Both LIS-1 and the dynein heavy-chain DHC-1 are required for integrity of the actin cytoskeleton in Caenorhabditis elegans. An RNAi screen revealed that knockdown of other actin regulators, including actin-capping protein genes and prefoldin subunit genes, suppresses dhc-1(or195ts)–induced lethality.
Abstract
Functional analysis of cytoplasmic dynein in Caenorhabditis elegans has revealed a wide range of cellular functions for this minus-end–directed motor protein. Dynein transports a variety of cargos to diverse cellular locations, and thus cargo selection and destination are likely regulated by accessory proteins. The microtubule-associated proteins LIS-1 and dynein interact, but the nature of this interaction remains poorly understood. Here we show that both LIS-1 and the dynein heavy-chain DHC-1 are required for integrity of the actin cytoskeleton in C. elegans. Although both dhc-1(or195ts) and lis-1 loss-of-function disrupt the actin cytoskeleton and produce embryonic lethality, a double mutant suppresses these defects. A targeted RNA interference screen revealed that knockdown of other actin regulators, including actin-capping protein genes and prefoldin subunit genes, suppresses dhc-1(or195ts)–induced lethality. We propose that release or relocation of the mutant dynein complex mediates this suppression of dhc-1(or195ts)--induced phenotypes. These results reveal an unexpected direct or indirect interaction between the actin cytoskeleton and dynein activity.
INTRODUCTION
Cytoplasmic dynein is a minus-end–directed microtubule motor protein that transports a wide range of cargos, including vesicles, organelles, and mRNAs (Mallik and Gross, 2004; Vallee et al., 2004). Dynein is also required for nuclear and cell migration and to reorient microtubules with respect to cellular architecture. Dynein is a large multisubunit motor protein that interacts with a large number of accessory proteins, including the multisubunit dynactin complex, and an assortment of dynactin and microtubule-binding proteins, to regulate its diverse interactions and functions (Vallee et al., 2004). In addition, in order to efficiently transport cargo to the minus end of microtubules, dynein must relocalize to a region near the plus end of microtubules. How this large and growing list of interacting proteins work together to regulate the spatial and temporal diversity of dynein function is not understood.
Cytoplasmic dynein is composed of two heavy chains (520 kDa) that contain ATPase and motor activities, two intermediate chains (74 kDa) thought to anchor dynein to its cargo, two light intermediate chains (53–59 kDa), and several light chains whose functions are poorly understood. The dynein heavy chain (DHC) is a member of the AAA+ (ATPases associated with diverse cellular activities) family of proteins. The C-terminus contains six AAA+ subunits (AAA1–AAA6) organized into a ring (Neuwald et al., 1999). The first four AAA modules (AAA1–AAA4) bind ATP, but only ATP hydrolysis by AAA1 has been linked to motor activity. A coiled-coil “stalk” responsible for microtubule binding is situated between domains AAA4 and AAA5 (Gee et al., 1997). The N-terminal “stem” region mediates interactions with the other heavy chain within the dynein complex and interactions with intermediate and light chains as well as the dynactin complex. Each of these dynein subunits and the multisubunit dynactin complex have been implicated in cargo binding and regulation of dynein activity (Gibbons et al., 1991; Ogawa, 1991; Koonce et al., 1992).
Cytoplasmic dynein function and localization has been extensively studied in the early Caenorhabditis elegans embryo. DHC localization by immunofluorescence shows dynamic cell cycle–dependent patterns in C. elegans embryos (Gonczy et al., 1999; Schmidt et al., 2005). DHC is distributed throughout the cytoplasm but is enriched at the nuclear envelope during prometaphase, at the spindle midzone during metaphase, and at the cell cortex in two-cell stage embryos. Small pools of dynein were also observed on the entire metaphase spindle. Dynein function in embryos has been investigated using RNA interference (RNAi) and conditional mutant alleles (Gonczy et al., 1999; Yoder and Han, 2001; Cockell et al., 2004). One of these conditional alleles, dhc-1(or195ts), is a mis-sense mutation substituting serine for leucine at amino acid 3200 (S3200L) in the DHC microtubule-binding stalk region. At the permissive temperature (15°C), dhc-1(or195ts) provides sufficient dynein activity for normal development, but growth at the nonpermissive temperature (25°C) results in embryonic lethality and adult sterility. The phenotype of dhc-1(or195ts) animals and their progeny grown continuously at the permissive temperature is similar to that of the wild type. However, at the nonpermissive temperature the dhc-1(or195ts) phenotype is indistinguishable from that of the strong loss-of-function phenotype produced by dhc-1 RNAi. Shifting these temperature-sensitive mutant animals and embryos to the nonpermissive temperature rapidly disrupts or reduces dynein function, revealing a wide range of defects, indicating that dynein is required for many microtubule-dependent processes, including pronuclear migration, spindle assembly, positioning and orientation, chromosome segregation, and cytokinesis (Gonczy et al., 1999; Yoder and Han, 2001; Schmidt et al., 2005). DHC-1(S3200L) localization at the nonpermissive temperature shows accumulation near the minus ends of centrosomal microtubules (Schmidt et al., 2005).
A variety of accessory proteins, including the dynactin complex and the microtubule-binding protein LIS-1, regulate cargo selection and dynein activity. The multisubunit dynactin complex attaches dynein to kinetochores and vesicular organelles and in vitro functions to increase dynein processivity (King, 2000). LIS-1, which acts to increase dynein ATPase activity, binds to two sites on dynein: the first site is located in the region also responsible for binding site of the intermediate chains involved in cargo binding, and the second binding site is the P-loop involved in motor activity (Sasaki et al., 2000; Tai et al., 2002; Mesngon et al., 2006). Coimmunoprecipitation studies show that LIS-1 and dynein can interact in vivo (Faulkner et al., 2000; Smith et al., 2000), but the character of this interaction remains elusive. LIS-1 also interacts with the dynamitin subunit of dynactin (Karki and Holzbaur, 1999; Tai et al., 2002). Because LIS-1 interacts with the dynein–dynactin complex, LIS-1 was thought to be a structural part of the motor complex (Liu et al., 1999; Faulkner et al., 2000; Smith et al., 2000). However, because LIS-1 increases dynein ATPase activity, it is more likely to perform a regulatory function (Mesngon et al., 2006).
In C. elegans, dynein localization is dependent on LIS-1, and LIS-1 localization, in turn, is dependent on dynein (Cockell et al., 2004). In the germline LIS-1 is expressed in the cytoplasm and is enriched at the nuclear envelope of oocytes (Buttner et al., 2007). In embryos LIS-1 localization is cell cycle-dependent. During late prophase LIS-1 localizes inside pronuclei in the one-cell stage embryo and at the nuclear periphery and around chromosomes in the two-cell stage embryo. During metaphase and anaphase LIS-1 localizes to the spindle, and in late anaphase and telophase LIS-1 is enriched at the ends of microtubules asters. LIS-1 also localizes along the microtubules (Cockell et al., 2004). In one-cell stage embryos, lis-1 loss of function phenocopies dhc-1 loss of function, causing defects in spindle assembly, pronuclear migration and centrosome separation (Cockell et al., 2004). Interestingly, in migrating neurons and in Dictiostylium, reduction in LIS-1 levels reduces filamentous (F)-actin content (Kholmanskikh et al., 2003; Rehberg et al., 2005). The reduction in F-actin is associated with reduced Cdc42 and Rac1 activity and altered actin dynamics. Although these observations link LIS-1 activity to regulation of the actin cytoskeleton, the nature of this interaction is still unknown.
Here we show that although C. elegans lis-1 is an essential gene, lis-1 knockdown by RNAi in dhc-1(or195ts) animals suppresses the lethality of both dhc-1(or195ts) and lis-1 RNAi, resulting in viable progeny. To investigate this surprising result, we used a genetic interactions database to assemble a list of 238 additional candidate DHC-1– and LIS-1–interacting proteins (Zhong and Sternberg, 2006) and then screened these genes for those that, like lis-1, when depleted by RNAi, suppress the temperature-sensitive lethality of dhc-1(or195ts). We found four genes, lis-1, cap-1, cap-2, and dli-1, that when depleted by RNAi significantly suppressed dhc-1(or195ts) lethality, cytoskeletal defects, and DHC localization in the germline and early embryo. Two of these genes, lis-1 and dli-1, are known dynein regulators, and lis-1, cap-1, and cap-2 are known or implicated regulators of F-actin dynamics. We propose that the unexpected rescue of dhc-1(or195ts) defects may be mediated by treatments that release or relocate the mutant dynein complex from the minus ends of microtubules.
MATERIALS AND METHODS
Nematode Strains and Culture Conditions
The following mutant strains were used in this study: C. elegans Bristol strain N2 (wild type), EU828, dhc-1(or195ts) I; MT12272, juIs73 III/lis-1(n3334) III; n3334 contains a deletion encompassing bases 4325–6342 of cosmid T03F6; HR10, dhc-1(ct42) dpy-5(e61)/unc-11(e47) bli-4(e937) I; KR332, dhc-1(h79) dpy-5(e61) unc-13(e450) I; sDp2(I;f). Nematodes were grown under standard culture conditions at 15–25°C. EU828was maintained at 15°C.
Gene Selection, Screen (RNAi), and Viability Assessment
To create a list of probable lis-1– and dhc-1–interacting genes, we used the computational data search system “Predictions of C. elegans Genetic Interactions” (http://tenaya.caltech.edu:8000/predict/; Zhong and Sternberg, 2006). As input genes, we used lis-1 and dhc-1 followed by cap-1 and -2. Candidate genes (n = 238) were screened using a genome-wide C. elegans RNAi library (Geneservice, Cambridge, United Kingdom). The library was constructed by J. Ahringer's group at the Wellcome CRC Institute, University of Cambridge, Cambridge, England. The primary screen was performed on 24-well plates (CorningCostar, Acton, MA) containing NG medium supplemented with 5 mM IPTG and 100 mg/ml carbenicillin. Plates were seeded with bacteria cultures and left overnight at room temperature to induce double-strand RNA (dsRNA) production. The next day two or three dhc-1(or195ts) L4 larvae were placed in each well, and the plates were incubated at 25°C. Seventy-two hours later each well was scored for viable progeny. All genes were tested at least three times. Genes for which knockdown resulted in any number of viable progeny were tested further.
For quantitative assessment of the suppression of dhc-1(or195ts) lethality, genes identified in the primary screen were retested on concentrated induced cultures, prepared as follows. Bacterial cultures were grown overnight and next diluted 1:3 followed by induction for 4 h at 37°C in LB media containing 1 mM IPTG and 100 mg/ml carbenicillin to produce dsRNA. Five milliliters of each culture was then concentrated and seeded into each well of a 24-well plate. Single animals were added to each well and after 24 h at 25° transferred to a similarly prepared fresh well for an additional 24 h. These plates were incubated an additional 24 h to allow all viable embryos to hatch. The average number of viable progeny per animal was calculated for each 24-h period from at least three separate trials. For experiments in which two genes were targeted (e.g., cap-1 and -2), the bacterial cultures were concentrated and mixed in a 1:1 ratio.
To assess the allele specific cosuppression of lis-1 and dhc-1, we grew strain KR332 on lis-1 RNAi food. Twenty animals produced no viable embryos.
Staining and Microscopy
F-actin and tubulin staining was performed on extruded gonad arms adhered to poly-l-lysine–coated slides and fixed in 4% formaldehyde in PBS for 40 min at room temperature. The slides were then rinsed in PBS, and the samples incubated with FITC-conjugated anti-α-tubulin mAb (1:50; Sigma-Aldrich, St. Louis, MO) overnight at room temperature, followed by incubation with rhodamine-conjugated phalloidin (0.164 μM; Invitrogen, Carlsbad, CA) for 2 h. Finally, slides were incubated in 1×x Hoechst (Invitrogen) for 5 min. Between incubations slides were washed in 1× PBST.
DHC-1 and microtubule staining of one-cell stage embryos was as described in Gonczy et al. (1999), with minor modifications. Anti-DHC-1 polyclonal antibodies (gift from Pierre Gonczy, ISREC, Epalinges, Switzerland) and secondary rhodamine-conjugated AffiniPure anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) were used at 1:100 dilution. FITC-conjugated anti-α-tubulin mAb was used at 1:50. Incubation time for all antibodies was 1 h.
F-actin, microtubules, and DHC-1 staining utilized a permeabilization protocol adapted from Huang et al. (2002) and then fixed in Cytofix solution (BD Biosciences, San Jose, CA) for 5 min at −20°C. Slides were first incubated with anti-DHC-1 polyclonal antibodies (gift from Pierre Gonczy or from Susan Strome, Indiana University; 1:100) and then rhodamine-conjugated AffiniPure anti-rabbit IgG (Jackson ImmunoResearch, 1:100). The slides were then incubated with FITC-conjugated anti-α-tubulin mAb (1:50), followed by incubation with AlexaFluor 405–conjugated phalloidin (Invitrogen).
All images were obtained using an Axiovert 200 spinning disk confocal microscope (Zeiss, Thornwood, NY) and analyzed with Axiovision software (v.4.6; Zeiss).
Drug Treatment
Jasplakinolide (Invitrogen) was kept as 1 mM solution in anhydrous methanol. To obtain 1 μM concentration, 1 μl of stock solution was added into 999 μl of M9. As a control we used M9 with the same volume of anhydrous methanol. Twenty-four-old adult animals were suspended in M9 medium containing either jasplakinolide or methanol and transferred onto poly-l-lysine–coated slides. Gonads were extruded from the animals, and slides were incubated in humid atmosphere for 30 min at room temperature, washed once in PBST, fixed with methanol, and then stained with anti-actin and anti-DHC-1 antibodies.
Densitometry
Densitometric analysis of lane profile plots was performed using ImageJ (http://rsb.info.nih.gov/ij/), according to guidelines in ImageJ documentation. The peak areas (corresponding to gel band intensities) of either LIS-1 or ZK858.3 (CTRL) were measured, and each peak was normalized to the appropriate peak area of the loading controls for the corresponding amount of RNA template used.
RESULTS
Loss of Function of Either dhc-1 or lis-1 Disrupts F-Actin Organization in the Pachytene Region of the Gonad
lis-1 and dhc-1 are required for similar microtubule-dependent cell cycle events in the early C. elegans embryo. In the one-cell embryo lis-1 and dhc-1 loss-of-function both produce defects in centrosome separation, pronuclear migration, and spindle assembly (Cockell et al., 2004). In addition, lis-1 is required for germline development; lis-1 loss of function reduces the proliferative capacity of the germline in part by disrupting the mitotic spindle (Buttner et al., 2007). To determine whether dhc-1 loss of function causes similar germline phenotypes, we compared the earliest effects of lis-1(n3334) and dhc-1(or195ts) on the development of the C. elegans germline, with a particular emphasis on the structure of the cytoskeleton.
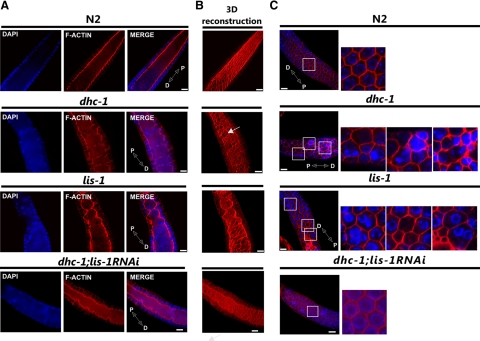
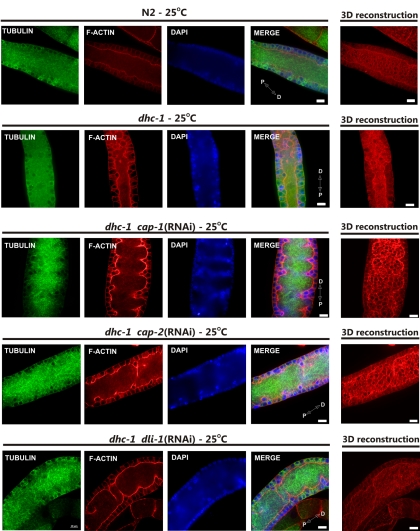
We used immunofluorescence microscopy to visualize and compare F-actin and microtubule cytoskeletal structures and organization in wild-type, lis-1(n3334), and dhc-1(or195ts) gonads and germlines (Figure 1). The C. elegans germline is syncytial, existing as proliferating and differentiating nuclei in a cytoplasmically continuous environment. The development of these nuclei is organized along the distal to proximal axis. At the distal end nuclei proliferate mitotically in a stem-cell-like niche. Proliferating nuclei exit this niche and begin meiosis in the transition zone, where chromosome pairs align and the synaptonemal complex begins to form. In the pachytene region, the nuclei are arranged around the cortex of the gonad creating a nuclei-free rachis. F-actin staining in wild type showed a cage-shaped structure surrounding each cortically localized nucleus (Figure 1C). This actin cage opens to the rachis through uniformly shaped “windows.” F-actin, visualized by three-dimensional reconstruction from serial optical sections, showed a straight smooth structure along the edge of the rachis (Figure 1B). In lis-1(n3334) and dhc-1(or195ts) animals the structure of F-actin in the pachytene region was disrupted. The structure of the rachis, rather than being straight and smooth, was ruffled and deformed (Figure 1A). Furthermore, a single actin cage often surrounded multiple nuclei, and many actin-free nuclei accumulated within the rachis (Figure 1C). Additionally, the windows connecting nuclei with the cytoplasm were larger and irregular (Figure 1B). The differences in the distal gonad were largely restricted to the pachytene region; few of these defects were evident in the most distal portion of the gonads (Supplementary Figure 1). These results suggest that both dhc-1 and lis-1 function in actin cytoskeleton organization. Alternatively, similar effects could result from the fact that LIS-1 and dynein interact with each other (Faulkner et al., 2000; Smith et al., 2000), or the defects in the actin cytoskeleton might be secondary to defects in microtubule organization.
Figure 1.
Mutations of dhc-1 and lis-1 disrupt F-actin organization in the pachytene region of the gonad. Phalloidin-rhodamine and Hoechst staining of extruded gonads to visualize F-actin (red) and DNA (blue), respectively. (A) A single optical section of the pachytene germline showing the F-actin structure of the cytoplasmic rachis in wild type (N2), dhc-1(or195ts), lis-1(n3334), and in dhc-1(or195ts); lis-1(RNAi). In wild-type gonads the germline rachis is straight with regularly positioned nuclei in the surrounding cortex, whereas in both mutants the F-actin lining the rachis is ruffled, and many cortical nuclei are displaced into the rachis. (B) 3D reconstruction of F-actin serial optical images to better visualize rachis abnormalities, deformation of the cytoskeleton, and irregularity of actin cages that connect nuclei to the cytoplasmic rachis (arrows). (C) Defects in cortical nuclear localization. Labeled as in A. Images in the leftmost column represent a single optical section near the gonad surface. Areas in white squares are magnified and displayed to the right to illustrate the regular actin network surrounding individual nuclei in wild type and the absence of nuclei or clusters of 2–3 nuclei within an actin ring in both mutants. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image. Scale bar, 10 μm.
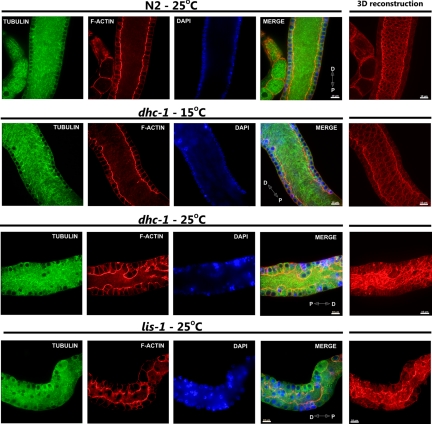
The microtubule cytoskeleton in wild-type animals was composed of long evenly distributed microtubules within the gonad rachis, creating a regular network (Figure 2). Each nucleus on the periphery of the rachis was surrounded by a regular network of microtubules. In lis-1(n3334) and dhc-1(or195ts) gonads the microtubule cytoskeleton was similar to that in wild-type gonads. We noticed some subtle changes in microtubule organization within the rachis; however, these changes were most likely caused by defects in the organization of the actin cytoskeleton surrounding the rachis (Figure 2). Thus, it is unlikely that the common actin cytoskeleton defects observed in lis-1 and dhc-1 are secondary to defects in microtubule organization.
Figure 2.
Microtubule organization in lis-1 and dhc-1 mutants is comparable to wild type. Anti-α-tubulin antibody, phalloidin-rhodamine, and Hoechst staining of extruded gonads (pachytene region) to visualize microtubules (green), F-actin (red), and nuclei (blue), respectively. The microtubule cytoskeleton of wild-type (N2) and mutant (dhc-1 and lis-1) gonads from animals cultured at indicated temperatures do not show any obvious abnormalities, whereas both mutants show obvious defects in the actin cytoskeleton. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image. Scale bar, 10 μm.
The Cytoskeletal Defects and Embryonic Lethality Associated with dhc-1(or195ts) and lis-1(RNAi) Are Cosuppressed in dhc-1(or195ts); lis-1(RNAi) Double “Mutant” Animals
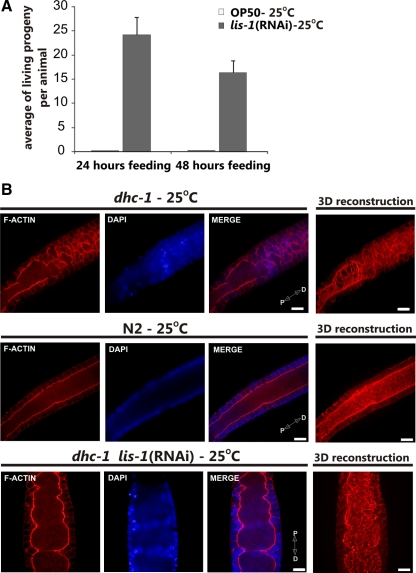
The above results indicated that the disruption of actin structures in lis-1(n3334) and dhc-1(or195ts) was not simply secondary to defects in microtubule disruption. Therefore, to determine whether depletion of both lis-1 and dhc-1 would produce a more severe phenotype and thereby reveal nonoverlapping requirements for actin organization, we used RNAi to deplete lis-1 in a dhc-1(or195ts) background at the nonpermissive temperature of 25°C. Surprisingly, codepletion of lis-1 and dhc-1 produced viable embryos. In contrast, depleting lis-1 in this background at the permissive temperature (15°C; data not shown) or simply maintaining the dhc-1(or195ts) animals at 25°C produced no viable embryos (Figure 3A). Because both lis-1(n3334) and dhc-1(or195ts) mutations produced similar dramatic changes in F-actin in the germline (Figure 1), we asked whether these abnormalities were present in the double knockdown animals. We found that the F-actin structures in the double lis-1(RNAi) dhc-1(or195ts) animals were restored to a level that functioned to maintain nuclei in the periphery (Figures 1 and 3B, and Figure S2). Likewise the microtubule cytoskeleton in the double mutants was comparable to wild type; we did not observe any changes in the length of microtubules or in microtubule organization (not shown). Thus, although the disruption of expression of either lis-1 or dhc-1 alone causes similar cytoskeletal defects and embryonic lethality, the codepletion of two genes cosuppress each other for both phenotypes.
Figure 3.
RNAi of lis-1 suppresses lethality of dhc-1(or195ts) animals. (A) Average number of viable hatched larvae laid by dhc-1(or195ts) animals in two consecutive 24-h periods on control bacteria or bacteria producing lis-1 dsRNA [lis-1(RNAi)]. Embryos were incubated an additional 24 h to assess hatching. n = 3; error bars, ±SEM. (B) Phalloidin-rhodamine and Hoechst staining of extruded gonads (pachytene region) to visualize F-actin (red) and DNA (blue), respectively, from dhc-1(or195ts) animals grown on control or lis-1 dsRNA-expressing bacteria at the nonpermissive temperature (25°C). 3D reconstruction of F-actin serial optical images. Depletion of lis-1 by RNAi greatly improved F-actin structure, comparable to N2 control (Figure 1A). Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image. Scale bar, 10 μm.
The Observed Cosuppression Depends on the dhc-1(or195ts) Allele and lis-1(RNAi)
The observed cosuppression was between a temperature-sensitive dhc-1(or195ts) allele that likely disrupts but does not eliminate dynein function and RNAi depletion of lis-1, which likely substantially reduces, but does not eliminate LIS-1 activity. Therefore, we determined whether other dhc-1alleles and null alleles of lis-1 can be similarly suppressed. We found that the dhc-1(h79) allele was similarly lethal, but that the lethality was not suppressed by lis-1 RNAi (data not shown). Similarly, the lis-1(n3334) null allele was not suppressed by dhc-1 RNAi (not shown). Further, we were unable to recover a viable dhc-1(or195ts); lis-1(n3334) double mutant strain. We conclude that the cosuppression is allele-specific and requires both the dhc-1(or195ts) mutant and reduced but not eliminated lis-1 function.
lis-1(RNAi) Rescues DHC-1(S3200L) Mislocalization
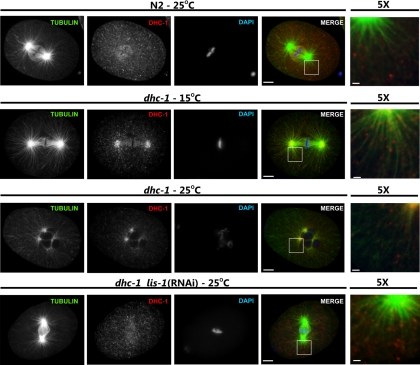
To investigate the nature of this allele-specific cosuppression we examined dynein localization in both one-cell embryos and the germline. As previously reported, at metaphase in wild-type one-cell embryos cytoplasmic and centrosome-associated DHC-1 localizes to the spindle on both sides of the metaphase plate (Gonczy et al., 1999). Although dhc-1(or195ts) embryos are fully viable at the permissive temperature, the localization of the mutant DHC-1(S3200L) was altered (Figure 4). In addition to the spindle localization at metaphase, DHC-1(S3200L) remained detectable both in the cytoplasm and at the centrosome throughout the cell cycle. DHC-1(S3200L) was also enriched in the spindle midzone. Similar to previous reports (Schmidt et al., 2005), at the nonpermissive temperature DHC-1(S3200L) localized primarily to the minus end of microtubules, close to or at the microtubule-organizing center (MTOC), which indicates defects in releasing dynein and its relocalization rather than in movement.(Figure 4). RNAi-mediated knockdown of lis-1 in wild-type animals, similar to the lis-1(null), caused DHC-1 to become cytoplasmic (Figure 5 and Cockell et al., 2004). However knockdown of lis-1 in dhc-1(or195ts) embryos altered DHC-1(S3200L) localization, such that a portion of DHC-1(S3200L) protein was distributed along microtubules (Figure 4). Compared with wild-type embryos, there was more dynein localized to microtubules surrounding centrosomes in the dhc-1(or195ts); lis-1(RNAi) embryos. Furthermore, while DHC-1(S3200L) was detected on microtubules at the metaphase plate, the level of staining was generally reduced compared with wild type. Finally, in some embryos DHC-1 was only localized along microtubules rather than on the metaphase plate (not shown).
Figure 4.
lis-1 knockdown restores DHC-1 localization in dhc-1(or195ts) embryos. One-cell stage embryos from wild-type animals (N2) or dhc-1(or195ts) animals (dhc-1) at permissive (15°C) or nonpermissive (25°C) temperatures and fed with either control or lis-1 dsRNA-producing bacteria [lis-1(RNAi)] were stained with anti-β-tubulin (green), anti-DHC-1 (red), and Hoechst (blue). Individual channels are shown in grayscale, and merged images are shown in color. To better visualize localization of dynein with respect to microtubules, the last column (5×) shows magnification of the white square–marked area of merged image; scale bar, 1 μm. lis-1 knockdown restored dynein localization to near wild type. Scale bar, 5 μm.
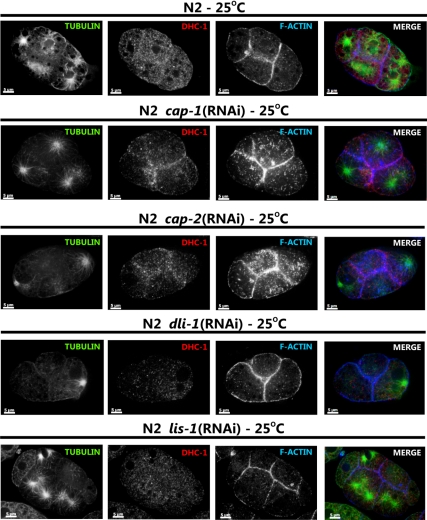
Figure 5.
Suppressors of dhc-1(or195ts) lethality affect dynein localization in wild-type embryos differently. Four-cell stage embryos from wild-type animals (N2) animals fed with either control bacteria or bacteria expressing the indicated dsRNA were stained with anti-β-tubulin (green), anti-DHC-1 (red), and rhodamine phalloidin (blue). Individual channels are shown in grayscale, and merged images are shown in color. Each image shows a single optical section of a four-cell stage embryo. RNAi knockdown of cap-1 and -2 causes DHC-1 localization to regions of cell-cell contact, an increase in F-actin levels, and the accumulation of short acting filaments. RNAi knockdown of dli-1 and lis-1 causes DHC-1 localization to the cytoplasm. Scale bar, 5 μm.
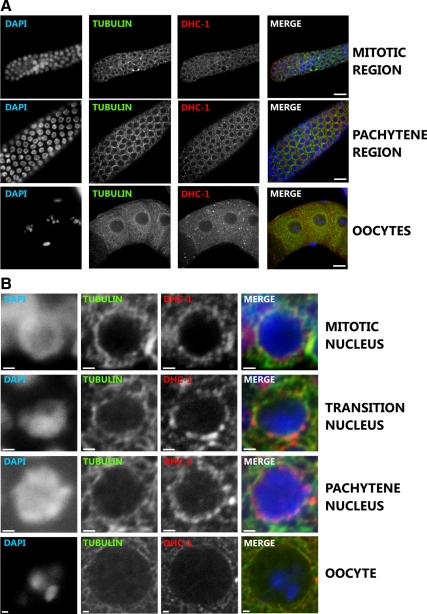
In the germline of wild-type animals, DHC-1 localization was perinuclear throughout the whole gonad, except in oocytes where the localization was cytoplasmic (Figure 6). In the transition zone the staining was more punctuated but remained perinuclear (Figure 6B). We failed to detect a significant overlap between dynein localization and microtubules in either wild-type or dhc-1(or195ts) gonads. We also did not detect any colocalization between dynein and F-actin. Similar to the observations in embryos, LIS-1 is required for DHC-1 localization in gonads, as knockdown of lis-1 caused the perinuclear-localized dynein to become cytoplasmic (Figure 7). This result suggests that LIS-1 may regulate dynein localization or be required for dynein attachment to microtubules.
Figure 6.
Cytoplasmic and perinuclear dynein localization in the wild-type germline. Anti-α-tubulin, anti-DHC-1, and Hoechst staining of wild-type gonads to visualize microtubules (green), DHC-1 (red), and nuclei (blue). (A) DHC-1 and tubulin localization in the mitotic, pachytene, and oocyte regions of the gonad. (B) Perinuclear localization of DHC-1 and tubulin in the indicated regions of wild-type germline. Scale bar, 1 μm. DHC-1 localization is perinuclear in the transition zone and pachytene region. DHC-1 and tubulin do not colocalize. Scale bar, 10 μm.
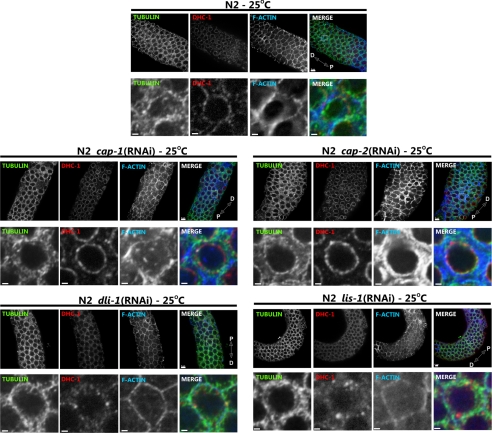
Figure 7.
Suppressors of dhc-1(or195ts) lethality differently affect dynein localization in the germline. Anti-β-tubulin (green), anti-DHC-1 (red), and rhodamine phalloidin (blue) staining of wild-type (N2) and the indicated RNAi-treated gonads. Individual channels are shown in grayscale, and merged images are shown in color. A fragment of the distal gonad is shown in the top row of each panel (scale bar, 5 μm) and a close-up of a single nucleus is shown in the bottom row (scale bar, 1 μm). cap-1 and -2 RNAi cause notable increase in F-actin levels and striking localization of DHC-1 to the perinuclear region. In contrast lis-1 and dli-1 RNAi cause an increase in cytoplasmic DHC-1 localization. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image.
Disruption of F-Actin Dynamics Suppresses dhc-1(or195ts) Embryonic Lethality
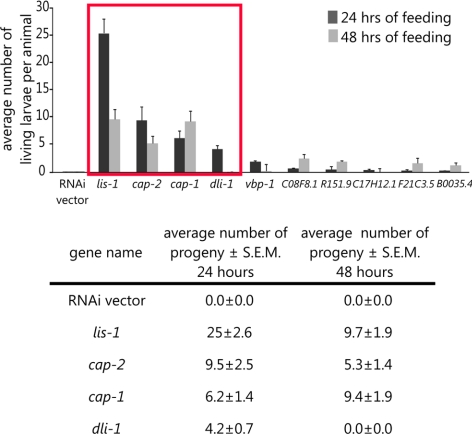
To gain insight into how reduction of lis-1 suppresses dhc-1(or195ts) embryonic lethality, we sought to identify additional genes that when depleted by RNAi also suppressed the temperature-sensitive embryonic lethality of dhc-1(or195ts). We first compiled a list of 238 candidate DHC-1– and LIS-1– interacting genes using the computational data search system “Predictions of C. elegans Genetic Interactions” (Table S1; Zhong and Sternberg, 2006; see Materials and Methods). We selected genes predicted to interact with dynein and LIS-1. Available bacterial clones for these genes were selected from an RNAi bacterial clone library, and dhc-1(or195ts) L4 animals were grown on these clones as the sole food source at 25°C. To score for suppression of dhc-1(or195ts) lethality, we determined the viability (hatching) of progeny produced in the subsequent 24 h. Bacterial clones that resulted in any viable dhc-1(or195ts) progeny were sequenced to verify the identity of the insert and retested in triplicate for suppression of dhc-1(or195ts). Overall, 36 clones produced viable progeny (Table S2). Among these genes, lis-1 RNAi produced the strongest effect, but the actin-capping proteins, cap-1 and -2, the dynein light chain, dli-1, and the prefoldin subunits (vbp-1, C08F8.1, R151.9, C17H12.1, F21C3.5, and B0035.4) produced significantly more progeny than other suppressing clones (Figure 8, boxed). The capping proteins and prefoldin subunits are directly or indirectly involved in F-actin assembly, suggesting that the actin cytoskeleton may have an important role in suppression of dhc-1-induced lethality.
Figure 8.
Suppressors of dhc-1(or195ts) lethality. Average number of viable hatched larvae laid by dhc-1(or195ts) animals in two consecutive 24-h periods on control bacteria (RNAi vector) or bacteria producing the indicated dsRNA (see Materials and Methods). The percent of fertile hermaphrodites is as follows: dhc-1; lis-1 RNAi, n = 29, 100%; dhc-1; cap-1 RNAi, n = 14, 85%; dhc-1; cap-2 RNAi, n = 14, 93%; and dhc-1; dli-1 RNAi, n = 14, 93%. Note that for single mutants for each produced no viable progeny. The graph shows data for select genes, and the table shows numerical data for the genes selected for further analysis (red rectangle). n = 3 different experiments. Error bars, ±SEM.
dhc-1(or195ts) Suppressors Have Distinct Effects on Cytoskeletal Organization
We next determined whether and how knockdown of these proteins disrupted F-actin organization in wild-type and dhc-1(or195ts) gonads and embryos. RNAi knockdown of the dynein light-IC (dli-1), like lis-1(RNAi), suppressed the actin abnormalities associated with dhc-1(or195ts) in the gonad and produced some viable progeny. The windows connecting the nuclei to the rachis were regular and small, resembling those observed in wild type (Figure 1), and few nuclei were present within the rachis (Figure 9). Interestingly, although dli-1 RNAi and lis-1 RNAi similarly improved the cytoskeleton structure and germline morphology, the level of embryonic rescue by dli-1 RNAi was much less than observed for lis-1 RNAi (Figure 8). That lis-1 is more dispensable than dli-1 in the dhc-1(or195ts) background likely reflects the requirement of dli-1 for dynein function.
Figure 9.
Suppression of dhc-1(or195ts) lethality is linked to the improvement of actin cytoskeleton structure. Anti-α-tubulin antibody, phalloidin-rhodamine, and Hoechst staining of gonads (pachytene region) extruded from dhc-1(or195ts) animals grown at the nonpermissive temperature (25°C) on bacteria expressing the indicated dsRNA shows that each of the RNAi knockdowns, cap-1, -2, and dli-1, improve F-actin (red) structure and nuclei (blue) positioning. Microtubule (green) structures are largely unaffected by the treatments. Note the apparent increase in F-actin content in cap-1 and -2 RNAi-treated animals. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image. Scale bar, 10 μm.
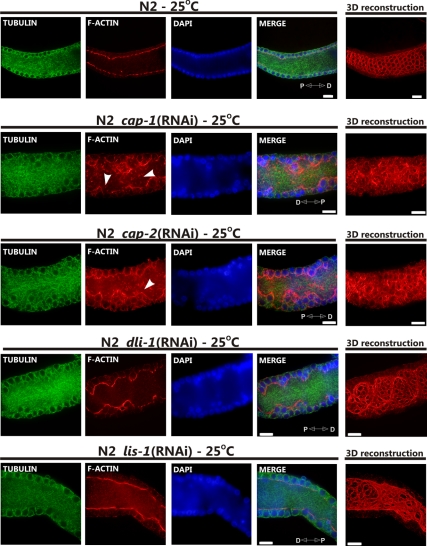
RNAi knockdown of the actin-capping proteins cap-1 and -2 produced indistinguishable results and hereafter will be referred to as cap(RNAi). cap(RNAi) dramatically increased the apparent level of F-actin in wild-type and dhc-1(or195ts) animals (Figures 9 and 10). In both wild-type and dhc-1(or195ts) animals the F-actin lining the rachis of the gonad was thickened and ruffled, but in dhc-1(or195ts); cap(RNAi) animals the windows connecting nuclei with the cytoplasm were more regular, suggesting that the dhc-1(or195ts) allele also partially suppresses cap(RNAi) defects. Capping proteins bind and stabilize the barbed plus end of actin, regulating the rate of assembly and disassembly (Hug et al., 1995; Hopmann and Miller, 2003). Thus, when G-actin levels are low, cap(RNAi) would be expected to reduce F-actin levels and when G-actin levels are high, cap(RNAi) would be expected to increase F-actin levels. The observed increase in F-actin thus leads us to conclude that there is abundant G-actin ready for polymerization in the gonad. We also noticed short actin filaments within the rachis of cap(RNAi) gonads, filaments which were not present in either wild-type gonads or in dhc-1(or195ts); cap(RNAi) gonads (Figure 10, arrowheads). The significance of these short actin filaments and their dependence on wild-type dhc-1 is currently unknown.
Figure 10.
Suppressors of dhc-1(or195ts) lethality have distinct effects on cytoskeletal organization. Anti-α-tubulin antibody, phalloidin-rhodamine, and Hoechst staining of gonads (pachytene region) extruded from wild-type (N2) animals grown at 25°C on bacteria expressing the indicated dsRNA. Each of the RNAi knockdowns, cap-1, -2, dli-1, and lis-1 affects F-actin (red) organization and the morphology of the rachis, including nuclei (blue) positioning. cap-1 and -2 RNAi treatments caused an apparent increase in F-actin content and the formation of distinct short actin filaments (arrowheads). Microtubule (green) structures are largely unaffected by the treatments. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image. Scale bar, 10 μm.
dhc-1(or195ts) Suppressors Have Distinct Effects on DHC-1(S3200L) Localization
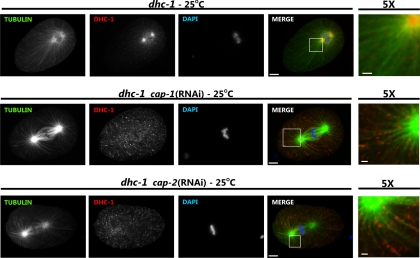
We showed above that in embryos lis-1 RNAi redistributed DHC-1(S3200L) protein (Figure 4) and in the germline caused wild-type DHC-1 to disassociate from perinuculear regions (Figure 7). To determine whether the mechanisms of dhc-1(or195ts) suppression were similar for the additional suppressors, we examined DHC-1(S3200L) localization in embryos and gonads of RNAi-treated animals. As shown previously (Yoder and Han, 2001), dli-1 RNAi did not affect DHC-1 localization in one-cell stage embryos. However, in the gonad dli-1 RNAi, like lis-1 RNAi, caused DHC-1(wt) and DHC-1(S3200L) protein to disassociate from the perinuclear region and become cytoplasmic (Figure 7). These results suggest that dli-1 and lis-1 suppress dhc-1(or195ts) by a similar mechanism. In contrast, cap RNAi caused DHC-1 to accumulate at F-actin–rich cell junctions in wild-type embryos and to localize very strongly along microtubules (Figures 5 and 11). Similarly, in the germline, DHC-1 localized very strongly to the perinuclear region of both meiotic germ cells and maturing oocytes, where F-actin also accumulated (Figure 7). However, we were unable to clearly demonstrate colocalization between F-actin and DHC-1 (Figure 7), and DHC-1(S3200L) did not accumulate on all F-actin surfaces (not shown). These observations suggest that DHC-1 localization near F-actin is likely indirect.
Figure 11.
cap-1 and -2 knockdowns restore DHC-1 localization in dhc-1(or195ts) embryos. One-cell stage embryos from dhc-1(or195ts) animals (dhc-1) at the nonpermissive temperature (25°C) and fed with either control or cap-1 or -2 dsRNA-producing bacteria were stained with anti-β-tubulin (green), anti-DHC-1 (red), and Hoechst (blue). Individual channels are shown in grayscale, and the merged images are shown in color. Scale bar, 5 μm. To better visualize localization of dynein with respect to microtubules, the last column (5×) shows magnification of the white square–marked area of merged image; scale bar, 1 μm. cap-1 and -2 knockdown restored dynein localization from centrosomal location to near wild-type localization along microtubules.
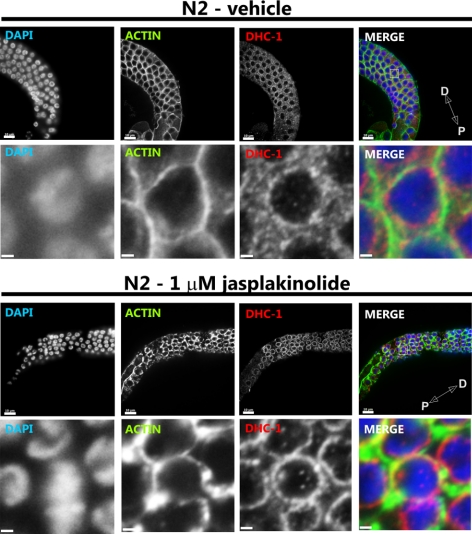
Jasplakinolide Increases F-Actin Levels and Relocalizes DHC-1 in the Gonad
To determine whether the relocalization of DHC-1 associated with suppression of dhc-1(or195ts) by cap(RNAi) is directly associated with the cap(RNAi)-induced increase in F-actin levels, we treated worms and isolated gonads with jasplakinolide. Jasplakinolide is a peptide isolated from the marine sponge Jaspis johnstoni that binds to and stabilizes F-actin in vitro (Bubb et al., 1994). In vivo jasplakinolide induces F-actin polymerization and aggregation (Senderowicz et al., 1995; Holzinger and Meindl, 1997). We found that the effects of a 30-min jasplakinolide treatment (1 μM) on the cytoskeleton of extruded wild-type gonads was similar to that of lis-1(RNAi) or dhc-1(RNAi) (Figure 12). The F-actin lining the rachis was ruffled, the windows connecting nuclei to the rachis were irregular, and many nuclei were within the rachis (not shown). Strikingly, the jasplakinolide-treated gonads showed very strong perinuclear DHC-1 localization (Figure 12). This result suggests that DHC-1 relocalization in cap(RNAi) animals is caused by changes in the actin cytoskeleton. However, we were unable to rescue the dhc-1(or195ts) defect by jasplakinolde treatment (see Materials and Methods). This could be due to the inability of dhc-1(or195ts) to suppress the toxicity of jasplakinolde or may indicate that cap(RNAi) rescues dhc-1(or195ts) lethality via a different mechanism. In this regard, capping proteins were also shown to be a part of the dynactin complex (Schafer et al., 1994a,b), so the rescue of dhc-1(or195ts) may be unrelated to the observed relocalization of DHC-1 to F-actin.
Figure 12.
Jasplakinolide treatment increases F-actin levels and causes dynein relocalization in the germline. Anti-actin antibody (green), anti-DHC-1 (red), and Hoechst staining of gonads (pachytene region) extruded from wild-type (N2) animals and incubated with either 0.1% MeOH (vehicle) or 1 μM jasplakinolide for 30 min at room temperature before fixation (see Materials and Methods). The pachytene region of the germline is shown in the top row of each panel (scale bar, 10 μm), and close-up of nuclei is shown in the bottom row (scale bar, 1 μm). The jasplakinolide-treated germline exhibits strong perinuclear localization of dynein, whereas the appearance of actin is similar to that of cap-1 and -2 RNAi germlines. Distal (D) and proximal (P) orientations of the gonad is indicated on the merged image.
DISCUSSION
Here we investigated the genetic and cellular interactions between dynein and LIS-1 in the C. elegans germline and early embryo. We found that a temperature-sensitive dynein heavy-chain mutant dhc-1(or195ts) and loss-of-function of lis-1 disrupted the F-actin cytoskeleton and cause similar developmental defects in the germline and early embryo. Unexpectedly, the dhc-1(or195ts); lis-1(RNAi) animals produced viable progeny. An RNAi screen of 238 genes predicted to interact with regulators of the cytoskeleton identified additional genes that when depleted suppress dhc-1(or195ts) lethality. We found that like the dhc-1(or195ts) mutant, depletion of these genes disrupts F-actin organization in the germline and early embryo. Furthermore, suppression of dhc-1(or195ts) lethality by depletion of these genes was associated with restoration of near normal F-actin structures as well as relocalization of the mutant DHC-1 protein. These results suggest that F-actin cytoskeleton organization may play an essential role in the suppression of dhc-1(or195ts) lethality.
Genetic studies previously suggested that lis-1 is required to maintain F-actin structure (Kholmanskikh et al., 2003; Rehberg et al., 2005). Similarly, we found that LIS-1 as well as its regulatory target, DHC-1, were required for F-actin cytoskeletal organization in the pachytene region of the C. elegans germline. In wild-type gonads F-actin surrounded each cortically localized nucleus, and the F-actin lining the rachis was straight and smooth. This highly regular F-actin organization was disrupted in a similar manner by RNAi that targeted the expression of genes encoding actin-capping proteins CAP-1 and -2, by treatment with the F-actin–stabilizing drug jasplakinolide, and by depletion of the microtubule-associated proteins LIS-1, DHC-1, and DLI-1. In each case the F-actin structure surrounding the rachis was deformed, resulting in a ruffled rachis lining, mislocalization of nuclei into the rachis, and empty and/or multiple nuclei within single irregularly shaped peripheral actin cages. These results suggest a role for dynein and the accessory proteins LIS-1 and DLI-1 in modulating actin dynamics.
We observed that treatments that alter F-actin levels or organization could suppress the temperature-sensitive dhc-1(or195ts) allele, suggesting that F-actin dynamics could modulate dynein activity. Importantly, treatments that suppress dhc-1(or195ts) did not simply bypass the F-actin defects, but restore the F-actin cytoskeleton to near wild-type patterns. This is particularly remarkable, because these treatments in a wild-type background resulted in similar actin-cytoskeleton defects. The suppression of dhc-1(or195ts) was also associated with changes in DHC-1 localization in both the germline and in embryos. However, these changes were different for lis-1, dli-1, and cap(RNAi). In the germline, depletion of the dynein-associated proteins DLI-1 and LIS-1 caused diffusion of DHC-1 into the cytoplasm, whereas depletion of CAP-1 and -2 caused strong accumulation of DHC-1 in the perinuclear region. Similar results were observed in one-cell stage C. elegans embryos. In lis-1(RNAi) and dli-1(RNAi) embryos DHC-1 localization became more cytoplasmic. In contrast, knockdown of the capping proteins shifted DHC-1 localization to the periphery of the embryo where the amount of F-actin also increased. We also observed clear localization of DHC-1 to microtubules in some embryos. These results demonstrate that changes and/or restoration of DHC-1 localization may bypass the dhc-1(or195ts) defect, restoring some dynein function and suppressing the embryonic lethality. These observations may explain both the allele specificity of the cosuppression and the wide variety of functions that can be disrupted to restore dhc-1 function (see below).
CAP-1 and -2 form heterodimers of actin-capping protein, and RNAi knockdown of each was nearly indistinguishable. Capping proteins bind the barbed end of actin with high affinity, inhibiting both assembly and disassembly of actin monomers (Wear and Cooper, 2004). In the gonad and at cell–cell junctions in early embryos, depletion of capping proteins caused an increase in F-actin levels, indicating that capping proteins were acting to limit actin assembly in these locations. Thus, the increase in DHC-1 localization to normal sites in response to increased F-actin content may represent either recruitment or redistribution of dynein. However treatment with the actin polymerization promoting peptide jasplakinolide caused an increase in F-actin levels and a redistribution of DHC-1, suggesting that F-actin levels and not an unknown effect of capping protein activity drive the redistribution of DHC-1.
Our screen revealed that the capping proteins, CAP-1 and -2, as well as the dynein light IC,*** DLI-1, suppress dhc-1 lethality, although not as strongly as lis-1 knockdown. In addition to these four moderately strong suppressors, we identified 32 additional suppressors that we have not further characterized. Interestingly, half of these are known or suspected to regulate actin or microtubule dynamics or function. These results supplement and expand those of O'Rourke et al. (2007), who performed a genome-wide RNAi screen for suppressors of dhc-1(or195ts)-induced lethality. None of the genes identified here were found in the screen by O'Rourke et al. (2007). This discrepancy might be due to different testing conditions. Our screen selected for fertile adults by exposing fourth-stage larvae to RNAi treatment at a completely nonpermissive 25°C growth temperature. In contrast, O'Rourke et al. (2007) exposed first-stage larvae to a semipermissive 23°C temperature resulting in substantially longer RNAi exposure covering many more developmental events. O'Rourke et al. (2007) may not have identified the same genes either because these genes fail to suppress dhc-1 at the earlier stages of development or because these genes have essential functions during the extended growth.
We imagine two mechanisms for suppression of dhc-1(or195ts). First, the DHC-1(or195ts) mutant protein localizes strongly to microtubules and MTOCs, and our RNAi knockdowns shift DHC-1(or195ts) to a more wild-type distribution. Thus, DHC-1(or195ts) may have normal motor functions but may maintain a persistent attachment to the minus ends of microtubules; perturbations that release DHC-1(or195ts) from microtubules may allow dynein to reengage in another round of normal motor function. Because LIS1 is thought to be a processivity factor (Coquelle et al., 2002; Mesngon et al., 2006), reducing LIS-1 levels may increase the release rate of DHC-1(or195ts) from microtubules. Similarly in Aspergillus nidulans a NUDF/LIS1 deletion mutant is suppressed by a dynein heavy-chain mutation that reduces ATPase activity and that causes an increase in dynein localization to microtubules (Zhuang et al., 2007). Thus the lack of a proposed processivity factor is suppressed by a mutation that increases the interaction between dynein and microtubules. This hypothesis is appealing, as it explains why both specific and nonspecific treatments restore dynein function. However, because different treatments had distinct effects on DHC-1 localization, a second proposed mechanism is that RNAi knockdowns redistribute DHC-1 by altering dynein targeting. LIS-1 is involved in targeting dynein to microtubule structures. Overexpression of LIS1 alters dynein distribution at the cell cortex (Faulkner et al., 2000). By analogy, disruption of lis-1 expression could impair dynein targeting, resulting in the observed increase of dynein pool in the cytoplasm. In contrast, knockdown of capping proteins resulted in strong localization of dynein to areas enriched with F-actin, as well as to the perinuclear region. Although dynein does not interact directly with F-actin, it could be anchored to cortical F-actin by other proteins, e.g., CLIP-170 or dynactin. An increase in the local concentration of F-actin and, thus, of dynein-anchoring proteins could explain the accumulation of dynein at sites with enriched F-actin. Indeed, our experiments with an actin polymerization–enhancing drug, jasplakinolide, showed similar effects to those caused by knockdown of capping proteins. We were unable to rescue dhc-1(or195ts) lethality by treatment with jasplakinolide, but this result was not surprising, as suppression of lethality likely requires both the restoration of localization and proper activity of DHC-1(S3200L).
In this study we found that the heat-sensitive allele of the dynein heavy chain, dhc-1(or195ts), disrupts the actin cytoskeleton in the C. elegans gonad. Surprisingly, depletion of factors that regulate dynein activity (lis-1and dli-1) and actin assembly (actin-capping protein and prefoldin) suppressed not only the temperature-sensitive lethality associated with this allele, but also restored the integrity of the actin network. Depletion of these factors in a wild-type background similarly disrupted actin structures, suggesting that despite the disparate processes mediated by these factors, a common mechanism is likely responsible for the suppression of dhc-1(or195ts) lethality. At the elevated temperature the mutant DHC-1 protein accumulated near the minus ends of microtubules; the above depletions caused DHC-1 to redistribute toward a more normal pattern. Therefore, we propose that the common bypass mechanism is decreased dynein processivity that allows the mutant DHC-1 protein to release from near microtubule minus ends and reinitiate a new functional interaction nearer the plus end of the microtubules. This work, thus illustrates a previously undescribed and likely indirect interaction between microtubule and actin cytoskeletal dynamics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pierre Gönczy and Susan Strome for generous gifts of antibodies, Hunter lab members for advice and discussions, and Konrad Krzewski for valuable comments. This study was supported in part by the Bernhard Foundation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0593) on June 16, 2010.
REFERENCES
- Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L., Korn E. D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Buttner E. A., Gil-Krzewska A. J., Rajpurohit A. K., Hunter C. P. Progression from mitotic catastrophe to germ cell death in Caenorhabditis elegans lis-1 mutants requires the spindle checkpoint. Dev. Biol. 2007;305:397–410. doi: 10.1016/j.ydbio.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M. M., Baumer K., Gonczy P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 2004;117:4571–4582. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Coquelle F. M., et al. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner N. E., Dujardin D. L., Tai C. Y., Vaughan K. T., O'Connell C. B., Wang Y., Vallee R. B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Gee M. A., Heuser J. E., Vallee R. B. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gibbons I. R., Gibbons B. H., Mocz G., Asai D. J. Multiple nucleotide-binding sites in the sequence of dynein beta heavy chain. Nature. 1991;352:640–643. doi: 10.1038/352640a0. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Pichler S., Kirkham M., Hyman A. A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A., Meindl U. Jasplakinolide, a novel actin targeting peptide, inhibits cell growth and induces actin filament polymerization in the green alga Micrasterias. Cell Motil. Cytoskelet. 1997;38:365–372. doi: 10.1002/(SICI)1097-0169(1997)38:4<365::AID-CM6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Hopmann R., Miller K. G. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol. Biol. Cell. 2003;14:118–128. doi: 10.1091/mbc.E02-05-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N. N., Mootz D. E., Walhout A. J., Vidal M., Hunter C. P. MEX-3 interacting proteins link cell polarity to asymetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- Hug C., Jay P. Y., Reddy I., McNally J. G., Bridgman P. C., Elson E. L., Cooper J. A. Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell. 1995;81:591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Karki S., Holzbaur E. L. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- Kholmanskikh S. S., Dobrin J. S., Wynshaw-Boris A., Letourneau P. C., Ross M. E. Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J. Neurosci. 2003;23:8673–8681. doi: 10.1523/JNEUROSCI.23-25-08673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. M. The dynein microtubule motor. Biochim. Biophys. Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- Koonce M. P., Grissom P. M., McIntosh J. R. Dynein from Dictyostelium: primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J. Cell Biol. 1992;119:1597–1604. doi: 10.1083/jcb.119.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xie T., Steward R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development. 1999;126:4477–4488. doi: 10.1242/dev.126.20.4477. [DOI] [PubMed] [Google Scholar]
- Mallik R., Gross S. P. Molecular motors: strategies to get along. Curr. Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Mesngon M. T., Tarricone C., Hebbar S., Guillotte A. M., Schmitt E. W., Lanier L., Musacchio A., King S. J., Smith D. S. Regulation of cytoplasmic dynein ATPase by Lis1. J. Neurosci. 2006;26:2132–2139. doi: 10.1523/JNEUROSCI.5095-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- O'Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K. Four ATP-binding sites in the midregion of the beta heavy chain of dynein. Nature. 1991;352:643–645. doi: 10.1038/352643a0. [DOI] [PubMed] [Google Scholar]
- Rehberg M., Kleylein-Sohn J., Faix J., Ho T. H., Schulz I., Graf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell. 2005;16:2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schafer D. A., Gill S. R., Cooper J. A., Heuser J. E., Schroer T. A. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 1994a;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. A., Korshunova Y. O., Schroer T. A., Cooper J. A. Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J. Cell Biol. 1994b;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D. J., Rose D. J., Saxton W. M., Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderowicz A. M., et al. Jasplakinolide's inhibition of the growth of prostate carcinoma cells in vitro with disruption of the actin cytoskeleton. J. Natl. Cancer Inst, 1995;87:46–51. doi: 10.1093/jnci/87.1.46. [DOI] [PubMed] [Google Scholar]
- Smith D. S., Niethammer M., Ayala R., Zhou Y., Gambello M. J., Wynshaw-Boris A., Tsai L. H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Tai C. Y., Dujardin D. L., Faulkner N. E., Vallee R. B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R. B., Williams J. C., Varma D., Barnhart L. E. Dynein: an ancient motor protein involved in multiple modes of transport. J. Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- Wear M. A., Cooper J. A. Capping protein: new insights into mechanism and regulation. Trends Bioch. Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Yoder J. H., Han M. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2921–2933. doi: 10.1091/mbc.12.10.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Sternberg P. W. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- Zhuang L., Zhang J., Xiang X. Point mutations in the stem region and the fourth AAA domain of cytoplasmic dynein heavy chain partially suppress the phenotype of NUDF/LIS1 loss in Aspergillus nidulans. Genetics. 2007;175:1185–1196. doi: 10.1534/genetics.106.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.