF-1 was cloned into the pMALc-2 expression vector (New England Biolabs, Beverly, MA) for generation of a maltose-binding protein (MBP)-tagged fusion protein. The subcutaneous injection of the fusion protein into the rabbits was performed by Bethyl Laboratories (Montgomery, TX). The collected antisera were affinity-purified using nitrocellulose-bound MBP-IFT70/CrDYF-1 fusion protein as bait.
This study also used antibodies against α-tubulin (clone B-5-1-2, ascites fluid; Sigma, St. Louis, MO), IFT57, IFT81, IFT139 (Cole et al., 1998), IFT74 (Qin et al., 2004), IFT46 (Hou et al., 2007), IFT27 (Qin et al., 2007), FLA10 (Cole et al., 1998), D1bLIC (Hou et al., 2004), and FMG-1 (Bloodgood et al., 1986).
Sucrose-Density Gradients
The method used for flagella isolation has been detailed elsewhere (Qin et al., 2004). The soluble flagellar proteins were fractionated through 12-ml 10–25% sucrose density gradients in HMDEK (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 25 mM KCl) in an SW41Ti rotor (Beckman Coulter, Fullerton, CA) for 14 h at 38,000 rpm. The gradients were typically fractionated into 24–26 0.5-ml aliquots. The standards used to calculate S values were bovine serum albumin (BSA; 4.4 S), aldolase (7.35 S), catalase (11.3 S), and thyroglobulin (19.4 S).
Immunoprecipitation
Before the immunoprecipitation experiments, protein A-Sepharose beads (GE Healthcare, Piscataway, NJ) were washed three times with HMDEK buffer plus 3% BSA. Flagellar soluble proteins (protein concentration approximates 3 mg/ml) were clarified by centrifugation at 100,000 × g for 10 min. The preparation was then incubated with antibodies for 1–2 h on ice. Immune complexes were recovered by incubation with pretreated protein A-Sepharose beads for 2–8 h at 4°C. After washing three times with 1 ml of HMDEK plus 0.05% NP-40 and then once with HMDEK plus 300 mM NaCl (each wash was for 10 min at room temperature), proteins were eluted from the resin by boiling in SDS-PAGE loading buffer and analyzed by SDS-PAGE, followed by immunoblotting.
Copurification of IFT70/CrDYF-1 and IFT46
IFT70/CrDYF-1 was expressed as a C-terminal fusion to MBP in pMAlc_2X (New England Biolabs). IFT46 was expressed with the N-terminal epitope Strep-II-Tag (W-S-H-P-Q-F-E-K; IBA Göttingen, Germany) in a vector derived from pRSFDuet-1 (Novagen, Madison, WI). The two expression plasmids were sequentially introduced into the Escherichia coli expression strain BL21(DE3) (Novagen). The dual-transformed strain grown in 50 ml of LB was induced at an OD600 of ∼ 0.4 with 1 mM IPTG and harvested after an additional 2.5 h of growth at 37°C. The cell pellet was suspended in amylose column buffer (ACB; 20 mM HEPES, 200 mM NaCl, pH 7.4) with protease inhibitors (10 μg/ml leupeptin, 0.1 μg/ml pepstatin-A, 1.7 μg/ml aprotinin, 5.0 μg/ml soybean trypsin inhibitor, 0.5 mM phenylmethylsulfonyl fluoride) and frozen. Frozen cell suspensions were thawed in ice-cold water and sonicated for 3 min on ice. Insoluble material was removed by centrifugation at 12,000 × g for 10 min at 4°C in a Sorvall SS34 rotor (Newton, CT). Clarified cell extract was applied to a 0.4-ml amylose column (New England Biolabs) equilibrated in ACB and subsequently washed with ACB. Bound protein was eluted into 0.4-ml aliquots using ACB + 10 mM maltose. Elution fractions 1 and 2 were pooled and applied to a 0.4-ml Streptactin (GE Healthcare) column equilibrated in ACB. After extensive washing with ACB, the bound proteins were eluted into 0.4-ml aliquots using 0.1M Tris-HCl, 0.15 M NaCl, and 2.5 mM desthiobiotin, pH 8.0. Aliquots (8 μl) from each step of the purification were resolved on 10% SDS-PAGE and visualized with Coomassie Blue staining.
Immunofluorescence Microscopy
Immunofluorescence staining of C. reinhardtii cells has been described in detail previously (Wang et al., 2009). After the staining, the cells were viewed with an Olympus IX-70 inverted fluorescence microscope (Melville, NY) at 100× magnification. The images were captured with an Image Point CCD camera (Photometrics, Woburn, MA) with an exposure time of 1–1.5 s and processed with the PCI software package.
Nucleic Acid Manipulations and Transformation
A cosilencing method (Rohr et al., 2004) was used to achieve the desired RNA interference (RNAi) knockdown of IFT70/CrDYF-1 expression. For the vector construction, two cDNA fragments including the forward partial IFT70/CrDYF-1 gene (1–650 base pairs) amplified by PCR using a set of primers 5′-GGAATTCTTCTTTCAGCAGCCCGCGAG-3′ and 5′-TCGAGGATCCGCCGTCCGTGTTGCTGCCCA-3′, and the inverted piece (1–450 base pairs) obtained by PCR using a set of primers 5′-CGACCATATGAATTCTTCTTTCAGCAGCCCGCGAG-3′ and 5′-TCGAGGATCCATGATGCAGCCGGTGTTGAC-3′ were inserted into the NE537 Maa7/X IR vector. The template used for PCR was the EST clone CL63d05 obtained from the Kazusa DNA Research Institute (Japan). The newly formed junctions in the resulting construct were confirmed by direct nucleotide sequencing.
The miRNA construct pchlamymiRNA3-INT-IFT70/CrDYF-1 was created as described previously (Molnar et al., 2009). The target sequence of IFT70/CrDYF-1 was scanned via http://wmd2.weigelworld/cgi-bin/mirnatools.pl for potential artificial miRNA target sites, and a 21-base pair sequence (ttagtagcaatatcctaggag) targeting the IFT70/CrDYF-1 3′-untranslated region (UTR) was chosen for further study. In detail, two 90-mer oligonucleotides (IFT70/CrDYF-1-ami-forward: CTAGTCTCCTAGGATATTGCTTCTAATCTCGCTGATCGGCACCATGGGGGTGGTGGTGATCAGCGCTATTAGTAGCAATATCCTAGGAGG and IFT70/CrDYF-1-ami-reverse: CTAGCCTCCTAGGATATTGCTACTAATAGCGCTGATCACCACCACCCCCATGGTGCCGATCAGCGAGATTAGAAGCAATATCCTAGGAGA, Invitrogen, Carlsbad, CA), containing the targeting sequence in opposite directions separated by a 42-base pair spacer sequence, were annealed in vitro and treated with T4 polynucleotide kinase (PNK, Fermentas, Hanover, MD) for phosphorylation. The miRNA vector pchlamymiRNA3int was digested with SpeI followed with CIAP (Fermentas) treatment for dephosphorylation. Thereafter, the 90-base pair DNA oligo was digested with SpeI and inserted into the pchlmaymiRNA3int vector. The correct miRNA construct was confirmed by direct nucleotide sequencing.
Transformation of C. reinhardtii cells with DNA was performed with glass beads as described previously (Kindle, 1990). Before the transformations, the cell wall of the wt cc125 cells was removed by autolysin treatment. The clones harboring the transgene IFT70/CrDYF-1 RNAi or miRNA constructs were selected in accordance with the previously described method (Rohr et al., 2004; Molnar et al., 2009).
Measurement of the Flagellar Length
Cells were fixed with 1% polyglutaraldehyde, mounted to slides, and viewed with an Olympus IX-70 inverted fluorescence microscope at 100× magnification. The phase-contrast images of the cells were captured with an Image Point CCD camera (Photometrics), and the flagellar length was measured with the software ImageJ 1.42 (http://rsb.info.nih.gov/ij/). The histogram showing the percentile distributions of flagellar length was created with Prism 5 (GraphPad Software, La Jolla, CA).
Transmission Electron Microscopy
IFT70/CrDYF-1 knockdown cells (strain miRNA-4) grown in TAP growth medium were fixed for 20 min at 20°C in TAP containing 2.5% glutaraldehyde and 1% formaldehyde, pH 7.2, followed by a fixation in 100 mM Na-cacodylate containing 2.5% glutaraldehyde and 1% formaldehyde, pH 7.2, at 4°C over night. The cells were rinsed three times in 100 mM Na-cacodylate, pH 7.2, and osmicated in 1% OsO4 in distilled water for 1 h at 4°C. The cells were washed three times in distilled water, embedded in 1% agar, stained en bloc with 1% aqueous uranyl acetate, rinsed three times in distilled water, and then dehydrated and embedded in Epon 812 (Serva, Heidelberg, Germany) as described by McFadden and Melkonian (1986). Ultrathin sections (∼50–60 nm) were cut with a diamond knife (type ultra 45°; Diatome, Biel, Switzerland) on an EM UC6 ultramicrotome (Leica, Wetzlar, Germany) and mounted on single-slot Pioloform-coated copper grids (Plano, Wetzlar, Germany). The sections were stained with uranyl acetate and lead citrate (Reynolds, 1963) and viewed with a JEM-2100 transmission electron microscope (JEOL, Tokyo, Japan) operated at 80 kV. Micrographs were taken using a 4000 × 4000 charge-coupled device camera (UltraScan 4000; Gatan, Pleasanton, CA) and Gatan Digital Micrograph software (version 1.70.16.).
RESULTS
DYF-1 Is the Most Conserved Protein among All the IFT Particle Subunits
By homologous sequence blast, a C. reinhardtii DYF-1 homologue was identified in the Joint Genome Institute (JGI) database and named as IFT70/CrDYF-1 (Figure 1A, also see Figure 3 for why the protein is named as IFT70). IFT70/CrDYF-1 is encoded by a single gene 128801 (JGI version 3, http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). Similar to other DYF-1 homologues, IFT70/CrDYF-1 contains three tetratricopeptide repeat (TPR) domains and a putative prenyltransferase domain (not shown). Phylogenetic analysis showed that IFT70/CrDYF-1 is more closely related to homologues of other ciliated protists than those of worms, insects, and vertebrates (Figure 1). Further sequence analysis revealed that DYF-1 has the highest percentage of conserved amino acids among all the identified IFT particle complex B subunits across the species (Table 1). Seventy-three percent of the amino acids of DYF-1 are conserved from the green alga C. reinhardtii to humans.
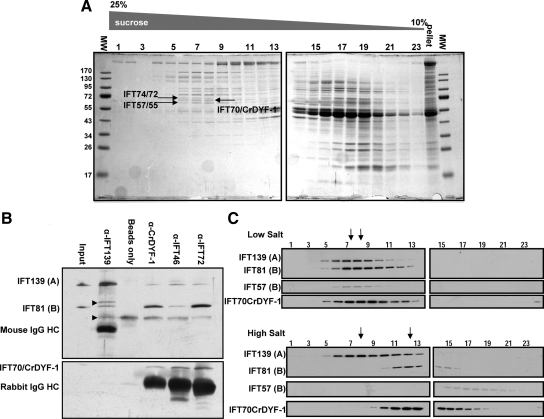
Figure 3.
IFT70/CrDYF-1 is a core component of the IFT particle complex B. (A) IFT70/CrDYF-1 comigrates with other IFT particle subunits at 16S. Flagellar matrix was fractionated through a 12-ml 10–25% sucrose density gradient. The gradient fractions were separated on 10% SDS-PAGE gels and stained with Coomassie Blue. IFT70/CrDYF-1 migrates between IFT72 and IFT57. The lane labeled “pellet” is collected from the bottom of the gradient. (B) IFT70/CrDYF-1 coimmunoprecipitates with other IFT particle complex B proteins. Immunoprecipitates with antibodies against IFT proteins from the flagellar membrane plus matrix were separated on 8% polyacrylamide gels and analyzed by Western blotting. The antibodies used for immunoprecipitation are listed above the Western blots. The antibodies used for Western blotting are indicated on the left. Nonspecific bands are indicated by arrowheads on the left. The band just above IFT81 may come from α-IFT139 antibody, because no such band exists in the starting membrane plus matrix material. The band just below IFT81 apparently comes from protein A beads, as it is present in the beads alone control. (C) Flagellar matrix was treated with or without high salt as described previously (Lucker et al., 2005) and fractionated through a 12-ml 10–25% sucrose density gradient. The sucrose density gradient fractions were separated by 10% SDS-PAGE and analyzed by Western blotting. The arrows mark the peaks of complexes A (left) and B (right).
IFT70/CrDYF-1 Has a Typical IFT Distribution Pattern and Its Flagellar Localization Is FLA10 Kinesin-II–dependent
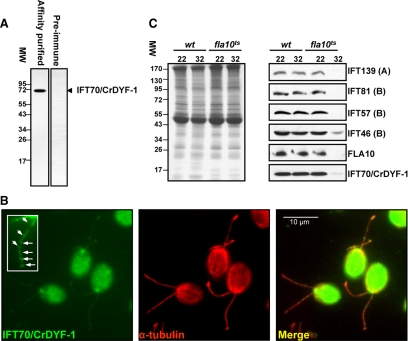
To confirm the flagellar localization of IFT70/CrDYF-1, a polyclonal antibody was raised against a recombinant MBP-IFT70/CrDYF-1 (1–380) and affinity-purified (see Materials and Methods). Immunoblot analysis of the flagellar proteins prepared from wt cc125 cells revealed that the antibody α-IFT70/CrDYF-1 specifically recognized a single protein band migrating at Mr ∼70,000 (Figure 2A). Immunofluorescent microscopy analysis with this affinity-purified antibody showed that IFT70/CrDYF-1 is concentrated at peri-basal body regions and is localized along the entire length of the flagellum as punctuated dots (Figure 2B), a cellular distribution pattern typical for IFT particle proteins (Cole et al., 1998; Deane et al., 2001).
Figure 2.
IFT70/CrDYF-1 has a typical localization pattern for IFT proteins, and its entrance into flagella is FLA10 dependent. (A) The wild-type (wt) flagella extract was probed with the affinity-purified α-IFT70/CrDYF-1 antibody with a single band detected. In contrast, preimmune serum does not recognize any specific band. (B) IFT70/CrDYF-1 is localized in the peri-basal body region and flagella. The wt cells were double-labeled with antibodies α-IFT70/CrDYF-1 (green) and α-tubulin (red). The staining with α-tubulin illustrates the position of the two flagella. IFT70/CrDYF-1 is localized primarily in the peri-basal body region as well as in dots along the flagella. The inset shows an enlargement of one of the flagella. (C) The entrance of IFT70/CrDYF-1 into the flagella is FLA10-dependent. Flagellar proteins were extracted from the wt and fla10ts cells after incubation for 50 min at either the permissive temperature (22°C) or the restrictive temperature (32°C), separated on an 8% polyacrylamide gel, transferred to nitrocellulose, and probed with antibodies against IFT70/CrDYF-1 and other IFT complex proteins, as indicated on the right of the Western blots. Equal amount of flagellar proteins were loaded for each sample, as shown by the Coomassie Blue–staining gel in the left panel. The labels A and B represent IFT complexes A and B, respectively.
The entrance of the IFT particle proteins into the flagella of C. reinhardtii is dependent on heterotrimeric FLA10 kinesin (Kozminski et al., 1995), the sole anterograde IFT motor (Huang et al., 1977; Walther et al., 1994; Kozminski et al., 1995; Cole et al., 1998). The temperature-sensitive mutant fla10ts harbors a mutation in the fla10 gene that encodes a motor subunit of FLA10 kinesin-II (Cole et al., 1998). The fla10ts cells have normal IFT at permissive temperature (22°C), whereas the anterograde IFT movement ceases within 1–2 h after the cells are shifted to the restrictive temperature (32°C; Kozminski et al., 1995; Iomini et al., 2001). Once anterograde IFT ceases, the IFT particle proteins are soon depleted from the fla10ts flagella, and thereafter the flagella gradually shorten. We found that, similarly to the known IFT particle proteins, IFT70/CrDYF-1 was dramatically reduced in the flagella of the fla10ts cells within 50 min after the cells were shifted to the restrictive temperature (Figure 2C). At the 50-min time point, flagella shortening was not observed, which was consistent with the previous observation that the disappearance of IFT precedes the shortening of flagella (Iomini et al., 2001). This result confirmed that the flagellar localization of IFT70/CrDYF-1 is FLA10 kinesin-II–dependent.
IFT70/CrDYF-1 Is a Subunit of the IFT Complex B Core
To determine whether IFT70/CrDYF-1 is an IFT particle subunit, sucrose density gradient analysis was utilized to determine its sedimentation pattern. The results clearly showed that IFT70/CrDYF-1 peaked in the 16S fractions with other IFT particle subunits (Figure 3, A and C). Based on the results of Coomassie Blue staining and Western blotting, the antibody α-IFT70/CrDYF-1 recognized a band between IFT74/72 and IFT57/55. After the bands corresponding to IFT74/72, IFT57/55 and IFT70/CrDYF-1 were cut off from the Coomassie Blue–stained gel, proteolytically digested and subjected to mass spectrometry, the identities of these proteins were confirmed. Compared with the staining intensity of IFT74/72 and IFT57 on the Coomassie Blue–stained gel (Figure 3A), IFT70/CrDYF-1 was present at a molar ratio of ∼1 relative to other IFT particle subunits, as expected for a bona fide IFT particle subunit (Cole et al., 1998). In concurrence with the IFT particle protein nomenclature, we named the C. reinhardtii DYF-1 homologue as IFT70/CrDYF-1 after the apparent size of the purified protein from isolated flagella. For clarity, in the rest of the report, IFT70 is used to represents orthologues in different organisms and IFT70/CrDYF-1 is used specifically for the C. reinhardtii protein.
Coimmunoprecipitation of flagellar soluble proteins was performed to determine if IFT70/CrDYF-1 belonged to IFT complex A or B. The immunoprecipitation assay with α-IFT70/CrDYF-1 showed that IFT70/CrDYF-1 and IFT81 (IFT complex B subunit), but not IFT139 (IFT complex A subunit), were enriched in the precipitates (Figure 3B), suggesting that IFT70/CrDYF-1 is an IFT complex B protein. Similar results were also previously observed when FLAG-tagged DYF-1 protein was immunoprecipitated from mouse IMCD3 cell extracts (Follit et al., 2009). Additionally, IFT70/CrDYF-1 was enriched in the precipitates when the assay was performed with α-IFT46 and α-IFT72 antibodies, both of which are against complex B subunits. In contrast, α-IFT139 (against the complex A protein IFT139) antibody which effectively precipitated complex A proteins (Cole et al., 1998; Qin et al., 2004) was unable to precipitate IFT70/CrDYF-1 (Figure 3B). Together, these data support that IFT70/CrDYF-1 is associated more strongly with IFT complex B than complex A.
At low ionic strength, both IFT complexes A and B sediment at approximately 16S on sucrose density gradient (Piperno and Mead, 1997; Cole et al., 1998). At higher ionic conditions, complex A remains intact, whereas complex B dissociates into an 11S core and a few free subunits (Lucker et al., 2005). Applying a similar analysis, we found that IFT70/CrDYF-1 cosedimented with IFT complex B proteins IFT81 and IFT57 on the low-salt sucrose density gradient and comigrated with IFT81 at 11S on the high-salt sucrose density gradient (Figure 3C), thus confirming that IFT70/CrDYF-1 is one of the core subunits of the IFT complex B.
IFT70/CrDYF-1 Binds Directly to IFT46
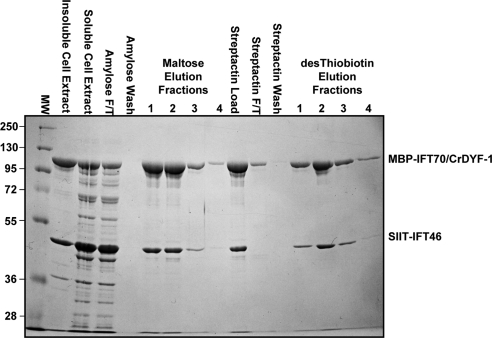
Until 2006, all the identified IFT complex B mutants in C. elegans shared a characteristic and distinct ciliary morphology: the ciliary axoneme of amphid cilia are highly stunted (Perkins et al., 1986; Haycraft et al., 2001, 2003; Qin et al., 2001). However the complex B mutant ift46 (Bell et al., 2006) was shown to have a different ciliary morphology from any of the previous identified complex B mutant. Similar to dyf-1, the mutant ift46 can assemble the middle segment, but fail to form the distal segment of the cilia. These observations prompted us to test if IFT70/CrDYF-1 and IFT46 interact directly. We therefore initiated a heterologous bacterial expression system (see Materials and Methods), which allowed coexpression of two proteins in a single host bacterium. Full-length MBP tagged IFT70/CrDYF-1 and Strep-II-Tag IFT46 were expressed simultaneously and then purified by tandem affinity chromatography. IFT70/CrDYF-1 and IFT46 were copurified with a 1:1 stoichiometric ratio (Figure 4), demonstrating that these two subunits interact directly. This result also strongly suggests that these two subunits should be capable forming a heterodimer in vivo.
Figure 4.
Coexpression and tandem purification of recombinant MBP-IFT70/CrDYF-1 and SIIT-IFT46. Shown here is the Coomassie Blue stained gel of samples from each step of the tandem purification; the first two lanes following the markers contain the insoluble and soluble fractions of bacterial cell lysates. Note that both proteins copurify after tandem purification with amylose and StrepTactin affinity resin. F/T stands for the flow-through proteins that did not bind to the resistive resins.
IFT Particle Proteins Are Partially Coisolated with the Axoneme
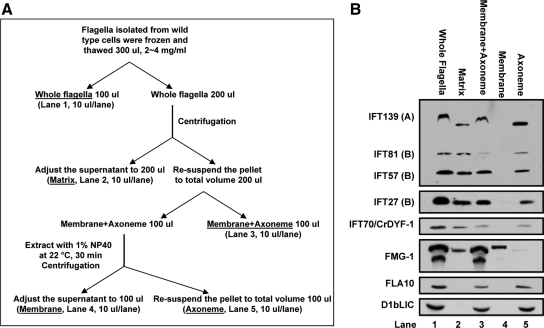
The zebrafish dyf-1/fleer mutant is missing a structural component of the axoneme, which leads to the hypothesis that DYF-1/Fleer is an integral axonemal protein (Pathak et al., 2007). Furthermore, Chlamydomonas flagellar proteomics analysis identified three IFT70/CrDYF-1–specific micropeptides with two in the detergent-soluble fraction and one in the axonemal fraction (Pazour et al., 2005). To investigate whether IFT70/CrDYF-1 is associated with the axoneme, the flagella isolated from wt cells was fractionated into the soluble flagellar matrix fraction and the insoluble membrane plus axoneme fraction by the freeze-thaw method (Figure 5A). Immunoblot assay showed that the majority of IFT70/CrDYF-1 protein was detected in the soluble matrix fraction; however, a small amount of IFT70/CrDYF-1 was also found in the insoluble membrane plus axoneme fraction (Figure 5B). To determine if this pool of IFT70/CrDYF-1 was associated with membrane or axoneme, the membrane was further separated from the axoneme fraction by applying the nonionic detergent NP-40 to the insoluble membrane plus axoneme fraction (Figure 5A). After this treatment, the flagellar transmembrane protein FMG-1 stayed in the membrane fraction, whereas the anterograde and retrograde motor proteins FLA10 and cytoplasmic dynein D1bLIC were exclusively detected in the axoneme fraction (Figure 5B). Therefore, the application of NP-40 had effectively stripped the membrane away from the axoneme, and this treatment did not dissolve IFT motor proteins. IFT70/CrDYF-1 together with other components of the IFT particle, including IFT139, IFT81, IFT57, and IFT27, were detected in both the flagellar matrix and the axonemal fractions but not the membrane fraction. The association of the IFT complex proteins with the axoneme was probably bridged through the IFT motors FLA10 kinesin-II and/or cytoplasmic dynein. The detailed mechanism, however, remains unknown.
Figure 5.
IFT70/CrDYF-1 is partially associated with the axoneme. (A) Procedure for the preparation of the flagellar fractions. This procedure is essentially the same as described in Huang et al. (2007). (B) Stoichiometrically equivalent levels of whole flagella, matrix, membrane plus axoneme, membrane proteins, and bare axoneme (labeled on the top) were isolated according to the procedure described in A. Note that, like other IFT particle proteins, a portion of IFT70/CrDYF-1 remains associated with the axoneme. The samples were separated by 10% SDS-PAGE and analyzed by Western blotting.
IFT70/CrDYF-1 Is Essential for Flagella Assembly
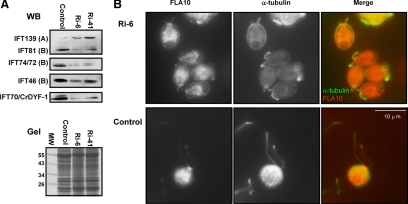
To investigate the role of IFT70/CrDYF-1 in flagellar assembly, vector-based RNAi was performed in wt cells. A total of 70 transformed colonies were obtained in two independent experiments. Among the 70 transformants, nine showed detectable reduction of the cellular IFT70/CrDYF-1 level as determined by immunoblot assay performed on the whole cell extracts (data not shown). For further phenotype analysis, we focused on two knockdown strains, Ri-6 and Ri-41, which both showed different levels of reduced IFT70/CrDYF-1. Although IFT70/CrDYF-1 was reduced, there were no obvious flagellar defects in Ri-41 cells, indicating that the reduction of IFT70 was too small to affect ciliogenesis. Strain Ri-6 had a much lower IFT70/CrDYF-1 expression level than strain Ri-41 (Figure 6A). Corresponding to the dramatic reduction of IFT70/CrDYF-1, most of the Ri-6 cells had extremely short flagella, indicated by α-tubulin staining (Figure 6B) and phase-contrast microscopy (data not shown). It was determined that the majority of the cells have a flagella length of ∼3–4 μm, although cells with full-length flagella (2∼5%) and flagella-less (bald) cells (<10%) also could be observed in the population. Immunofluorescence microscopy assay was performed on Ri-6 cells with α-IFT72 and α-FLA10 antibodies, which showed that both IFT72 (data not shown) and FLA10 were localized to the basal body region and flagella (Figure 6B). Therefore, the IFT70/CrDYF-1 depletion had no effect on the cellular localization of other IFT proteins or FLA10.
Figure 6.
The IFT70/CrDYF-1 knockdown cells have reduced levels of IFT complex B proteins and shortened flagella. (A) Immunoblots (top panels, WB) of the whole cell extracts isolated from the knockdown cells Ri-6 and Ri-41 and control (wild-type) cells. Letters A and B represent the IFT particle complexes A and B, respectively. The bottom panel is part of a Coomassie Blue stained gel (Gel) to show the equal loading of all the samples. (B) Dual-staining of Ri-6 and control cells with antibodies against α-tubulin (red) and FLA10 (green). Ri-6 cells have much shorter flagella than the control cells. The FLA10 localization pattern in Ri-6 cells appears normal.
To confirm that the short flagella were caused by the lack of IFT70/CrDYF-1 rather than accidental insertional mutagenesis or off-targeting of the RNAi vector, artificial miRNA that targeted the IFT70/CrDYF-1 3′-UTR region was used to knock down the expression of IFT70/CrDYF-1. The construct was transformed into the cell wall–deficient but otherwise normal cw92 cells and a total of 40 transformants were obtained in two independent experiments. Determined by immunoblot assays, two transformed strains, miRNA-1 and miRNA-4, showed reduced cellular levels of IFT70/CrDYF-1 (Figure 7A).
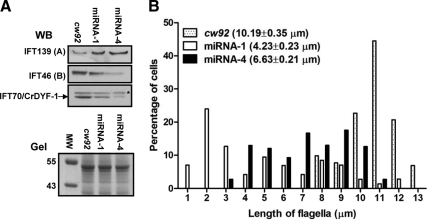
Figure 7.
IFT70/CrDYF-1 is required for flagellar assembly. (A) Levels of cellular IFT particle complex A proteins are elevated while Complex B proteins are reduced when IFT70/CrDYF-1 is reduced. The upper panels show the Western-blotting (WB) results of a few IFT particle proteins of whole cell extracts isolated from the knockdown strains miRNA-1 and miRNA-4, and the control cw92 cells. The IFT70/CrDYF-1 protein is indicated by an arrow on the left. The α-IFT70/CrDYF-1 antibody also recognized a non-specific band indicated by an asterisk (*) on the right. The lower panel is part of a Coomassie Blue-staining gel (Gel) to show the equal loading of all the samples. (B) IFT70/CrDYF-1 reduced miRNA cells assemble short flagella. The plot shows flagellar length distribution of cw92 (n = 116), miRNA-1 (n = 116), and miRNA-4 (n = 108) cells. The mean lengths of the flagella are listed.
Examination by phase-contrast microscopy showed that miRNA-1 and miRNA-4 cells had shorter flagella than the control cw92 cells. miRNA-1 and miRNA-4 cells showed a average flagella length of 6.63 and 4.23 μm, respectively, whereas cw92 cells had a average flagella length of 10.19 μm (Figure 7B). These results confirmed that the depletion of IFT70/CrDYF-1 resulted in the formation of short flagella.
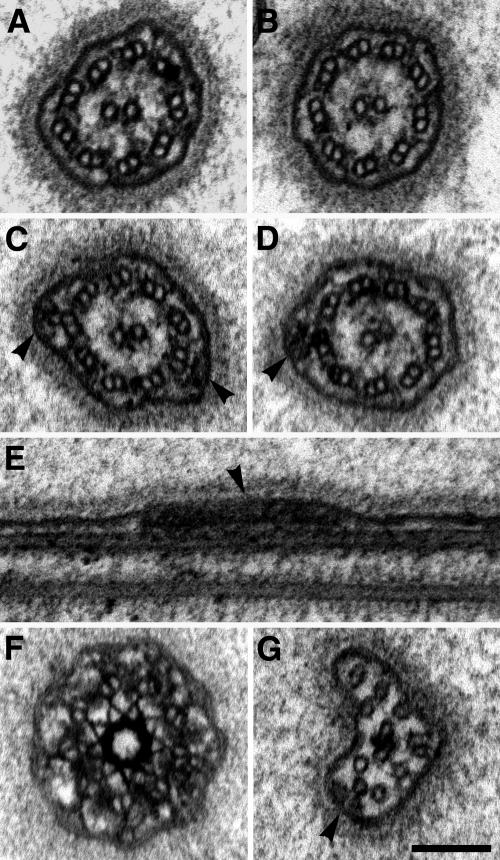
Ultrastructural analysis by transmission electron microscopy (EM) did not reveal any abnormal structure in the basal bodies and flagella of miRNA-4 cells (Figure 8). The doublet axonemal microtubules, along with the attached dynein arms, and radial spokes appeared normal. The structure of the very distal tip of the flagellum, which contains only A subfibers also appeared to be normal (Figure 8G). In addition, the IFT-trains in the miRNA-4 flagella were observable along the axoneme in both cross and longitudinal sections, and at the distal tip (Figure 8, B–G). Taken together, these data strongly support that the remaining IFT particle complex B in the miRNA-4 cells were still functional, but insufficient to assemble full-length flagella.
Figure 8.
The flagella of IFT70/CrDYF-1 knockdown cells display a normal ultrastructure. (A) Cross section of a flagellum of a C. reinhardtii cw92 cell showing the 9 + 2 microtubule architecture of the axoneme. (B–G) Electron micrographs of flagella of IFT70/CrDYF-1 knockdown cells (strain miRNA-4). (B–D) Cross sections of flagella. IFT-trains (arrowheads) attached to the B-subfiber of outer-doublets are visible. (E) Longitudinal section of a flagellum with a visible IFT-train (arrowhead). (F) Cross section through a transition zone showing a normal ultrastructure. (G) Cross section through the distal tip of a flagellum. An IFT-train is visible (arrowhead). Scale bar, (A–G), 100 nm.
Partial Depletion of IFT70/CrDYF-1 Causes the Reduced Cellular Levels of the IFT Complex B Proteins
As previously reported, complex B mutants ift46 (Hou et al., 2007) and bld1/ift52 (Qin et al., 2007) have elevated cellular levels of the complex A proteins but substantially reduced levels of the complex B subunits. Therefore, some complex B subunits are critical in maintaining the stability of the B complex. Immunoblot assays of whole cell extracts showed that the complex A protein IFT139 was increased, whereas complex B proteins were dramatically decreased in all the examined IFT70/CrDYF-1 knockdown cell lines including Ri-6, Ri-41 (Figure 6A), miRNA-1 and miRNA-4 (Figure 7A). This result is consistent with IFT70/CrDYF-1 serving as an integral component of the B complex and its loss results in the destabilization of the other B subunits in C. reinhardtii.
DISCUSSION
Through rigorous biochemical analysis, this study firmly establishes that IFT70/CrDYF-1, the DYF-1 homologue in C. reinhardtii, is an essential, stoichiometric component of IFT complex B. Previously, based on the motility profile of green fluorescent protein (GFP)-tagged proteins in wt and bbs mutants in C. elegans, DYF-1 was predicted to be a complex B–associated protein (Ou et al., 2007). BBS proteins are important in maintaining an intact IFT particle (Blacque et al., 2004) because in bbs-7 and bbs-8 mutants complex A and B are moved separately at different speeds by kinesin-II and OSM-3, respectively (Ou et al., 2005, 2007). In these mutants, DYF-1 moves at the same velocities as complex B, but not complex A; thus DYF-1 must be a protein associated with complex B, not complex A. The current study confirms this prediction and further shows unequivocally that IFT70/CrDYF-1 is a core component of complex B.
The addition of IFT70 as an integral IFT complex B subunit also indicates that the current inventory of IFT particle subunits is unlikely to be complete. The identification of IFT particle proteins has relied primarily on biochemical purifications (Piperno and Mead, 1997; Cole et al., 1998). Although powerful, these approaches have their own imperfections. In C. reinhardtii for example, putative IFT particle proteins have to be visible on the stained gels (Cole et al., 1998). We believe that IFT 70 CrDYF-1 evaded early detection because it was disguised by other proteins that co-migrated on early protein gels. Since the electrophonetic mobility of the IFT proteins can be affected substantially by the various reagents used in both the SDS-PAGE gel and the buffer, such as SDS (Wang et al., 2009), we changed conditions to maximize separation of IFT 70/CrDYF-1 from other proteins such as IFT 74/72 (Figure 3A). In addition, we also show that yield of individual IFT complex B subunits extracted from flagella varies (Figure 5B), indicating that the strength of which different IFT complex B subunits interact with the axoneme is variable. It is thus possible that IFT70/CrDYF-1 was not extracted effectively from the flagella in the previous purifications.
Accumulating evidence in recent years clearly demonstrates that IFT directly transports flagellar precursors to balance the continuous turnover at the flagellar tip (Qin et al., 2004; Hou et al., 2007; Ahmed et al., 2008). Because no ultrastructural defects were identified, the inability to assemble full-length flagella likely results from insufficient amount of IFT particles in IFT70/CrDYF-1 knockdown cells. However, neither the RNAi nor miRNA methods could completely deplete IFT70/CrDYF-1, thus we were unable to address if IFT70/CrDYF-1 is important for a particular flagellar structure or carries a specific precursor. On the other hand, interfering with precursor transport alone may not solely account for the shorter flagella phenotype in IFT70/CrDYF-1–depleted cells. The fact that IFT70, an IFT particle subunit, functions specifically in regulating OSM-3 activity in C. elegans (Ou et al., 2005) and affects tubulin polyglutamylation in both zebrafish (Pathak et al., 2007) and Tetrahymena (Dave et al., 2009), raises a tantalizing possibility that the role of this IFT particle protein in ciliogenesis could be multifaceted. Almost certainly, IFT particle proteins are used as more than just a scaffold to bridge flagellar precursors to the motors. They might facilitate transport of other nonaxonemal structural proteins, such as tubulin polyglutamylase, to indirectly impact flagellar assembly or stability.
Clearly, the IFT machinery likely has species- and tissue-specific variations with functional ramifications. In C. reinhardtii (this report), zebrafish (Pathak et al., 2007), and Tetrahymena (Dave et al., 2009), IFT70/CrDYF-1 is essential for maintaining the entire axoneme structure of cilia and flagella. In zebrafish, Fleer/DYF-1 is an essential regulator of cilia tubulin polyglutamylation, which is important in stabilizing the axonemes. The cause for the dyf-1 mutant of Tetrahymena failing to assemble axonemes or only assemble extremely short remnants is still unresolved. In the axoneme remnants of the DYF1p knockout Tetrahymena, in contrast to the results with the fleer zebrafish, the level of tubulin glutamylation was increased (Dave et al., 2009). However, further investigation is needed to address whether the increased level of tubulin glutamylation is the cause or the consequence of the absence of DYF-1 protein. In C. reinhardtii IFT70/CrDYF-1 appears to be involved in the stability of IFT complex B, as the cellular levels of IFT complex B proteins were reduced proportionally to that of IFT70/CrDYF-1 in the IFT70/CrDYF-1 knockdown cells (Figures 6 and 7). This result is consistent with IFT70/CrDYF-1 being an IFT particle complex B subunit. Therefore, in this study, results from both biochemical purifications and functional analysis all point to an unambiguous conclusion: IFT70/CrDYF-1 is a canonical subunit of IFT particle complex B.
In contrast, DYF-1 is essential only for assembly of the distal segment of sensory cilia in C. elegans (Ou et al., 2005). In this organism, DYF-1 clearly has unique functions that are not possessed by many complex B proteins. Mutations affecting existing known proteins that function as part of IFT particle complex A or B in C. elegans display characteristic and distinct morphologies. The ciliary axoneme of most complex B mutants is much shorter compared with that of complex A mutants (Perkins et al., 1986; Haycraft et al., 2001, 2003; Qin et al., 2001). Mutant dyf-1 nematodes, however, possess an intact ciliary middle segment, which is different from the complete loss of cilia observed in most complex B mutants. In addition, many complex B proteins are essential for the stability or intraflagellar transport of complex B (Haycraft et al., 2003; Hou et al., 2007; Qin et al., 2007). This is not the case in the dyf-1 mutant in which IFT movement persists along the remaining middle segment (Ou et al., 2005); therefore, the DYF-1 protein is not essential for nematode IFT complex B formation and function. Furthermore, DYF-1 serves as an essential OSM-3 regulator (Ou et al., 2005), a function that has not been revealed for any complex B subunit yet. Based on these observations, it appears that in C. elegans DYF-1 is no longer an essential component of complex B, but has gained specialized roles to sustain the species specific ciliary structures.
It is well established that IFT particles serve as scaffolds to bridge flagellar precursors to IFT motors (Qin et al., 2004; Hou et al., 2007; Ahmed et al., 2008). However, subunits within IFT particles clearly function beyond that spectrum. At least one IFT particle complex B protein, DYF-1, regulates the activity of the IFT motor OSM-3 (Ou et al., 2005). Moreover, recent studies in C. elegans showed that similar to dyf-1 mutant, the cilia of complex B mutants ift46 (Bell et al., 2006), ift81 and ift72 (Kobayashi et al., 2007) assemble the middle segment, but fail to form the complete distal segment. Furthermore, here we showed that IFT70/CrDYF-1 directly interacts with IFT46 (Figure 4); thus, these two subunits may function cooperatively to regulate the activity of OSM-3. On the basis of these observations, we speculate that the activity of OSM-3, and possibly all of the IFT motors, is subject to regulation by the subunits within the IFT complexes. Presently, little is known about how IFT motor activity is activated and deactivated. Future efforts in understanding the role of IFT particle proteins in regulating the activity of IFT motors will provide insight into this important problem.
ACKNOWLEDGMENTS
We deeply appreciate Dr. Duan Liu (Texas A&M University) for his technical assistance, Dr. George Witman (University of Massachusetts Medical School) for generously sharing antibodies, the Kazusa DNA Research Institute, Japan, for providing the EST clones, and Chlamydomonas Genetics Center for sending us strains. We thank the Laboratory of Biological Mass Spectrometry at TAMU for protein sequencing. We also thank other members of the Qin lab, Dr. Dennis Diener (Yale University), and Drs. Sachs Matthew, and Brian Perkins (Texas A&M University) for careful reading of the manuscript. This work was supported by Polycystic Kidney Disease Foundation Grant 173G08a and National Science Foundation Grant MCB-0923835 to H.Q and a grant from the Natioanl Institutes of Health, GM 61920 to D.G.C.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0191) on June 9, 2010.
REFERENCES
- Ahmed N. T., Gao C., Lucker B. F., Cole D. G., Mitchell D. R. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J. Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L. R., Stone S., Yochem J., Shaw J. E., Herman R. K. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood R. A., Woodward M. P., Salomonsky N. L. Redistribution and shedding of flagellar membrane glycoproteins visualized using an anti-carbohydrate monoclonal antibody and concanavalin A. J. Cell Biol. 1986;102:1797–1812. doi: 10.1083/jcb.102.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L. M., Rieder C. L., Edde B., Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D., Wloga D., Sharma N., Gaertig J. DYF-1 Is required for assembly of the axoneme in Tetrahymena thermophila. Eukaryot. Cell. 2009;8:1397–1406. doi: 10.1128/EC.00378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Follit J. A., Xu F., Keady B. T., Pazour G. J. Characterization of mouse IFT complex B. Cell Motil. Cytoskelet. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Schafer J. C., Zhang Q., Taulman P. D., Yoder B. K. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Swoboda P., Taulman P. D., Thomas J. H., Yoder B. K. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- Hou Y., Pazour G. J., Witman G. B. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., Witman G. B. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Rifkin M. R., Luck D. J. Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J. Cell Biol. 1977;72:67–85. doi: 10.1083/jcb.72.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Diener D. R., Mitchell A., Pazour G. J., Witman G. B., Rosenbaum J. L. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C., Besharse J. C. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev. Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C., Pathak N., Perkins B., Drummond I., Besharse J. C. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev. Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Babaev-Khaimov V., Sassaroli M., Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I., Mitani S. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells. 2007;12:593–602. doi: 10.1111/j.1365-2443.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Beech P. L., Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L., Cole D. G. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- McFadden G. I., Melkonian M. The use of HEPES buffer for microalgal culture media and fixation for electron microscopy. Phycologia. 1986;25:551–557. [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Ou G., Koga M., Blacque O. E., Murayama T., Ohshima Y., Schafer J. C., Li C., Yoder B. K., Leroux M. R., Scholey J. M. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Obara T., Mangos S., Liu Y., Drummond I. A. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D. R., Rosenbaum J. L., Lupetti P. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Rosenbaum J. L., Barr M. M. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- Qin H., Wang Z., Diener D., Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker V., Levilliers N., Vinolo E., Rossier J., Jaillard D., Burnette D., Gaertig J., Bre M. H. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of alpha- and beta-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 2005;280:596–606. doi: 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- Reynolds E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr J., Sarkar N., Balenger S., Jeong B. R., Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Snow J. J., Ou G., Gunnarson A. L., Walker M. R., Zhou H. M., Brust-Mascher I., Scholey J. M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Walther Z., Vashishtha M., Hall J. L. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fan Z. C., Williamson S. M., Qin H. Intraflagellar transport (IFT) protein IFT25 is a phosphoprotein component of IFT complex B and physically interacts with IFT27 in Chlamydomonas. PLoS One. 2009;4:e5384. doi: 10.1371/journal.pone.0005384. [DOI] [PMC free article] [PubMed] [Google Scholar]